16.8 million SARS-CoV-2 infections in the US went undiagnosed in the first 6 months of the pandemic compared to 3.5 million diagnosed infections.
Elucidating seroprevalence in COVID-19
Symptoms of SARS-CoV-2 infection range from completely asymptomatic, to those of a common cold, to a drop in oxygen saturation and lung function, and death in some patients. To evaluate the proportion of the U.S. population who had an undiagnosed infection during the first wave of the COVID-19 pandemic, we measured antibody prevalence in study participants who had not previously been diagnosed with a SARS-CoV-2 infection. By mid-July of 2020, 16.8 million people had an undiagnosed SARS-CoV-2 infection, almost five times the rate of diagnosed infections.
Abstract
Asymptomatic SARS-CoV-2 infection and delayed implementation of diagnostics have led to poorly defined viral prevalence rates in the United States and elsewhere. To address this, we analyzed seropositivity in 9089 adults in the United States who had not been diagnosed previously with COVID-19. Individuals with characteristics that reflected the U.S. population (n = 27,716) were selected by quota sampling from 462,949 volunteers. Enrolled participants (n = 11,382) provided medical, geographic, demographic, and socioeconomic information and dried blood samples. Survey questions coincident with the Behavioral Risk Factor Surveillance System survey, a large probability-based national survey, were used to adjust for selection bias. Most blood samples (88.7%) were collected between 10 May and 31 July 2020 and were processed using ELISA to measure seropositivity (IgG and IgM antibodies against SARS-CoV-2 spike protein and the spike protein receptor binding domain). The overall weighted undiagnosed seropositivity estimate was 4.6% (95% CI, 2.6 to 6.5%), with race, age, sex, ethnicity, and urban/rural subgroup estimates ranging from 1.1% to 14.2%. The highest seropositivity estimates were in African American participants; younger, female, and Hispanic participants; and residents of urban centers. These data indicate that there were 4.8 undiagnosed SARS-CoV-2 infections for every diagnosed case of COVID-19, and an estimated 16.8 million infections were undiagnosed by mid-July 2020 in the United States.
INTRODUCTION
Coronavirus disease 2019 (COVID-19), the disease caused by severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) infection, presents with a spectrum of illness ranging from asymptomatic to severe disease. As with most respiratory viral diseases, it is difficult to estimate the true prevalence of the disease during a pandemic and the extent of its spread is only known after extensive study (1–3). Most patients infected with SARS-CoV-2 develop robust antibody responses against the viral spike protein, nucleocapsid protein, and the envelope protein that can be detected by serological testing (4–8). Antibodies against spike protein persist for months and can neutralize SARS-CoV-2 (9). Frequently, these neutralizing antibodies bind to the receptor binding domain (RBD) of the spike protein, but antibodies against the spike protein S2 domain have also been observed (10–15).
To characterize the spread of SARS-CoV-2 infection in the United States, we evaluated seropositivity in a national survey of participants who had not previously been diagnosed with SARS-CoV-2 infection. We used quota sampling from a large pool of volunteers (n = 462,949) to obtain a representative sample (n = 9089) and performed statistical weighting to generate prevalence estimates that revealed the extent of SARS-CoV-2 infection in the general population. To ensure accurate classification of seropositivity, we used our dual-antigen enzyme-linked immunosorbent assay (ELISA) protocol that evaluated immunoglobulin G (IgG) and IgM antibodies against both the full viral spike protein ectodomain and the RBD (8, 16).
RESULTS
Enrollment and demographic representation
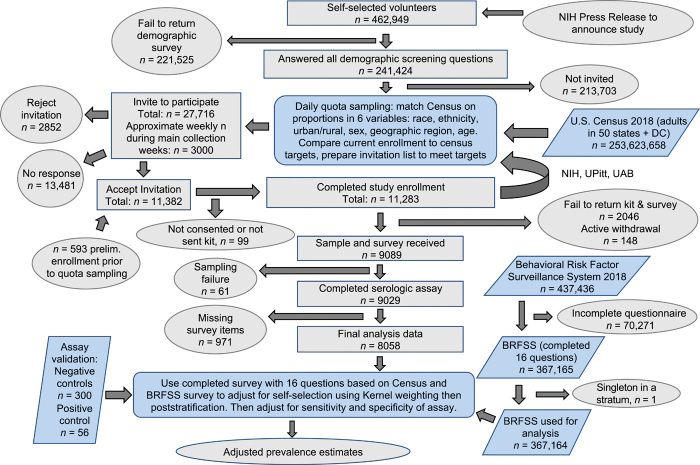
Recruitment took place from 1 April 2020 to 4 August 2020. During that time, 11,283 participants were enrolled from a pool of 241,424 volunteers in the United States (50 states and the District of Columbia). Of these participants, 214 had blood collected via venipuncture and 11,069 were sent volumetric dried blood microsamplers (absorbent polymer, 20-μl collection volume). More than 80% of the microsamplers were returned (9089 participants). Ultimately, 9028 participant blood samples were analyzed using ELISA for the presence of anti–SARS-CoV-2 spike protein antibodies. Of those, 8058 participants had a complete clinical questionnaire and were included in the weighted analysis (Fig. 1). Most blood sample collection (>88%) occurred within the 11-week period between 10 May and 31 July 2020 (figs. S1 and S2). The six major demographic factors used in participant selection are summarized in Table 1. Participant sampling was representative of the U.S. population. When expanded to include the additional 10 demographic or health-related factors captured by the Behavioral Risk Factor Surveillance System (BRFSS), many factors were well matched, but there were some differences, for example, our sample population was more highly educated, had higher employment rates, and had better access to health care compared to the general U.S. population (Table 1).
Fig. 1. SARS-CoV-2 serosurvey study overview and statistical workflow.
A flow chart of participant recruitment through data analysis displays steps in data acquisition and lists participant attrition. Ovals show the start and end of data analysis or data acquisition; gray rectangles indicate subsets of participants in this study; blue parallelograms represent individuals from outside data sets that contributed to adjusted prevalence estimates; blue rounded rectangles present analysis processes.
Table 1. Characteristics of the serosurvey population compared to the U.S. population.
Census and BRFSS (2018) data on selection criteria were used for quota-based sampling in our SARS-CoV-2 serosurvey. Other values from BRFSS were used for statistical weighting. The table shows comparisons between the estimated proportion of the U.S. population in each category according to weighted BRFSS data compared to our sample population in the SARS-CoV-2 serosurvey. NLF, not in the labor force (student, retired, unable to work, refused to answer, not asked/missing).
| U.S. population (BRFSS, census survey) | SARS-CoV-2 serosurvey population | ||||
| n | % |
Weighted
(%) |
n | % | |
| Selection criteria | |||||
| Region | |||||
| North East | 91,307 | 21.19 | 17.6 | 1,508 | 16.7 |
| Midwest | 67,110 | 15.57 | 16.97 | 1,445 | 16.01 |
| Mid-Atlantic | 80,979 | 18.79 | 16.91 | 1,833 | 20.3 |
| South/Central | 60,482 | 14.03 | 15.35 | 1,293 | 14.32 |
| Mountain/Southwest | 86,204 | 20 | 15.89 | 1,392 | 15.42 |
| West/Pacific | 44,866 | 10.41 | 17.27 | 1,557 | 17.25 |
| Age group | |||||
| 18–45 | 125,081 | 28.59 | 46 | 3,837 | 42.51 |
| 45–70 | 207,749 | 47.49 | 39.84 | 3,783 | 41.91 |
| 70–95 | 104,605 | 23.91 | 14.17 | 1,407 | 15.59 |
| Sex | |||||
| Male | 197,411 | 45.24 | 48.66 | 4,318 | 47.83 |
| Female | 238,911 | 54.76 | 51.34 | 4,710 | 52.17 |
| Urban/rural | |||||
| Urban | 365,714 | 84.9 | 93.48 | 8,550 | 94.78 |
| Rural | 65,234 | 15.1 | 6.52 | 471 | 5.22 |
| Race | |||||
| White only | 345,710 | 81 | 73.41 | 6,986 | 77.4 |
| Black only | 37,862 | 8.87 | 12.9 | 830 | 9.2 |
| Others | 43,219 | 10.13 | 13.69 | 1,210 | 13.41 |
| Ethnicity | |||||
| Hispanic | 36,941 | 8.53 | 17.06 | 1,495 | 16.56 |
| Not Hispanic | 395,931 | 91.47 | 82.94 | 7,532 | 83.44 |
| Additional weighting criteria | |||||
| Children | |||||
| Yes | 113,408 | 26.21 | 35.81 | 2,943 | 32.88 |
| No | 319,281 | 73.79 | 64.19 | 6,009 | 67.12 |
| Education | |||||
| <=HS | 151,606 | 34.79 | 41.07 | 240 | 2.68 |
| College | 119,979 | 27.53 | 30.88 | 1,284 | 14.35 |
| >=College | 164,229 | 37.68 | 28.05 | 7,422 | 82.96 |
| Homeowner | |||||
| Own | 305,545 | 70.36 | 66.49 | 6,635 | 74.12 |
| Rent | 107,208 | 24.69 | 27.32 | 1,861 | 20.79 |
| Other | 21,535 | 4.96 | 6.19 | 456 | 5.09 |
| Employment | |||||
| Employed | 219,493 | 50.75 | 57.74 | 6,364 | 71.09 |
| NLF | 174,920 | 40.45 | 31.38 | 2,129 | 23.78 |
| Unemployed | 38,053 | 8.8 | 10.88 | 459 | 5.13 |
| Health insurance | |||||
| Yes | 400,028 | 91.86 | 87.85 | 8,697 | 97.31 |
| No | 35,433 | 8.14 | 12.15 | 240 | 2.69 |
| Flu vaccinated | |||||
| Yes | 234,727 | 59 | 50.62 | 6,198 | 73.73 |
| No | 163,124 | 41 | 49.38 | 2,208 | 26.27 |
| Cardiovascular disease | |||||
| Yes | 52,284 | 12.07 | 9.07 | 354 | 3.98 |
| No | 380,985 | 87.93 | 90.93 | 8,541 | 96.02 |
| Pulmonary disease | |||||
| Yes | 84,102 | 19.33 | 18.53 | 1,671 | 18.96 |
| No | 350,913 | 80.67 | 81.47 | 7,140 | 81.04 |
| Immune disease | |||||
| Yes | 170,115 | 39.14 | 29.29 | 2,039 | 23.1 |
| No | 264,571 | 60.86 | 70.71 | 6,787 | 76.9 |
| Diabetes | |||||
| Yes | 60,703 | 13.9 | 11.41 | 482 | 5.41 |
| No | 375,876 | 86.09 | 88.59 | 8,430 | 94.59 |
Estimates of seroprevalence
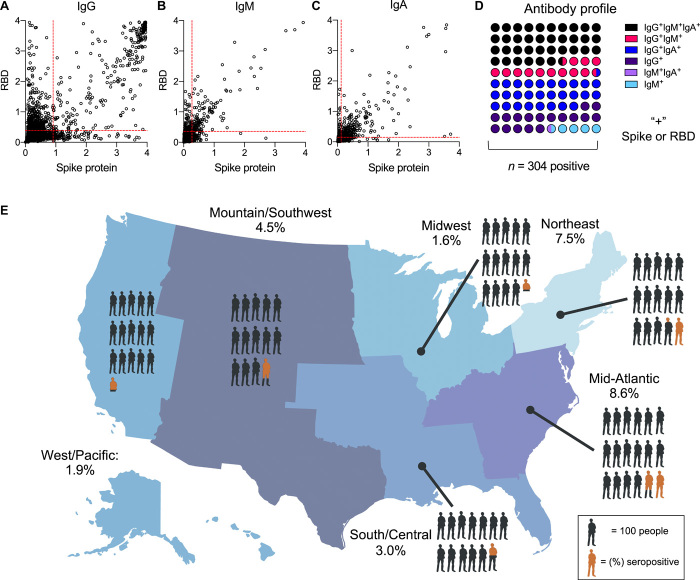
There were 304 seropositive participants in the analysis set (Fig. 2). This gave a weighted estimate of 4.6% of the undiagnosed adults in the U.S. population who were seropositive for SARS-CoV-2 infection [95% confidence interval (CI), 2.6% to 6.5%, n = 8058 complete testing and survey]. Using this average rate over the study period, we estimated that there were 4.8 undiagnosed SARS-CoV-2 infections for each diagnosed case over the course of the study (95% CI, 2.8 to 6.8). Among seropositive participants, 36.51% were IgG+IgM+IgA+, 28.29% were IgG+IgM−IgA+, 17.11% were IgG+IgM−IgA−, 13.16% were IgG+IgM+IgA−, 4.28% were IgG−IgM+IgA−, and 0.66% were IgG−IgM+IgA+ (Fig. 2, A to D, and fig. S3). There were variations in antibody profiles across different demographic groups, specifically anti-spike protein and anti-RBD IgG antibodies (figs. S4 and S5).
Fig. 2. Geographic distribution of undiagnosed seropositivity in the United States from May to July 2020.
Raw serology data for (A) IgG, (B) IgM, and (C) IgA against SARS-CoV-2 spike protein and the receptor binding domain (RBD) of spike protein are displayed. Cut points for positivity are shown as red dashed lines; data are optical density (OD). (D) Serologic phenotype of antibody presence in 304 seropositive participants. (E) The map of the United States displays seropositivity in the six regions surveyed: Northeast: ME, NH, VT, MA, NY, CT, RI, PA, and NJ, 7.5% (95% CI, 3.9% to 12.4%); Midwest: MN, IA, WI, IL, IN, MI, and OH, 1.6% (95% CI, 0.3% to 2.4%); Mid-Atlantic: MD, DE, DC, VA, WV, KY, TN, NC, SC, and GA, 8.6% (95% CI, 2.6% to 18.9%); South/Central: FL, MS, AL, LA, AR, MO, KS, and OK, 3.0% (95% CI, 1.2% to 5.0%); Mountain/Southwest: TX, NM, AZ, CO, UT, WY, NE, SD, ND, MT, and ID, 4.5% (95% CI, 1.3% to 9.5%); West/Pacific: WA, OR, NV, CA, AK, and HI, 1.9% (95% CI, 0.2% to 3.8%). Each person in (E) represents 100 participants; orange represents weighted prevalence estimate within the geographic region.
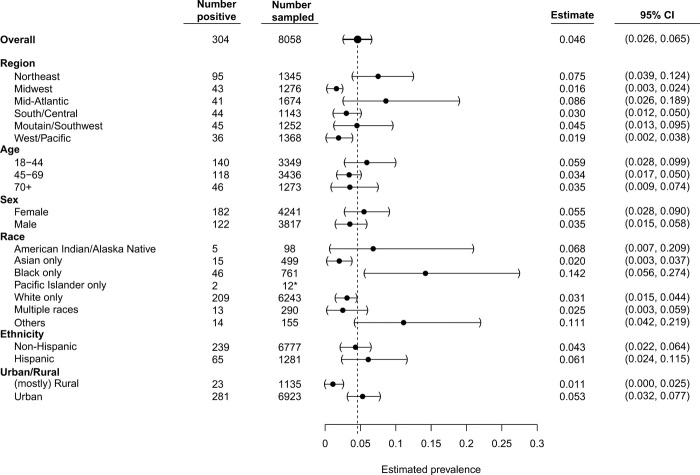
We found regional variations in seroprevalence estimates across the United States (Figs. 2E and 3). The Northeast and Mid-Atlantic regions showed the highest rates of seropositivity, whereas the lowest seropositivity was in the Midwest. Urban areas were estimated to have higher point estimates of seropositivity (5.3%) compared to rural areas (1.1%) at the time blood samples were collected. Estimates of seroprevalence were calculated for other demographic subgroups (Fig. 3). The youngest age group, 18 to 44 years, had the highest estimated seropositivity (5.9%). Estimated seroprevalence for females was 5.5% and was 3.5% for males. The seroprevalence estimate for African Americans was highest at 14.2% followed by participants who self-identified as other/unlisted race (11.1%), American Indian/Alaska Native (6.8%), followed by White/Caucasian (3.1%), whereas those identifying as Asian displayed the lowest seroprevalence estimate (2.0%).
Fig. 3. Undiagnosed SARS-CoV-2 seroprevalence in the main demographic categories.
Six main categories were used during quota-based sampling: region, age, sex, race, ethnicity, and urban/rural. Seropositivity estimates of blood samples that had a full clinical questionnaire completed and successful sampling are shown. Data are weighted estimates ± 95% CIs. Black dashed vertical line, weighted national seroprevalence estimate; *, n value too low to make a proper weighted estimate so raw positivity is displayed.
Participants who reported a known exposure to a SARS-CoV-2–infected individual had a higher seroprevalence estimate (15.6%) compared to those who did not (2.7%). In comparison to the national average (4.6%), those who worked from home had a lower seropositivity estimate of 3.0%. Those who reported previous vaccination (for influenza 3.2% or pneumonia 2.3%) had a lower likelihood of undiagnosed seropositivity. Those who had health conditions associated with poor outcomes in SARS-CoV-2 infection, including coronary heart disease, asthma, and diabetes, displayed lower rates of seropositivity (Fig. 4). Other health conditions were also correlated with a decreased seropositivity rate such as skin cancer, stroke, or arthritis.
Fig. 4. Seroprevalence estimates according to socioeconomic and health characteristics.
(A to C) Evaluation of the effect of nondemographic traits on seroprevalence estimates for blood samples that had a full clinical questionnaire completed and successful sampling. Nondemographic traits included (B) socioeconomic and (C) health characteristics. Data are weighted estimates ± 95% CIs. Gray dashed vertical line, weighted national seroprevalence estimate.
Our results estimate that as of July 2020, there were about 4.79 undiagnosed infections (95% CI, 2.76 to 6.82; fig. S6) for every identified case of COVID-19, suggesting a potential 16.8 million undiagnosed infections by July 2020 in addition to the reported 3.5 million diagnosed cases in the United States. These data suggest that a higher level of infection-induced immunity exists in the U.S. population than previously predicted.
DISCUSSION
These results, including the subgroup analysis, provide us a previously undescribed view into the spread of the COVID-19 pandemic by more clearly identifying the large numbers of individuals with undiagnosed infections during the initial months of the pandemic. These data are of great importance as we consider the impact vaccination may have on the future course of the pandemic and plan for current and future available vaccines to be administered. In addition, these data can also help us to better assess the public health measures taken during the pandemic and how to take the best approaches forward during any future public health emergencies.
This study demonstrates that spread of the SARS-CoV-2 virus in the United States during the first 6 months of the pandemic was more widespread than has been suggested by data reporting diagnostic test-confirmed cases. Similar to responses to other respiratory viruses, such as influenza, many individuals develop asymptomatic or mild disease that is not medically attended and therefore never diagnosed. Our findings indicate that there are nearly five individuals with a previous asymptomatic infection for every diagnosed case of COVID-19. Furthermore, patterns of our seroprevalence data match well with those of diagnosed cases reported during a similar time frame (17). For example, the greater seropositivity estimated in densely populated urban areas follows the observed initial spread of SARS-CoV-2. In comparison to the national average, we found that the Midwest, South, and West had lower seroprevalence rates during the study time frame, which preceded a substantial increase in SARS-CoV-2 infections in these regions detected by viral testing.
Our data suggest that the youngest age group had the highest undiagnosed seroprevalence, which is consistent with observations that they display less severe symptoms than older patients (18). We also found higher undiagnosed seroprevalence in females, possibly suggesting a higher risk for asymptomatic disease. Participants with chronic diseases that are more likely to be associated with severe clinical manifestations of COVID-19, including diabetes, heart disease, and asthma, had a lower prevalence of asymptomatic SARS-CoV-2 infection in comparison to the national average. Those with known exposure to SARS-CoV-2–infected individuals had a higher estimated incidence of undiagnosed seropositivity. We also found that African American and Hispanic participants had higher undiagnosed seropositivity, correlating with national data on disease burden in these subgroups.
Our study reports a representative population sample across the United States and evaluated regional, demographic, and socioeconomic differences in the prevalence of asymptomatic SARS-CoV-2 infection. In contrast, other reports of seroprevalence data focus on specific groups of individuals or geographic locations, such as dialysis patients or individuals who reported for blood draws that may be biased toward those needing medical care during the pandemic (19–36). These previous studies came within the range of our estimate of undiagnosed cases when considering the additional diagnosed cases within the same time frame. Our results provide new insight into the spread of SARS-CoV-2, estimating the national undiagnosed exposure rate to illuminate the scope of infection during the first 6 months of the pandemic. As expected, given delayed arrival in different geographic areas such as the Midwest and rural South, undiagnosed infection estimates varied by region, with the Mid-Atlantic region having the largest proportion of undiagnosed infections in comparison to diagnosed cases. Given the high point estimate of undiagnosed seropositivity in younger participants, lower point estimates in individuals with preexisting conditions such as diabetes, and the vaccine rollout starting with older persons and those at risk, we could see a faster onset of herd immunity due to these undiagnosed infections in populations that are in lower priority groups for vaccination. Young and healthy individuals, such as those under the age of 16 who were not eligible for the first wave of vaccines in the United States and those under 12 who are still ineligible, could serve as an asymptomatic reservoir for viral mutations leading to increased transmissibility or vaccine escape mutations, which has been shown in unvaccinated children and adults with viral persistence (37). Further long-term studies of immunity in the population will be necessary to understand durability of the immune response to the vaccine versus infection, how infection-induced immunity affects vaccine response and performance, and whether herd immunity can play a role in controlling the spread of SARS-CoV-2. In addition, further subgroup analysis of these data will be useful in clarifying the spread of disease in the presence of public health measures and how we may be able to refine and further target those measures in the future.
Our study has several limitations. First, although extensive statistical adjustments were made, our study cohort is based on a nonrandom volunteer sample, which can have selection bias. Traditional random sampling studies using probability sampling design may have low response rates, calling into question the advantages of that practice (38, 39). Our study population also exhibited some differences from the general U.S. population, such as higher education level and access to health care that had to be adjusted for with statistical weighting. Larger sample sizes would allow us to make more detailed estimates, although potentially at the cost of how representative the population is. We used both census and behavioral data to weight our results, although it is possible that there are variables associated with disease transmission that were not accounted for in our weighting. Although we used extensive validation methods on our ELISA (8) for seropositivity designations, we used historical serum samples and convalescent post-infection samples because dried blood was unavailable from historical samples on the collection devices. Future cross-verification with an independent analyte, such as the nucleocapsid protein, could prove useful, although antibodies to nucleocapsid fade and would require correction for antibody decay.
Our data suggest a larger spread of the COVID-19 pandemic in the United States during the first 6 months than originally thought. Our findings have implications for understanding SARS-CoV-2 spread, epidemiological characteristics of spread, and prevalence in different communities and could have a potential impact on decisions involved in vaccine rollout. Continued large-scale surveillance of SARS-CoV-2 immunity is in progress, discriminating infection-based and vaccine-induced antibody responses. Mathematical models are being generated to understand the pandemic, vaccine performance, and public health measure efficacy and to provide insight into the best approach for handling the next virus with pandemic potential.
MATERIALS AND METHODS
Study design
This study was designed to determine the seroprevalence of anti–SARS-CoV-2 antibodies in adults 18 years of age or older in the United States who had not been previously diagnosed with COVID-19. The primary endpoint was the weighted estimate of seroprevalence in the United States. Secondary endpoints were weighted estimates for subgroups categorized by demographics or risk factors. An initial period enrolled a convenience sample of 593 volunteers before the quota sample. Participants across the United States (50 states and District of Columbia) were then enrolled through telephone consent from a pool of volunteers who provided basic demographic data in response to the study announcement. Recruitment calls were made from three sites: National Institute of Allergy and Infectious Diseases (NIAID) Laboratory of Infectious Diseases Clinical Studies Unit, the University of Pittsburgh Clinical and Translational Science Institute (CTSI), and the University of Alabama at Birmingham Center for Clinical and Translational Science (CCTS). The selection of participants is described below. Selected participants were contacted by the study team, consented, and sent a blood microsampling kit and questionnaire in the online REDCap platform (project-redcap.org). For a small subset of participants (n = 214) working on the National Institutes of Health (NIH) campus, serum was collected by venipuncture.
This serosurvey clinical study (ClinicalTrials.gov NCT04334954) is ongoing and will follow the same cohort of participants over time to evaluate seroprevalence and antibody profiles in comparison to the demographic, health, and socioeconomic data provided by each participant. This study was approved by the NIH Institutional Review Board and conducted in accordance with the provisions of the Declaration of Helsinki and Good Clinical Practice guidelines. All participants provided verbal informed consent before enrollment.
Participant selection
The study was advertised online through an official NIH Press Release that linked to an email address to volunteer for selection in the study (www.niaid.nih.gov/news-events/nih-begins-study-quantify-undetected-cases-coronavirus-infection). This press release was subsequently publicized by local and national news outlets and covered via broadcast television news, print news, and internet news articles. All volunteers were emailed an initial survey to collect basic demographic characteristics. Survey responses were de-identified and aggregated by subcategory of state, type of locality approximated from zip codes, age, sex, race, and ethnicity (Fig. 1). Target sample sizes for these subcategories were determined from the U.S. census and were updated every evening based on the characteristics of people who had already enrolled to assure that individuals in each subcategory were enrolled evenly over time. Within each subcategory, participants were initially assigned a selection probability calculated from the target number as a proportion of the available pool. Specific subcategories that had insufficient numbers were aggregated to estimate their impact on the overall distribution of the six main characteristics. If a particular characteristic had insufficient numbers, sample probabilities were boosted for volunteers who had the characteristic. For each day’s call list, the most representative of 20,000 randomly generated lists was used, each list drawn without replacement from the volunteer pool based on the sampling probabilities previously defined. Representativeness was assessed by estimating a weighted sum of squared differences from the desired targets and picking the list with the lowest deviation. Unselected participants were eligible to be called at a later date. This algorithm is designed such that each cohort of invited participants is representative of the diversity of the U.S. population with respect to the six sampling variables (see section S4).
Blood sample collection
Participants provided blood samples by mail using a Mitra microsampling kit (Neoteryx, Torrance, CA) or standard venipuncture. Microsampling kits contained visual instructions on the sampling process, bandages, gauze, lancets, and four 20-μl microsampling devices for a total collection of 80 μl of whole blood. Participants used the lancet to draw blood from their fingertip and collect blood onto each of the four microsamplers. Participants returned the dried microsamplers with desiccant via overnight shipping. Those who underwent venipuncture did so in the NIH Clinical Center phlebotomy laboratory, where 18 ml of blood was collected in a serum separator and whole blood tube. Once received in the laboratory, serum samples were processed, and microsamplers were stored dry at −80°C until elution and analysis.
Serologic assays
Antibodies from samples were analyzed using ELISA as previously described (8, 40–42). To maintain longitudinal quality control and ensure that the assays remained stable across multiple months of assay implementation, positive and negative controls were included on each assay plate and monitored for stability (fig. S7). Seropositivity cut points were defined by evaluating 300 true-negative samples and 56 true-positive samples. Positivity thresholds were based on the mean optical density (absorbance) plus 3 SDs (see the Supplementary Materials for details). The final criterion of a Spike+ and RBD+ for any combination of IgG or IgM gave estimated sensitivity and specificity of 1, with raw values for recombinant antibody results reported in fig. S8 and table S1. In addition, IgA was evaluated via previously described ELISA to further phenotype the participant’s serologic status. Raw sample positivity data by state can be found in fig. S9.
Statistical analysis
The iterative quota sampling (described in the “Participant selection” section) that we used continuously matched the proportion of people in the study with the census estimated proportion of people in the United States on six variables (Table 1 and Fig. 1). This ensured that each periodic sample of participants over the course of the study was representative, and the time effects of the pandemic were approximately independent of those six variables (fig. S2). Each participant was asked demographic and health-related questions that matched those on the BRFSS survey, a large probability-based national survey (43). Responses to those matching questions were used with BRFSS survey data to adjust estimators to account for important criteria that may be related to both selection probability and seropositivity but were not accounted for in our quota sampling. Those adjusted estimators used weighting based on the propensity of being a quota sample versus a BRFSS sample participant and poststratification to U.S. census data. Weighting additionally accounted for sensitivity and specificity. CIs were calculated for the final seroprevalence estimates accounting for both the variability of the weighting and of the sensitivity and specificity adjustment. The ratio of undiagnosed SARS-CoV-2 infections to diagnosed cases of COVID-19 was estimated as the final seroprevalence estimate times a factor calculated from the daily national population and diagnosed cases. Detailed statistical methods are provided in the Supplementary Materials. The main computer code used in this study is available at: https://zenodo.org/record/4958017#.YMkzYpNKh26. Sources used for analysis can be found in (8, 38, 39, 43–56).
Acknowledgments
We thank all of the participants in this study. We acknowledge K. Corbett and B. Graham of the NIAID VRC for their generous donation of coronavirus spike expression plasmid, and A. Schmidt, J. Feldman, B. M. Hauser, and T. M. Caradonna of the Ragon Institute of MGH, MIT, and Harvard for their donation of their RBD expression plasmid. We thank J. Crissey and the CRIMSON team (NIAID, NIH) for their assistance and data management. We thank J. McLellan for scholarly discussions regarding the spike S-2P construct. We thank the CCTS Clinical Research Support Program (A. Delbridge and L. Dukes), UAB School of Public Health and its Survey Research Unit (L. Battle, J. Carson, M. DeRamus, T. Fields, T. Graham, T. Jackson, E. Pruitt, A. Underwood, and P. Wolff), the University of Pittsburgh CTSI staff and leadership (J. Avolio, L. Bash, S. Clayton, M. Cristinziano, K. Daw, C. Fascetti, E. Gyurisin, J. Huwe, N. Jones, D. Mathias, S. Mathias, A. Mykita, B. Petersen, M. Phillips, C. Rush, E. Shepherd, S. Shetty, A. Socci, L. Stearns, S. Ugbomah, K. Underwood, and L. Yasko), and University of Pittsburgh Information Technology (D. McGaughey, S. Ritzman, and T. Smith) for their contributions to this study. We thank D. Bernard for his assistance with sample delivery, and S. Ford-Scheimer for organizational assistance. Funding: This research was supported, in part, by the Intramural Research Program of the NIH, including the National Institute for Biomedical Imaging and Bioengineering, the National Institute of Allergy and Infectious Disease, and the National Center for Advancing Translational Sciences. This project has been funded, in part, with Federal funds from the National Cancer Institute, NIH, under contract number HHSN261200800001E, 75N91019D00024, Task Order No. 75N91019F00130, Clinical and Translational Science Awards Program grants UL1TR003096 (UAB) and UL1TR001857 (University of Pittsburgh). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. The NIH, its officers, and employees do not recommend or endorse any company, product, or service. Author contributions: H.K., C.K.-T., J.H., J.T., K.P., J.S., T.N., K.M.A., M.K., A.S., R.L., and K.S. processed blood samples and performed serology experiments. J.M., M.D., K.R.S., P.F., V.W., J.-P.D., S.M., W.G., and D.E. synthesized antigen constructs and produced protein for serology. H.K., C.K.-T., and K.S. performed seropositivity and quality control analysis on serology data. H.A.B., J.A.C., L.C., O.B., C.C., A.H., L.T.G., L.A.R., R.B., R.A., A.C.-M., M.G., B.H., S.V., R.C., M.K.M., A.K., R.S., S.S., S.R., E.W.F., S.G.M., R.P.K., S.E.R., and M.J.M. recruited and enrolled participants. S.H., M.P.F., N.S., J.W., Y.L., and B.I.G. performed statistical weighting and survey analyses. S.H., M.P.F., R.P.K., S.E.R., M.D.H., M.J.M., and K.S. interpreted seroprevalence results. M.D.H., D.E., M.J.M., S.H., and K.S. designed the study. H.K., C.K.-T., S.H., R.P.K., S.E.R., M.D.H., D.E., M.J.M., and K.S. wrote and edited the manuscript. Competing interests: K.S. and M.J.M. are co-inventors on a provisional U.S. patent application no. 63/092,350 entitled “Antibody specific for SARS-CoV-2 receptor binding domain and therapeutic methods.” The other authors declare no competing interests. Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. Serologic data have been deposited in the NCI SeroNet hub (https://covid19serohub.nih.gov/). Protein and antigen sequences are available in (8). Statistical code is available at: https://zenodo.org/record/4958017#.YMkzYpNKh26. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using this material.
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Fauci A. S., Lane H. C., Redfield R. R., Covid-19—navigating the uncharted. N. Engl. J. Med. 382, 1268–1269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taubenberger J. K., Morens D. M., The pathology of influenza virus infections. Annu. Rev. Pathol. 3, 499–522 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taubenberger J. K., Morens D. M., 1918 influenza: The mother of all pandemics. Rev. Biomed. 17, 69–79 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang A. T., Garcia-Carreras B., Hitchings M. D. T., Yang B., Katzelnick L. C., Rattigan S. M., Borgert B. A., Moreno C. A., Solomon B. D., Rodriguez-Barraquer I., Lessler J., Salje H., Burke D., Wesolowski A., Cummings D. A. T., A systematic review of antibody mediated immunity to coronaviruses: Kinetics, correlates of protection, and association with severity. Nat. Commun. 11, 4704 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., Wang D.-Q., Hu Y., Ren J.-H., Tang N., Xu Y.-Y., Yu L.-H., Mo Z., Gong F., Zhang X.-L., Tian W.-G., Hu L., Zhang X.-X., Xiang J.-L., Du H.-X., Liu H.-W., Lang C.-H., Luo X.-H., Wu S.-B., Cui X.-P., Zhou Z., Zhu M.-M., Wang J., Xue C.-J., Li X.-F., Wang L., Li Z.-J., Wang K., Niu C.-C., Yang Q.-J., Tang X.-J., Zhang Y., Liu X.-M., Li J.-J., Zhang D.-C., Zhang F., Liu P., Yuan J., Li Q., Hu J.-L., Chen J., Huang A.-L., Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 26, 845–848 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Ainsworth M., Andersson M., Auckland K., Baillie J. K., Barnes E., Beer S., Beveridge A., Bibi S., Blackwell L., Borak M., Bown A., Brooks T., Burgess-Brown N. A., Camara S., Catton M., Chau K. K., Christott T., Clutterbuck E., Coker J., Cornall R. J., Cox S., Crawford-Jones D., Crook D. W., D’Arcangelo S., Dejnirattsai W., Dequaire J. M. M., Dimitriadis S., Dingle K. E., Doherty G., Dold C., Dong T., Dunachie S. J., Ebner D., Emmenegger M., Espinosa A., Eyre D. W., Fairhead R., Fassih S., Feehily C., Felle S., Fernandez-Cid A., Fernandez Mendoza M., Foord T. H., Fordwoh T., Fox McKee D., Frater J., Gallardo Sanchez V., Gent N., Georgiou D., Groves C. J., Hallis B., Hammond P. M., Hatch S. B., Harvala H. J., Hill J., Hoosdally S. J., Horsington B., Howarth A., James T., Jeffery K., Jones E., Justice A., Karpe F., Kavanagh J., Kim D. S., Kirton R., Klenerman P., Knight J. C., Koukouflis L., Kwok A., Leuschner U., Levin R., Linder A., Lockett T., Lumley S. F., Marinou S., Marsden B. D., Martinez J., Martins Ferreira L., Mason L., Matthews P. C., Mentzer A. J., Mobbs A., Mongkolsapaya J., Morrow J., Mukhopadhyay S. M. M., Neville M. J., Oakley S., Oliveira M., Otter A., Paddon K., Pascoe J., Peng Y., Perez E., Perumal P. K., Peto T. E. A., Pickford H., Ploeg R. J., Pollard A. J., Richardson A., Ritter T. G., Roberts D. J., Rodger G., Rollier C. S., Rowe C., Rudkin J. K., Screaton G., Semple M. G., Sienkiewicz A., Silva-Reyes L., Skelly D. T., Sobrino Diaz A., Stafford L., Stockdale L., Stoesser N., Street T., Stuart D. I., Sweed A., Taylor A., Thraves H., Tsang H. P., Verheul M. K., Vipond R., Walker T. M., Wareing S., Warren Y., Wells C., Wilson C., Withycombe K., Young R. K., Performance characteristics of five immunoassays for SARS-CoV-2: A head-to-head benchmark comparison. Lancet Infect. Dis. 20, 1390–1400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G. A., Tan J., Bhavsar D., Capuano C., Kirkpatrick E., Meade P., Brito R. N., Teo C., Mahon M. M., Simon V., Krammer F., SARS-CoV-2 seroconversion in humans: A detailed protocol for a serological assay, antigen production, and test setup. Curr. Protoc. Microbiol. 57, e100 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klumpp-Thomas C., Kalish H., Drew M., Hunsberger S., Snead K., Fay M. P., Mehalko J., Shunmugavel A., Wall V., Frank P., Denson J. P., Hong M., Gulten G., Messing S., Hicks J., Michael S., Gillette W., Hall M. D., Memoli M. J., Esposito D., Sadtler K., Standardization of ELISA protocols for serosurveys of the SARS-CoV-2 pandemic using clinical and at-home blood sampling. Nat. Commun. 12, 113 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. M. Dan, J. Mateus, Y. Kato, K. M. Hastie, C. E. Faliti, S. I. Ramirez, A. Frazier, E. D. Yu, A. Grifoni, S. A. Rawlings, B. Peters, F. Krammer, V. Simon, E. O. Saphire, D. M. Smith, D. Weiskopf, A. Sette, S. Crotty, Immunological memory to SARS-CoV-2 assessed for greater than six months after infection. bioRxiv 2020.11.15.383323 [Preprint]. 16 November 2020. 10.1101/2020.11.15.383323. [DOI]
- 10.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang J., Li Y., Song X., Chen Y., Xia L., Fu L., Hou L., Xu J., Yu C., Li J., Zhou Q., Chen W., A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 369, 650–655 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., Jiang S., Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: Implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 324, 773–781 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L., Wang P., Nair M. S., Yu J., Rapp M., Wang Q., Luo Y., Chan J. F. W., Sahi V., Figueroa A., Guo X. V., Cerutti G., Bimela J., Gorman J., Zhou T., Chen Z., Yuen K. Y., Kwong P. D., Sodroski J. G., Yin M. T., Sheng Z., Huang Y., Shapiro L., Ho D. D., Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Pinto D., Park Y.-J., Beltramello M., Walls A. C., Tortorici M. A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J. B., Chen R. E., Havenar-Daughton C., Snell G., Telenti A., Virgin H. W., Lanzavecchia A., Diamond M. S., Fink K., Veesler D., Corti D., Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Shi R., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W. J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.-S., Wang Q., Gao G. F., Yuan Z., Yan J., A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature 584, 120–124 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Z. Wang, J. C. C. Lorenzi, F. Muecksch, S. Finkin, C. Viant, C. Gaebler, M. Cipolla, H.-H. Hoffman, T. Y. Oliveira, D. A. Oren, V. Ramos, L. Nogueira, E. Michailidis, D. F. Robbiani, A. Gazumyan, C. M. Rice, T. Hatziioannou, P. D. Bieniasz, M. Caskey, M. C. Nussenzweig, Enhanced SARS-CoV-2 neutralization by secretory IgA in vitro. bioRxiv 2020.09.09.288555 [Preprint]. 9 September 2020. 10.1101/2020.09.09.288555. [DOI]
- 16.Hicks J., Klumpp-Thomas C., Kalish H., Shunmugavel A., Mehalko J., Denson J.-P., Snead K. R., Drew M., Corbett K. S., Graham B. S., Hall M. D., Memoli M. J., Esposito D., Sadtler K., Serologic cross-reactivity of SARS-CoV-2 with endemic and seasonal betacoronaviruses. J. Clin. Immunol. , 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong E., Du H., Gardner L., An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533–534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehra M. R., Desai S. S., Kuy S., Henry T. D., Patel A. N., Cardiovascular disease, drug therapy, and mortality in Covid-19. N. Engl. J. Med. 382, e102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Caini S., Bellerba F., Corso F., Díaz-Basabe A., Natoli G., Paget J., Facciotti F., de Angelis S. P., Raimondi S., Palli D., Mazzarella L., Pelicci P. G., Vineis P., Gandini S., Meta-analysis of diagnostic performance of serological tests for SARS-CoV-2 antibodies up to 25 April 2020 and public health implications. Euro Surveill. 25, 2000980 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kontou P. I., Braliou G. G., Dimou N. L., Nikolopoulos G., Bagos P. G., Antibody tests in detecting SARS-CoV-2 infection: A meta-analysis. Diagnostics 10, 319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C. Y.-P., Lin R. T. P., Renia L., Ng L. F. P., Serological approaches for COVID-19: Epidemiologic perspective on surveillance and control. Front. Immunol. 11, 879 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bastos M. L., Tavaziva G., Abidi S. K., Campbell J. R., Haraoui L., Johnston J. C., Lan Z., Law S., MacLean E., Trajman A., Menzies D., Benedetti A., Khan F. A., Diagnostic accuracy of serological tests for covid-19: Systematic review and meta-analysis. BMJ 370, m2516 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biggs H. M., Harris J. B., Breakwell L., Dahlgren F. S., Abedi G. R., Szablewski C. M., Drobeniuc J., Bustamante N. D., Almendares O., Schnall A. H., Gilani Z., Smith T., Gieraltowski L., Johnson J. A., Bajema K. L., David K. M., Schafer I. J., Sullivan V., Punkova L., Tejada-Strop A., Amiling R., Mattison C. P., Cortese M. M., Ford S. E., Paxton L. A., Drenzek C., Tate J. E.; CDC Field Surveyor Team , Estimated community seroprevalence of SARS-CoV-2 antibodies—Two Georgia counties, April 28–May 3, 2020. Morb. Mortal. Wkly. Rep. 69, 965–970 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryan A., Pepper G., Wener M. H., Fink S. L., Morishima C., Chaudhary A., Jerome K. R., Mathias P. C., Greninger A. L., Performance characteristics of the Abbott Architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J. Clin. Microbiol. 58, e00941-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.M. L. Dietrich, E. B. Norton, D. Elliott, A. R. Smira, J. A. Rouelle, N. G. Bond, K. Aime-Marcelin, A. Prystowsky, R. Kemnitz, A. Sarma, S. T. Himmelfarb, N. Sharma, A. E. Stone, R. Craver, A. R. Lindrose, L. A. Smitley, R. B. Uddo, L. Myers, S. S. Drury, J. S. Schieffelin, J. E. Robinson, K. J. Zwezdaryk, SARS-CoV-2 seroprevalence rates of children in Louisiana during the state stay at home order. medRxiv 2020.07.07.20147884 (2020).
- 26.Feehan A. K., Fort D., Garcia-Diaz J., Price-Haywood E. G., Velasco C., Sapp E., Pevey D., Seoane L., Seroprevalence of SARS-CoV-2 and infection fatality ratio, Orleans and Jefferson Parishes, Louisiana, USA, May 2020. Emerg. Infect. Dis. 26, 2765–2768 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Havers F. P., Reed C., Lim T., Montgomery J. M., Klena J. D., Hall A. J., Fry A. M., Cannon D. L., Chiang C.-F., Gibbons A., Krapiunaya I., Morales-Betoulle M., Roguski K., Rasheed M. A. U., Freeman B., Lester S., Mills L., Carroll D. S., Owen S. M., Johnson J. A., Semenova V., Blackmore C., Blog D., Chai S. J., Dunn A., Hand J., Jain S., Lindquist S., Lynfield R., Pritchard S., Sokol T., Sosa L., Turabelidze G., Watkins S. M., Wiesman J., Williams R. W., Yendell S., Schiffer J., Thornburg N. J., Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, March 23-May 12, 2020. JAMA Intern. Med. 180, 1576–1586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.C. C. McLaughlin, M. K. Doll, K. T. Morrison, W. L. Mc Laughlin, T. O’Connor, A. M. Sholukh, E. L. Bossard, K. Phasouk, E. S. Ford, K. Diem, A. M. Klock, K. R. Jerome, L. Corey, High community SARS-CoV-2 antibody seroprevalence in a ski resort community, Blaine County, Idaho, US. Preliminary results. medRxiv 2020.07.19.20157198 (2020).
- 29.Menachemi N., Yiannoutsos C. T., Dixon B. E., Duszynski T. J., Fadel W. F., Wools-Kaloustian K. K., Needleman N. U., Box K., Caine V., Norwood C., Weaver L., Halverson P. K., Population point prevalence of SARS-CoV-2 infection based on a statewide random sample—Indiana, April 25–29, 2020. Morb. Mortal. Wkly. Rep. 69, 960–964 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng D. L., Goldgof G. M., Shy B. R., Levine A. G., Balcerek J., Bapat S. P., Prostko J., Rodgers M., Coller K., Pearce S., Franz S., Du L., Stone M., Pillai S. K., Sotomayor-Gonzalez A., Servellita V., Sanchez San Martin C., Granados A., Glasner D. R., Han L. M., Truong K., Akagi N., Nguyen D. N., Neumann N. M., Qazi D., Hsu E., Gu W., Santos Y. A., Custer B., Green V., Williamson P., Hills N. K., Lu C. M., Whitman J. D., Stramer S. L., Wang C., Reyes K., Hakim J. M. C., Sujishi K., Alazzeh F., Pham L., Thornborrow E., Oon C.-Y., Miller S., Kurtz T., Simmons G., Hackett J. Jr., Busch M. P., Chiu C. Y., SARS-CoV-2 seroprevalence and neutralizing activity in donor and patient blood. Nat. Commun. 11, 4668 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberg E. S., Tesoriero J. M., Rosenthal E. M., Chung R., Barranco M. A., Styer L. M., Parker M. M., John Leung S. Y., Morne J. E., Greene D., Holtgrave D. R., Hoefer D., Kumar J., Udo T., Hutton B., Zucker H. A., Cumulative incidence and diagnosis of SARS-CoV-2 infection in New York. Ann. Epidemiol. 48, 23–29.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sood N., Simon P., Ebner P., Eichner D., Reynolds J., Bendavid E., Bhattacharya J., Seroprevalence of SARS-CoV-2–specific antibodies among adults in Los Angeles County, California, on April 10-11, 2020. JAMA 323, 2425–2427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutton M., Cieslak P., Linder M., Notes from the field: Seroprevalence estimates of SARS-CoV-2 infection in convenience sample—Oregon, May 11–June 15, 2020. Morb. Mortal. Wkly. Rep. 69, 1100–1101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anand S., Montez-Rath M., Han J., Bozeman J., Kerschmann R., Beyer P., Parsonnet J., Chertow G. M., Prevalence of SARS-CoV-2 antibodies in a large nationwide sample of patients on dialysis in the USA: A cross-sectional study. Lancet 396, 1335–1344 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bajema K. L., Wiegand R. E., Cuffe K., Patel S. V., Iachan R., Lim T., Lee A., Moyse D., Havers F. P., Harding L., Fry A. M., Hall A. J., Martin K., Biel M., Deng Y., Meyer W. A. III, Mathur M., Kyle T., Gundlapalli A. V., Thornburg N. J., Petersen L. R., Edens C., Estimated SARS-CoV-2 seroprevalence in the US as of September 2020. JAMA Intern. Med. 181, 450–460 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rostami A., Sepidarkish M., Leeflang M. M. G., Riahi S. M., Shiadeh M. N., Esfandyari S., Mokdad A. H., Hotez P. J., Gasser R. B., SARS-CoV-2 seroprevalence worldwide: A systematic review and meta-analysis. Clin. Microbiol. Infect. 27, 331–340 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Truong T. T., Ryutov A., Pandey U., Yee R., Goldberg L., Bhojwani D., Aguayo-Hiraldo P., Pinsky B. A., Pekosz A., Shen L., Boyd S. D., Wirz O. F., Röltgen K., Bootwalla M., Maglinte D. T., Ostrow D., Ruble D., Han J. H., Biegel J. A., Li M., Huang C. H., Sahoo M. K., Pannaraj P. S., O’Gorman M., Judkins A. R., Gai X., Bard J. D., Increased viral variants in children and young adults with impaired humoral immunity and persistent SARS-CoV-2 infection: A consecutive case series. EBioMedicine 67, 103355 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elliott M. R., Valliant R., Inference for nonprobability samples. Stat. Sci. 32, 249–264 (2017). [Google Scholar]
- 39.D. Hill, “The dirty little secret pollsters need to own up to,” Washington Post, 2020.
- 40.Hicks J., Klumpp-Thomas C., Kalish H., Shunmugavel A., Mehalko J., Denson J.-P., Snead K., Drew M., Corbett K., Graham B., Hall M. D., Memoli M. J., Esposito D., Sadtler K., Serologic cross-reactivity of SARS-CoV-2 with endemic and seasonal Betacoronaviruses. J. Clin. Immunol. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esposito D., Mehalko J., Drew M., Snead K., Wall V., Taylor T., Frank P., Denson J.-P., Hong M., Gulten G., Sadtler K., Messing S., Gillette W., Optimizing high-yield production of SARS-CoV-2 soluble spike trimers for serology assays. Protein Expr. Purif. 174, 105686 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehalko J., Drew M., Snead K., Denson J.-P., Wall V., Taylor T., Sadtler K., Messing S., Gillette W., Esposito D., Improved production of SARS-CoV-2 spike receptor-binding domain (RBD) for serology assays. Protein Expr. Purif. 179, 105802 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Centers for Disease Control and Prevention (CDC) (2018). 2018 BRFSS Questionnaire. Available at https://www.cdc.gov/brfss/questionnaires/index.htm.
- 44.C. C. Chow, J. C. Chang, R. C. Gerkin, S. Vattikuti, Global prediction of unreported SARS-CoV2 infection from observed COVID-19 cases. medRxiv 2020.04.29.20083485 (2020).
- 45.Fay M. P., Proschan M. A., Brittain E., Combining one-sample confidence procedures for inference in the two-sample case. Biometrics 71, 146–156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ingram D. D., Franco S. J., NCHS urban-rural classification scheme for counties. Vital Health Stat. , 1–65 (2012). [PubMed] [Google Scholar]
- 47.Klumpp-Thomas C., Kalish H., Hicks J., Mehalko J., Drew M., Memoli M. J., Hall M. D., Esposito D., Sadtler K., D614G Spike variant does not alter IgG, IgM, or IgA spike seroassay performance. J. Infect. Dis. 223, 802–804 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korn E. L., Graubard B. I., Confidence intervals for proportions with small expected number of positive counts estimated from survey data. Surv. Methodol. 24, 193–201 (1998). [Google Scholar]
- 49.Leeb R. T., Price S., Sliwa S., Kimball A., Szucs L., Caruso E., Godfred-Cato S., Lozier M., COVID-19 trends among school-aged children—United States, March 1–September 19, 2020. Morb. Mortal. Wkly. Rep. 69, 1410–1415 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lumley T., Analysis of complex survey samples. J. Stat. Softw. 9, 1–19 (2004). [Google Scholar]
- 51.Rogan W. J., Gladen B., Estimating prevalence from the results of a screening test. Am. J. Epidemiol. 107, 71–76 (1978). [DOI] [PubMed] [Google Scholar]
- 52.United States Census Bureau (2020) downloaded files: Data counts by county, age, race, ethnicity (as of 2018; accessed April 16, 2020). FIPS Codes (accessed April 16, 2020). Urban/rural codes (as of the 2010 census; accessed April 15, 2020). U.S. and World Population Clock (accessed on December 12, 2020). Estimates of the Total Resident Population and Resident Population Age 18 Years and Older for the United States, Regions, States, and the District of Columbia: July 1, 2020; (accessed on March 25, 2021).
- 53.Valliant R., Poststratification and conditional variance estimation. J. Am. Stat. Assoc. 88, 89–96 (1993). [Google Scholar]
- 54.Valliant R., Dever J. A., Estimating propensity adjustments for volunteer web surveys. Sociol. Methods Res. 40, 105–137 (2011). [Google Scholar]
- 55.L. Wang, B. I. Graubard, H. A. Katki, Y. Li, Efficient and robust propensity-score-based methods for population inference using epidemiologic cohorts. arXiv:2011.14850 [stat.ME] (30 November 2020).
- 56.L. Wang, R. Valliant, Y. Li, Adjusted logistic propensity weighting methods for population inference using nonprobability volunteer-based epidemiologic cohorts. arXiv:2007.02476 [stat.AP] (6 July 2020). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.