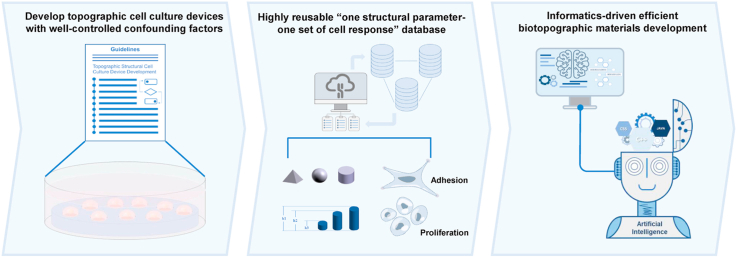
Abstract
Micro/nano topographic structures have shown great utility in many biomedical areas including cell therapies, tissue engineering, and implantable devices. Computer-assisted informatics methods hold great promise for the design of topographic structures with targeted properties for a specific medical application. To benefit from these methods, researchers and engineers require a highly reusable “one structural parameter – one set of cell responses” database. However, existing confounding factors in topographic cell culture devices seriously impede the acquisition of this kind of data. Through carefully dissecting the confounding factors and their possible reasons for emergence, we developed corresponding guideline requirements for topographic cell culture device development to remove or control the influence of such factors. Based on these requirements, we then suggested potential strategies to meet them. In this work, we also experimentally demonstrated a topographic cell culture device with controlled confounding factors based on these guideline requirements and corresponding strategies. A “guideline for the development of topographic cell culture devices” was summarized to instruct researchers to develop topographic cell culture devices with the confounding factors removed or well controlled. This guideline aims to promote the establishment of a highly reusable “one structural parameter – one set of cell responses” database that could facilitate the application of informatics methods, such as artificial intelligence, in the rational design of future biotopographic structures with high efficacy.
Keywords: Biotopographic materials, Cell culture device, Confounding factor, Database, Informatics
Graphical abstract
1. Introduction
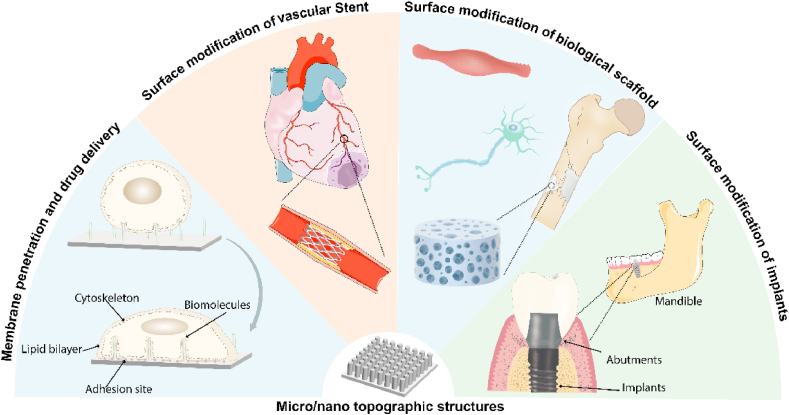
Micro/nano topographic structures have extensive applications in many medical fields, such as oral implants, orthopedic implants, and cardiovascular stents (Fig. 1) [[1], [2], [3]]. The underlying mechanisms are very complicated, including changing the conformation of adsorbed proteins, modulating the focal adhesion activity and cytoskeleton dynamics, etc [4,5]. So far empirical “Trial and Error” is the most widely adopted approach to develop micro/nano topographic structures that can trigger desired cell responses [6]. In this manner, scientists fabricated many micro/nano topographic structures and then tested their influences on cell responses one by one to screen out an ideal candidate. Although this approach has made a significant contribution to the development of biomaterials, serious problems such as low efficiency still exist. Informatics-driven methods such as quantitative structure-activity relationships (QSAR) have been proposed to address this issue [7]. QSAR is a method to find the quantitative mathematical relationship (often represented as “biological response = function (property1, property2, property3 …)” between biological responses and physicochemical properties (e.g., shape, dimension, density, stiffness, and functional groups) through data mining based on a large-scale experimental dataset. With informatics methods, the biological responses of numerous complicated micro/nano topographic structures with different structural parameters can be rapidly predicted, and a selected set of highly promising candidates can be efficiently screened out to be tested in the laboratory for following application in real medical devices.
Fig. 1.
Wide applications of biomicro/nano topographic structures in medical fields. Micro/nano topographic structures have an extensive application in many medical fields, such as dentistry, orthopedics, cardiovascular, neurology, gastroenterology, hepatobiliary surgery, tumor therapy, etc.
Despite the promising prospects of informatics methods, the lack of suitable high-quality databases significantly impedes their application [8]. To establish high-quality databases of micro/nano topographic cell responses, sufficient experimental data linking “one structural parameter” with “one set of cell responses” (named “micro/nano topographic cell responses”) should be extracted and integrated from published literature. However, contrary to the rapidly increasing number of published papers, the reusability of micro/nano topographic structures to guide cell responses seems low. For example, Oh et al. reported that nanotubes with 70 nm–100 nm diameters showed better osteoblastic differentiation than nanotubes with smaller diameters [9]. While Park et al. found opposite results: nanotubes approximately 15 nm in diameter were more favorable for osteoblastic differentiation than larger tubes [10]. Prineas et al. noted that few studies on nanotoxicology provide consistent results due to insufficient nanomaterial characterizations and poorly defined experimental conditions [11]. Low reusability has made it extremely difficult to establish a high-quality database and seriously reduces the prediction capacity and accuracy of the informatics-driven methods.
The highly variable reporting components of experimental results is one of the reasons for the low reusability, and Minimum Information Reporting in Bio-Nano Experimental Literature (MIRIBEL) has been proposed to solve this issue [12]. However, even with MIRIBEL, reusability is still unsatisfactory because there are many confounding factors involved in the establishment and application of topographic cell culture devices. For example, autoclaving, which is the most widely used sterilization method in biological researches, was reported to probably cause surface alteration of titanium nanotubes [13]; thus, the cell responses tested after sterilization actually cannot be precisely linked to the structural parameters characterized before sterilization. In addition, it was reported that some substrate materials for generating topographic structures may be partially degraded by mRNA or protein extraction reagents, which would cause degradation of experimental samples [14] and then affect the accuracy of the gene expression or protein synthesis results.
Therefore, it is necessary to carefully control or remove confounding factors when developing and applying topographic cell culture devices for the establishment of a highly reusable “one structural parameter – one set of cell responses” database. Many potential confounding factors need to be taken into consideration. After careful dissection, we found that these factors were derived from three major processes: 1) structure generation, 2) cell culture, and 3) cell response analysis (Table 1). It then came to us that a corresponding guideline based on removing these three types of confounding factors may need to be proposed to guide researchers to correctly achieve this purpose.
Table 1.
Confounding factors existing in topographic cell culture devices that impede the establishment of high-quality topographic cell responses database.
| Steps | Confounding factors derived from cell culture device | Example |
|---|---|---|
| Structure Generation | 1. Structural inconsistency in a surface | Random topographic structures generated by surface roughening [37]. |
| 2. Inconsistent physical properties | Inconsistent stiffness between control and topographies [38]. | |
| 3. Additional bioactive chemicals | Fluorine residual on the topographic structure after hydrofluoric acid treatment [39]. | |
| Cell Culture Application | 1. Physicochemical instability during sterilization | Autoclaving sterilization process probably causes alteration of surface properties of titanium nanotube [13]. |
| 2. Physicochemical instability during immersion | During immersion, the topographies on biodegradable materials, such as PLGA, might deform due to swelling or degradation [40]. | |
| 3. Moveable or non-fixable during immersion | Due to the low density, some cell culture devices, such as collagen membranes, cannot keep immobility in the culture medium. | |
| 4. Cytotoxicity | Many toxic reagents might be introduced during the fabrication process, such as the etching process in the photolithography method. | |
| Cell Response Analysis | 1. Disturbed signal transmission and collection during the morphological assay | The opaqueness of some metal-made cell culture devices hinders the cell density and cell status assessment before cell response detection. |
| 2. Non-fixable during cell adhesion force assay | During hydrodynamic shear assay by shear force [41], the force may not be normally applied to cells on non-fixable substrates. | |
| 3. Structural change during wound-healing assay | During wound-healing assay, physical scratch using pipette tip [28] might destroy the topographic structures. | |
| 4. Interference with transmittance during absorbance test | Some topographic substrates change the transmission of visible light [[42], [43]], thus interfering with the detection of absorbance in CCK-8 or MTT assays. | |
| 5. Interference with mRNA or protein extraction | The topographic substrates may react to the mRNA or protein extraction reagent, thus causing the degradation of samples [14]. | |
| 6. Interference with the relative quantification of mRNA or protein expression | Topographic substrates can regulate cytoskeletal dynamic [31] so that may influence the expression of some housekeeping genes, such as β-actin or α-tubulin, of which the constant expression in other cases was helpful to correct sample loading deviation in RT-qPCR or WB. |
Developing biotopographic cell culture devices with controlled confounding factors should become a preliminary requirement before initiating micro/nano topographic cell response researches. It is the authors’ ambition that the proposed guideline can greatly improve the reusability of “one structural parameter – one set of cell responses” research outcomes. After that, a high-quality database could be established through data extraction and integration, and informatics-driven methods such as QSAR can be applied in the efficient prediction and rational design of biomicro/nano topographic structures, effectively facilitating the development of advanced topographic biomaterials with shorter research cycles, lower costs and ultimately wide applicability.
In this work, the classification and detailed reasons for the emergence of confounding factors were first carefully dissected, and the corresponding guideline requirements for topographic cell culture device development were proposed to remove or control them. Based on these requirements, we then suggested potential strategies to meet them. We attempted to develop a topographic cell culture device with controlled confounding factors based on these guideline requirements and corresponding strategies. During practice, some strategies were confirmed to be effective, while others were proven to require further optimization. After careful optimization, we successfully developed a topographic cell culture device meeting most of the guideline requirements, thus controlling or removing the unwanted confounding factors.
2. Materials and methods
Fabrication and characterization of topographic structures with well-defined parameters: Replica molding and polydimethylsiloxane (PDMS) were chosen as fabrication method and substrate material respectively. Briefly, the templates were thoroughly cleaned in milli-Q H2O and ethanol by ultrasonic cleaning. The PDMS kit (Dow Corning, USA) was mixed in the mass ratio of 10:1 (base: crosslinker) with mechanical stirring, which was then spread on the templates under vacuum (−0.1 MPa) for 30min. After being cured in the oven for 1 h at 100 °C, solidified PDMS films with defined topographic structures were peeled off from the templates gently. To improve the resolution of topographic structures, n-hexane was added at different volume ratios to decrease the viscosity of PDMS. The geometric morphologies of generated topographic structures were characterized by scanning electron microscope (SEM, SU8220, Hitachi, Japan).
Assessment of chemical properties and structure-independent differences in physical properties: The chemical composition was measured through X-ray photoelectron spectroscopy (XPS, ESCALab250, Thermo Fisher Scientific, USA), and the chemical bond and functional group distribution were measured through Fourier transform infrared spectroscopy analyzer (FTIR, Nicolet/Nexus 670, Thermo Fisher Scientific, USA). The surface stiffness was measured by atomic force microscopy (AFM, Dimension FastScan, Bruker, Germany). To identify the experimental conditions that affect surface stiffness, different ratios of coupling agent (5:1, 10:1, 15:1, 20:1, 25:1) and different heating temperatures (80 °C, 100 °C, 120 °C, 140 °C, 160 °C) were applied to synthesize the substrates.
Assessing the geometric and physicochemical stability of generated topographic structures during sterilization conditions: In ethanol sterilization, substrates were immersed in 75% ethanol for 30min then dried in air at room temperature; In UV sterilization, substrates were treated with UV light for 30min (254 nm, 100 μW/cm2) in a laboratory hood (about 50 cm from UV source). For autoclaving, substrates were placed in an autoclave at 121 °C for 30min. In γ-ray sterilization, substrates were exposed to γ-ray irradiation at 75 kGy with a dose rate of 10 kGy/h. After sterilization processes, the geometric morphologies, surface stiffness, chemical properties of sterilized and unsterilized topographic structures were characterized respectively by SEM, AFM, XPS, and FTIR.
Assessing the geometric and physicochemical stability of generated topographic structures under immersion environment: PDMS substrates were immersed in Dulbecco's Modified Eagle Medium (DMEM, Thermo Fisher Scientific, USA) or deionized water under a humidified environment (37 °C, 5% CO2) for 0, 7, 14, 21, 28 days. Then the geometric morphologies, surface stiffness, chemical properties of soaked and unsoaked topographic structures were further characterized as aforementioned.
Assessing the immobility of the cell culture devices during immersion: PDMS substrates were adhered to the cell culture plate and immersed in DMEM under a humidified environment (37 °C, 5%CO2) for 1, 3, 5, 7 days. Then the interfaces between the substrates and the bottom of the cell culture plate were observed. The side view pictures were taken by a camera (D610, Nikon, Japan).
Assessing the structure-independent cytotoxicity: After being sterilized, the cell culture devices were immersed by DMEM supplied with 10% fetal bovine serum (FBS, Thermo Fisher Scientific, USA) and 1% penicillin/streptomycin (Thermo Fisher Scientific, USA) for 24 h. Raw264.7 cells were seeded on a 96-well culture plate at a density of 2000 and cultured with the collected immersion medium, then incubated in a humidified environment (37 °C, 5%CO2). After 1, 3, and 5 days, cell viability was detected by a CCK-8 assay.
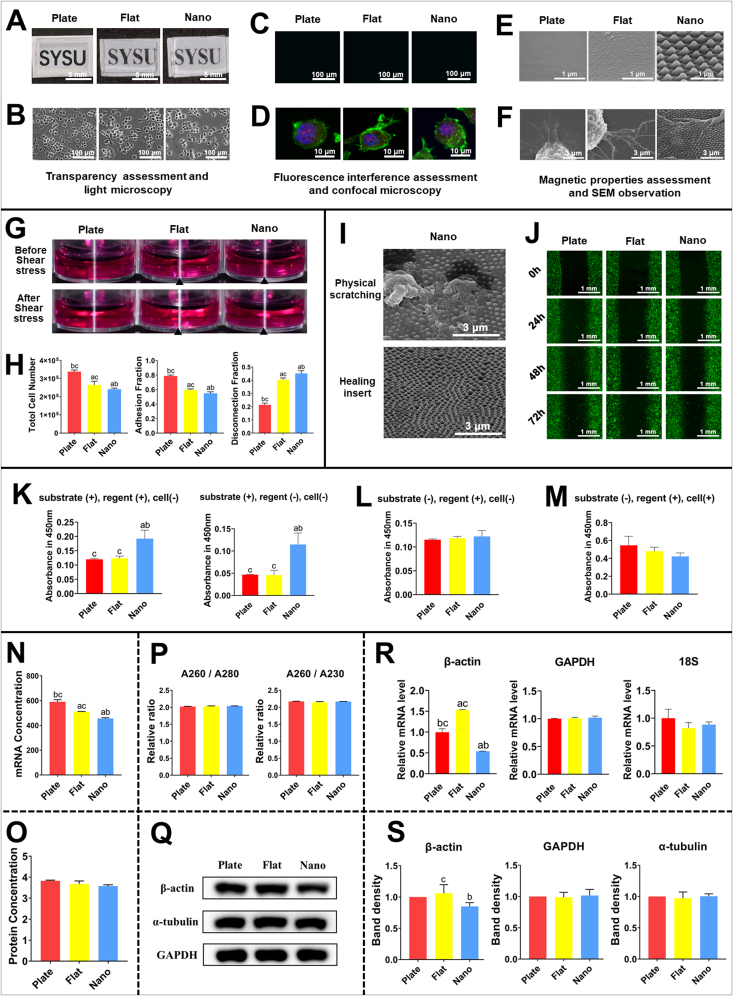
Transparency, fluorescence interference, magnetic property assessment, and cell morphology evaluation: The transparency of PDMS substrates were recorded by a camera. The magnetic property was assessed by direct SEM observation, XPS, and FTIR evaluation. Fluorescence interference was assessed by visualizing PDMS substrates under different fluorescence channels. Then Raw264.7 cells were seeded on the substrates at a density of 104. Cell density and cell status were observed by light microscopy (Axio vert.A1, Zeiss, Germany). Subcellular structures, including nuclear, cytoskeleton, and focal complex were observed by confocal microscopy (FV3000, Olympus, Japan) through visualizing DAPI (Blue, Beyotime, China), Actin-Tracker (Green, Beyotime, China), and Vinculin (Red, Abcam, USA). The direct interaction between the cells and the topographic structures was observed by SEM.
Immobility assessment during shear stress and hydrodynamic flow assay: DMEM was added into each well of the 12-well plate with or without substrates. Then the plate was placed on an Orbital Shaker (Essenscien, USA) at 200 rpm/min for 5 min, and the interfaces were observed as aforementioned. After confirming the immobility during shear stress, Raw264.7 cells were seeded on the substrates and cultured for 30 min at 37 °C with 5%CO2, then the plate was placed on the oscillator at 200 rpm/min for 5 min. The supernatant was collected for further calculation of detached cells. The residual cells were scraped off to calculate the adhered cells. The cell number was detected by a cellometer (Nexcelom Bioscience, USA).
Geometric stability assessment after wound preparation and wound-healing assay: To assess the geometric stability after wound preparation, Raw264.7 cells were seeded onto the substrates and incubated at 37 °C to obtain a confluent monolayer. Wounds were created on the surfaces by scratching with a 200 μL pipette tip and were observed by SEM. To avoid the geometric change after physical scratching, healing inserts were used to prepare the wounds, and the geometric stability was testified by SEM. After that, the wound-healing process was captured at 0, 24, 48, and 72 h by a fluorescence microscope.
Confounding absorbance assessment and CCK-8 assay: To assess whether the substrates would introduce confounding absorbance, the sterilized substrates were adhered to the bottom of the 96-well plates. Detection reagents were added to each well and incubated at 37 °C with 5%CO2 for 1h at dark. Then the absorbance at 450 nm was measured by a microplate reader (BioTek, USA). To remove the confounding absorbance, the reacted solution was transferred to a new 96-well plate and was measured again under the same condition. After that, Raw264.7 cells were seeded on the substrates with a density of 2000 and incubated for 24h, and a CCK-8 assay was performed in this way.
Assessment of mRNA quality and housekeeping genes expression: Total mRNA was extracted by mRNA-Quick Purification Kit (ES Science, China). The quantity and quality of the extracted mRNA were analyzed by a spectrophotometer (NanoDrop One, Thermo Fisher Scientific, USA). The expression of commonly used housekeeping genes (GAPDH, 18-S, β-actin, primers were outlined in Tab. S1) was assessed by RT-qPCR. The melting curves were recorded to further assess the quality of samples. CT (threshold cycle) values for each sample were recorded to detect the expression of housekeeping genes.
Protein quality and housekeeping proteins expression assessment: The cells were lysed by the mixture of RIPA (CWBIO, China) and Protease inhibitor cocktail (CWBIO, China) for 30 min at low temperature. The protein concentration was subsequently measured by BCA protein quantitative kit (CWBIO, China). The expression of commonly used housekeeping proteins was detected by western blot (α-tubulin 1: 5000; β-actin 1:1000; GAPDH 1:50000). The semi-quantitative analysis was carried out as previously described [15].
Statistical analysis: Data were analyzed using GraphPad Prism 7 (GraphPad Software, USA). Results were expressed in the form of mean ± standard deviation (SD). Statistical analyses were performed according to one-way ANOVA. Statistical significance was confirmed at P < 0.05.
3. Results and discussion
3.1. Removing and controlling confounding factors derived from structure generation
Ideally, we would expect the generated topographic structures to possess only one variant parameter, i.e. structure difference, so that we can study the cell responses toward the pure structural parameter effect without any side interference factors. However, the structural features are sometimes poorly controlled because the size and shape controllability of some fabrication methods are unsatisfactory. For example, the surface roughening methods cannot well control the size and shape of topographic structures, so the random topographies generated by these methods have multiple variable geometric parameters, and we cannot precisely link the cell responses to “one structural parameter”. In addition, the chosen substrate materials are sometimes not compatible with the chosen fabrication method. For example, when choosing substrate materials with low mechanical strength, such as hydrogels, for the model transfer methods, topographic structures cannot maintain integrity during mold release. Well-controlled structural features thus become the first requirement of topographic devices.
However, it is almost impossible to achieve only one variant parameter due to the substrate materials having their physicochemical properties. Hence, it is needed to carefully manage the physicochemical properties of the substrates to avoid interference when studying topographic cell responses. Bioactive molecules can elicit significant effects on cell responses no matter in planar or topographic surfaces [16], which may lead to some side effects, affecting the investigation of sole topographic structures-mediated cell responses. Hence, we would expect that the surface chemistry is biologically inert and consistent among different micro/nano structures so that it will bring minimal side effects on cell responses when investigating the topographic structures-mediated cell responses.
For physical properties (stiffness wettability, roughness, charge, etc.), the structure-independent differences and structure-dependent differences should be distinguished. The former ones are not elicited by the topographic structures, and will trigger confounding cell responses, for which should be kept consistent. The latter ones are elicited by the topographic structures, it is part of the mechanism by which the micro/nano topographies regulate cell responses, and it is difficult and inappropriate to eliminate these differences.
Therefore, to control this type of confounding factor, we need to generate micro/nano topographic structures with well-controlled “one structural parameter”, achieve biologically inertness and consistent surface chemistry, and ensure minimum structure-independent differences of physical properties.
-
1)
How to achieve a well-controlled “one structural parameter”?
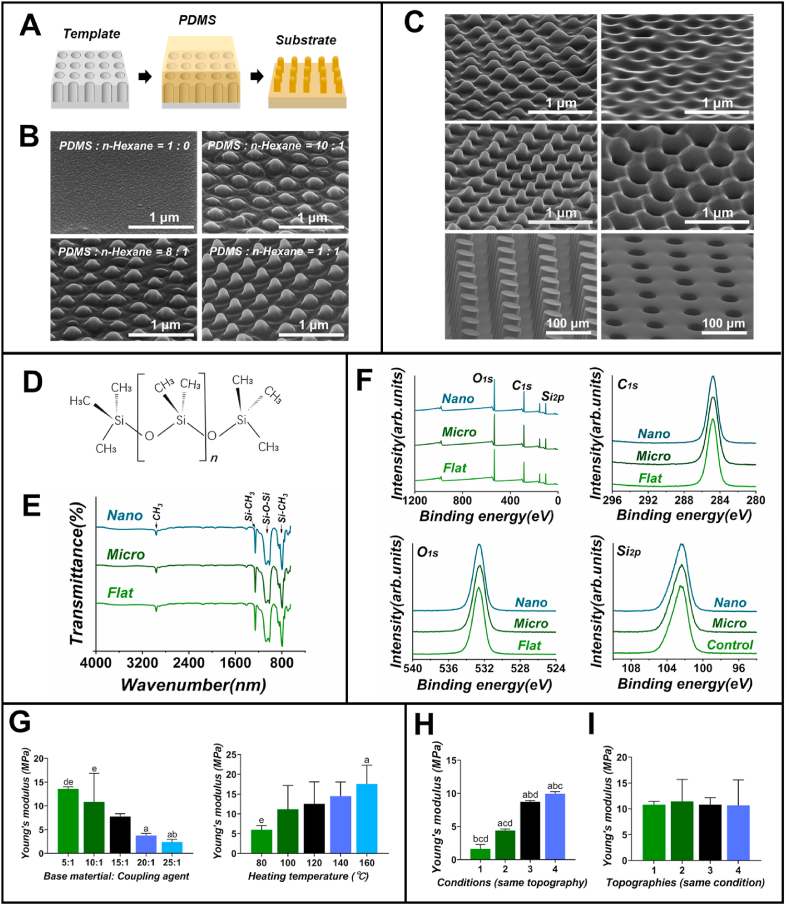
To achieve this aim, researchers should first choose an appropriate fabrication method, which has been thoroughly discussed in a review paper [17]. Many fabrication or synthesis methods have been reported to generate micro/nano topographic surfaces. Generally, those methods can be divided into top-down methods including photolithography, electron beam and X-ray lithography, reactive ion etching (RIE), chemical etching, phase separation and anodization, and bottom-up methods ranging from colloidal lithography, electrospinning to pattern transfer (nanoimprinting and replica molding). The fabrication methods should be chosen in accordance with the desired surface geometries. For example, photolithography, electron beam and X-ray lithography, and pattern transfer are suitable for generating surfaces with regular micro/nano-patterns, including gratings, grooves, pillars, and nanopit arrays [[18], [19], [20], [21]], while electrospinning and phase separation are more preferred when generating fibers in nanoscale [17]. To achieve one structural parameter, fabrication accuracy and resolution should be relatively high to avoid extra or not-intended topographies. In addition, to fabricate numbers of different well-controlled topographic surfaces and perform various biological characterizations, throughput should also be focused. Replica molding is the method that can both generate various topographic surfaces and possess high shape and size controllability, as well as good throughput [22]; therefore it well meets the mentioned requirements. Thus, the replica molding (Fig. 2A) method was chosen to generate topographic structures.
Fig. 2.
Removing or controlling confounding factors derived from structure generation. (A-C) Achieve a well-controlled “one structural parameter” difference: (A) Schematic of generating topographic structures choosing replica molding as method and PDMS as substrate material. (B) SEM images of nanostructures generated by PDMS diluted with n-hexane in different volume ratios, and (C) different micro/nano topographic structures generated after optimizing the viscosity of PDMS. (D-E) Ensure consistent chemical properties: (D) Chemical formula of PDMS. (E) FTIR showed the chemical bonds of generated topographic structures. (F) XPS showed the chemical composition and structure of generated topographic structures. (G-I) Achieve consistent structure-independent physical properties: (G) Young's modulus of PDMS in different experimental conditions. (H) Young's modulus of the same topographic structure that prepared under different conditions. (I) Young's modulus of different topographic structures that prepared under the same condition.
Next, a substrate material that is compatible with the chosen method should be selected. For replica molding, the substrate material should finely fit into the mold in the liquid state, be able to transform into a solid substrate, and can be easily released from the mold. PDMS possesses sufficient elasticity, thermal curability, mechanical strength, and superior mold-releasing properties [23]. It can thus finely fit the mold and form a solid topographic substrate, and maintain structural integrity during mold release, which is compatible with the replica molding method. Hence, we chose PDMS as the substrate material to generate topographic structures.
However, after choosing an appropriate fabrication method and compatible substrate material, a well-controlled structural parameter still cannot be achieved (Fig. 2B). We speculated that this might be due to the high viscosity of PDMS, which might impedes its deformation. Therefore, we diluted PDMS with n-hexane at different dilution ratios to reduce the viscosity, and the results showed that the viscosity decreased significantly with the increase in n-hexane in PDMS (Tab. S2). The SEM images confirmed that diluting PDMS with n-hexane can effectively improve the resolution of the generated nano-structures (Fig. 2B). In this way, we successfully prepared a variety of micro/nano topographies with high resolution that meet the requirements of cell response testing (Fig. 2C).
Therefore, after determining the appropriate method and material for generating topographic structures, researchers should also optimize the technique details and material parameters to improve the controllability of feature parameters.
-
2)
How to ensure consistent chemical properties?
To further ensure minimum confounding chemical properties, biologically inert substrate materials should be chosen, and the additional chemicals used during the fabrication process should be thoroughly removed. Although bioactive components, such as protein, ions, or functional groups, are powerful for enhancing biological performance, they are not recommended to be used for this application as the chemical signals can trigger compounding cell responses.
PDMS is a chemically and biologically inert substrate material (Fig. 2D); thus, choosing this material and excluding any bioactive molecule coating is the first consideration to avoid chemical confounding factors. The n-hexane used during the fabrication process may form a residue that can impede the chemical consistency among the generated topographic structures, but it was expected that high-temperature conditions during PDMS curing can promote the volatilization of n-hexane. To determine whether n-hexane can be thoroughly removed under this condition, we utilized gas chromatography. The results showed that there were no n-hexane peaks in our test samples (Fig. S1), indicating that n-hexane was completely removed from the substrates. To further assess the chemical properties, FTIR and XPS characterizations were applied. The FTIR spectra of the three different topographies showed similar peaks representing the chemical groups of “Si–CH3”, “Si–O–Si”, and “-CH3” (Fig. 2E). XPS spectra also showed similar peaks representing the elements of “O1s”, “C1s”, and “Si2p” (Fig. 2F). The atomic ratios of C, O, and Si were also similar in different topographic structures (Fig. S2A). They collectively indicated that the generated topographic structures were only composed of pristine, biologically inert PDMS without other chemical impurities, and they were chemically consistent.
-
3)
How to ensure minimum structure-independent differences of physical properties?
It could have more than one type of confounding physical properties (stiffness, wettability, roughness, and charge, etc.), depending on the type of topographic structure and the way it is fabricated. All these properties may influence cell responses, which should theoretically be completely controlled or removed. Surface stiffness is easily influenced in the chosen replica molding method, we thus took stiffness as an example here to show the process of identifying and unifying the factors that can affect physical properties.
During the generation of micro/nano structures through replica molding using PDMS, we first assessed the effect of experimental details such as heating temperature, and the ratio of coupling agent on the surface stiffness of PDMS. The results showed that the heating temperature and ratio of the coupling agent significantly affected Young's modulus of PDMS (Fig. 2G). The same topographic structure generated by PDMS under different conditions showed a distinct Young's modulus (Fig. 2H). These results indicated that the difference in the heating temperature and the ratio of the coupling agent would cause inconsistent surface stiffness and thus should be carefully controlled during structure generation. After finely unifying these experimental details, the generated substrates with different topographic structures showed similar Young's modulus (Fig. 2I), indicating that we successfully achieved consistent surface stiffness among the generated micro/nano topographic structures.
In summary, we found that inaccurate topographic structures, bioactive or inconsistent surface chemistry, and structure-independent differences in physical properties are the common confounding factors during the structure generation stage. The application of the replica molding method and PDMS substrate seemed to be feasible for controlling these confounding factors, after optimizing the substrate material parameters and technique details. It should be noted that stiffness is not the only side physical property. Many more confounding physical properties may come up in different types of topographic structures or fabrication methods. Researchers can try to manage all of them by referring to the way we control stiffness in this study. Other methods, substrate materials, or strategies can also be utilized if they meet the mentioned guideline requirements.
3.2. Controlling or removing the confounding factors derived from topographic structures in cell culture
To prepare for cell culture studies, topographic structures should undergo sterilization and immersion environments and be co-cultured with cells. The common confounding factors include alterations of topographic structures or their physicochemical properties during sterilization or immersion, uncontrollable contact status between cells and topographic structures due to the floating of substrate material upon immersion, and the cytotoxicity of substrates. Hence, the topographic structures should be able to maintain geometric and physicochemical stability during sterilization and immersion. Further, immobility during immersion also needs to be taken into consideration. For cytotoxicity, structure-related cytotoxicity and structure-independent cytotoxicity should be distinguished. Structure-related cytotoxicity is part of the entire cell response triggered by topographic structures, which does not need to be monitored. Structure-independent cytotoxicity is mainly derived from toxic chemical components, which damage normal cell responses toward topographic structures and thus should be assessed and eliminated. Collectively, to control confounding factors during the cell culture process, the generated micro/nanostructures should act as immobile cell culture devices with geometric and physicochemical stability during sterilization and immersion and no structure-independent cytotoxicity.
-
1)
How to achieve geometric and physicochemical stability during the sterilization process?
Widely used sterilization methods include autoclaving (AU), ultraviolet irradiation (UV) or γ-ray irradiation (γ-ray), and ethanol immersion (EI), etc. Different sterilizing methods may elicit distinct effects on the model topographic structures, resulting in the damage of structural signals. Therefore, we need to test the geometric and physicochemical stability of candidate substrate materials under the chosen sterilization methods before the formal experiments and determine the appropriate sterilization method.
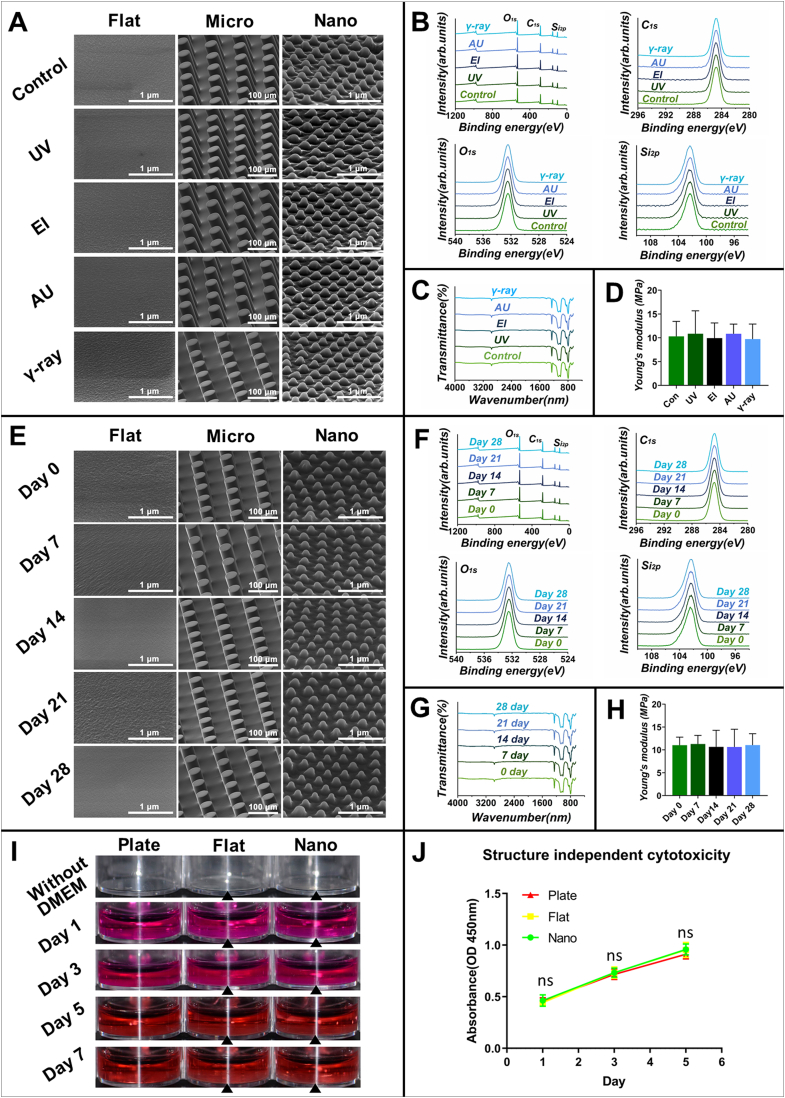
After glass transition, PDMS possesses superior thermostability and irradiation stability and is also stable in ethanol. Hence, we hypothesize that the micro/nano structures generated by PDMS can maintain geometric and physicochemical stability during commonly used sterilization methods. We then performed EI, UV, AU, and γ-ray treatment of the PDMS substrates and characterized their geometric morphologies, stiffness, and chemical properties. Compared with unsterilized substrates, the geometric morphologies of the substrates disinfected by the four methods were unaltered (Fig. 3A). Both the XPS and infraed spectra of the substrates disinfected by the four sterilization methods were the same as those of unsterilized substrates (Fig. 3B and C), and the atomic ratios were also similar in different sterilization groups (Fig. S2B). In addition, Young's modulus of substrates disinfected by the four sterilization methods was similar to that of unsterilized substrates (Fig. 3D). These results indicated that generated PDMS substrate material-based topographic structures can keep geometric and physicochemical stability during different sterilization processes.
-
2)
How to achieve geometric and physicochemical stability during immersion?
Fig. 3.
Controlling or removing the confounding factors derived from micro/nano topographic cell culture. (A-D) Achieve geometric and physicochemical stability during sterilization process: (A) SEM images, (B) XPS spectra, (C) FTIR spectra, and (D) Young's modulus of generated micro/nano topographic structures before and after sterilized by ethanol (EI), ultraviolet radiation (UV), autoclaving (AU), gamma-irradiation (γ-ray), respectively. (E-H) Ensure geometrical and physicochemical stability during immersion: (E) SEM images, (F) XPS spectra, (G) FTIR spectra, and (H) Young's modulus of generated micro/nano topographic structures immersed in deionized water for 0, 7, 14, 21, 28 days. (I) Achieve immobility during immersion: pictures of the generated cell culture devices immersed in DMEM for 1, 3, 5, 7 days, respectively. (J) Avoid structure independent toxicity: cell viability when cultured with the immersion medium of different cell culture devices assessed by CCK-8.
To maintain geometric and physicochemical stability during immersion, the swellability and degradability of substrate materials should be carefully considered. PDMS is chemically inert and does not swell or degrade when in contact with water; therefore, we hypothesized that it can maintain geometric and physicochemical stability during immersion. To confirm this assumption, we immersed the PDMS substrates with nanotopography in Dulbecco's Modified Eagle Medium (DMEM) and then characterized their geometric and physicochemical stability. Interestingly, we found that the geometric morphology changed gradually over time (Fig. S3), which was different from our assumption. This was probably due to the organic substances in DMEM adsorbing to the surface, which is one of the structure-dependent effects but interfered with assessing the structural change mediated by swelling or degradation. This indicated that DMEM might be not appropriate for using as the assessment medium when evaluating the stability of topographic structures during immersion, and another assessment medium should be selected to avoid this problem.
It has been reported that the mineral salt in PBS can adsorb to the surface of some substrate materials, such as magnesium [24] and nacre [25], so that it may cover the topographic structures. Hence, we thought that deionized water would be the most suitable candidate for the assessment medium because it can avoid solution components adsorbing to topographic structures so that the geometric change mediated by swelling or degradation can be assessed without interference. Thus, the geometric and physicochemical stability of PDMS substrates was tested by immersion in deionized water. The results showed that the geometric morphologies (Fig. 3E), chemical properties (Fig. 3F and G; Fig. S2C), and surface stiffness (Fig. 3H) of different substrates were stable during 28 days of soaking.
Therefore, in addition to carefully choosing non-swellable and non-degradable substrate materials, researchers should also select an appropriate assessment medium when evaluating the geometric and physicochemical stability during immersion. Generally, deionized water is recommended as the assessment medium.
-
3)
How to achieve immobility during immersion?
To maintain immobility during immersion, it is recommended to choose a substrate material with a relatively high density. The density of PDMS is approximately 965 kg/m³, lower than the density of water and culture medium, so PDMS substrates may not maintain immobility during immersion. To verify this assumption, we added DMEM to the culture plate and different substrates for 1, 3, 5, and 7 days. Interestingly, the results showed that the substrates still adhered well to the bottom of the cell culture plate (Fig. 3I). This indicated that the cell culture devices generated by PDMS can maintain immobility during immersion. This is because of the binding force between PDMS substrates and the culture plate, which can finely compensate for its relatively low density.
Therefore, high density is recommended but is not a necessity when choosing substrate materials. For substrate materials with a relatively lower density, such as some synthetic or natural polymers, there should be other retention mechanisms, such as adhesive ability or artificial retention design, to maintain the immobility of the cell culture devices during immersion.
-
4)
How to avoid structure-independent cytotoxicity?
To avoid structure-independent cytotoxicity, the cell culture devices should not contain toxic chemical substances, including chemical residues or toxic substances released during degradation.
The PDMS substrate is biologically inert, without chemical coating, and n-hexane was thoroughly removed during the fabrication process. Additionally, PDMS is not degradable in the cell culture medium. Hence, it is anticipated that the cell culture devices generated by PDMS do not exhibit structure-independent cytotoxicity during the cell culture process. To further confirm this assumption, the cell culture devices with different topographic structures were immersed in the cell culture medium to collect the soaking medium, which contains the releasable components of the substrates. Then cells were co-cultured with the soaking medium, and the cell vitality was evaluated. The results demonstrated that the cell vitality levels in the three groups on days 1, 3, and 5 were similar (Fig. 3J), which confirmed that the generated topographic structure did not exhibit structure-independent cytotoxicity and can be used for cell culture without introducing confounding factors.
Collectively, we determined that the common confounding factors in the cell culture application stage include the geometric or physiochemical alteration during sterilization and immersion, floating of substrate material, and structure-independent cytotoxicity. Our results indicated that the generated topographic surfaces are free from these confounding factors. It should be noted that we only examined some common confounding factors, while the confounding factors derived from the sterilization and immersion processes could be many more. Researchers should try to eliminate or control all of them following their project requirements.
3.3. Controlling or removing the confounding factors derived from topographic cell response analysis
Cell response analysis is the core step in determining cellular responses to surface topographic structures. In this process, cell responses such as morphology, adhesion, migration, proliferation, and differentiation are assessed by different experimental methods. Ideally, we would expect that the detection processes would be carried out without the participation of the substrate material to minimize confounding factors. However, many experimental methods must be carried out directly on the substrate material, such as cell morphology observation through microscopy, cell adhesion evaluation, and migration evaluation. Hence, for all cell response analysis methods of interest, researchers should carefully analyze the possible confounding factors and then try to remove or control them.
To show the process of identifying and controlling confounding factors in different cell response evaluations, we need a model cell for further investigation. Immune cells can actively interact with the topographic bio-interfaces and regulate the outcome and long-term performance of the implanted biomaterials. In this interaction, macrophages are the main effector among them due to their high plasticity. Raw 264.7 is a widely used cell line of macrophage [26,27]. Therefore, we selected raw 264.7 cells as the model cells.
-
1)
Controlling or removing confounding factors during cell morphology evaluation.
Cell morphology evaluation includes observing the overall cell density and cell status, cell shape, and subcellular structures. To comprehensively assess the cell morphology toward biomicro/nano topographic structures, different experimental methods can be used, such as optical microscopy, confocal microscopy, and scanning electron microscopy. Because the cell morphology is adhesion-dependent, they cannot be detected without the existence of cell culture devices. Generally, the devices should not disturb signal transmission and collection during cell morphology evaluation.
Optical microscopy provides a convenient way to observe the overall cell density and cell status in real-time before formal cell response analysis, which is important for cell responses toward biomicro/nano topographic structures. Because most of the optical microscopes equipped in the laboratory are transmission light microscopes, it is better for the cell culture devices to be transparent. It is thus recommended to choose substrate materials with high translucency. PDMS is a substrate material with high translucency (Fig. 4A), so we can conveniently observe the overall cell density and cell status with the direct existence of substrates (Fig. 4B).
Fig. 4.
Controlling or removing the confounding factors derived from micro/nano topographic structural cell response detection. (A-F) Ensuring precise cell morphology evaluation: (A) Translucency assessment. (B) Observe cell density and status without confounding factors through optical microscopy. (C) Fluorescence interference assessment. (D) Observe subcellular structures without confounding factors. (E) Magnetic properties assessment. (F) Observe the interaction of cell and topographic structure without confounding factors. (G-H) Ensuring precise cell adhesion affinity evaluation: (G) Pictures of the generated cell culture devices after undergoing shear stress. (H) Evaluating cell adhesion affinity without confounding factors. (I-J) Ensuring precise cell migration evaluation: (I) Assessing the integrity of topographic structures after preparing wound by pipette tip scratching or healing inserts through SEM. (J) Evaluating cell migration without confounding factors. (K-M) Ensuring precise cell proliferation evaluation: (K–L) The influence of topographic structures on absorbance with or without the detection reagents of CCK-8 assay, and removing the confounding absorbance by detecting without substrate materials. (M) Evaluating cell proliferation without confounding factors by the optimized protocol. (N–S) Ensuring precise gene expression and protein synthesis evaluation: Assessing the concentration (N, O) and the quality (P, Q) of the mRNA or the protein extracted from the cells cultured on the substrate materials. (R, S) Evaluating the expression of housekeeping molecules in RT-qPCR and WB assays.
Confocal fluorescence microscopy can greatly improve the resolution and contrast of traditional microscopy, which makes it a great tool for studying cell shape and subcellular structure observation. In this method, subcellular structures are observed by recording the fluorescence emitted from an object. It is thus recommended to choose substrate materials without autofluorescence or the capacity to adsorb fluorescence molecules. PDMS does not have autofluorescence and will not swell or adsorb fluorescent molecules (Fig. 4C); thus, we successfully observed cell shape and subcellular structures such as filopodia and cytoskeleton, without any interference (Fig. 4D).
Scanning electron microscopy is a useful tool to study the direct interaction between cells and micro/nano topographic structures because of its nanoscale resolution. Magnetic components (such as Fe, Co, and Ni) would seriously disturb the observation through SEM. Hence, it is recommended to choose substrate materials without magnetic components. PDMS contains no magnetic components (Fig. 4E); thus, we successfully observed the direct interaction between the cell and the geometric topographic structures through SEM without interference (Fig. 4F).
-
2)
Controlling or removing confounding factors during cell adhesion affinity evaluation.
Hydrodynamic flow assay is a widely used method to assess cell adhesion affinity. In this assay, the cell culture devices should undergo shear stress. Stress may not be normally applied to cells when the cell culture devices cannot maintain immobility under shear stress. Thus, the selected substrate material should possess sufficient immobility to undergo the hydrodynamic flow assay.
As mentioned above, PDMS can maintain immobility by adhering to culture plates. However, whether it is enough to undergo shear stress is unknown. We examined the immobility of the cell culture devices under the condition of hydrodynamic flow assay. The results showed that the cell culture devices would not float into the culture medium (Fig. 4G), indicating that they meet the requirement of cell adhesion evaluation. Then, cells were seeded into the devices to formally carry out the hydrodynamic flow assay, and the total cell number, adhesion fraction, and disconnection fraction were successfully acquired without interference (Fig. 4H), which can indicate the cell adhesion affinity to the generated topographic structures.
-
3)
Controlling or removing confounding factors during cell migration evaluation.
Wound-healing assays are widely applied to evaluate the cell migration response toward micro/nano structures. In this assay, a wound is prepared after the cells form a confluent monolayer to expose the pristine topographic structures and then observe the cell migration response. To ensure that the acquired cell migration response is precisely linked to the designed topographic structures, the topographic structures should be geometrically stable after wound preparation.
Despite the convenience of preparing wounds by scratching using a pipette tip [28], the changes in topographic structures after scratching were often neglected. To assess the geometric stability of our cell culture devices in this process, the topographic structures after wound preparation by physical scratching were observed through SEM. The results showed that the structures changed significantly (Fig. 4I), with cell debris seemingly covering the topography. This indicated that physical scratching would cause confounding factors in the wound-healing assay. Alternatively, healing inserts are available for generating wounds without physical scratching [29,30], which occupy the position of the designed wound during cell seeding and are removed after cells confluent. We thus used healing inserts to generate wounds and then assessed the geometric stability. It can be seen that the geometric topography did not change (Fig. 4I), indicating that the above confounding factor had been effectively removed. Then, a wound-healing assay was successfully conducted without confounding factors (Fig. 4J).
Therefore, when evaluating the cell migration response toward micro/nano topographies by wound-healing assay, it is recommended to prepare the wound using healing inserts rather than by physical scratching.
-
4)
Controlling or removing confounding factors during cell proliferation evaluation.
Cell proliferation is usually evaluated by CCK-8 or MTT assays by measuring the absorbance of the detection reagent after interacting with cell metabolic enzymes. In this process, the detection reagent and cell culture devices are all located between the optical transmitter and the optical receiver and are transmitted by the light of a defined wavelength. Thus, the effect of topographic substrates on absorbance should be carefully assessed.
We carried out a CCK-8 assay on substrates without cells. In this case, the absorbance should be similar among different groups. However, the cell culture devices with nanotopography showed an increased absorbance (Fig. 4K). There were no cells on the devices, implying the increased absorbance was probably derived from the devices themselves. Thus, we further investigated the absorbance of the topographic cell culture devices themselves without the reagents of the CCK-8 assay. The results showed a similar trend (Fig. 4K), which verified that the increased absorbance was caused by the topographic substrate. These results indicated that topographic substrates may absorb the detecting light, which might becomes a confounding factor for the absorbance assays.
To remove this confounding factor, the protocol should be optimized. We transferred the detection reagent mentioned above to a new cell culture plate without substrates to detect the absorbance, and the results showed that the absorbance was similar in different groups (Fig 4L), indicating that confounding factors were successfully eliminated. After that, cells were seeded directly onto the cell culture devices, and the direct cell proliferation response on different topographic structures was evaluated by the optimized protocol (Fig 4M).
Hence, in absorbance assays, such as evaluating cell proliferation through CCK-8 or MTT assays, it is recommended to detect the absorbance without substrate materials after the reaction between the cell and the detection reagent.
-
5)
Controlling or removing confounding factors while evaluating the expression of genes or proteins.
RT-qPCR (Real-time quantitative polymerase chain reaction) and WB (Western blot) are the most widely used methods to assess the expression of genes and proteins, respectively. However, topographic substrates may react to mRNA or protein extraction reagents, thus causing sample degradation [14]. In addition, it was reported that topographic structures can regulate cytoskeletal dynamics [31] and may influence the expression of some cytoskeleton-related housekeeping molecules, of which the constant expression in other cases is utilized to correct sample loading deviation in RT-qPCR or WB assays. These confounding factors will impede the reusability of the results. Hence, during RT-qPCR and WB assays, the substrates should not affect sample quality and the expression of internal references. To meet these requirements, the chosen substrate material should not be obviously dissolved by the lysis reagents, and it is necessary to choose the stably expressed internal references.
PDMS was reported to possess low solubility [32] in many organic solvents, such as phenol, which is the main component of the lysis reagents. We thus hypothesized that it will not be obviously degraded by the lysis reagents and affect the sample quality. To verify this assumption, we extracted mRNA and protein samples from the cells cultured on our cell culture devices and characterized their quality, the results showed that sufficient mRNA and protein could be extracted (Fig 4N, 4O). The absorbance ratio of mRNA at 260 nm/280 nm was approximately 2.0, while that at 260 nm/230 nm was approximately 2.05 (Fig 4P). When RT-qPCR was performed using the extracted mRNA, the melting curves of housekeeping genes (GAPDH, β-actin, 18S) were similar in each group (Fig. S4). In addition, when WB was performed using the extracted protein, the protein bands were clean and sharp (Fig 4Q). These results indicated that the extracted samples were integrated and not contaminated by impurities.
To choose the stably expressed internal references, the expression of housekeeping molecules on different topographic structures should be assessed before formal experiments. We examined the expression levels of commonly used housekeeping genes (GAPDH, β-actin, 18S) and proteins (GAPDH, α-tubulin, β-actin) on PDMS substrates with different topographic structures. The results showed that the expression level of β-actin varied among groups (Fig 4R, 4S), suggesting that it is a confounding factor when choosing β-actin as the internal reference in this situation. However, the expression of GAPDH, 18S, and α-tubulin was stable in each group (Fig 4R, 4S). Thus, β-actin is not recommended as an internal reference when assessing gene and protein expression on topographic structures.
In this section, we revealed several confounding factors in different cell response assays, including geometric alteration during physical scratching in wound-healing assay, increased absorbance of the topographic substrate in absorbance assay, and unstably-expressed β-actin on different topographic surfaces in RT-qPCR and WB assays. Through preparing wounds with healing inserts, detecting the absorbance without the participation of substrate material, and choosing the stably-expressed internal reference, we successfully controlled these confounding factors. Notably, there are a large number of methods for analyzing cell responses. The management of confounding factors should be carried out following our guideline, when other cell response detecting methods are performed.
4. Summary and perspective
As mentioned in the Introduction section of this paper, the development of biomicro/nano topographic structures is about to enter a new era, in which scientists can efficiently predict the cell responses towards topographic structures using informatics. Before this can happen, the establishment of a high-quality database is a prerequisite. Sets of highly reusable data consistent with the “one structural parameter – one set of cell responses” are needed to establish this type of database. However, many published results are not suitable for this intention due to confounding factors.
From the point of controlling confounding factors, this work is to establishing guidelines to instruct the development and application of topographic cell culture devices. After summarizing the aforementioned requirements, we proposed the “Guideline for the development of topographic cell culture devices with controlled or removed confounding factors” (Table 2). It clearly defines the criteria for topographic cell culture devices with controlled confounding factors. The strategies for meeting these guideline requirements are also included, which will guide researchers to fabricate qualified devices with controlled confounding factors. In addition, they can also instruct researchers to correctly apply the generated devices in cell culture and analysis processes to avoid confounding factors.
Table 2.
Guideline for the development of topographic cell culture devices with controlled or removed confounding factors.
| Stage | Requirements | Strategies | Notes |
|---|---|---|---|
| Structure generation related assessments | “One structural parameter” difference | 1. Select a fabrication method with high accuracy, high throughput in accordance with the desired surface geometry | Replica molding may be a feasible method, others can also be selected if meeting these requirements |
| 2. Select an appropriate substrate material that compatible with the chosen method | Material selection should meet other requirements | ||
| 3. Optimize the technique details and material parameters | The details need to be optimized vary with methods | ||
| Minimum and consistent chemical properties | 1. Choose a biologically inert material | Because the chemical molecules-induced cellular effects will interfere with investigating the sole topographic structures-mediated cell responses | |
| 2. Avoid bioactive coatings | |||
| 3. Thoroughly remove the chemical residues | |||
| Consistent structure-independent differences in physical properties | 1. Unify the fabrication conditions to control the surface stiffness | Its main affecting factors may be various in different methods | |
| 2. Other types of confounding physical properties (wettability, roughness, and charge, etc.) should also be managed by referring to the way we control stiffness. | |||
| Cell culture related assessments | Geometrically and physicochemically stable under sterilization | Choose a material that is stable under sterilization condition, and select its compatible sterilization method | Stability in high temperature, irradiation, and ethanol, etc. should be considerate |
| Geometrically and physicochemically stable during immersion | Choose an un-swellable and un-degradable material | Deionized water is suggested to be the assessment medium | |
| Immobile during immersion | Choose a material with high density or fixable property | Material selection should meet other requirements | |
| No structure-independent toxicity | 1. Thoroughly remove the chemical residues | ||
| 2. Choose an un-degradable material | |||
| Cell response analysis related assessments | For morphological assay: Not disturb signal transmission and collection |
Choose a material that is transparent, non-magnetic, non-autofluorescence | Material selection should meet other requirements |
| For hydrodynamic flow assay: Immobile during shear stress | Choose a material with high density or fixable property | Material selection should meet other requirements | |
| For wound-healing assay: Geometrically stable after wound preparation | Prepare wounds by healing inserts rather than physical scratching | Commercial or self-designed healing inserts both can meet the requirement | |
| For CCK-8, MTT assays: Not to bring absorbance difference |
Detect the absorbance without the substrate materials | Ensure the same volume reaction reagent being transferred to the new plate | |
| For RT-qPCR, WB assays: Not affect sample quality and internal reference |
1. Choose a material with low solubility in the cell lysis reagents. | Material selection should meet other requirements | |
| 2. Carefully choose internal reference | β-actin is not stably expressed on different topographies | ||
| There are a large number of methods for analyzing cell responses. The management of confounding factors should be carried out following our guideline, when other cell response detecting methods are performed. | |||
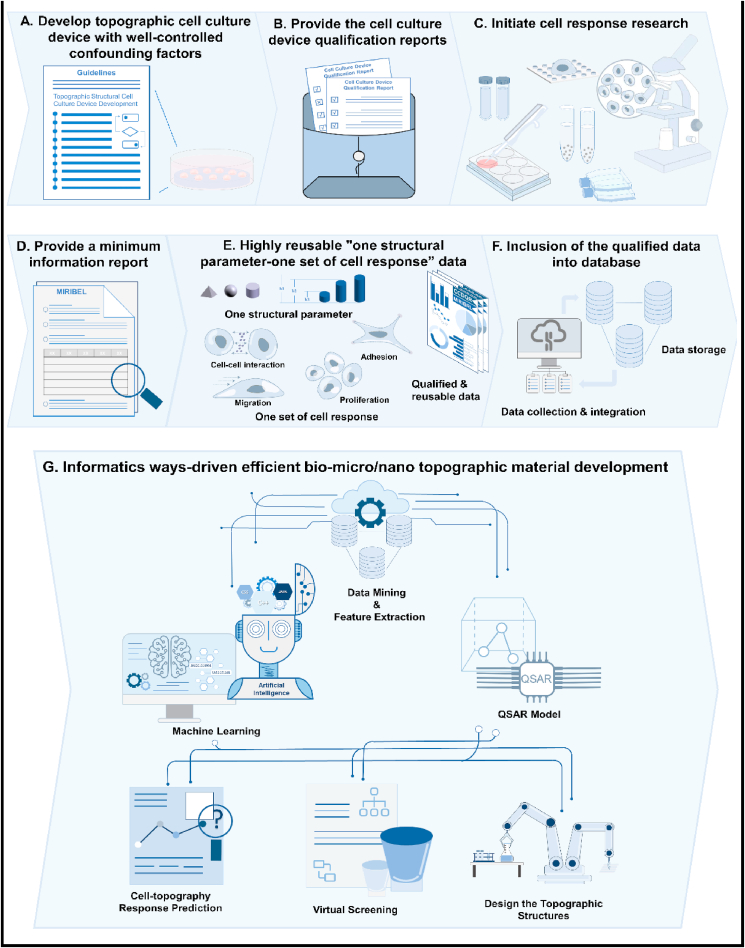
There may be many confounding factors existing in topographic cell response researches. It is difficult to identify all of them. Even to those identified confounding factors, we may not be able to completely remove them. Nevertheless, with this thoughtful guideline, we can still prepare a topographic cell culture device with minimum confounding factors. Cell culture and analysis can then be carried out to obtain the “one structure parameter – one set of cell response” data. To enter these data in a database, we suggest labeling them with two reports: 1) a “Cell culture device qualification report” (Table 3) reporting the information of structural parameters and controlling of all the related confounding factors and 2) mimicking MIRIBEL, a “Minimal information report” recording all the conditions when carrying out cell culture and analysis. The benefit of this approach is that we can clearly know how and under what kind of circumstances the data were generated, and whether the confounding factors have been well-controlled or still exist. This can give us significant confidence in reusing them. Based on these proposals, we summarized a detailed pathway for the generation and enrollment of the “one structure parameter – one set of cell response” data for constructing a high-quality database (Fig. 5).
Table 3.
Cell culture devices qualification report. After developing topographic cell culture devices based on the guideline, researchers should report the controlling of confounding factors.
| 1. Structures Fabrication Related Confounding Factors Testing ☐ Well-controlled “one structural parameter” difference Supporting testing results: e.g., SEM demonstrated that the shapes of nanopores are similar, pore size is the only one variable structural parameter (50 nm, 100 nm, 200 nm), and other structural parameters are consistent between different topographies (pore depth: 50 nm; pore interval: 100 nm). ☐ Minimum and consistent chemical properties Supporting testing results: e.g., XPS and FTIR spectra showed no unwanted chemical elements or bioactive functional groups, and similar peaks between different topographic structures; ☐ The consistent structure-independent difference in physical properties Supporting testing results: e.g., AFM show similar Young's modulus between different substrates |
| 2. Cell Culture Related Confounding Factors Testing ☐ Geometrical and physicochemical stability during the sterilization process Supporting testing results: e.g., SEM showed no obvious change in topography after sterilization. XPS, FTIR, and AFM proved no physicochemical alteration after sterilization. ☐ Geometrical and physicochemical stability during immersion Supporting testing results: e.g., SEM showed no obvious change in topography after immersion. XPS, FTIR, and AFM proved no physicochemical alteration after immersion. ☐ Immobile during immersion Supporting testing results: e.g., It can be visible that the substrates wound not float during immersion ☐ No structure-independent cytotoxicity Supporting testing results: e.g., CCK-8 showed good cell viability in the soaking medium of substrates |
| 3. Cell Response Analysis Related Confounding Factors Testing ☐ For light microscopy: Transparency Supporting testing results: e.g., It can be visible that the substrates were transparent ☐ For confocal microscopy: No fluorescent interference Supporting testing results: e.g., Confocal microscopy exhibited no fluorescent interference ☐ For SEM: Non-magnetic Supporting testing results: e.g. XPS showed no magnetic elements in the cell culture device ☐ For hydrodynamic flow assay: Immobile during shear stress Supporting testing results: e.g., It can be visible that the substrates wound not float under shear stress ☐ For wound-healing assay: Geometrical stable after wound preparation Supporting testing results: e.g., SEM showed no change of the topographies after wound preparation. ☐ For CCK-8, MTT assays: Not to bring absorbance difference Supporting testing results: e.g., CCK-8 showed no confounding absorbance ☐ For RT-qPCR, WB assays: Not affect sample quality Supporting testing results: e.g., Spectrophotometry demonstrated a good quality of the mRNA extracted from the devices (A260/A280 > 1.8, A260/A230 > 1.8). ☐ For RT-qPCR, WB assays: Not affect internal reference Supporting testing results: e.g., we chose GAPDH as an internal reference, which stably expressed on different topographic structures. |
Fig. 5.
Schematic figure. Efficient biomicro/nano topographic material development procedure. (A–B) Develop cell culture devices based on this guideline to remove or control confounding factors before initiating biotopographic cell response research projects, then provide a “cell culture devices qualification report” to report the information of structural parameters and controlling for all the related confounding factors. (C–D) Initiate cell response research then provide a “minimum information report” that recording all the conditions when carrying out cell culture and analysis. (E–F) Obtain highly reusable “one structural parameter – one set of cell response” data and establish a high-quality database. (G) Virtual topographic structures prediction, screening and design based on the high-quality database.
The successful establishment of this database is of vital significance because it can break through the bottleneck of the currently lacking highly reusable database in topographic structure efficient development. It may advance biotopographic structure development into the “Materials Genome Initiative” era, which has played a vital role in advanced material development in the fields of electronics, energy, transportation, and health care [33]. In this era, scientists can quickly predict the cell responses of topographic structures through informatics methods such as QSAR and artificial intelligence, profoundly saving time and costs (Fig. 5). For example, if we want to develop titanium implant surfaces with anti-inflammatory effects, we can first search for possible topographic structures with “anti-inflammatory effects” from the database with the aid of informatics-driven methods like QSAR and artificial intelligence. Two reports (Cell culture device qualification report and Minimal information report) of each data should be noticed before formal application. This will also help to further narrow down the number of candidate topographies according to the requirements of the projects. After obtaining effective topographies, we can then apply proper fabrication techniques to generate titanium implant topographic surfaces. Further cell and animal studies can be carried out to testify the desired inflammatory effects.
In addition to surface topographic structures, surface chemistry is also effective and powerful in regulating cell response. For example, the –NH2 and –SH groups promoted the osteogenic differentiation of bone marrow-derived mesenchymal stem cells (BMSCs) [34]. Hence, many promising chemical modification strategies were proposed to optimize biological performance, such as via cell-adhesive modifications and the antifouling surfaces to minimize nonspecific cell adhesion [35,36]. Our study can also shed light on the establement of “surface chemistry – cell responses” database. Researhers can also propose a guideline for the development of cell culture devices with specific surface chemistry and confounding factors controlled. The “surface chemistry – cell responses” database may be subsequently built up, to promote informatics-driven efficient bioactive surface chemistry development.
CRediT authorship contribution statement
Yuanlong Guo: Methodology, Investigation, Data curation, Formal analysis, Writing – original draft. Jiaomei Mi: Investigation, Data curation, Formal analysis, Writing – original draft. Chen Ye: Investigation, Data curation, Formal analysis, Writing – original draft. Yong Ao: Investigation, Writing – original draft. Mengru Shi: Investigation. Zhengjie Shan: Investigation. Bingzhi Li: Visualization. Zetao Chen: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. Zhuofan Chen: Conceptualization, Writing – original draft. Krasimir Vasilev: Conceptualization, Writing – original draft. Yin Xiao: Conceptualization, Writing – original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (82071167), Natural Science Foundation of Guangdong Province (2018B030306030), International Team for Implantology (ITI) Research Grant (1536_2020), Guangdong Financial Fund for High-Caliber Hospital Construction, Special Funds for the Cultivation of Guangdong College Students' Scientific and Technological Innovation (“Climbing Program” Special Funds, pdjh2020b0011).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.06.013.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary data to this article had been submitted as a separated file, which includes the following messages: Primers used in this study; Viscosity of PDMS diluted by n-hexane at different volume ratios; Gas chromatography of n-hexane and the PDMS diluted by n-hexane; Atomic ratios of the substrates with different topographic structures, different sterilization methods, and different immersion time; Nano-structures immersed by DMEM for 28 days; The melting curve of different housekeeping genes of different mRNA samples detected by RT-qPCR.
References
- 1.Gulati K., Moon H.J., Li T., Sudheesh Kumar P.T., Ivanovski S. Titania nanopores with dual micro-/nano-topography for selective cellular bioactivity. Mater Sci Eng C Mater Biol Appl. 2018;91:624–630. doi: 10.1016/j.msec.2018.05.075. [DOI] [PubMed] [Google Scholar]
- 2.Long E.G., Buluk M., Gallagher M.B., Schneider J.M., Brown J.L. Human mesenchymal stem cell morphology, migration, and differentiation on micro and nano-textured titanium. Bioact Mater. 2019;4:249–255. doi: 10.1016/j.bioactmat.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schieber R., Lasserre F., Hans M., Fernandez-Yague M., Diaz-Ricart M., Escolar G., Ginebra M.P. Direct laser interference patterning of CoCr alloy surfaces to control endothelial cell and platelet response for cardiovascular applications. Adv Healthc Mater. 2017;6:1700327. doi: 10.1002/adhm.201700327. [DOI] [PubMed] [Google Scholar]
- 4.Visalakshan R.M., Cavallaro A.A., MacGregor M.N., Lawrence E.P., Koynov K., Hayball J.D., Vasilev K.J.A.F.M. vol. 29. 2019. Nanotopography‐Induced Unfolding of Fibrinogen Modulates Leukocyte Binding and Activation; p. 1807453. [DOI] [Google Scholar]
- 5.Yim E.K., Darling E.M., Kulangara K., Guilak F., Leong K.W. Nanotopography-induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomaterials. 2010;31:1299–1306. doi: 10.1016/j.biomaterials.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalby M.J., Gadegaard N., Oreffo R.O. Harnessing nanotopography and integrin-matrix interactions to influence stem cell fate. Nat. Mater. 2014;13:558–569. doi: 10.1038/nmat3980. [DOI] [PubMed] [Google Scholar]
- 7.Puzyn T., Rasulev B., Gajewicz A., Hu X., Dasari T.P., Michalkova A., Hwang H.M. Using nano-QSAR to predict the cytotoxicity of metal oxide nanoparticles. Nat. Nanotechnol. 2011;6:175–178. doi: 10.1038/nnano.2011.10. [DOI] [PubMed] [Google Scholar]
- 8.Feng R., Yu F., Xu J., Hu X. Knowledge gaps in immune response and immunotherapy involving nanomaterials: databases and artificial intelligence for material design. Biomaterials. 2021;266:120469. doi: 10.1016/j.biomaterials.2020.120469. [DOI] [PubMed] [Google Scholar]
- 9.Oh S., Brammer K.S., Li Y.S., Teng D., Engler A.J., Chien S., Jin S. Stem cell fate dictated solely by altered nanotube dimension. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2130–2135. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park J., Bauer S., Schlegel K.A., Neukam F.W., von der Mark K., Schmuki P. TiO2 nanotube surfaces: 15 nm--an optimal length scale of surface topography for cell adhesion and differentiation. Small. 2009;5:666–671. doi: 10.1002/smll.200801476. [DOI] [PubMed] [Google Scholar]
- 11.Join the dialogue. Nat. Nanotechnol. 2012;7:545. doi: 10.1038/nnano.2012.150. [DOI] [PubMed] [Google Scholar]
- 12.Faria M., Bjornmalm M., Thurecht K.J., Kent S.J., Parton R.G., Kavallaris M., Johnston A.P.R. Minimum information reporting in bio-nano experimental literature. Nat. Nanotechnol. 2018;13:777–785. doi: 10.1038/s41565-018-0246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao L., Mei S., Wang W., Chu P.K., Wu Z., Zhang Y. The role of sterilization in the cytocompatibility of titania nanotubes. Biomaterials. 2010;31:2055–2063. doi: 10.1016/j.biomaterials.2009.11.103. [DOI] [PubMed] [Google Scholar]
- 14.Gasparian A., Daneshian L., Ji H., Jabbari E., Shtutman M. Purification of high-quality RNA from synthetic polyethylene glycol-based hydrogels. Anal. Biochem. 2015;484:1–3. doi: 10.1016/j.ab.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z., Ni S., Han S., Crawford R., Lu S., Wei F., Chang J. Nanoporous microstructures mediate osteogenesis by modulating the osteo-immune response of macrophages. Nanoscale. 2017;9:706–718. doi: 10.1039/c6nr06421c. [DOI] [PubMed] [Google Scholar]
- 16.Li H.-y., Huang D.-n., Ren K.-f., Ji J. Inorganic-polymer composite coatings for biomedical devices. Smart Materials in Medicine. 2021;2:1–14. doi: 10.1016/j.smaim.2020.10.002. [DOI] [Google Scholar]
- 17.Chen W., Shao Y., Li X., Zhao G., Fu J. Nanotopographical surfaces for stem cell fate control: engineering mechanobiology from the bottom. Nano Today. 2014;9:759–784. doi: 10.1016/j.nantod.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bettinger C.J., Zhang Z., Gerecht S., Borenstein J.T., Langer R. Enhancement of in vitro capillary tube formation by substrate nanotopography. Adv. Mater. 2008;20:99–103. doi: 10.1002/adma.200702487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalby M.J., Gadegaard N., Tare R., Andar A., Riehle M.O., Herzyk P., Wilkinson C.D.W. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mater. 2007;6:997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 20.Yang K., Jung K., Ko E., Kim J., Park K.I., Kim J., Cho S.W. Nanotopographical manipulation of focal adhesion formation for enhanced differentiation of human neural stem cells. ACS Appl. Mater. Interfaces. 2013;5:10529–10540. doi: 10.1021/am402156f. [DOI] [PubMed] [Google Scholar]
- 21.Yim E.K., Pang S.W., Leong K.W. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Exp. Cell Res. 2007;313:1820–1829. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z., Bachhuka A., Wei F., Wang X., Liu G., Vasilev K., Xiao Y. Nanotopography-based strategy for the precise manipulation of osteoimmunomodulation in bone regeneration. Nanoscale. 2017;9:18129–18152. doi: 10.1039/c7nr05913b. [DOI] [PubMed] [Google Scholar]
- 23.Sereni J.G.R. 2015. Reference Module in Materials Science and Materials Engineering.http://scitechconnect.elsevier.com/reference-module-material-science/ [Google Scholar]
- 24.Wang Y., Ding B.H., Gao S.Y., Chen X.B., Zeng R.C., Cui L.Y., Li S.J. In vitro corrosion of pure Mg in phosphate buffer solution-Influences of isoelectric point and molecular structure of amino acids. Mater Sci Eng C Mater Biol Appl. 2019;105:110042. doi: 10.1016/j.msec.2019.110042. [DOI] [PubMed] [Google Scholar]
- 25.Ni M., Ratner B.D. Nacre surface transformation to hydroxyapatite in a phosphate buffer solution. Biomaterials. 2003;24:4323–4331. doi: 10.1016/s0142-9612(03)00236-9. [DOI] [PubMed] [Google Scholar]
- 26.Kagan V.E., Tyurina Y.Y., Tyurin V.A., Konduru N.V., Potapovich A.I., Osipov A.N., Kisin E.R. Direct and indirect effects of single walled carbon nanotubes on RAW 264.7 macrophages: role of iron. Toxicol. Lett. 2006;165:88–100. doi: 10.1016/j.toxlet.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Barth K.A., Waterfield J.D., Brunette D.M. The effect of surface roughness on RAW 264.7 macrophage phenotype. J. Biomed. Mater. Res. 2013;101:2679–2688. doi: 10.1002/jbm.a.34562. [DOI] [PubMed] [Google Scholar]
- 28.Jonkman J.E., Cathcart J.A., Xu F., Bartolini M.E., Amon J.E., Stevens K.M., Colarusso P. An introduction to the wound healing assay using live-cell microscopy. Cell Adhes. Migrat. 2014;8:440–451. doi: 10.4161/cam.36224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lv X., Geng Z., Fan Z., Wang S., Pei W., Chen H. A PDMS device coupled with culture dish for in vitro cell migration assay. Appl. Biochem. Biotechnol. 2018;186:633–643. doi: 10.1007/s12010-018-2737-z. [DOI] [PubMed] [Google Scholar]
- 30.Liu R.H., Chen S.C., Huang P.N., Liu G.Q., Luo P., Li Z.P., Xiao Y. Immunomodulation-based strategy for improving soft tissue and metal implant integration and its implications in the development of metal soft tissue materials. Adv. Funct. Mater. 2020;30:1910672. doi: 10.1002/adfm.201910672. [DOI] [Google Scholar]
- 31.Teo B.K., Wong S.T., Lim C.K., Kung T.Y., Yap C.H., Ramagopal Y., Romer L.H. Nanotopography modulates mechanotransduction of stem cells and induces differentiation through focal adhesion kinase. ACS Nano. 2013;7:4785–4798. doi: 10.1021/nn304966z. [DOI] [PubMed] [Google Scholar]
- 32.Lee J.N., Park C., Whitesides G.M. Solvent compatibility of poly(dimethylsiloxane)-based microfluidic devices. Anal. Chem. 2003;75:6544–6554. doi: 10.1021/ac0346712. [DOI] [PubMed] [Google Scholar]
- 33.Olson G.B. Genomic materials design: the ferrous frontier. Acta Mater. 2013;61:771–781. doi: 10.1016/j.actamat.2012.10.045. [DOI] [Google Scholar]
- 34.Curran J.M., Chen R., Hunt J.A. The guidance of human mesenchymal stem cell differentiation in vitro by controlled modifications to the cell substrate. Biomaterials. 2006;27:4783–4793. doi: 10.1016/j.biomaterials.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Chen Q., Yu S., Zhang D., Zhang W., Zhang H., Zou J., Mao Z. Impact of antifouling PEG layer on the performance of functional peptides in regulating cell behaviors. J. Am. Chem. Soc. 2019;141:16772–16780. doi: 10.1021/jacs.9b07105. [DOI] [PubMed] [Google Scholar]
- 36.Chen Q., Zhang D., Zhang W., Zhang H., Zou J., Chen M., Li J. Dual mechanism β-amino acid polymers promoting cell adhesion. Nat. Commun. 2021;12:562. doi: 10.1038/s41467-020-20858-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strickstrock M., Rothe H., Grohmann S., Hildebrand G., Zylla I.M., Liefeith K. Influence of surface roughness of dental zirconia implants on their mechanical stability, cell behavior and osseointegration. BioNanoMaterials. 2017;18 doi: 10.1515/bnm-2016-0013. [DOI] [Google Scholar]
- 38.Wood M.A., Wilkinson C.D., Curtis A.S. The effects of colloidal nanotopography on initial fibroblast adhesion and morphology. IEEE Trans. NanoBioscience. 2006;5:20–31. doi: 10.1109/tnb.2005.864015. [DOI] [PubMed] [Google Scholar]
- 39.Lamolle S.F., Monjo M., Rubert M., Haugen H.J., Lyngstadaas S.P., Ellingsen J.E. The effect of hydrofluoric acid treatment of titanium surface on nanostructural and chemical changes and the growth of MC3T3-E1 cells. Biomaterials. 2009;30:736–742. doi: 10.1016/j.biomaterials.2008.10.052. [DOI] [PubMed] [Google Scholar]
- 40.Zong X., Ran S., Kim K.S., Fang D., Hsiao B.S., Chu B. Structure and morphology changes during in vitro degradation of electrospun poly(glycolide-co-lactide) nanofiber membrane. Biomacromolecules. 2003;4:416–423. doi: 10.1021/bm025717o. [DOI] [PubMed] [Google Scholar]
- 41.Zhou D.W., Garcia A.J. Measurement systems for cell adhesive forces. J. Biomech. Eng. 2015;137 doi: 10.1115/1.4029210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn J.S., Hammond P.T., Rubner M.F., Lee I. Self-assembled particle monolayers on polyelectrolyte multilayers: particle size effects on formation, structure, and optical properties. Colloid. Surface. Physicochem. Eng. Aspect. 2005;259:45–53. doi: 10.1016/j.colsurfa.2005.02.008. [DOI] [Google Scholar]
- 43.Zhang M., Li C., Yang S., Hirte J., Zhao W., Wei Q., Diao Z. Ultra-transparent slippery surface. Smart Materials in Medicine. 2021;2:38–45. doi: 10.1016/j.smaim.2020.10.001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.