Abstract
Circular RNA (circRNA) is a new type of long-sequence RNA formed by a noncanonical form of alternative splicing called back-splicing. Emerging evidence has revealed that circRNAs are involved in cancer progression, regulating cancer-related genes through sponging microRNAs (miRNAs). In our study, we identified a novel upregulated circRNA, circSERPINE2, through analyzing circRNAs microarray data of glioblastoma from GEO datasets (GSE146463). Quantitative real-time PCR was used to further confirm the upregulation of circSERPINE2 in glioblastoma cell lines and tissues. Silencing circSERPINE2 inhibits glioblastoma proliferation in vivo and in vitro through cell counting kit-8 (CCK-8) assay, colony formation assay, flow cytometry analysis, and western blot analysis and xenograft tumor model. Mechanistically, circSERPINE2 could directly sponge miR-324-5p and miR-361-3p. BCL2, known as a novel anti-apoptosis gene, is a target gene both of miR-324-5p and miR-361-3p. Thus, circSERPINE2 promotes BCL2 expression through sponging miR-324-5p and miR-361-3p. In conclusion, our study revealed the biological function and mechanism of circSERPINE2 in glioblastoma progression and that circSERPINE2 could be a potential therapeutic target for glioblastoma.
Keywords: circSERPINE2, glioma, miR-361-3p, miR-324-5p, BCL2
Graphical abstract
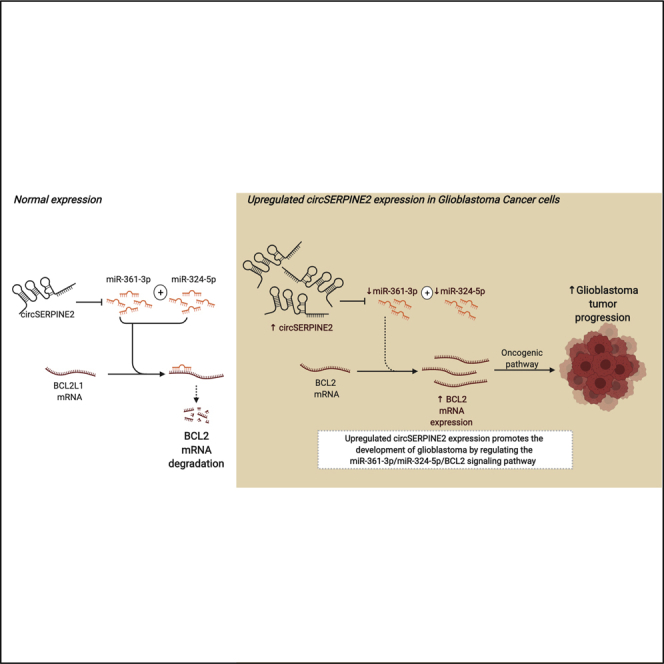
Li et al. revealed a new oncogenic circular RNA, circSERPINE2, in glioblastoma progression. circSERPINE2 is upregulated in glioblastoma and is related with poor prognosis of glioblastoma. This study found that circSERPINE2 promotes proliferation of glioblastoma through inhibiting BCL2 expression via sponging miR-324-5p and miR-361-3p.
Introduction
Glioma is one of the most prevalent and aggressive primary tumors of the brain and spinal cord.1 Accumulating studies show that its 5-year survival rate has been poor due to the aggressive growth and the difficulty of complete resection.2 Despite the gradual optimization of the treatment plan, the recurrence rate and mortality rate are still high. In recent years, many studies have explored the occurrence and development of gliomas by revealing the molecular mechanisms of tumors, so that targeted drugs can be used to accurately treat tumors to achieve good progress.3, 4, 5 However, the molecular mechanisms of glioma formation and development are still unclear. Therefore, the deep pathogenesis of gliomas remains to be further studied, including the abnormal activation of proto-oncogenes and the inactivation of tumor suppressor factors.
Circular RNA (circRNA) is a new type of long sequence RNA formed by a noncanonical form of alternative splicing called back-splicing.6 circRNA has been a subject of study in recent years due to its significant role in the pathogenesis of many human diseases such as cancer. For instance, a study by Fan et al.7 had revealed that the circRNA circ_POLA2 contributes to the increased stemness of lung cancer cells by endogenously regulating the miR-326/GNB1 axis while Dong et al.8 reported that the circRNA ACVR2A acts as a tumor suppressor gene that inhibits the proliferation and metastasis of bladder cancer cells via the regulation of the miR-626/EYA4 axis. Specifically, the circ-SERPINE2 has been demonstrated to promote the progression of gastric carcinoma by downregulating miR-375 expression through sponging and by regulating the YWHAZ gene.9 However, no study has reported its role in glioma.
Therefore, in this study, we aimed to understand the role of circ-SERPINE2 in the development and progression of glioma and also elucidate its molecular mechanism of regulation. To achieve this, we analyzed microarray chip data (GSE146463) containing 8 groups of malignant glioma cells (G cells) and 3 groups of neural precondition cells (N) and found that circSERPINE2 (has-circ-0001103) is highly expressed in malignant glioma cells. This result was further confirmed in collected clinical samples and clinicopathological data. We found that circSERPINE2 is highly expressed in the tissues of glioma patients. Through the circRNA interactome, we found that circSERPINE2 can target miR-361-3p and miR-324-5p. The miR-361-3p is involved in the development of pancreatic cancer10 and cervical cancer,11 and its low expression in cancer cells has been reported to functionally regulate multiple signal pathways and inhibit tumor growth.10,12,13 Similarly, miR-324-5p has been shown to inhibit tumor progression in gallbladder cancer14 and also glioma.15 Through starBase, we predicted that both miR-361-3p and miR-324-5p can jointly target the 3′ UTR of BCL2. BCL2 is an important regulatory hub in the apoptosis signaling pathway.16 Mechanistically, we showed that the aberrant expression of circSERPINE2 contributes to the pathogenesis of glioma by regulating the miR-361-3p/miR-324-5p/BCL2 signaling axis, which might be an effective drug target for the treatment of glioma.
Results
circSERPINE2 is upregulated in the glioma tissue and cells
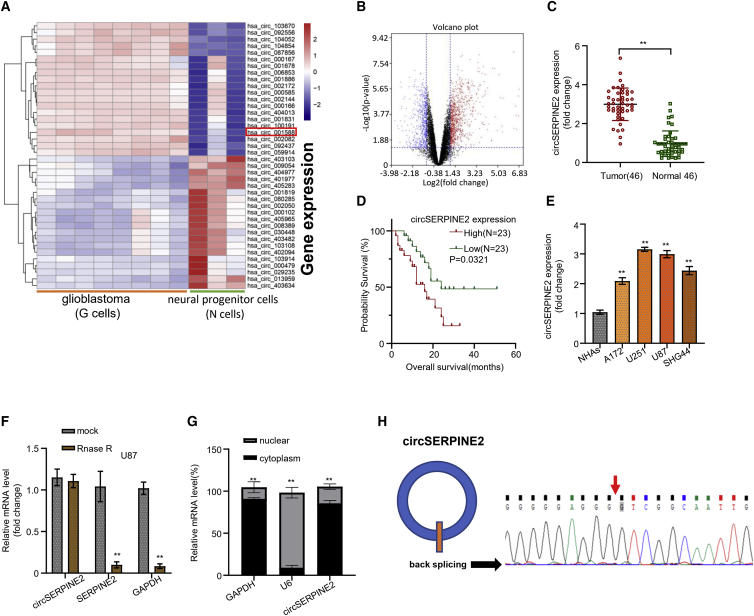
At the start of this study, we downloaded microarray chip data (GSE146463), containing 8 glioblastoma (G) cells and 3 neural progenitor (N) cells, from the GEO website and conducted a differential expression analysis on the data. The heatmap analysis result showed the distribution of the 1,361 differentially expressed circRNAs in the samples, after the filtration criteria (|log2FC| > 1, p value < 0.05; Figure 1A). A volcano plot was subsequently plotted, which showed that a total of 822 and 539 circRNAs were significantly upregulated and downregulated, respectively (Figure 1B). Notably, the result showed that the circSERPINE2 was significantly overexpressed in the malignant G cells. To confirm the expression of circSERPINE2 in malignant G cells, we conducted quantitative real-time PCR analysis in 46 glioblastoma (GM) cancer tissues collected from patients and 46 normal brain (N) tissues. We found that the circSERPINE2 expression in GM tissues was significantly higher than the one observed in the normal brain (N) tissues (Figure 1C, Figure S1A, p < 0.001). We also detected expression of other dysregulated circRNAs, including circ_103670, circ_087856, and circ_002172), and found that there is no significance between tumor and normal tissues (data not shown). Furthermore, we sought to determine the effect of circSERPINE2 expression level on the overall survival rate of GM patients. Using the median expression value of circSERPINE2 in our initial result, we determined the expression level boundary and divided the 46 GM patients into a high expression group and a low expression group. Afterward, a KM-plotter was used to analyze and plot the survival curve of the GM patients in the high and low circSERPINE2 expression group. From the result, we found that high circSERPINE2 expression level was significantly associated with the poor overall prognosis of GM patients in this study (Figure 1D, p = 0.0321). Combining this expression level data with the recorded clinicopathological data of the GM patients, we observed that high circSERPINE2 expression level had no significant correlation with gender and age, but with staging, grade, and lymph node (Table S1, p < 0.001).
Figure 1.
circSERPINE2 is upregulated in glioblastoma tissue and cells
Heatmap produced from hierarchical clustering of 1,361 differentially expressed circRNAs in 8 GM cells and 3 neural progenitor (N) cells; the most upregulated and downregulated circRNAs are represented in red and purple respectively (A). Volcano plot of differentially expressed circRNAs showed that a total of 822 and 539 circRNAs were significantly upregulated and downregulated, respectively; black is the no differential expression, blue is significantly downregulated, and red is upregulated (B). Quantitative real-time PCR detects that circSERPINE2 expression is markedly upregulated in GM cancer tissues compared to normal brain tissues (C). Survival curve of GM patients in the high- and low circSERPINE2 expression group showed that high circSERPINE2 expression level was significantly associated with the poor prognosis of GM patients (D). Quantitative real-time PCR analysis of circSERPINE2 expression level in normal human astrocytes (NHAs) and GM cell lines (A172, U251, U87, and SHG44); circSERPINE2 was markedly upregulated in the GM cell lines with the highest expression level in the U251 and U87 cell lines (E). RNase R treatment to determine the circular nature circSERPINE2 in GM cell line; Rnase R had no significant effect on the expression level of circSERPINE2 mRNA when compared to the blank control mock (F). Detection of the subcellular localization of circSERPINE2; quantitative real-time PCR analysis showed that circSERPINE2 was significantly expressed in the cytoplasmic fraction of the GM cell line compared to the nuclear fraction (result is comparable to that of the nuclear-enriched U6 and cytoplasmic-enriched GAPDH) (G). Sequencing of junction sites of circSERPINE2 (H). ∗∗p < 0.01.
To further determine the molecular function of the circSERPINE2 in GM cancer, we examined the expression level of circSERPINE2 in normal human astrocytes (NHAs) and GM cell lines (A172, U251, U87, and SHG44) through quantitative real-time PCR analysis. As shown in Figure 1E, circSERPINE2 was markedly upregulated in the GM cell lines with the highest expression level observed in the U251 and U87 cell lines, which were chosen for further experiments. We also provided the mutational status of cancer-related genes (including PIK3CA, PTEN, KRAS, AKT1, and mTOR) of the glioblastoma cell lines and speculated that somatic mutations of cancer-related genes have no effects on function and expression of circ-SERPINE2 (Table S2). We also confirmed the structure of the studied circSERPINE2 and its subcellular localization. Figure 1F shows that Rnase R had no significant effect on the expression level of circSERPINE2 mRNA compared to the blank control mock, while it effectively reduced the mRNA expression level of the linear form of SERPINE2 and also GAPDH, confirming the circular structure of circSERPINE2 (p < 0.001). We also observed that the circSERPINE2 was significantly expressed in the cytoplasmic fraction of the GM cell line compared to the nuclear fraction, suggesting that the circSERPINE2 is subcellularly located in the cytoplasm of GM cancer cells (Figure 1G, p < 0.001). Besides, sequencing analysis revealed the back junction sites of circSERPINE2 (Figure 1H). The result is comparable to the one obtained for the nuclear-enriched U6 and cytoplasmic-enriched GAPDH, which were used as controls.
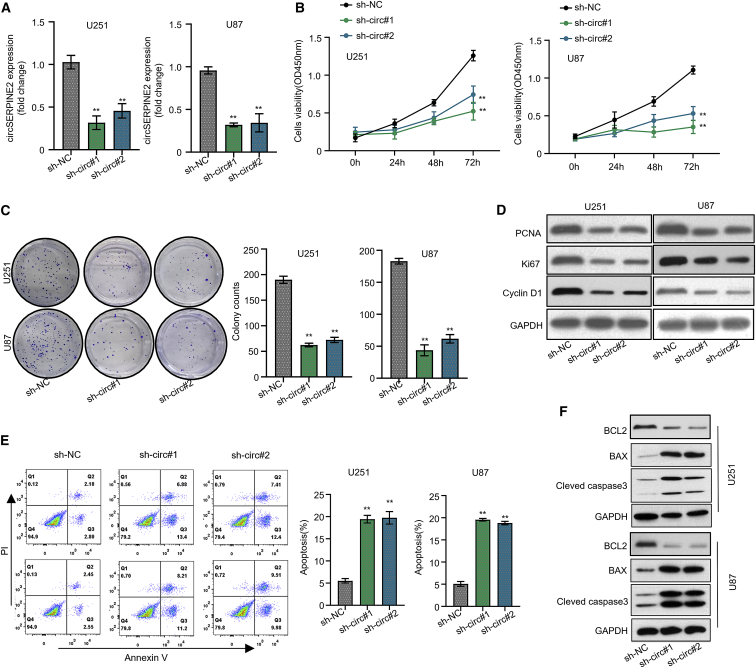
Knockdown of circSERPINE2 represses the glioblastoma cell proliferation and promotes its apoptosis in vitro
To determine the molecular function of circSERPINE2 in GM cells, we conducted a loss-of-function experiment by knocking down the expression of circSERPINE2 in the cell lines using two interfering RNAs constructed to target the circSERPINE2 sequence (and are hereafter referred to as short hairpin (sh)-circ#1 and sh-circ#2). As shown in Figure 2A, sh-circ#1 and sh-circ#2 significantly reduced circSERPINE2 expression level to less than 50% in both U251 and U87 GM cell lines, confirming the efficacy of the interfering RNAs used (p < 0.001). Furthermore, we observed the function of circSERPINE2 in the proliferation of GM cells via cell counting kit-8 (CCK-8) and colony formation assay. The CCK-8 result showed that the sh-circ#1 and sh-circ#2 significantly reduced the viability of the GM cell lines, after 72 h of transfection, especially when compared to the sh-NC (negative control transfection; Figure 2B, p < 0.001). Our colony formation assay also revealed that the sh-circ#1 and sh-circ#2 could significantly reduce the number of GM cell colonies formed after transfection compared to the sh-NC (Figure 2C, p < 0.001). Besides, the expression level of the proliferative proteins (proliferating cell nuclear antigen (PCNA), Ki67, and cyclin D), measured by western blot analysis, after circSERPINE2 knockdown in the U251 and U87 GM cell lines was drastically reduced (Figure 2D). Our apoptosis analysis data suggested that silencing circSERPINE2 markedly enhanced the apoptosis ability of both GM cell lines (Figure 2E, p < 0.001). Similarly, further data obtained from western blot analysis suggested that circSERPINE2 knockdown repressed the protein expression level of the anti-apoptotic gene (BCL2) but enhanced the expression of the pro-apoptotic genes (Bax and cleaved caspase-3; Figure 2F).
Figure 2.
Knockdown of circSERPINE2 represses the glioblastoma cell proliferation and promote its apoptosis in vitro
Quantitative real-time PCR detects the expression level of circSERPINE2 in U251 and U87 after knockdown; circSERPINE2 expression was significantly downregulated (A). CCK-8 assay detects the viability of GM cell lines after circSERPINE2 knockdown and at different intervals; the viability of the GM cell lines was significantly reduced (B). Colony formation assay showed that sh-circ#1 and sh-circ#2 significantly reduced the colony formation ability of the GM cells (C). Western blot experiment detects PCNA, Ki67, and cyclin D1 protein expression level in U251 and U87 cells after circSERPINE2 knockdown, which was drastically reduced (D). Flow cytometry analysis detects apoptosis of U251 and U87 cells treated with sh-NC, sh-circ#1, and sh-circ#2; silencing circSERPINE2 markedly enhanced the apoptosis ability of both GM cell lines (E). Western blot analysis detects BCL2, BAX, and cleaved caspase-3 protein expression level in U251 and U87 cells treated with different groups of sh-NC, sh-circ#1, and sh-circ#2; the expression level of the BCL2 was enhanced while that of Bax and cleaved caspase-3 was repressed (F). ∗∗p < 0.01.
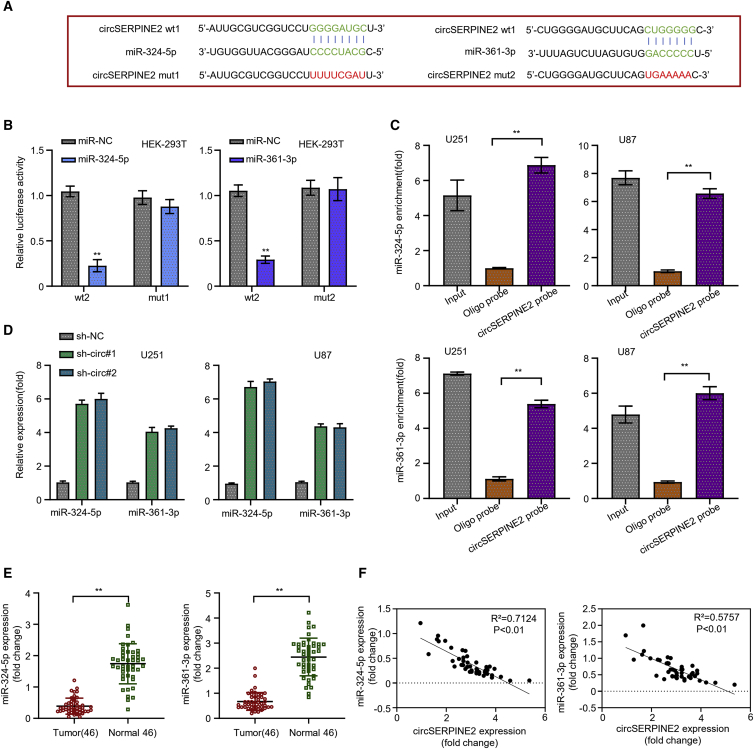
circSERPINE2 endogenously sponges miR-361-3p and miR-324-5p and accelerates their degradation
Based on the circRNA interactome (https://circinteractome.nia.nih.gov/) bioinformatics analysis, miR-361-3p and miR-324-5p were predicted as the putative interacting microRNA (miRNAs) of circSERPINE2, due to the presence of its complementary binding site sequence in circSERPINE2 (Figure 3A). This analysis result was later confirmed through a dual-luciferase reporter assay experiment. In the experiment, the normal human cell lines (HEK293T) were co-transfected with circSERPINE2 wild-type (WT) or mutant-type (mut) reporter vector, together with miR-361-3p and miR-324-5p mimics (or their negative miR-NC control). The result in both experimental groups showed that miR-361-3p and miR-324-5p mimics significantly inhibited the luciferase activity of the HEK293T cells transfected with circSERPINE2 WT sequence, but no significant inhibiting effect was observed in those transfected with the circSERPINE2 mut sequence (Figure 3B). Furthermore, a conducted biotinylated RNA pull-down assay revealed that more miR-361-3p and miR-324-5p mRNA were enriched in the circSERPINE2 oligonucleotide probe in both U251 and U87 GM cell lines, than the control oligo probe (Figure 3C, p < 0.001), confirming, to some extent, the ability of circSERPINE2 to interact with both miRNAs. To further understand this molecular mechanism of regulation, we measured the relative expression of miR-361-3p and miR-324-5p mRNA in the cell lines after silencing circSERPINE2 through quantitative real-time PCR analysis. We found that silencing circSERPINE2 lead to a significant upregulation of the miR-361-3p and miR-324-5p mRNA expression levels in both cell lines (Figure 3D, p < 0.001). Besides, miR-361-3p and miR-324-5p mRNA was found to be significantly downregulated in the 46 GM cancer tissues compared to the normal brain tissues and their expression level was negatively correlated to that of circSERPINE2 (Figure 3E, p < 0.001; Figure 3F, p < 0.01).
Figure 3.
circSERPINE2 sponges miR-361-3p and miR-324-5p and accelerates their degradation
The circular RNA interactome predicted the complementary sequence of circSERPINE2 as miR-361-3p and miR-324-5p (A). Luciferase reporter assay experiment in HEK293T cells co-transfected with miR-361-3p and miR-324-5p (mut and WT) and miR-NC; the luciferase activity of the HEK293T cells transfected with circSERPINE2 WT sequence was significantly reduced but no significant inhibiting effect was observed in those transfected with the circSERPINE2 mut sequence (B). Biotinylated RNA pull-down assay revealed that more miR-361-3p and miR-324-5p mRNA were enriched in the circSERPINE2 oligonucleotide probe in the GM cell lines compared to the control oligo probe (C). Quantitative real-time PCR analysis showed that circSERPINE2 knockdown significantly upregulated miR-361-3p and miR-324-5p expression level in GM cell lines (D). Differential expression analysis of miR-361-3p and miR-324-5p mRNA in 46 GM cancer tissues compared to the normal brain tissues; miR-361-3p and miR-324-5p was significantly downregulated in the GM tissues (E). Correlation analysis of miR-361-3p or miR-324-5p expression level and circSERPINE2 expression level; their expression was negatively correlated (F). ∗∗p < 0.01.
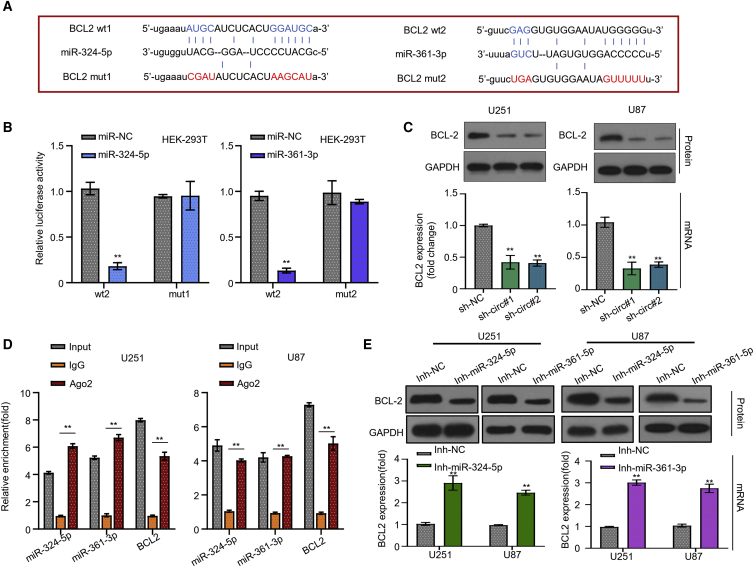
BCL2 gene is the common target of miR-361-3p and miR-324-5p
Furthermore, starBase prediction analysis showed that the miR-361-3p and miR-324-5p can co-target the 3′ UTR region of BCL2 gene to regulate its expression level (Figure 4A). And the confirmatory luciferase reporter gene experiment revealed that miR-361-3p and miR-324-5p mimics significantly reduced the luciferase activity of the HEK293T cell group transfected with WT BCL2 luciferase reporter vector compared to the miR-NC. On the contrary, we found that miR-361-3p and miR-324-5p mimics could not repress the luciferase activity of the HEK293T cell group transfected with the mut BCL2 luciferase reporter vector (Figure 4B, p < 0.001). We observed that silencing circSERPINE2 significantly reduced BCL2 mRNA and protein expression level in both U251 and U87 GM cell lines (Figure 4C, p < 0.001). RNA immunoprecipitation (RIP)-quantitative real-time PCR analysis revealed that more miR-361-3p, miR-324-5p, and BCL2 mRNA were enriched in the precipitated Ago2 group after transfection, compared to the immunoglobulin G (IgG) control group, suggesting that the miRNAs and BLC2 mRNA could bind through the Agos2 complex (Figure 4D, p < 0.001). Moreover, transfecting the U251 and U87 GM cell lines with miR-361-3p and miR-324-5p inhibitor notably reduced the expression level of miR-361-3p and miR-324-5p mRNA in both experimental cell lines (Figure S1B, p < 0.001). BCL2 expression was increased when miR-361-3p and miR-324-5p reduced by miRNA inhibitors (Figure 4E, p < 0.001). These results indicated that BCL2 was the target gene of miR-324-5p and miR-362-3p.
Figure 4.
BCL2 gene is the common target of miR-361-3p and miR-324-5p
StarBase predicts miR-361-3p and miR-324-5p target on BCL2 (A), transfected into HEK293T cells (B). RIP-quantitative real-time PCR experiments confirm the interaction between miR-361-3p and miR-324-5p and BCL2 mRNA and protein expression level (C), quantitative real-time PCR, and western blot experiments detect BCL2 expression in U251, U87 cells treated with sh-NC, sh-circ#1, sh-circ#2 (D), and inhibitors of miR-361-3p and miR-324-5p (E). ∗∗p < 0.01.
circSERPINE2 promotes the proliferation and inhibits the apoptosis of glioblastoma cell by sponging miR-361-3p and miR-324-5p and modulating BCL2 gene
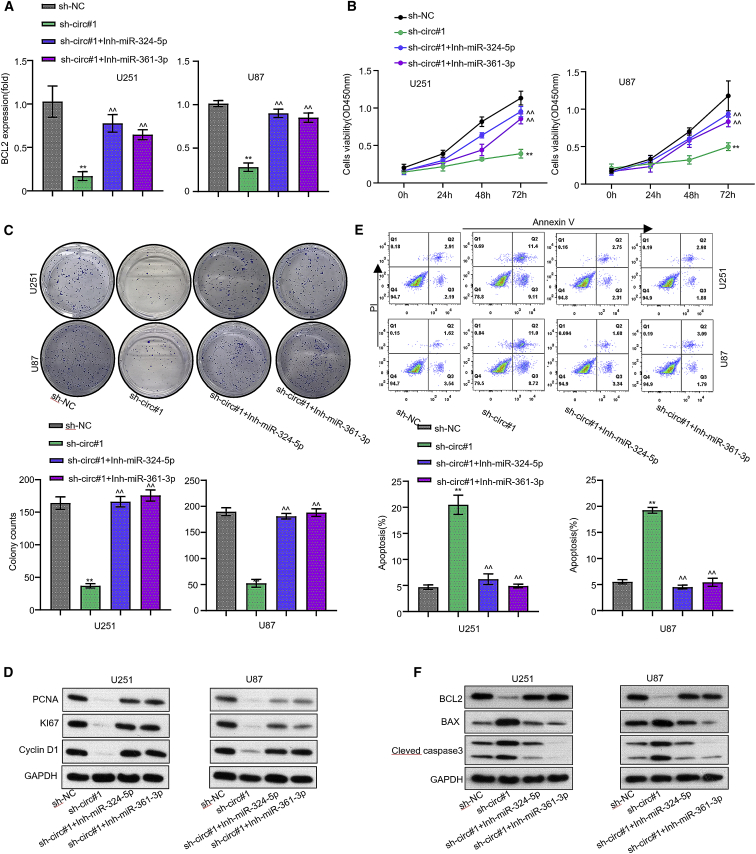
To confirm that circSERPINE2 could promote the proliferation of glioblastoma cancer cell and inhibit the apoptosis through the miR-361-3p/miR-324-5p/BCL2 signaling axis, we assessed the corresponding cell phenotype after silencing circSERPINE2 and inhibiting the miR-361-3p and miR-324-5p expression in the U251 and U87 GM cell lines. Quantitative real-time PCR result showed that silencing circSERPINE2 (with sh-circ#1) in the cell lines inhibited the BCL2 mRNA expression. This inhibiting effect was later subsided after co-transfecting the cell lines with miR-361-3p inhibitor or miR-324-5p inhibitor, significantly restoring BCL2 expression in both cell lines (Figure 5A, p < 0.001). The functional effect of this treatment on the proliferation of the glioma cell lines was also assessed using CCK-8 assay. The result showed that inhibiting miR-361-3p and miR-324-5p could restore the viability of the two circSERPINE2-silenced GM cell lines (Figure 5B, p < 0.001). Similarly, colony formation assay revealed that the number of cell colonies formed in the two circSERPINE2-silenced GM cell lines was significantly restored after co-transfecting the cell lines with miR-361-3p or miR-324-5p inhibitor (Figure 5C, p < 0.001). Besides, we found that the increased apoptosis rate of the circSERPINE2-silenced GM cell lines was markedly reduced after co-transfecting the cells with either miR-361-3p inhibitor or miR-324-5p inhibitor (Figure 5D, p < 0.001). Additionally, western blot analysis provided us with evidence that miR-361-3p or miR-324-5p inhibition in the circSERPINE2-knockdown cell lines could restore the expression level of the proliferative (PCNA, Ki67, and cyclin D) and anti-apoptotic (BCL2) proteins but reduced the expression level of the pro-apoptotic proteins (Bax and cleaved caspase-3) in the GM cell (Figures 5E and 5F), suggesting that the overexpression of circSERPINE2 promotes the proliferation of GM cell and increased its apoptosis by inhibiting the miR-361-3p and miR-324-5p expression and upregulating BCL2 expression.
Figure 5.
circSERPINE2 promotes the proliferation and inhibit the apoptosis of glioblastoma cell by sponging miR-361-3p and miR-324-5p and modulating BCL2 gene
Expression of BCL2 in transfected U251 and U87 cells detected by quantitative real-time PCR (A) and cell viability (B). Clonal formation ability of U251 transfected cells (C). Protein levels of PCNA, Ki67, and cyclin D1 via western blot (D). Apoptosis detection (E) and protein levels of BCL2, BAX, and cleaved caspase-3 (F) were evaluated in cells transfected with sh-NC, sh-circ#1, sh-circ#1+miR-361-3p inhibitor, and sh-circ#1+miR-324-5p inhibitor. ∗∗p < 0.01, and ^^p < 0.01.
Knockdown of circSERPINE2 represses the development of glioma in vivo
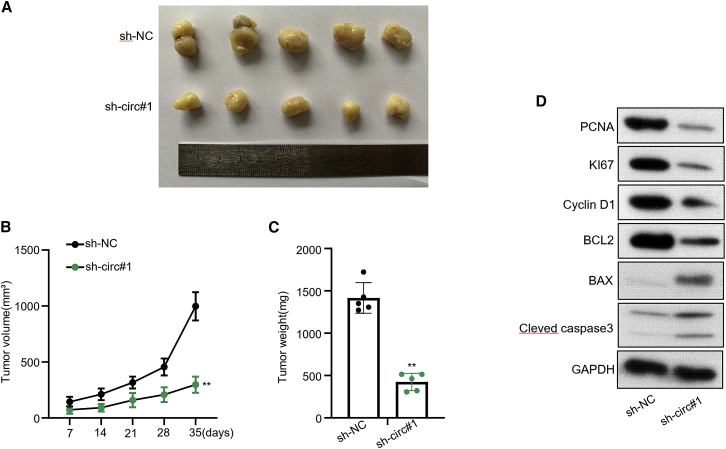
Lastly, an in vivo tumor assay was conducted to further observe and validate the functional effect of circSERPINE2 knockdown. Our result demonstrated that silencing circSERPINE2 resulted in a substantial reduction in the sizes of tumors formed in the xenograft mice model after 35 days of injection (Figure 6A). Also, the tumor volume measure was significantly lower in the circSERPINE2-knockdown mice compared to the negative control (sh-NC) mice, especially after 35 days of injection (Figure 6B, p < 0.001). At the end of the experiment, we observed the volume of the tumor formed in both groups of mice (sh-NC group and sh-circ#1, i.e., circSERPINE2-knockdown group). The measured tumor volume was significantly lower in the circSERPINE2-knockdown mice compared to the sh-NC mice (Figure 6C, p < 0.001). Finally, we examined the protein expression level of some specific biomarkers in the excised tumor after the experiment, using western blot analysis. The result showed that circSERPINE2 knockdown markedly reduced the protein expression level of the proliferative (PCNA, Ki67, and cyclin D) and anti-apoptotic (BCL2) genes in the tumor but on the contrary, reduced that of the pro-apoptotic (Bax and cleaved caspase-3) genes (Figure 6D).
Figure 6.
Knockdown of circSERPINE2 represses development of glioma in vivo
Tumor weight and volume were evaluated in cells transfected with sh-NC and sh-circ#1 (A–C). Protein levels of PCNA, Ki67, cyclin D1, BCL2, BAX, and cleaved caspase-3 were detected (D). ∗∗p < 0.01.
Discussion
Glioblastoma remains a leading brain disease with invariably poor prognosis and high recurrence rate, which eventually results in more aggressive, therapy-resistant relapses in patients.17,18 However, its molecular mechanism of development and progression has not been well-documented. Thus, in the present study, we demonstrated that the upregulation of circSERPINE2 significantly contributed to the development and progression of glioblastoma by downregulating miR-361-3p and miR-324-5p through sponging and upregulating BCL2 gene expression. Specifically, we found that the circSERPINE2 expression was significantly higher in GM tissues compared to the normal brain tissues, and its high expression level was associated with poor prognosis of patients, suggesting that the circSERPINE2 might be actively involved in the progression of GM in patients and might be an important biomarker for the diagnosis, prognosis, and treatment of the disease. Several studies had already shown the importance of circRNAs in the progression of cancers and their potential as biomarkers.19, 20, 21 A study by Jin et al.22 had revealed that the circRNA circHIPK3 might be a prospective therapeutic target and prognostic marker for the effective treatment of glioma. Besides, the downregulation of circRNAs has been observed to have different implications in tumor progress in carcinoma. For instance, a study by Sun et al.23 showed that the downregulation of hsa_circ_0000520 enhances the progress of gastric cancer. In contrast, the downregulation of circ-SERPINE2 inhibits the progress of gastric carcinoma.9 Dysregulation of circRNAs also leads to drug resistance in glioma, which indicated that circRNAs may be potential targets to overcome drug-insensitive. Exosomal circNFIX confers temozolomide-resistance in glioma cells by regulating cell migration, invasion, half maximal inhibitory concentration (IC50) value to temozolomide, and apoptosis.19 circASAP1 could enhance temozolomide-resistance in glioma cells via NRAS/MEK1/ERK-2 signaling.24 Therefore, this study further explored the role of circSERPINE2 in GM and reveals that circSERPINE2 expression reduction was almost halved after the knockdown. Silencing of circ-SERPINE2 in glioma cells promotes cell apoptosis, restrained cell viability, and proliferation by reducing the expression of proliferative protein markers. These suggest that circ-SERPINE2 plays the role of an oncogene in glioma cells.
Furthermore, accumulating evidence has demonstrated that circRNAs sequester with several miRNAs to influence the post-transcriptional actions of miRNAs.25,26 In most cases, they play a role as competing endogenous RNA that binds miRNAs to regulate their expression.25 circRNA hsa_circ_0000670 functions as a tumor promoter by binding with miR-384 to upregulate sine occulis-related homeobox 4 (SIX4) gene and enhance the progress of gastric carcinoma tumor.27 The upregulated expression of circPRKCI glioma tissues has been reported to sponge miR-545.28 Besides, Qu et al.29 demonstrated that the upregulation of circ-0079593 was associated with increased glioma tumor size, which was possibly a result of its ability to sponge miR-182 and miR-433 in the cancer cell. Generally, the miRNA is known to post-transcriptionally regulate gene expression by targeting the 3′ UTR region of a protein-coding gene.30 They have been recognized as important molecular biomarkers in cancer based on their pervasive cancer-associated aberrant expression.31 For example, Li et al.32 showed that the aberrant expression of miR-188-5p promotes the migration and invasion of gastric carcinoma by significantly reducing tumor suppressor gene PTEN. The downregulated expression level of miR-505 has been implicated in breast cancer progression.33 Also, the restored expression of miR-324-5p significantly mitigated gallbladder carcinoma cell migration, invasion, and epithelial-mesenchymal transit (EMT) in vitro,14 while the overexpression of miR-361-3p fosters migration and invasion of pancreatic cancer cell in vitro.10 In our study, the predicted circSERPINE2 target miRNAs, miR-361-3p and miR-324-5p, were found to be remarkably downregulated in GM tumor tissues. Interestingly, we found that the upregulation of miR-361-3p and miR-324-5p could repress the expression of their common target gene: BCL2, which consequently inhibited the progression of glioma, suggesting that miR-361-3p and miR-324-5p functions as a tumor suppressor in glioma cells. The deregulation of BCL2 family members has been implicated in human malignant diseases.16 Repression of BCL2 by miR-15 and miR-16 has been demonstrated to induce apoptosis in chronic lymphocytic leukemia (CLL).34 Our observation from this study reveals that miR-361-3p inhibitor or miR-324-5p inhibitor significantly restores BCL2 expression and also cell proliferation, indicating that BCL2 could promote the progression of glioma cells.
Consequently, this report is the foremost to elucidate the molecular mechanism and functional role of circSERPINE2 in glioma cells. And although there might be some limitations in the current study, we believe that our results would help to further explore the miR-361-3p/miR-324-5p/BCL2 upstream pathway and the association of circSERPINE2 upregulation with pathological differentiation in glioma cells. In summary, the upregulation of circSERPINE2 in human glioma cells and tissues leads to the progression of glioma by sponging miR-361-3p and miR-324-5p mRNA and regulating the BCL2 expression. The miR-361-3p/miR-324-5p/BCL2 signaling axis could be a novel therapeutic target for the effective treatment of glioblastoma. Besides, further experiments, which could determine whether circSERPINE2 is involved in drug resistance in glioma cells, are worthy to be performed in the future.
Conclusions
The circSERPINE2 promotes the progression of glioblastoma by regulating the miR-361-3p/miR-324-5p/BCL2 signaling pathway and could be a promising biomarker for the early diagnosis and effective prognosis of the disease. Also, the miR-361-3p/miR-324-5p/BCL2 signaling axis could be a novel therapeutic target for the treatment of glioblastoma.
Materials and methods
Heatmap analysis
The heatmap analysis was done by analyzing the GEO (GSE146463) chip data using the R package Limma R.35 The significantly upregulated and downregulated circRNA was retrieved and a volcano plot was used to present the results using R language.
Human tissue sample collection
46 pairs of human glioma samples and normal healthy brain samples surgically resected from glioma patients and volunteers that undertook surgery at Fudan University Shanghai Cancer Center were used for this study. The excised samples were kept in liquid nitrogen at −80°C immediately after collection. This study was permitted by the Ethics Committee of Fudan University Shanghai Cancer Center.
Cell culture
The human glioma cells (A172, U251, U87, and SHG44) were obtained from American Type Culture Collection (ATCC, USA) together with the NHAs, and cultured in Dulbecco’s modified Eagle’s medium (DMEM, USA) complimented with 10% fetal bovine serum (GIBCO, USA), penicillin (100 unit ml/L, GIBCO, USA), and streptomycin (100 μg ml/L, GIBCO, USA) with 5% CO2 at 37°C. The mutational status of cancer-related genes of the glioblastoma cell lines A172, U251, U87, and SHG44 was shown in Table S2.
Cell transfection
miR-361-3p/324-5p inhibitors and the NC inhibitor were acquired from RiboBio (China). shRNA targeting circSERPINE2 sequence (sh-circ#1 and sh-circ#2) was used for silencing the expression of circSERPINE2 in this study (GeneChem, China). The transfection was done using Lipofectamine 2000 reagent (Invitrogen, USA). After, the cellular effect of this transfection was investigated through various molecular experiments such as cell viability and apoptosis. The sequence of small interfering RNAs (siRNAs) was shown in Table S1.
Quantitative real-time PCR
Isolation of the RNA was performed with the TRIzol reagent (Takara, Japan), and the isolated RNA sample was exposed to RNase R (Geneseed, China) for 30 min to obtain high-quality circRNAs. The RNA was transcribed reversely to cDNA with a specific reverse transcription kit (Thermo Fisher Scientific, USA). The transcribed RNA (cDNA) was mixed with SYBR Green (Takara, Japan), and specific primers were used to perform quantitative real-time PCR analysis using ABI 7900 system (ABI, USA). GAPDH and U6 acted as internal controls, while the relative gene expression was estimated using the 2–ΔΔCt method.2 The sequence of primers was shown in the Table S1.
CCK-8 assay
Cells transfected were cultured in a 96-well plate (2 × 103 cells per well). 10 μL of CCK-8 reagent (Dojindo, Japan) was added into the medium and then incubated for 2 h. After, the microplate reader (BioTek, USA) was used to measure the absorbance of the cell at 450 nm after 0, 24, 48, 72, and 96 h of transfection. These results were then used to estimate cell viability.
Cell apoptosis assay
Cell apoptosis was determined by a flow cytometer. The cell lines were suspended in 400 μL of 1× binding buffer comprising 10 mM HEPES/NaOH, NaCl (140 mM), and CaCl2 (2.5 mM) at pH 7.4 and were kept in annexin V-fluorescein isothiocyanate (5 μL) and propidium iodide (5 μL; PI, KeyGEN, China) for 10 mins in the dark. The fluorescence expression was evaluated by a flow cytometer (BD Biosciences, USA).
Colony formation assay
U251 and U87 cells were trypsinized. Then, 1 × 103 of the cells were placed into 6-well plates, transfected with sh-NC, sh-circ#1, and sh-circ#2, and later incubated for 10 days at 37°C. A dyeing solution consisting of methanol (20%) and crystal violet (0.1%) was used on the colonies, which was counted and analyzed afterward.
Western blot assay
Lysis of the glioma cells was done with lysis buffer and proteinase inhibitor (Invitrogen, USA), and the protein quantification was carried out using the bicinchoninic acid (BCA) protein assay (Bio-Rad, USA). Subsequently, the same lysed protein was loaded onto sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, Thermo Fisher Scientific, USA), and moved onto polyvinylidene difluoride (PVDF; Thermo Fisher Scientific, USA). Membranes were masked with skim milk solution (5%), reacted with the primary antibodies (PCNA, Ki67, cyclin D1, BCL2, Bax, cleaved-caspase-3; 1:1,000, Abcam, USA) and GADPH (1:3,000, Abcam, USA) as a reference control. After washing, the membranes were probed with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:2,000 dilution, USA) at room temperature. Then, visualization of the antibody binding was done and evaluated with enhanced chemiluminescence (ECL) Western Blotting Detection Kit (Solarbio, China) and ImageJ software (National Institutes of Health, USA), respectively.
Luciferase reporter assay
Amplification of the WT and mut sequences of circSERPINE2 3′ UTR was done by PCR using human genomic DNA of the HEK293T cells line and cloned into the fluorescein reporter vector plasmid (Promega, USA). U251 and U87 cells (5 × 104 cells/well) were placed in 24-well plates and co-transfected with miR-361-3p mimics, miR-324-5p mimics, and WT or mut 3′ UTR of circSERPINE2 (has-circ-0001103) with Lipofectamine 3000 reagent (Invitrogen, USA) in accordance with the manufacturer’s instruction. The Renilla plasmid (RL-SV40) was employed as the reference. After 48 h of transfection, the luciferase activity of the cell was estimated with the dual-luciferase reporter assay system (Promega, USA) following the producer’s guide.
Biotinylated RNA pull-down assay
The RNA precipitation assay was performed to investigate the coupling ability of circSERPINE2 with the RNA-induced silencing complex (RISC), using synthesized has-circ-0001103 as a probe. Lysates of U251 and U87 were incubated with synthesized biotin-labeled sense and anti-sense DNA probes for 3 h. Then, streptavidin-coupled agarose beads (Invitrogen, USA) were added to the binding reaction mixture for RNA complex isolation and incubated at room temperature for 1 h. The expression levels of the miR-361-3p and miR-324-5p were determined using quantitative real-time PCR.
RIP assay
The interaction between miR-361-3p and miR-324-5p and mRNA was confirmed with RIP assay through the binding of miR-361-3p and miR-324-5p via Ago2. RIP assay was performed with a Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore, USA) according to the producer’s protocol. RIP assays antibodies against AGO2 and IgG were obtained from Abcam (ab5072, rabbit polyclonal antibody, USA).
In vivo assays for tumor growth
To investigate tumor growth through in vivo method, we developed a xenograft model with 6- to 8-week-old nude mice (Shanghai Laboratory Animal Center, China). Approximately 1 × 106 suspended transfected cells with sh-NC and sh-circ#1 were inoculated subcutaneously into each mouse. Tumor growth was estimated every 7 days, and tumor volumes were calculated (0.5 × length × width). After the inoculation, the mice were sacrificed, and tumors were excised and weighed. Proteins from the tumor specimens were extracted and the expression of PCNA, Ki67, cyclin D1, BCL2, BAX, and cleaved caspase-3 was detected.
Target gene prediction
Prediction of circSERPINE2 (has-circ-0001103) target miRNA (miR-361-3p and miR-324-5p) was done using circRNA interactome (https://circinteractome.nia.nih.gov/). StarBase v2.0 (http://starbase.sysu.edu.cn/) was used to predict the miRNAs target mRNA (gene).
Statistical analysis
All the experiments were carried out in triplicate. The data are shown as mean ± standard deviation (SD), and the comparisons between the two-group data were examined by the Pearson’s correlation, while chi-square/ANOVA analysis was done to compare the differences in multiple groups. All the data were analyzed by SPSS 17.0 and Graphpad Prism 7. The significant difference was taken to be p < 0.05.
Acknowledgments
This work was supported by the National Nature Science Foundation of China (grant number 81802494), Shanghai Anticancer Association EYAS PROJECT (grant number SACA-CY20B03), and Shanghai Municipal Commission of Health and Family Planning Foundation, China (grant number 20184Y0107).
Author contributions
D.L. and L.L. designed the project, collected data, analyzed the data, and drafted the manuscript. X.C. and W.Y. did almost all the experiments. Y.C. was involved in data collection and analysis and revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2021.07.010.
Contributor Information
Wentao Yang, Email: yangwt2000@163.com.
Yiqun Cao, Email: yiqun_fduscc@163.com.
Supplemental information
References
- 1.Chen R., Smith-Cohn M., Cohen A.L., Colman H. Glioma Subclassifications and Their Clinical Significance. Neurotherapeutics. 2017;14:284–297. doi: 10.1007/s13311-017-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batash R., Asna N., Schaffer P., Francis N., Schaffer M. Glioblastoma Multiforme, Diagnosis and Treatment; Recent Literature Review. Curr. Med. Chem. 2017;24:3002–3009. doi: 10.2174/0929867324666170516123206. [DOI] [PubMed] [Google Scholar]
- 3.Le Rhun E., Preusser M., Roth P., Reardon D.A., van den Bent M., Wen P., Reifenberger G., Weller M. Molecular targeted therapy of glioblastoma. Cancer Treat. Rev. 2019;80:101896. doi: 10.1016/j.ctrv.2019.101896. [DOI] [PubMed] [Google Scholar]
- 4.Vastrad B., Vastrad C., Godavarthi A., Chandrashekar R. Molecular mechanisms underlying gliomas and glioblastoma pathogenesis revealed by bioinformatics analysis of microarray data. Med. Oncol. 2017;34:182. doi: 10.1007/s12032-017-1043-x. [DOI] [PubMed] [Google Scholar]
- 5.Sasmita A.O., Wong Y.P., Ling A.P.K. Biomarkers and therapeutic advances in glioblastoma multiforme. Asia Pac. J. Clin. Oncol. 2018;14:40–51. doi: 10.1111/ajco.12756. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhary R., Muys B.R., Grammatikakis I., De S., Abdelmohsen K., Li X.L., Zhu Y., Daulatabad S.V., Tsitsipatis D., Meltzer P.S. Levels; 2020. A Circular RNA from the MDM2 Locus Controls Cell Cycle Progression by Suppressing; p. 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan Z., Bai Y., Zhang Q., Qian P. CircRNA circ_POLA2 promotes lung cancer cell stemness via regulating the miR-326/GNB1 axis. Environ. Toxicol. 2020;35:1146–1156. doi: 10.1002/tox.22980. [DOI] [PubMed] [Google Scholar]
- 8.Dong W., Bi J., Liu H., Yan D., He Q., Zhou Q., Wang Q., Xie R., Su Y., Yang M. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol. Cancer. 2019;18:95. doi: 10.1186/s12943-019-1025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J., Song S., Lin S., Zhang M., Du Y., Zhang D., Xu W., Wang H. Circ-SERPINE2 promotes the development of gastric carcinoma by sponging miR-375 and modulating YWHAZ. Cell Prolif. 2019;52:e12648. doi: 10.1111/cpr.12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu J., Li L., Chen H., Zhang G., Liu H., Kong R., Chen H., Wang Y., Li Y., Tian F. MiR-361-3p regulates ERK1/2-induced EMT via DUSP2 mRNA degradation in pancreatic ductal adenocarcinoma. Cell Death Dis. 2018;9:807. doi: 10.1038/s41419-018-0839-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J., Yang B., Wang L., Zhu Y., Zhu X., Xia Z., Zhao Z., Xu L. LncRNA BBOX1-AS1 upregulates HOXC6 expression through miR-361-3p and HuR to drive cervical cancer progression. Cell Prolif. 2020;53:e12823. doi: 10.1111/cpr.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W., Wang J., Liu S., Wang S., Cheng Y., Zhou W., Duan C., Zhang C. MicroRNA-361-3p suppresses tumor cell proliferation and metastasis by directly targeting SH2B1 in NSCLC. J. Exp. Clin. Cancer Res. 2016;35:76. doi: 10.1186/s13046-016-0357-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao D., Cui Z. MicroRNA-361-3p regulates retinoblastoma cell proliferation and stemness by targeting hedgehog signaling. Exp. Ther. Med. 2019;17:1154–1162. doi: 10.3892/etm.2018.7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X., Zhang L., Chen M., Liu D. miR-324-5p inhibits gallbladder carcinoma cell metastatic behaviours by downregulation of transforming growth factor beta 2 expression. Artif. Cells Nanomed. Biotechnol. 2020;48:315–324. doi: 10.1080/21691401.2019.1703724. [DOI] [PubMed] [Google Scholar]
- 15.Xu H.S., Zong H.L., Shang M., Ming X., Zhao J.P., Ma C., Cao L. MiR-324-5p inhibits proliferation of glioma by target regulation of GLI1. Eur. Rev. Med. Pharmacol. Sci. 2014;18:828–832. [PubMed] [Google Scholar]
- 16.Frenzel A., Grespi F., Chmelewskij W., Villunger A. Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis. 2009;14:584–596. doi: 10.1007/s10495-008-0300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campos B., Olsen L.R., Urup T., Poulsen H.S. A comprehensive profile of recurrent glioblastoma. Oncogene. 2016;35:5819–5825. doi: 10.1038/onc.2016.85. [DOI] [PubMed] [Google Scholar]
- 18.Davis M.E. Glioblastoma: Overview of Disease and Treatment. Clin. J. Oncol. Nurs. 2016;20(5, Suppl):S2–S8. doi: 10.1188/16.CJON.S1.2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding C., Yi X., Wu X., Bu X., Wang D., Wu Z., Zhang G., Gu J., Kang D. Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 2020;479:1–12. doi: 10.1016/j.canlet.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y., Liu H., Li W., Yu J., Li J., Shen Z., Ye G., Qi X., Li G. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY) 2017;9:1585–1594. doi: 10.18632/aging.101254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su Y., Lv X., Yin W., Zhou L., Hu Y., Zhou A., Qi F. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY) 2019;11:8183–8203. doi: 10.18632/aging.102312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin P., Huang Y., Zhu P., Zou Y., Shao T., Wang O. CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem. Biophys. Res. Commun. 2018;503:1570–1574. doi: 10.1016/j.bbrc.2018.07.081. [DOI] [PubMed] [Google Scholar]
- 23.Sun H., Tang W., Rong D., Jin H., Fu K., Zhang W., Liu Z., Cao H., Cao X. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21:299–306. doi: 10.3233/CBM-170379. [DOI] [PubMed] [Google Scholar]
- 24.Wei Y., Lu C., Zhou P., Zhao L., Lyu X., Yin J., Shi Z., You Y. EIF4A3-induced circular RNA ASAP1 promotes tumorigenesis and temozolomide resistance of glioblastoma via NRAS/MEK1/ERK1-2 signaling. Neuro-oncol. 2021;23:611–624. doi: 10.1093/neuonc/noaa214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mumtaz P.T., Taban Q., Dar M.A., Mir S., Haq Z.U., Zargar S.M., Shah R.A., Ahmad S.M. Deep Insights in Circular RNAs: from biogenesis to therapeutics. Biol. Proced. Online. 2020;22:10. doi: 10.1186/s12575-020-00122-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou S.Y., Chen W., Yang S.J., Xu Z.H., Hu J.H., Zhang H.D., Zhong S.L., Tang J.H. The emerging role of circular RNAs in breast cancer. Biosci. Rep. 2019;39 doi: 10.1042/BSR20190621. BSR20190621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu P., Cai S., Li N. Circular RNA-hsa-circ-0000670 promotes gastric cancer progression through the microRNA-384/SIX4 axis. Exp. Cell Res. 2020;394:112141. doi: 10.1016/j.yexcr.2020.112141. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X., Yang H., Zhao L., Li G., Duan Y. Circular RNA PRKCI promotes glioma cell progression by inhibiting microRNA-545. Cell Death Dis. 2019;10:616. doi: 10.1038/s41419-019-1863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu Y., Zhu J., Liu J., Qi L. Circular RNA circ_0079593 indicates a poor prognosis and facilitates cell growth and invasion by sponging miR-182 and miR-433 in glioma. J. Cell. Biochem. 2019;120:18005–18013. doi: 10.1002/jcb.29103. [DOI] [PubMed] [Google Scholar]
- 30.Cannell I.G., Kong Y.W., Bushell M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- 31.Lan H., Lu H., Wang X., Jin H. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. BioMed Res. Int. 2015;2015:125094. doi: 10.1155/2015/125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Yan X., Shi J., He Y., Xu J., Lin L., Chen W., Lin X., Lin X. Aberrantly expressed miR-188-5p promotes gastric cancer metastasis by activating Wnt/β-catenin signaling. BMC Cancer. 2019;19:505. doi: 10.1186/s12885-019-5731-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J., Liu H., Li M. Downregulation of miR-505 promotes cell proliferation, migration and invasion, and predicts poor prognosis in breast cancer. Oncol. Lett. 2019;18:247–254. doi: 10.3892/ol.2019.10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cimmino A., Calin G.A., Fabbri M., Iorio M.V., Ferracin M., Shimizu M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.