Graphical abstract
Abbreviations: PCa, prostate cancer; mCRPC, metastatic castration-resistant prostate cancer; TCGA, The Cancer Genome Atlas; miRNAs, microRNAs; MIRcor, miRNA-correlated; SCNAs, somatic copy number alterations; k-NN, K-nearest neighbor; GEO, Gene Expression Omnibus; DFS, disease-free survival; OS, overall survival; BCR, biochemical recurrence; NMF, non-negative matrix factorization; FDR, false discovery rate; NTP, Nearest template prediction; SubMap, Subclass mapping; DEmiRs, differentially expressed miRNAs; GSEA, Gene Set Enrichment Analysis; GEP, gene expression profile; TNAs, tumor neoantigens; TMB, tumor mutation burden; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; ssGSEA, single-sample gene set enrichment analysis; CCLs, cancer cell lines; AUC, Area under the dose-response curve; CTLA-4, cytotoxic T-lymphocyte associated protein-4; PD-1, programmed cell death protein-1; PD-L1, programmed death-ligand 1; TGFβ, transforming growth factor β; AR, androgen receptor; NEPC, neuroendocrine prostate cancer; EMT, epithelial-mesenchymal transition; IFN, interferon; MDSCs, myeloid-derived suppressor cells; Tregs, regulatory T cells; CAFs, cancer-associated fibroblasts; ADT, androgen deprivation therapy; ICB, immune checkpoint blockade
Keywords: Prostate cancer, miRNAs, Molecular subtypes, Heterogeneity
Abstract
MicroRNA (miRNA) deregulation plays a critical role in the heterogeneous development of prostate cancer (PCa) by tuning mRNA levels. Herein, we aimed to characterize the molecular features of PCa by clustering the miRNA-regulated transcriptome with non-negative matrix factorization. Using 478 PCa samples from The Cancer Genome Atlas, four molecular subtypes (S-I, S-II, S-III, and S-IV) were identified and validated in two merged microarray and RNAseq datasets with 656 and 252 samples, respectively. Interestingly, the four subtypes showed distinct clinical and biological features after comprehensive analyses of clinical features, multiomic profiles, immune infiltration, and drug sensitivity. S-I is basal/stem/mesenchymal-like and immune-excluded with marked transforming growth factor β, epithelial-mesenchymal transition and hypoxia signals, increased sensitivity to olaparib, and intermediate prognosis. S-II is luminal/metabolism-active and responsive to androgen deprivation therapy with frequent TMPRSS2-ERG fusion and a good prognosis. S-III is characterized by moderate proliferative and metabolic activity, sensitivity to taxane-based chemotherapy, and intermediate prognosis. S-IV is highly proliferative with moderate EMT and stemness, frequent deletions of TP53, PTEN and RB, and the poorest prognosis; it is also immune-inflamed and sensitive to anti-PD-L1 therapy. Overall, based on miRNA-regulated gene profiles, this study identified four distinct PCa subtypes that could improve risk stratification at diagnosis and provide therapeutic guidance.
1. Introduction
Prostate cancer (PCa) is the second-most (14.1%) commonly diagnosed cancer and fifth (6.8%) leading cause of cancer death among men [1]. Despite the high incidence rate, a proportion of localized PCa is relatively indolent with good prognosis and therefore, these patients should be spared from invasive treatments and concomitant complications [2], [3]. On the other hand, a subset of PCa is clinically significant and aggressive, and tends to progress into metastatic castration-resistant prostate cancer (mCRPC) with limited therapeutic options [4], [5]. Early identification of this subset of patients will allow for more active interventions and close follow-up, which could theoretically reduce relapse and prolong survival. Attributed to the inherent genetic heterogeneity of PCa [6], widely used risk stratification tools including serum prostate-specific antigen level, TNM and Gleason score often fail to pinpoint PCa patients with either aggressive or indolent forms of the disease [7], [8], [9]. As a result, these patients are at risk of undertreatment or overtreatment [2]. To address this challenge, genomic profiling has been exploited to characterize the molecular heterogeneity of PCa and has been proven to increase the accuracy of risk predictions in clinical practice [10], [11]. Recently, The Cancer Genome Atlas (TCGA) used somatic mutations or transcript fusions to define PCa subtypes [12]. However, this classification fails to discriminate patients with distinct prognoses, and therefore, its use in routine clinical practice is severely compromised. Additionally, most published molecular classifications did not investigate whether different molecular subtypes respond differently to conventional therapies [12], [13], [14], [15], [10], which limited their application into clinical practice.
MicroRNAs (miRNAs) are a group of small non-coding regulatory RNA molecules that repress mRNA translation or promote degradation by binding to targeted mRNAs [16]. Mounting evidence suggests that miRNA aberrations occur frequently in PCa and play key roles in PCa initiation and progression [17], [18]. Nevertheless, it remains unclear to what extent miRNA deregulation affects PCa heterogeneity, and thus molecular subtyping and therapeutic response.
Herein, using miRNA-correlated (MIRcor) genes, we sought to develop and validate a novel molecular classification that could accurately identify PCa patients with differential prognoses and therapeutic responses. We intended to characterize the identified PCa subtypes at the epigenetic, genomic, transcriptional, immune, and clinical levels. We also sought to determine potential subtype-driven miRNAs in each subtype.
2. Materials and methods
2.1. Data collection and preprocessing
Mature miRNA expression (level 3, Illumina miRNA-Seq), processed mRNA expression (level 3, Illumina RNASeq), 450 K methylation data (level 3, Illumina HumanMethylation450 BeadChip), somatic copy number alterations (SCNAs, level 3, Affymetrix SNP 6.0 array), somatic mutation (level 4, MAF files), and clinical information of PCa patients up to November 2020 were obtained from TCGA database (http://cancergenome.nih.gov/). We filtered out miRNA, mRNA and CpG sites with>25% missing values across all samples and imputed the remaining missing values using the K-nearest neighbor (k-NN) approach in the ‘impute’ R package. We used log2 (normalized count + 1) as the expression levels of mRNA and miRNA. DNA methylation probes were filtered and normalized using the R ‘ChAMP’ package. We only selected CpG sites residing in the promoter region (TSS1500, TSS200, 5′UTR, and 1stExon). Somatic variants were detected using the TCGA MuTect2 pipeline and further analyzed by the R ‘maftools’ package. GISTIC2.0 (Gene Pattern) was used to infer recurrent SCNAs [19]. After removing ineligible samples, 478 treatment-naïve PCa patients with complete prognostic information were selected for subsequent analyses (Table S1).
For external validation, we retrieved 8 independent datasets with available GEPs and matched prognostic information, including GSE116918 (n = 248), GSE70769 (n = 92), GSE70768 (n = 111), GSE107299 (n = 65), and GSE54460 (n = 91) from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/), and DKFZ-PRAD (n = 81), SU2C-PRAD (n = 80) and MSKCC-PRAD (n = 140) from the cBioPortal dataset (https://www.cbioportal.org/). Raw expression data from Affymetrix were background adjusted and normalized using the robust multiarray average algorithm (‘affy’ R package). The fragments per kilobase per million values in RNA sequencing cohorts were also transformed into transcripts per million values that were more comparable with the microarray data [20]. Next, the RNAseq- and microarray-based cohorts were merged into two meta-cohorts (Table S2 and Table S3), respectively. Batch effects were removed by ‘combat’ functions in the ‘sva’ R package (Fig. S1). Disease-free survival (DFS), overall survival (OS), and biochemical recurrence (BCR) rates were all considered as endpoint events but were analyzed separately.
2.2. Identification of miRNA-mRNA pairs
A two-step approach was adopted to identify miRNA-correlated genes as in other miRNA studies [21], [22]. First, three miRNA databases were used to predict potential mRNA targets of miRNAs in the TCGA-PRAD dataset. Second, we computed the Pearson correlation coefficients of the intersecting miRNA-mRNA pairs, with Benjamini and Hochberg corrections (q-values) to control for multiple testing. We selected only those with q-values < 0.05 and correlation coefficients < -0.3 for downstream analysis because miRNAs have been widely shown and validated to reduce mRNA levels through complementary interactions between their seeding sequences and the 3′ untranslated regions of targeted mRNAs [16].
2.3. Development and validation of molecular subtypes
Non-negative matrix factorization (NMF, R ‘NMF’ package) was applied to 570 MIRcor gene expression profiles with the setting of 30 iterations for k = 2 to k = 6. The optimal number of clusters was determined by taking into account the cophenetic coefficient and consensus maps from 2 to 6 clusters.
To validate the classification, we first identified differentially expressed genes among the four subtypes using the R ‘limma’ package. A false discovery rate (FDR) < 0.05 and log2 |fold change| >1 were used as the cutoff criteria when comparing each subtype with the rest. The gene signature for each subtype was defined as the unique and upregulated mRNAs in the corresponding subtype. Next, using subtype-specific signatures, the nearest template prediction (NTP) algorithm [23] (Gene Pattern) was employed to classify PCa patients in two merged datasets. To test the robustness of class assignment, we conducted subclass mapping (SubMap) analyses [24] (Gene Pattern) to evaluate the similarity of molecular subtypes between validation cohorts and the TCGA-PRAD cohort.
2.4. Associations of PCa subtypes with multiomics data in TCGA-PRAD
We first performed differential expression analysis of miRNAs by comparing each subtype to other tumor samples. FDR < 0.05 and log2 |fold change| >0.5 were used to define differentially expressed miRNAs (DEmiRs). We also computed and compared the mean expression of miRNAs of each subtype and adjacent normal samples. Furthermore, to search for DEmiRs potentially driving PC subtypes, we defined miRNA gene sets using MIRcor genes (correlation coefficients < 0 and q values < 0.05) and conducted Gene Set Enrichment Analysis (GSEA) to determine contributions of candidate miRNAs to gene expression profiles (GEPs) of PCa subtypes. Normalized enrichment score > 1.5 and adjusted P values < 0.05 were used as the cutoff points.
For DNA methylation analysis, we calculated and compared the mean levels of promoter CpG methylation in each subtype. Differentially altered and mutated genes were detected by comparing their distributions among the subtypes. The focal and arm level changes of SCNAs were determined as the total number of altered mRNAs with absolute CNA values>0.2. The predicted aneuploidy score, tumor neoantigens (TNAs) and tumor mutation burden (TMB) data were extracted from a previous study by Thorsson et al. [25].
2.5. Biological characterization of PC subtypes
Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) were analyzed via the ‘clusterProfiler’ R package. Single-sample gene set enrichment analysis (ssGSEA, ‘GSVA’ R package) was performed on molecular pathways from the Molecular Signature Database and curated gene sets representing lineage plasticity, TMPRSS-ERG fusion, various biological processes and the immune microenvironment (Table S4). Meanwhile, we used the R ‘estimate’ package to infer tumor purity and estimate the degree of immune and stromal infiltration.
2.6. Exploration of therapeutic vulnerabilities across PC subtypes
Sequencing data of human cancer cell lines (CCLs) and drug sensitivity data against 482 CCLs were retrieved from the Cancer Cell Line Encyclopedia project (https://portals.broadinstitute.org/ccle/) and the secondary PRISM Repurposing dataset (https://depmap.org/portal/prism/), respectively. Area under the dose–response curve (AUC) values were used to measure drug sensitivity with lower AUC values indicating increased sensitivity. We applied ridge regression models (R ‘pRRophetic’ package) to estimate the AUC values of all PCa patients. Moreover, we used SubMap analysis to infer the efficacy of enzalutamide, anti-cytotoxic T-lymphocyte associated protein-4 (CTLA-4), anti-programmed cell death protein-1 (PD-1) and anti-programmed death-ligand 1 (PD-L1) therapies in PCa subtypes by evaluating the similarity of GEPs between the PCa subtypes and patients receiving corresponding therapies in previous studies [26], [27], [28].
2.7. Statistical analysis
All statistical and bioinformatics analyses were conducted with R software version 4.0.2 and Gene Pattern. Associations between PCa subtypes and categorical data were assessed by Fisher’s exact or chi-square tests. For continuous variables, the Student’s t-test or Wilcoxon rank-sum test was used to compare two groups while the Kruskal–Wallis test or ANOVA was used for more than two groups. Survival analyses were carried out by Kaplan–Meier curves and log-rank tests. A two-sided P value < 0.05 was recognized as statistically significant.
3. Results
3.1. Identifying the miRNA-regulated transcriptional subtypes in PCa
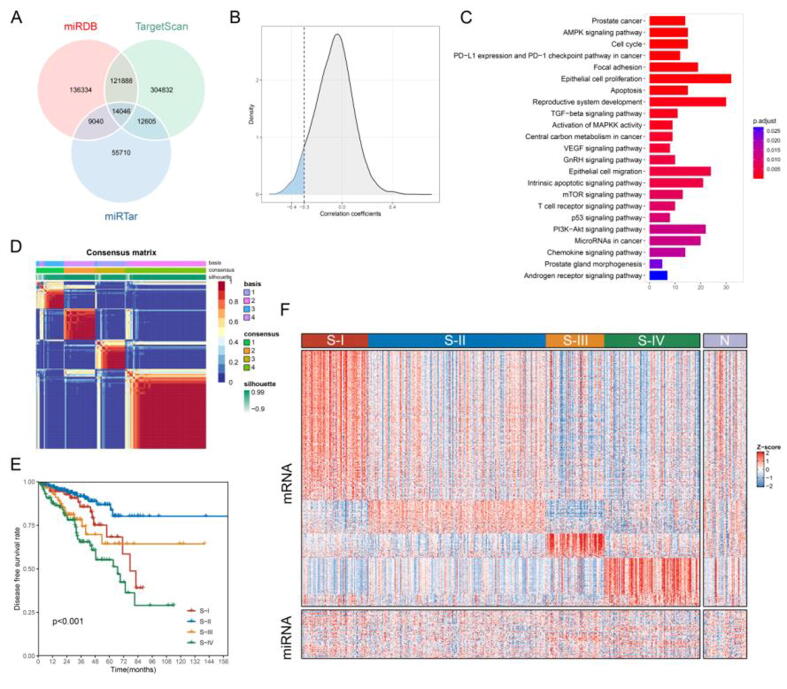
A flow chart was produced to summarize this study (Fig. 1). First, we used three miRNA databases to predict potential mRNA targets of 494 miRNAs among 16,766 mRNAs in 478 PCa samples of the TCGA-PRAD dataset. As shown in Fig. 2A, we obtained 281,308 miRNA-mRNA pairs from 487 miRNAs and 15,186 mRNAs in the miRDB database [29], 91,401 miRNA-mRNA pairs from 494 miRNAs and 12,791 mRNAs in the miRTar database [30], and 453,371 miRNA-mRNA pairs from 489 miRNAs and 16,178 mRNAs in the TargetScan v7.2 database [31], respectively. We then selected 14,046 pairs of miRNAs (n = 461) and mRNAs (n = 4565) that coexisted in the three databases. Eventually, using Pearson correlation coefficients (<-0.3; Fig. 2B) and q-values (<0.05; Table S5), we identified 570 MIRcor genes. KEGG and GO analyses revealed that these MIRcor genes were significantly enriched in epithelial cell proliferation and migration, cell cycle, reproductive system development, PI3K-Akt signaling pathway, p53 signaling pathway, T cell receptor signaling pathway, and transforming growth factor β (TGFβ) signaling pathway (Fig. 2C and Table S6). These results suggested that MIRcor genes might play important and diverse roles in regulating the biological functions of both prostate tumor cells and the tumor microenvironment.
Fig. 1.
The flow chart of the present study. The prostate cancer classification is developed by the TCGA-PRAD cohort (n = 478) and then validated by two merged datasets, i.e. the microarray cohort (6 public datasets, n = 656) and RNAseq cohort (3 public datasets, n = 252).
Fig. 2.
Identification of miRNA-correlated mRNAs and prostate cancer (PCa) subtypes with distinct prognoses and gene expression profiles. (A) Overlap of the miRNA-mRNA pairs identified in three miRNA databases. (B) Distribution of Pearson correlation coefficients for intersecting miRNA-mRNA pairs in the TCGA-PRAD cohort. Pearson correlation coefficients less than −0.3 with q values < 0.05 were used to define miRNA-correlated (MIRcor) genes. (C) KEGG and GO analyses of MIRcor genes. (D) Consensus map (k = 4) of non-negative matrix factorization classification based on MIRcor genes. Four PCa subtypes (S-I, S-II, S-III, and S-IV) were identified in the TCGA-PRAD cohort. (E) Kaplan-Meier curves show distinct prognoses among four PCa subtypes. (F) Heatmap of differentially expressed mRNAs and miRNAs among four subtypes and adjacent normal samples in the TCGA-PRAD cohort.
Next, we applied NMF to 570 MIRcor genes and chose k = 4 as the optimal number of clusters according to the cophenetic correlation coefficients and consensus maps (Fig. 2D and Fig. S2). Four subclasses were therefore identified in the TCGA-PRAD cohort (S-I, n = 80; S-II, n = 213; S-III, n = 70; S-IV, n = 115). Kaplan–Meier analysis revealed that the four subclasses had distinct prognostic differences (DFS, p < 0.001; Fig. 2E). S-IV (24.1%) was associated with the poorest prognosis while the largest subtype, S-II (44.6%) showed the best prognosis. The prognoses of S-I (16.7%) and S-III (14.6%) were somewhere in between. These results indicated that the MIRcor gene expression profile could stratify PCa patients into distinct prognostic groups.
3.2. Validation of PCa subtypes across independent datasets
To verify the robustness of the clustering method, differential expression analysis was first conducted to reveal 443 significantly upregulated genes (Table S7) with 260 for S-I, 57 for S-II, 42 for S-III, and 84 for S-IV, as shown in Fig. 2F. Pathway analysis of subtype-specific genes (Fig. S3 and Table S8) showed that oxidative phosphorylation and mitochondrial protein complexes were significantly enriched in S-III and that mesenchymal-related pathways such as extracellular matrix modeling, focal adhesion and muscle contraction were overrepresented in S-I. S-IV-specific genes were more involved in cell division-relevant pathways while S-II-specific genes were significantly associated with secretory and transport processes. These results highlighted differentially active pathways among the four PCa subtypes and further supported the rationale of using MIRcor genes for class discovery.
Next, subtype-specific genes were used to assign class labels to each sample in two merged datasets by the NTP algorithm. SubMap analyses showed that the subtypes in both merged datasets were specifically and significantly correlated with the corresponding subtypes in TCGA-PRAD (all p < 0.01, Fig. 3A). Moreover, similar prognostic differences were verified in the two merged datasets (BCR, p < 0.001; Fig. 3B). All independent cohorts demonstrated similar results (GSE54460: BCR, p = 0.001; GSE70768: BCR, p < 0.001; GSE70769: BCR, p < 0.001; GSE107299: BCR, p = 0.047; GSE116918: BCR, p = 0.002; DKFZ-PRAD: BCR, p < 0.001; MSKCC-PRAD: DFS, p < 0.001; SU2C-PRAD: OS, p = 0.002; Fig. 3C).
Fig. 3.
Independent validation of the prognostic significance of PCa subtypes across eight external cohorts. (A) Subclass mapping analyses demonstrated that corresponding subtypes were specifically and significantly correlated among the TCGA-PRAD, Microarray, and RNAseq cohorts. (B) Kaplan-Meier curves demonstrated distinct biochemical recurrence rates (BCR) among four PCa subtypes in six public datasets. (C) Kaplan-Meier curves for each public dataset showing similar and significant differences of BCR, DFS, or OS among PCa subtypes.
3.3. Associations of the PCa subtypes with miRNA, DNA methylation, somatic copy number alterations and mutations
We first analyzed the miRNA patterns among four PCa subtypes. Compared with normal samples, the four subtypes showed higher levels of mean miRNA expression (Fig. 4A). No significant difference was observed among subtypes except that S-IV had a higher level of miRNA expression than S-II (p < 0.001). Differential analyses revealed that S-II exhibited fewer DEmiRs (n = 9) than other subtypes (S-I, n = 29; S-III, n = 17; S-IV, n = 30; Fig. 4B and Table S9). Next, GSEA was exploited to select DEmiRs that were overrepresented in the corresponding subtypes (Fig. 4C and Table S10). The results showed that four upregulated and two downregulated DEmiRs were significantly enriched in S-III and S-II, respectively. Hsa-miR-103a-3p enriched in S-III belongs to the miR-103 family, which is responsible for targeting genes involved in oxidative phosphorylation in PCa [18]. Of seven DEmiRs enriched in S-IV, upregulated hsa-miR-106-5p and downregulated hsa-miR-135a-5p had been found significantly associated with very high-risk aggressive prostate cancer at circulating levels [32]. S-IV was also overrepresented by downregulated hsa-miR-30d-5p, which inhibited PCa cell proliferation and invasion by targeting Ecto-5′-nucleotidase [33]. In contrast, overexpressed hsa-miR-30d-5p was significantly enriched in S-I and was correlated with low androgen receptor (AR) activity and inhibition of PCa cell growth [34], [33]. Altogether, these results suggested that S-II was less affected by miRNA aberrations, whereas S-IV and S-I were heavily regulated. Furthermore, the identified subtype-driven miRNAs have been extensively linked to the distinct biological features of PCa subtypes (Fig. S4 and Table S11) with some experimentally validated to act as tumor-suppressive or oncogenic miRNAs.
Fig. 4.
The genomic and epigenetic landscape of the PCa subtypes in TCGA-PRAD. (A) Comparison of mean miRNA expression level among four PCa subtypes and normal samples. (B) A Venn diagram shows the number of unique and overlapping differentially expressed miRNAs (DEmiRs) among subtypes. (C) GSEA of subtype-specific DEmiRs identifies a subset of miRNAs that significantly contribute to the mRNA expression profiles of corresponding subtypes. (D) The levels of promoter methylation, somatic mutation, tumor neoantigen and aneuploidy were compared across subtypes. (E) The burden of copy number gain and loss at focal and arm levels in the four PCa subtypes. Oncoprints of top 20 somatic mutations (F) and somatic recurrent copy number alterations (G) among subtypes. The proportion of alteration of listed genes was shown on the right side. “ns” denotes no statistical significance, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To elucidate epigenetic and genomic alterations among PCa subtypes, we calculated methylation levels, somatic mutations, and SCNAs. Promoter methylation levels were comparable among S-II, S-III and S-IV except that S-I showed a lower level of methylation (p < 0.01; Fig. 4D). S-IV had the highest aneuploidy score and TMB, followed by S-III (p < 0.05; Fig. 4D). Both S-IV and S-III showed higher TNA loads than S-I and S-II (p < 0.05; Fig. 4D). Compared with other subtypes, S-IV had the highest burden in copy number gain and loss at both focal and arm levels (p < 0.001; Fig. 4E). Specifically, several tumor-suppressive genes (i.e. TP53, RB1, and PTEN) were more frequently deleted in S-IV (p < 0.001; Fig. 4F and Table S12). MYC and CCND1 amplifications were significantly observed in S-IV and least seen in S-II (p < 0.001). We also identified the top 20 frequently mutated genes in PCa (Fig. 4G and Table S13) and found that TP53 (31.30%, p < 0.001), FOXA1 (13.04%, p < 0.01), and LRP1B (8.70%, p < 0.05) were most frequently mutated in S-IV, followed by S-III.
3.4. Associations of PCa subtypes with clinical and biological features
We then compared the available clinicopathologic features among the four PCa subtypes (Tables S1–S3): elderly patients (>60 years, p = 0.004), surgical margins (p < 0.001) and lymph node invasion (p < 0.001) were significantly associated with S-IV in the TCGA-PRAD cohort. TMPRSS2-ERG fusion was significantly observed in S-II, S-III and S-IV when compared with S-I in both TCGA-PRAD (p = 0.035) and RNAseq cohorts (p = 0.031). Advanced ISUP (4 or 5, all p < 0.001) was significantly associated with S-I, S-III and S-IV in both TCGA-PRAD and microarray cohorts. Moreover, advanced pathological stage (T3 or T4, all p < 0.001) was remarkably correlated with S-IV in three cohorts. Notably, four subtypes were identified in each independent cohort, suggesting widespread intertumoral heterogeneity of PCa.
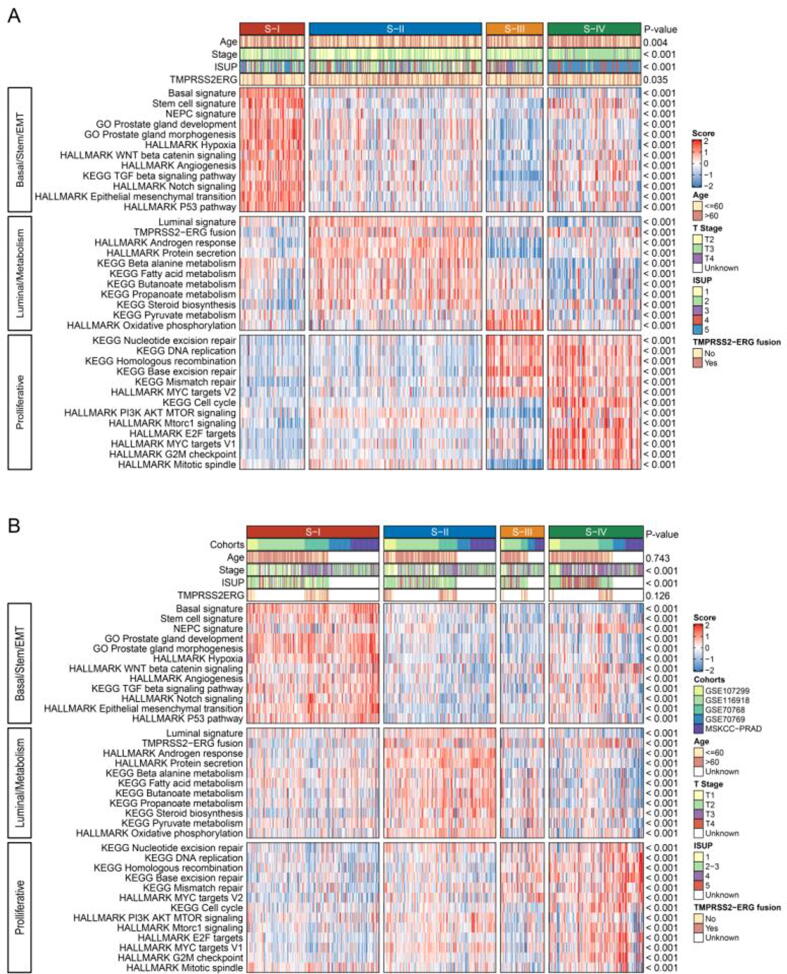
Using single-sample gene set enrichment analysis (ssGSEA), we next identified and validated distinct biological characteristics among the four subtypes in the TCGA-PRAD (Fig. 5A and Fig. S5A), microarray (Fig. 5B, Fig. S5B), and RNAseq cohorts (Fig. S6). S-I was significantly correlated with basal cell, stemness, neuroendocrine prostate cancer (NEPC), prostate gland development and mesenchymal-related (epithelial-mesenchymal transition [EMT], hypoxia, angiogenesis, Wnt, Notch, and TGFβ) signatures whereas S-II was enriched in luminal cells, ERG fusion, androgen response and metabolism-related (such as fatty acid metabolism, butanoate metabolism, steroid biosynthesis, and protein secretion) signatures. S-III exhibited elevated oxidative phosphorylation and moderate levels of proliferation as indicated by enriched DNA repair pathways (including base excision repair, nucleotide excision repair, homologous recombination, and mismatch repair). S-IV was characterized by moderate levels of NEPC, stemness, EMT and angiogenesis, as well as marked activities of DNA repair and cell cycle-related pathways such as E2F, G2M, and MYC targets.
Fig. 5.
Associations of clinicopathologic and biological features with the PCa subtypes. Single-sample gene set enrichment analysis (ssGSEA) identified distinct biological characteristics among subtypes in the TCGA-PRAD (A) and Microarray (B) cohorts. Red indicates enrichment of signatures while blue suggests the opposite. NEPC, neuroendocrine prostate cancer. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. Associations of PCa subtypes with immune infiltration
Next, we characterized the immune landscape of PCa subtypes in the TCGA-PRAD and microarray cohorts. The ESTIMATE algorithm revealed that S-I and S-IV had relatively higher immune and stromal scores and lower tumor purity than S-II and S-III (p < 0.05; Fig. 6A and 6C). Hence, we suspected that S-I and S-IV were likely to be immune-related. To decode the immune microenvironments, ssGSEA was performed and revealed that S-IV was significantly enriched in immune-inflamed signatures (such as human leukocyte antigen signature, CD8 T effector cell, natural killer cell cytotoxicity, and T cell inflamed GEP; Fig. 6B and 6D, Fig. S7A and S7B). Although S-I displayed enrichment of immune-inflamed signatures similar to S-IV, it was found to have higher levels of myeloid-derived suppressor cells (MDSCs), regulatory T cells (Tregs), T-cell exhaustion, TGFβ family members and receptors and cancer-associated fibroblasts (CAFs) than other subtypes. Thus, S-I showed an increased tendency toward an immune-excluded subtype. Overall, PCa subtypes in the RNAseq cohort demonstrated similar results (Fig. S8A-S8C).
Fig. 6.
The immune landscape of the PCa subtypes. The immune score, stromal score and tumor purity were compared across subtypes in the TCGA-PRAD (A) and Microarray (C) cohorts. “ns” denotes no statistical significance, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.Heatmaps of ssGSEA scores of curated immune-related gene sets in the TCGA-PRAD (B) and Microarray (D) cohorts. Red indicates enrichment of signatures while blue suggests the opposite. HLA, human leukocyte antigen; TCR, T cell receptor; IFN, interferon; GEP, gene expression profile; MDSC, myeloid-derived suppressor cells; Treg cells, regulatory T cells; TGFb, transforming growth factor β; Pan F TBRs, pan tissue fibroblast TGF-β response signature. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Associations of PCa subtypes with therapeutic efficacy
To explore whether PCa subtypes responded differentially to conventional PCa therapies, we computed the AUC values of abiraterone, olaparib and taxane-based chemotherapy (Fig. 7A-7C) in PCa cohorts. As a result, S-II was significantly associated with increased sensitivity to abiraterone whereas S-I was more sensitive to olaparib than other subtypes. Although inconsistent results were demonstrated across PCa cohorts, S-III and S-IV patients were, in general, more susceptible to docetaxel and cabazitaxel. We also used subMap to compare GEPs between PCa subtypes and 34 mCRPC patients treated with enzalutamide (160 mg/d). Data showed that S-I was significantly correlated with the enzalutamide-resistant group (all p < 0.01; Fig. 7D). In contrast, the GEP of S-II was more similar to that of the enzalutamide-sensitive group (TCGA-PRAD, p = 0.172; RNAseq cohort, p = 0.067; Microarray cohort, p = 0.018). Since PCa subtypes correlated with different immune phenotypes, we asked whether they responded differently to immunotherapy. We first investigated the expression levels of PD-1, PD-L1 and CTLA-4, and found that they did not differ significantly between S-I and S-IV (except that S-I had slightly higher PD-L1 levels in the RNAseq cohort [p < 0.05]; Fig. S7C and S7D, Fig. S8D). SubMap analysis showed that S-I was significantly associated with anti-PD-L1 and anti-PD-1nonresponders. In contrast, S-IV shared similar GEP with anti-PD-L1 responders (all p = 0.001; Fig. 7D), indicating that S-IV may benefit from anti-PD-L1 therapy. Additionally, there were no significant associations between PCa subtypes and anti-CTLA-4 patients.
Fig. 7.
Associations of the efficacy of conventional therapy with the PCa subtypes. Sensitivity results of abiraterone, docetaxel, cabazitaxel and olaparib in the Microarray (A), RNAseq (B) and TCGA-PRAD (C) cohorts. “ns” denotes no statistical significance, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. The similarity of gene expression profiles between PCa subtypes and patients receiving enzalutamide, anti-PD-1, anti-PD-L1 or anti-CTLA-4 therapies were shown in three cohorts (C). “R” denotes responders while “NR” represents nonresponders.
4. Discussion
The present study is the first to establish a robust PCa molecular classification based on miRNA-regulated GEPs, which offers new insights into the role of miRNAs in PCa heterogeneity and subtyping. We identified and validated four PCa subtypes exhibiting distinct epigenetic, genomic, transcriptional, and clinicopathologic features. Of the four subtypes, S-I and S-IV exhibit immune-excluded and immune-inflamed phenotypes, respectively. Moreover, we uncovered subtype-driven miRNAs that could potentially serve as circulating diagnostic biomarkers and therapeutic targets for corresponding subtypes. Finally, possible treatment options for each subtype were suggested based on drug sensitivity analysis.
Whether prostate adenocarcinoma and NEPC originate from basal cells or luminal cells or both is constantly debated with conflicting results reported by previous studies [35], [36], [37], [38]. Differences between S-I and S-II in our study support the hypothesis that PCa could be derived from either luminal or basal cell lineages, and that different cell origins give rise to distinct subtypes with divergent biological behaviors and drug responses. The identification of basal- and luminal-like subtypes has also been reported by previous studies [10], [11]. Zhao et al. used PAM50 classifiers of breast cancer to assign PCa patients into luminal A, luminal B and basal-like subtypes and found that the luminal B subtype had worse prognosis than other subtypes [11]. Nevertheless, the direct application of this breast cancer classifier to PCa subtyping poses the risk of neglecting important genetic features of PCa. Spratt et al. studied the heterogeneity of AR activity in primary treatment naïve PCa and identified a low AR-active and androgen deprivation therapy (ADT)-insensitive subclass that is in accordance with S-I characterized by overexpression of basal and NEPC signatures and decreased DNA repair [39]. These biological features also support the results of drug sensitivity in S-I. NEPC and low AR activity were extensively demonstrated to confer castration resistance [40]. Deficient DNA repair mechanisms, especially homologous recombination repair were associated with the antitumor activity of PARP inhibitors in patients with mCRPC [41]. In contrast to S-I, S-II was highly enriched in the androgen response and therefore, showed favorable response to ADTs. Although neither luminal nor basal signatures were overrepresented in S-III and S-IV, TMPRSS2-ERG fusion at both genomic and transcriptional levels was more enriched in S-II, S-III, and S-IV than in S-I, favoring the assumption that S-III and S-IV shared luminal cell origin with S-II. However, S-IV was also enriched in NEPC signatures, suggesting a possible role of lineage plasticity during S-IV evolution. This was supported by Dong et al. who used single-cell RNA sequencing and found that luminal-like adenocarcinoma cells could transdifferentiate into NE cells with loss of luminal markers [42].
Our data showed that basal-like S-I exhibited stem- and mesenchymal-like properties, which was consistent with a previous study uncovering intrinsic stem cell and EMT features in prostatic basal cells [43]. However, our study does not support the linkage between the basal cell signature and aggressive PCa [43]. Instead of S-I, S-IV was characterized as a more aggressive subtype of PCa with worse prognosis, significant enrichment in cell cycle-related pathways, and moderate levels of EMT and stemness. In support of this observation, lineage analysis by Wang et al. revealed that PCa of luminal origin was more aggressive than basal origin prostate cancer and exhibited more active cell cycle-related pathways [38]. The most likely explanation for this is that although S-I is more migratory and mesenchymal-like, it is strikingly less proliferative than S-IV. Reasonably, proliferative capacity is essential to generate PCa macrometastasis once tumor cells reach distant sites with the help of EMT [44]. Increased CNV burden and aneuploidy also contributed to the worse prognosis of S-IV, as these factors were reported to promote aggressive PCa development and earlier biochemical relapse [45], [46]. Specifically, we identified higher frequencies of TP53 and RB1 functional loss in S-IV. The biallelic loss of TP53 and RB1 has been associated with attenuated AR activity, increased cell proliferation and DNA repair activity, as well as exhibition of stemness and NE differentiation in LNCaP cells [47]. Similar observations in S-IV support these associations, which highlight the important roles of TP53 and RB1 loss in the development of aggressive PCa.
Immune checkpoint blockade (ICB) treatments have been proven to be effective against several cancers including melanoma and bladder cancer. However, the use of ICB alone has thus far shown modest efficacy in advanced PCa as most PCa has insufficient immunogenicity and few tumor-infiltrating T cells [48], [49], [50]. It is, therefore, necessary to select a subset of PCa that may benefit from ICB. Our study showed that S-I was unlikely to respond to anti-PD-1 and anti-PD-L1 therapies because it was mostly immune-excluded, with enrichment in CAFs, MDSCs, and T cell exhaustion. Furthermore, increased TGFβ activity in S-I was reported to promote T cell exclusion and concomitant resistance to ICB [51]. Consequently, ICB resistance in S-I patients may be overcome by combining anti-TGFβ treatment. S-IV was mostly immune-inflamed and therefore, may benefit from anti-PD-L1 therapy. The higher TMB and TAN loads in S-IV could also increase the efficacy by driving infiltrating T cell responses, as evidenced by previous studies [27], [52]. Taken together, these findings suggest that the current PCa subtypes offer the potential to provide ICB guidance.
Our study has several limitations that need to be optimized in the future. First, since we only used miRNA-regulated GEPs to classify PCa patients, our classification system is biased and may obscure important features determined by other omics data. Second, due to the lack of miRNA expression data, the classification pipeline used in the TCGA-PRAD dataset was not repeated in the validation datasets. Last, prospective cohorts and functional experiments are needed to validate the proposed classification and confirm our findings.
5. Conclusions
In summary, findings from the present study expand our understanding of PCa heterogeneity from the perspective of miRNAs regulation. While the proposed classification has the potential to predict clinical outcomes and inform personalized treatments for PCa patients, further experimental studies are required to verify this conclusion.
CRediT authorship contribution statement
Bing-Biao Lin: Conceptualization, Methodology, Software, Investigation, Visualization, Writing – original draft. Han-Qi Lei: Methodology, Investigation, Writing – original draft. Hai-Yun Xiong: Methodology, Investigation, Writing – original draft. Xing Fu: Methodology, Investigation, Visualization, Writing - review & editing. Fu Shi: Investigation, Writing - review & editing. Xiang-Wei Yang: Investigation, Writing - review & editing. Ya-Fei Yang: . Guo-Long Liao: Writing - review & editing. Yu-Peng Feng: Writing - review & editing. Dong-Gen Jiang: Conceptualization, Writing - review & editing, Funding acquisition, Supervision. Jun Pang: Conceptualization, Writing - review & editing, Funding acquisition, Supervision.
Acknowledgments
Acknowledgments
We would like to thank every author that contributes to TCGA, GEO, cBioPortal, CCLE, and PRISM Repurposing datasets.
Funding
This work was supported by the National Natural Science Foundation of China (81772754 and 81902613), Major Basic Research and Cultivation Program of Natural Science Foundation of Guangdong Province (2017A03038009), National Key R&D Program of China (2018YFA0902800), Shenzhen Basic Science Research (JCYJ20190809164617205), Sanming Project of Medicine in Shenzhen (SZSM202011011) and the Hospital Research Fund of SAHSYSU (ZSQYLCKYJJ202019).
Data availability
The data analyzed in this study are publicly available in TCGA, GEO, cBioPortal, CCLE, and PRISM Repurposing datasets.
Footnotes
Figure S1. Identification and removal of batch effects during datasets merging. The principal component analysis demonstrated significant batch effects in the microarray-based (A) and RNA-sequencing (B) cohorts, respectively. Batch effects were removed in the microarray-based (C) and RNA-sequencing (D) cohorts.
Figure S2. Results of non-negative matrix factorization based on miRNA-correlated mRNAs. Consensus maps (A) and cophenetic coefficients (B) of PCa classifications with clustering numbers ranging from 2 to 6.
Figure S3. KEGG and GO analyses of subtype-specific upregulated mRNAs. Distinct pathway enrichments were shown for S-I (A), S-II (B), S-III (C) and S-IV (D).
Figure S4. KEGG and GO analyses of subtype-driven miRNAs. Active pathways determined by correlated mRNAs were only observed in S-I (A), S-II (B) and S-IV (C).
Figure S5. Biological features associated with the PCa subtypes. Boxplots showing ssGSEA scores of biological pathways across subtypes in the TCGA-PRAD (A) and Microarray (B) cohorts.
Figure S6. Clinicopathologic and biological features associated with the PCa subtypes in the RNAseq cohort. The heatmap (A) and boxplots (B) showing ssGSEA scores of biological pathways across subtypes. Red indicates enrichment of signatures while blue suggests the opposite. “ns” denotes no statistical significance, *p< 0.05, **p<0.01, ***p<0.001, ****p<0.0001. NEPC, neuroendocrine prostate cancer.
Figure S7. Immune characterization of the PCa subtypes. Boxplots showing ssGSEA scores of immune-related pathways for the PCa subtypes in the TCGA-PRAD (A) and Microarray (B) cohorts. The levels of immune checkpoint expression in the TCGA-PRAD (C) and Microarray (D) cohorts. “ns” denotes no statistical significance, *p< 0.05, **p<0.01, ***p<0.001, ****p<0.0001. HLA, human leukocyte antigen; TCR, T cell receptor; IFN, interferon; GEP, gene expression profile; MDSC, myeloid-derived suppressor cells; Treg cells, regulatory T cells; TGFb, transforming growth factor β; Pan F TBRs, pan tissue fibroblast TGF-β response signature. CTLA-4, cytotoxic T-lymphocyte associated protein-4; PD-1, programmed cell death protein-1; PD-L1, programmed death-ligand 1.
Figure S8. Immune characterization of the PCa subtypes in the RNAseq cohort. The immune score, stromal score and tumor purity were compared across subtypes (A). The heatmap (B) and boxplots (C) show ssGSEA scores of immune-related pathways in the four PCa subtypes. Red indicates enrichment of signatures while blue suggests the opposite. Boxplots (D) showing the levels of immune checkpoint expression. “ns” denotes no statistical significance, *p< 0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.08.046.
Contributor Information
Dong-Gen Jiang, Email: jiangdg5@mail.sysu.edu.cn.
Jun Pang, Email: pangjun2@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.v71.310.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Cooperberg M.R., Broering J.M., Carroll P.R. Time trends and local variation in primary treatment of localized prostate cancer. J Clin Oncol. 2010;28(7):1117–1123. doi: 10.1200/JCO.2009.26.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanda M.G., Dunn R.L., Michalski J., Sandler H.M., Northouse L., Hembroff L. Quality of life and satisfaction with outcome among prostate-cancer survivors. N Engl J Med. 2008;358(12):1250–1261. doi: 10.1056/NEJMoa074311. [DOI] [PubMed] [Google Scholar]
- 4.Basch E., Loblaw D.A., Oliver T.K., Carducci M., Chen R.C., Frame J.N. Systemic therapy in men with metastatic castration-resistant prostate cancer: American Society of Clinical Oncology and Cancer Care Ontario clinical practice guideline. J Clin Oncol. 2014;32(30):3436–3448. doi: 10.1200/JCO.2013.54.8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith M.R., Kabbinavar F., Saad F., Hussain A., Gittelman M.C., Bilhartz D.L. Natural history of rising serum prostate-specific antigen in men with castrate nonmetastatic prostate cancer. J Clin Oncol. 2005;23(13):2918–2925. doi: 10.1200/JCO.2005.01.529. [DOI] [PubMed] [Google Scholar]
- 6.Haffner M.C., Zwart W., Roudier M.P., True L.D., Nelson W.G., Epstein J.I. Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev Urol. 2021;18(2):79–92. doi: 10.1038/s41585-020-00400-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conti S.L., Dall'Era M., Fradet V., Cowan J.E., Simko J., Carroll P.R. Pathological outcomes of candidates for active surveillance of prostate cancer. J Urol. 2009;181(4):1628–1634. doi: 10.1016/j.juro.2008.11.107. [DOI] [PubMed] [Google Scholar]
- 8.Cooperberg M.R., Pasta D.J., Elkin E.P., Litwin M.S., Latini D.M. The University of California, San Francisco Cancer of the Prostate Risk Assessment score: a straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J Urol. 2005;173(6):1938–1942. doi: 10.1097/01.ju.0000158155.33890.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gleason D.F., Mellinger G.T. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J Urol. 1974;111(1):58–64. doi: 10.1016/S0022-5347(17)59889-4. [DOI] [PubMed] [Google Scholar]
- 10.You S., Knudsen B.S., Erho N., Alshalalfa M., Takhar M., Al-deen Ashab H. Integrated classification of prostate cancer reveals a novel luminal subtype with poor outcome. Cancer Res. 2016;76(17):4948–4958. doi: 10.1158/0008-5472.CAN-16-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao S.G., Chang S.L., Erho N., Yu M., Lehrer J., Alshalalfa M. Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA Oncol. 2017;3(12):1663. doi: 10.1001/jamaoncol.2017.0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research N (2015) The Molecular Taxonomy of Primary Prostate Cancer. Cell 163, 1011-1025, doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed]
- 13.Cuzick J., Swanson G.P., Fisher G., Brothman A.R., Berney D.M., Reid J.E. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12(3):245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamoun A., Cancel-Tassin G., Fromont G., Elarouci N., Armenoult L., Ayadi M. Comprehensive molecular classification of localized prostate adenocarcinoma reveals a tumour subtype predictive of non-aggressive disease. Ann Oncol. 2018;29(8):1814–1821. doi: 10.1093/annonc/mdy224. [DOI] [PubMed] [Google Scholar]
- 15.Klein E.A., Cooperberg M.R., Magi-Galluzzi C., Simko J.P., Falzarano S.M., Maddala T. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol. 2014;66(3):550–560. doi: 10.1016/j.eururo.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Macfarlane L.A., Murphy P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eringyte I., Zamarbide Losada J.N., Powell S.M., Bevan C.L., Fletcher C.E. Coordinated AR and microRNA regulation in prostate cancer. Asian J Urol. 2020;7(3):233–250. doi: 10.1016/j.ajur.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton M.P., Rajapakshe K.I., Bader D.A., Cerne J.Z., Smith E.A., Coarfa C. The Landscape of microRNA Targeting in Prostate Cancer Defined by AGO-PAR-CLIP. Neoplasia. 2016;18(6):356–370. doi: 10.1016/j.neo.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4) doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner G.P., Kin K., Lynch V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131(4):281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 21.Holt J., Walter V., Yin X., Marron D., Wilkerson M.D., Choi H.Y. Integrative Analysis of miRNAs Identifies Clinically Relevant Epithelial and Stromal Subtypes of Head and Neck Squamous Cell Carcinoma. Clin Cancer Res. 2021;27(3):831–842. doi: 10.1158/1078-0432.CCR-20-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Liu J, Wang D, Feng M & Wu X (2020) Epigenetically regulated gene expression profiles reveal four molecular subtypes with prognostic and therapeutic implications in colorectal cancer. Brief Bioinform, doi: 10.1093/bib/bbaa309. [DOI] [PubMed]
- 23.Hoshida Y., Tan P. Nearest template prediction: a single-sample-based flexible class prediction with confidence assessment. PLoS ONE. 2010;5(11):e15543. doi: 10.1371/journal.pone.0015543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoshida Y., Brunet J.-P., Tamayo P., Golub T.R., Mesirov J.P., Hofmann O. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS ONE. 2007;2(11):e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.-H. The Immune Landscape of Cancer. Immunity. 2018;48(4):812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alumkal J.J., Sun D., Lu E., Beer T.M., Thomas G.V., Latour E. Transcriptional profiling identifies an androgen receptor activity-low, stemness program associated with enzalutamide resistance. Proc Natl Acad Sci U S A. 2020;117(22):12315–12323. doi: 10.1073/pnas.1922207117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balar A.V., Galsky M.D., Rosenberg J.E., Powles T., Petrylak D.P., Bellmunt J. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. The Lancet. 2017;389(10064):67–76. doi: 10.1016/S0140-6736(16)32455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen P.-L., Roh W., Reuben A., Cooper Z.A., Spencer C.N., Prieto P.A. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer Discov. 2016;6(8):827–837. doi: 10.1158/2159-8290.CD-15-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, et al. (2018) miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 46, D296-D302, doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed]
- 31.Agarwal V, Bell GW, Nam JW & Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4, doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed]
- 32.Alhasan A.H., Scott A.W., Wu J.J., Feng G., Meeks J.J., Thaxton C.S. Circulating microRNA signature for the diagnosis of very high-risk prostate cancer. Proc Natl Acad Sci U S A. 2016;113(38):10655–10660. doi: 10.1073/pnas.1611596113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song Y., Song C., Yang S. Tumor-Suppressive Function of miR-30d-5p in Prostate Cancer Cell Proliferation and Migration by Targeting NT5E. Cancer Biother Radiopharm. 2018;33(5):203–211. doi: 10.1089/cbr.2018.2457. [DOI] [PubMed] [Google Scholar]
- 34.Kumar B., Khaleghzadegan S., Mears B., Hatano K., Kudrolli T.A., Chowdhury W.H. Identification of miR-30b-3p and miR-30d-5p as direct regulators of androgen receptor signaling in prostate cancer by complementary functional microRNA library screening. Oncotarget. 2016;7(45):72593–72607. doi: 10.18632/oncotarget.v7i4510.18632/oncotarget.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi N., Zhang B., Zhang L.i., Ittmann M., Xin L.i. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21(2):253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldstein A.S., Huang J., Guo C., Garraway I.P., Witte O.N. Identification of a cell of origin for human prostate cancer. Science. 2010;329(5991):568–571. doi: 10.1126/science:1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X.i., Julio M.-d., Economides K.D., Walker D., Yu H., Halili M.V. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461(7263):495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z.A., Mitrofanova A., Bergren S.K., Abate-Shen C., Cardiff R.D., Califano A. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat Cell Biol. 2013;15(3):274–283. doi: 10.1038/ncb2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spratt D.E., Alshalalfa M., Fishbane N., Weiner A.B., Mehra R. Transcriptomic Heterogeneity of Androgen Receptor Activity Defines a de novo low AR-Active Subclass in Treatment Naive Primary Prostate Cancer. Clin Cancer Res. 2019;25:6721–6730. doi: 10.1158/1078-0432.CCR-19-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carceles-Cordon M., Kelly W.K., Gomella L., Knudsen K.E., Rodriguez-Bravo V., Domingo-Domenech J. Cellular rewiring in lethal prostate cancer: the architect of drug resistance. Nat Rev Urol. 2020;17(5):292–307. doi: 10.1038/s41585-020-0298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mateo J., Porta N., Bianchini D., McGovern U., Elliott T., Jones R. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21(1):162–174. doi: 10.1016/S1470-2045(19)30684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong B., Miao J., Wang Y., Luo W., Ji Z., Lai H. Single-cell analysis supports a luminal-neuroendocrine transdifferentiation in human prostate cancer. Commun Biol. 2020;3(1) doi: 10.1038/s42003-020-01476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang D., Park D., Zhong Y.i., Lu Y., Rycaj K., Gong S. Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat Commun. 2016;7(1) doi: 10.1038/ncomms10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruscetti M., Quach B., Dadashian E.L., Mulholland D.J., Wu H. Tracking and Functional Characterization of Epithelial-Mesenchymal Transition and Mesenchymal Tumor Cells during Prostate Cancer Metastasis. Cancer Res. 2015;75(13):2749–2759. doi: 10.1158/0008-5472.CAN-14-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hieronymus H., Schultz N., Gopalan A., Carver B.S., Chang M.T., Xiao Y. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A. 2014;111(30):11139–11144. doi: 10.1073/pnas.1411446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stopsack K.H., Whittaker C.A., Gerke T.A., Loda M., Kantoff P.W., Mucci L.A. Aneuploidy drives lethal progression in prostate cancer. Proc Natl Acad Sci U S A. 2019;116(23):11390–11395. doi: 10.1073/pnas.1902645116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nyquist M.D., Corella A., Coleman I., De Sarkar N., Kaipainen A., Ha G. Combined TP53 and RB1 Loss Promotes Prostate Cancer Resistance to a Spectrum of Therapeutics and Confers Vulnerability to Replication Stress. Cell Rep. 2020;31(8):107669. doi: 10.1016/j.celrep.2020.107669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cha H.-R., Lee J.H., Ponnazhagan S. Revisiting Immunotherapy: A Focus on Prostate Cancer. Cancer Res. 2020;80(8):1615–1623. doi: 10.1158/0008-5472.CAN-19-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon E.D., Drake C.G., Scher H.I., Fizazi K., Bossi A., van den Eertwegh A.J.M. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. 2014;15(7):700–712. doi: 10.1016/S1470-2045(14)70189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madan R.A., Gulley J.L. Finding an Immunologic Beachhead in the Prostate Cancer Microenvironment. J Natl Cancer Inst. 2019;111:219–220. doi: 10.1093/jnci/djy145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mariathasan S., Turley S.J., Nickles D., Castiglioni A., Yuen K. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma P., Pachynski R.K., Narayan V., Fléchon A., Gravis G., Galsky M.D. Nivolumab Plus Ipilimumab for Metastatic Castration-Resistant Prostate Cancer: Preliminary Analysis of Patients in the CheckMate 650 Trial. Cancer Cell. 2020;38(4):489–499.e3. doi: 10.1016/j.ccell.2020.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed in this study are publicly available in TCGA, GEO, cBioPortal, CCLE, and PRISM Repurposing datasets.