Abstract
We report a PCR–enzyme-linked immunosorbent assay which identifies Campylobacter species by capture hybridization of a single-stranded 16S rRNA gene amplicon with species-specific probes in a microtiter plate format. Specificities were confirmed for both reference and field strains, but the type strain of Campylobacter coli was atypical. The assay was rapid, informative, and usable with stool-extracted DNA.
We have previously described PCRs which identify individual Campylobacter species from pure cultures or fecal DNA extracts (3–5). In routine clinical laboratory practice, a further advantage would be to avoid multiple PCRs by generating one amplicon containing sequence polymorphisms suitable for species-specific identification.
DNA was extracted (8) from the reference strains listed in Table 1, 49 Campylobacter jejuni and 14 C. coli Penner serotype reference strains, 10 field isolates of C. jejuni, 8 of C. coli, 4 of C. lari, 4 of C. upsaliensis, 4 of C. helveticus, 2 of C. hyointestinalis, and 5 of C. fetus. These were used to analyze probe specificities.
TABLE 1.
Reference strains of Campylobacter, Arcobacter, and Helicobacter species
| Species | Sourcea |
|---|---|
| Campylobacter spp. | |
| C. jejuni subsp. jejuni | NCTC 11351T |
| C. jejuni subsp. doylei | NCTC 11951T |
| C. coli | NCTC 11366T |
| C. lari | NCTC 11352T |
| C. upsaliensis | NCTC 11541T |
| C. helveticus | NCTC 12470T |
| UPTC | NCTC 11845 |
| C. fetus subsp. fetus | NCTC 10842T |
| C. fetus subsp. venerealis | NCTC 10354T |
| C. hyointestinalis | NCTC 11608T |
| C. sputorum subsp. sputorum | NCTC 11528T |
| C. sputorum subsp. fecalis | NCTC 11415T |
| C. sputorum subsp. bubulus | NCTC 11367T |
| C. gracilis | NCTC 12738T |
| [Bacteroides] ureolyticusb | NCTC 10941T |
| C. concisus | NCTC 11485T |
| C. rectus | NCTC 11489T |
| C. curvus | NCTC 11649T |
| C. showae | NCTC 12843T |
| C. mucosalis | NCTC 11001 |
| Helicobacter spp. | |
| H. felis | NCTC 12436T |
| H. canis | NCTC 12739T |
| H. pullorum | NCTC 12824T |
| H. rappini | NCTC 12461T |
| H. acinonyx | NCTC 12686T |
| H. cinaedi | NCTC 12423T |
| CLO-3c | NCTC 12462 |
| H. fennelliae | NCTC 11612T |
| H. mustelae | NCTC 12198T |
| H. pylori | NCTC 11637T |
| H. hepaticus | ATCC 51448T |
| H. muridarum | NCTC 12714T |
| H. bilis | ATCC 51630T |
| H. pamatensis | ATCC 51478T |
| H. nemestriae | NCTC 12491T |
| Arcobacter spp. | |
| A. cryaerophilus | NCTC 11885T |
| A. skirrowii | NCTC 12713T |
| A. butzleri | NCTC 12481T |
| A. nitrofigilis | NCTC 12551T |
| Wollinella succinogenes | NCTC 11488T |
NCTC, National Collection of Type Cultures; ATCC, American Type Culture Collection; T denotes a type strain.
Species incertae sedis which is genotypically Campylobacter (7).
CLO-3, Campylobacter-like organism.
First-round symmetric PCR was performed with genus-specific primers, C412F and C1228R (5). Second-round asymmetric PCR was performed with 10 μl of the first-round reaction mixture in a 50-μl volume, with a single 5′ end fluorescein-labelled primer, C690F (see below). Conditions were as described previously (5), except that the annealing temperature was raised to 60°C and the primer concentration of 400 nM gave a >5:1 ratio of C690F/C1228R. The PCR–enzyme-linked immunosorbent assay (ELISA) was performed as described previously (6), except that 100 ng of capture probe was immobilized, denaturation of second-round PCR product was unnecessary, and the conjugate was diluted 1:1,000. The optical density (OD) of the final yellow product was measured at 450/620 nm (see Fig. 1): signals were considered negative if the absorbance was below 0.1 OD unit and positive if it was above 0.2 OD unit. Certain first-round PCR products were investigated by sequencing with a Prism DNA cycle sequencing kit, a 373A automated DNA sequencer (Applied Biosystems), and the primers C1228R (5) and C641F (see below).
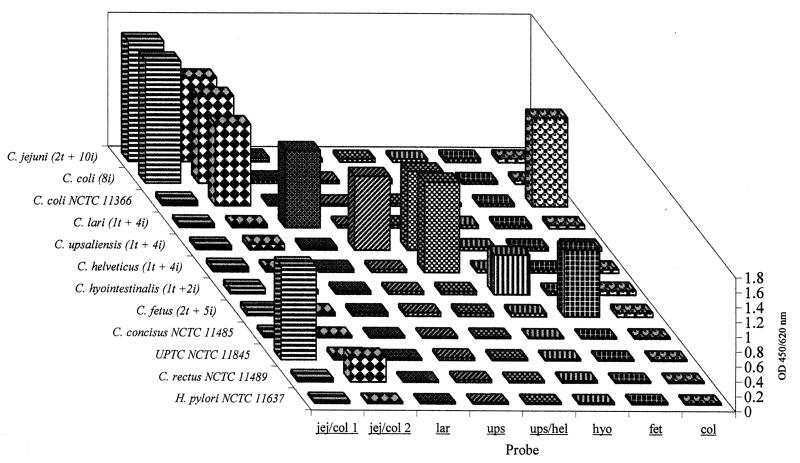
FIG. 1.
PCR-ELISA results for 13 strains and 37 field isolates of Campylobacter. Values shown for C. jejuni, C. lari, C. upsaliensis, C. helveticus, C. hyointestinalis, and C. fetus are the average for type strains (t), described in Table 1, and field isolates (i). C. coli field isolates behaved differently from the type strain. Values are shown for type strains of C. concisus and C. rectus, a reference strain of the UPTC group, and a negative control, Helicobacter pylori.
16S rRNA gene sequences of the following species were aligned by using the CLUSTAL program (2): C. gracilis L37787 and L04320, [Bacteroides] ureolyticus L04321, Campylobacter sp. strain CLO L14632, Campylobacter sp. strain L04318, C. coli L04312 and M59073, C. concisus L06977 and L04322, C. curvus L06976 and L04313, C. fetus subsp. venerealis L14633, C. fetus subsp. fetus L04314, C. helveticus U03022, C. hyointestinalis M65010 and M65009, C. jejuni Z29326 and M59298, C. jejuni subsp. jejuni L04315, C. lari L04316, C. mucosalis-like L14629, C. mucosalis L06978, C. rectus L06973 and L04317, C. showae L06975, L06974, and UPTC L14631, C. sputorum subsp. bubulus L04319, C. sputorum subsp. sputorum X67775, and C. upsaliensis L14628. Two Campylobacter genus-specific primers, C641F (5′-GAGAGGCAGATGGAATTGG-3′; sequencing primer) and C690F (5′-AGATACCCTGGTAGTCCACG-3′; second-round primer), were thereby designed.
Eight capture probes were designed to target variable regions; accession numbers of sequences bearing perfect matches to these probes are given in Table 2. Four probes (lar, fet, hyo, and ups) were designed to capture amplicons from C. lari, C. fetus, C. hyointestinalis, and C. upsaliensis, respectively. Probe ups/hel was designed to capture amplicons from C. upsaliensis and C. helveticus. Thus, capture with both ups and ups/hel would indicate C. upsaliensis; capture with the latter alone would indicate C. helveticus. A positive reaction with probes jej/col 1 and jej/col 2 should indicate C. jejuni, that with col and jej/col 2 should indicate C. coli, that with jej/col 1 alone should indicate the urease-positive thermophilic campylobacter group (UPTC), and that with jej/col 2 alone should indicate oral campylobacters (Table 2).
TABLE 2.
Campylobacter capture probes, their sequences, and homologies
| Capture probe | Sequencea | Sequence identity with: | EMBL/GenBank accession no.b |
|---|---|---|---|
| jej/col 1 | tttGCGGTACACTTAATGCGTT | C. jejuni | Z29326, M59074, L04315, L14630 |
| UPTC | L14631 | ||
| jej/col 2 | taaGCTCGGCCGAACCGTTA | C. coli | L04312 |
| C. jejuni | Z29326, M59074, L04315, L14630 | ||
| C. rectus | L06973 | ||
| C. showae | L06974 | ||
| C. curvus | L06976 | ||
| C. gracilis | L04320, L37787 | ||
| col | aaaCCCTGACTAGCAGAGCAA | C. coli | L04312 |
| lar | taaGCTCACCCGAAGTGTTAG | C. lari | L04316, M92310 |
| ups | aaaCTACAGAATTTGTTGGATATC | C. upsaliensis | L14628 |
| ups/hel | taaGCTCGACCGAATCGTTAG | C. helveticus | U03022 |
| C. upsaliensis | L14628 | ||
| hyo | ataCTCTAAGATGTTATTAGGATAT | C. hyointestinalis | M65009, M65010 |
| fet | aaaCTAAGAGATTAGTTGGATATC | C. fetus | M65011, M65012L04314, L14633 |
Bases in lowercase letters represent nonhybridizing spacer regions.
Species identified only to genus level have not been included.
To evaluate the probes, amplicons from the 21 reference strains of Campylobacter, the 16 reference strains of Helicobacter, the 4 reference strains of Arcobacter (Table 1), 63 Penner serotype reference strains (see above), and 37 Campylobacter field isolates (speciated by phenotype) were hybridized with the eight probes in a microtiter plate format. All capture probes except col hybridized with both their respective type strains and the corresponding field isolates (Table 1; Fig. 1). No capture was observed for the remaining species of Campylobacter, Helicobacter, and Arcobacter. Probe col reacted with the type strain but not with the serotype reference strains or field isolates of C. coli. Probe jej/col 1 reacted with all strains and isolates of C. jejuni and also C. coli (except NCTC 11366T).
To further analyze these two apparent anomalies, we sequenced amplicons from 52 of 63 Penner serotype reference strains of C. coli and C. jejuni. All had a similar sequence between positions 732 and 1198, falling into three groups which varied only at positions 837, 1010, and 1019. Nineteen C. jejuni reference strains (Penner serotypes HS 1 to 6, 10, 13 to 18, 22, 31, 37, 40, 44, 62, and 64) and 12 C. coli reference strains (HS 5, 14, 25, 26, 28, 30, 34, 39, 47, 48, 51, and 54) had G, T, and A at positions 837, 1010, and 1019, respectively (EMBL accession no. AJ006477). Nineteen C. jejuni strains (HS 7 to 9, 12, 19, 21, 23, 27, 29, 32, 33, 36, 38, 41, 42, 53, 58, 60, and 63) and one C. coli strain (HS 20) had A at positions 837 and 1010 and T at position 1019 (accession no. AJ006478). C. jejuni HS 43 uniquely had G at position 837, A at position 1010, and T at position 1019 (accession no. AJ006479).
The new assay format (without probe col) was tested on DNA extracted from 40 clinical fecal samples and 10 fecal samples from healthy volunteers as described previously (3). The clinical samples comprised 28 from which presumptive C. jejuni or C. coli had been cultured by standard methods (4), 10 Campylobacter culture-negative samples (5 positive for Salmonella, 1 for Shigella, 1 for Aeromonas, 1 for Cryptosporidium, and 2 for Giardia), and 2 samples in which no pathogenic organisms were detected. Amplicons from the 28 Campylobacter culture-positive samples hybridized with jej/col 1 and jej/col 2, indicating C. jejuni or C. coli, and were assigned to one of these species as previously described (4). The 10 samples positive for other enteropathogens showed no hybridization. One culture-negative sample reacted with ups and ups/hel, and the other reacted with hyo. Results for these two samples were confirmed with individual species-specific PCR assays for C. upsaliensis and C. hyointestinalis (4, 5). For the 10 samples from healthy volunteers, 6 negative results were obtained with all probes, while 2 were weakly positive with jej/col 1 alone and 2 were weakly positive with jej/col 2 alone, consistent with the presence of nonpathogenic campylobacters.
The data for probes jej/col 1, jej/col 2, and col and the sequencing studies described above indicate that the C. coli type strain is not representative of that species. This and the fact that there are no clear-cut differences between C. jejuni and C. coli explain why probe col did not hybridize with any other C. coli strains and highlight the need for caution when using sequence data derived from type strains alone (see also reference 1).
This PCR-ELISA is fast, informative, and can be customized by the addition or subtraction of probes to identify species of particular interest. Its microtiter plate format and nonisotopic detection are readily suited to automation. It should find use both in diagnostic laboratories and in molecular ecological studies of campylobacter prevalence in human disease.
Acknowledgments
This work was funded by a grant from the Department of Health, London, United Kingdom (DH220B).
We thank the Campylobacter Reference Unit, Central Public Health Laboratory, for provision of field isolates.
REFERENCES
- 1.Clayton R A, Sutton G, Hinkle P S, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–599. doi: 10.1099/00207713-45-3-595. [DOI] [PubMed] [Google Scholar]
- 2.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 3.Lawson A J, Linton D, Stanley J, Owen R J. Polymerase chain reaction detection and speciation of Campylobacter upsaliensis and C. helveticus in human faeces and comparison with culture techniques. J Appl Microbiol. 1997;83:375–380. doi: 10.1046/j.1365-2672.1997.00240.x. [DOI] [PubMed] [Google Scholar]
- 4.Linton D, Lawson A J, Owen R J, Stanley J. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J Clin Microbiol. 1997;35:2568–2572. doi: 10.1128/jcm.35.10.2568-2572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linton D, Owen R J, Stanley J. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res Microbiol. 1996;147:707–718. doi: 10.1016/s0923-2508(97)85118-2. [DOI] [PubMed] [Google Scholar]
- 6.Patel S, Yates M, Saunders N A. PCR–enzyme-linked immunosorbent assay and partial rRNA gene sequencing: a rational approach to identifying mycobacteria. J Clin Microbiol. 1997;35:2375–2380. doi: 10.1128/jcm.35.9.2375-2380.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vandamme P, Daneshvar M I, Dewhirst F E, et al. Chemotaxonomic analyses of Bacteroides gracilis and Bacteroides ureolyticus and reclassification of B. gracilis as Campylobacter gracilis comb. nov. Int J Syst Bacteriol. 1995;45:145–152. doi: 10.1099/00207713-45-1-145. [DOI] [PubMed] [Google Scholar]
- 8.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. pp. 2.4.1–2.4.5. [Google Scholar]