Abstract
Kinase fusions represent an important type of somatic alterations that promote oncogenesis and serve as diagnostic markers in lung cancer. We aimed to identify the landscape of clinically relevant kinase fusions in Chinese lung cancer and to explore rare kinase rearrangements; thus, providing valuable evidence for therapeutic decision making. We performed genomic profiling of 425 cancer-relevant genes from tumor/plasma biopsies from a total of 17,442 Chinese lung cancer patients using next generation sequencing (NGS). Patients’ clinical characteristics and treatment histories were retrospectively studied. A total of 1162 patients (6.66%; 1162/17,442) were identified as having kinase fusions, including 906 adenocarcinomas (ADCs) and 35 squamous cell carcinomas (SCCs). In ADC, 170 unique gene fusion pairs were observed, including rare kinase fusions, SLC12A2-ROS1, NCOA4-RET, and ANK3-RET. As for SCC, 15 unique gene fusions were identified, among which the most frequent were EML4-ALK and FGFR3-TACC3. Analyses of oncogenic mutations revealed a dual role for the gene fusions, CCDC6-RET and FGFR3-TACC3, in driving oncogenesis or serving as acquired resistance mechanisms to kinase inhibitors. In addition, our real-world evidence showed that patients with recurrent kinase fusions with low frequency (two occurrences) could benefit from treatment with kinase inhibitors’ off-label use. Notably, patients with stage IV ADC who had novel RORB-ALK or AFF2-RET fusions, but no other known oncogenic driver mutations, demonstrated favorable clinical outcomes on tyrosine kinase inhibitors. Our data provide a comprehensive overview of the landscape of oncogenic kinase fusions in lung cancer, which assist in recognizing potentially druggable fusions that can be translated into therapeutic applications.
Subject terms: Cancer, Molecular biology
Introduction
Lung cancer is the leading cause of cancer-related death worldwide, with a 5-year survival rate of less than 21%1. There are two major histological groups of lung cancer, including small cell lung cancer (SCLC) and non-SCLC (NSCLC). NSCLC accounts for approximately 85% of all lung cancers1 and can be further divided into different subtypes, among which, the most common is adenocarcinoma (ADC), followed by squamous cell carcinoma (SCC), adeno-squamous cell carcinoma (ASC), and large cell carcinoma (LCC). However, the molecular features of these subtypes are significantly distinct. For that reason, the identification of oncogenic driver genes and novel therapeutic targets is highly important for lung cancer treatment.
Kinases activated by gene fusions have been reported to be major classes of oncogenic drivers in lung cancer, which are produced by translocation or structural chromosome rearrangements, and function as potential targets of anticancer drugs2,3. Advances in next-generation sequencing (NGS) technologies have enabled the characterization of kinase fusions among different lung cancer subtypes, the identification of concurrent cancer-relevant alterations, and the identification of novel driver fusions.
In this study, we comprehensively analyzed DNA-seq data from 17,442 Chinese lung cancer patients, described the clinical and pathological characteristics of patients with different kinase fusions, and distinguished kinase fusion types between ADC and SCC. Additionally, we deeply studied the fusion breakpoint preferences of kinases and analyzed treatment-relevant mutations that co-occurred with primary recurrent kinase fusions. Furthermore, we identified multiple novel druggable fusions, which benefited from tyrosine kinase inhibitor (TKI) treatment. These studies facilitated a deep understanding of the unique clinical and molecular features, and the outcomes of lung cancer patients with kinase fusions.
Results
The landscape of kinase fusions in lung cancer
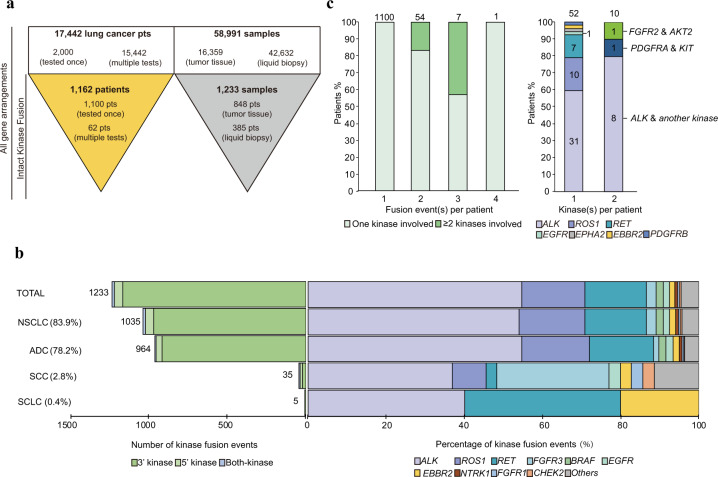
DNA-seq data from 17,442 lung cancer patients were analyzed, among which 2000 were unique cases while 15,442 patients had been tested more than once (Fig. 1a). After a series of filters and identifications, 1233 fusions with intact kinase domains were observed in 1162 patients with various histological subtypes, including ADC (78%; 906/1162), SCC (3.0%; 35/1162), adeno-squamous carcinoma (ASC; 0.8%; 9/1162), LCC (0.3%; 4/1162), and SCLC (0.4%; 5/1162) (Table 1). Sixty-one patients had bone metastasis. The median age of patients with ADC was 53 years (range: 21–95 years), which was significantly younger than that of other NSCLC subtypes (p < 0.001). In addition, the prevalence of males was higher than females (65.7% vs. 34.3%, respectively) in SCC patients.
Fig. 1. Overview of kinase fusions in lung cancer patients.
a Pipeline for kinase fusion calling and filtering. b The left bar plot shows the distribution of the three kinase fusion categories in different lung cancer subtypes. The number of events in each condition is indicated to the left of each bar. The right bar plot shows the percentage of the kinase genes involved across different lung cancer subtypes. c The left bar graph shows the number of kinase fusions in one patient. The right bar graph shows the kinase genes of patients who carried more than one kinase fusion. The number of patients in each condition is indicated on the top of each bar.
Table 1.
Summary of the demographic and clinical characteristics of the kinase fusion cohort in this study.
| Total | NSCLC | SCLC | Unknown | |||||
|---|---|---|---|---|---|---|---|---|
| ADC | SCC | ASC | LCC | Unknown | ||||
| Cases | 17,442 | 10,932 | 1353 | 162 | 63 | 217 | 549 | 4166 |
| Cases with kinase fusions | 1162 | 906 | 35 | 9 | 4 | 22 | 5 | 181 |
| Median age (range), years | 53 (21–95) | 53 (21–95) | 60 (31–82) | 60 (37–70) | 61 (44–64) | 60 (39–82) | 59 (46–64) | 54 (26–86) |
| <60, n (%) | 713 (61.4) | 585 (64.6) | 16 (45.7) | 4 (44.4) | 1 (25.0) | 10 (45.4) | 3 (60.0) | 94 (51.9) |
| ≥60 years, n (%) | 366 (31.5) | 277 (30.6) | 17 (48.6) | 5 (55.6) | 2 (50.0) | 11 (50.0) | 2 (40.0) | 52 (28.7) |
| Unknown | 83 (7.1) | 44 (4.8) | 2 (5.7) | 0 | 1 (25.0) | 1 (4.6) | 0 | 35 (19.4) |
| Sex, n (%) | ||||||||
| Male | 602 (51.8) | 434 (47.9) | 23 (65.7) | 4 (44.4) | 1 (25.0) | 14 (63.6) | 3 (60.0) | 81 (44.7) |
| Female | 560 (48.2) | 472 (52.1) | 12 (34.3) | 5 (55.6) | 3 (75.0) | 8 (36.4) | 2 (40.0) | 100 (55.3) |
| Stage, n (%) | ||||||||
| I | 21 (1.8) | 19 (2.1) | 0 | 1 (11.1) | 0 | 0 | 0 | 1 (0.5) |
| II | 20 (1.7) | 18 (2.0) | 2 (5.7) | 0 | 0 | 0 | 0 | 0 |
| III | 68 (5.9) | 60 (6.6) | 2 (5.7) | 0 | 0 | 1 (4.6) | 0 | 5 (2.8) |
| IV | 535 (46.0) | 457 (50.4) | 14 (40.0) | 5 (55.6) | 2 (50.0) | 10 (45.4) | 3 (60.0) | 44 (24.3) |
| Unknown | 518 (44.6) | 352 (38.9) | 17 (48.6) | 3 (33.3) | 2 (50.0) | 11 (50.0) | 2 (40.0) | 131 (72.4) |
Abbreviations: NSCLC non-small-cell lung cancer, SCLC small cell lung cancer, ADC adenocarcinoma, SCC squamous cell carcinoma, ASC adeno-squamous carcinoma, LCC large cell carcinoma.
We classified the 1233 kinase fusion events into three categories, including 1165 with a kinase at the 3′ end (3′ kinase), 57 with a kinase at the 5′ end (5′ kinase) and 11 fusions in which both partners were kinases (both-kinase) (Fig. 1b). In both-kinase patients, TRIM24-BRAF fusions were the most frequent (54.4%; 6/11; data not shown) and reported to be sensitive to a MEK inhibitor4. ALK (54.8%; 674/1233), ROS1 (16.2%; 200/1233), RET (15.8%; 195/1233), and FGFR3 (2.5%; 31/1233) were the most frequent recurring kinases identified, but with different frequencies in ADC and SCC (Fig. 1b and Supplementary Table 1). ALK fusions were the most common among all histologic subtypes (Supplementary Table 1 and Supplementary Table 2). Compared to ADC, the frequencies of ROS1 and RET were lower in SCC, and accounted for 8.5% and 2.9%, respectively. The prevalence of FGFR3 was much higher in SCC than ADC (SCC vs. ADC: 28.6% vs. 1.5%, respectively).
We also summarized the number of kinase fusions carried by each patient (Fig. 1c). As expected, 94.7% of patients contained only one fusion, while 5.3% of patients harbored at least two fusions. The ALK gene rearrangement was most frequently observed in patients with either single fusion event or multiple fusions (59.6% and 80%, respectively). We also identified one patient with four kinase fusions that all belonged to KIF5B-RET, but with different fusion breakpoints.
We further analyzed the gender distribution in patients with different fusions (a sample size of more than 15 cases was required for each fusion) (Supplementary Fig. 1a). Most patients with CCDC6-RET and FGFR3-TACC3 fusions were male, while the frequency of females was higher among patients with VCL-ALK and SDC4-ROS1 fusions. Age was also statistically associated with fusion type. The ages of patients with ROS1 fusions involving different partners varied significantly, as patients with EZR-ROS1 were older than patients with CD74-, SLC34A2-, and SDC4-ROS1 fusions (Supplementary Fig. 1b).
Characterizing recurrent kinase fusions in lung cancer patients
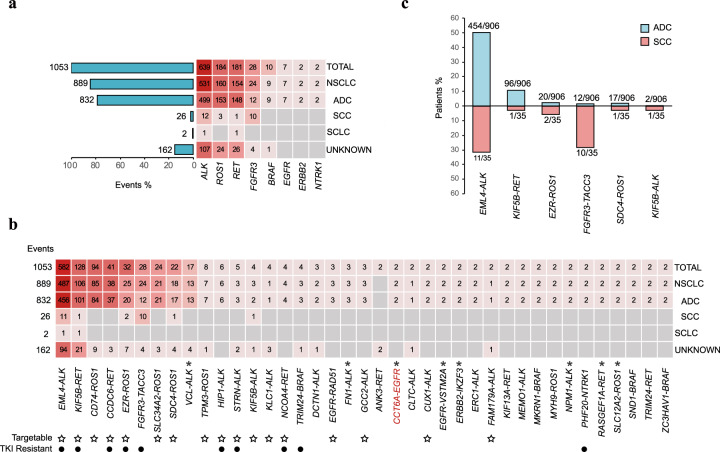
Recurrent kinase fusions have been of great interest and regarded as potential therapeutic targets for cancer treatment. Among 219 kinase fusion types, we observed 39 recurrent fusions involving 8 kinases, including ALK, ROS1, RET, FGFR3, BRAF, EGFR, ERBB2, and NTRK1 (Fig. 2a, b). The most frequently occurring kinase was ALK (60.7%; 639/1053) and included EML4-ALK (91.1%; 582/639), HIP1-ALK (0.9%; 6/639), and STRN-ALK (0.8%; 5/639) fusions, and has been well documented in NSCLC5–7 (Fig. 2b). We also found several recurrent ALK fusions which have not been reported in lung cancer, but have been identified in other cancer types, such as VCL-ALK, FN1-ALK, and NPM1-ALK. Those data indicated that such rarely-reported driver fusions may also play crucial roles in the carcinogenesis of lung cancer. For the recurrent kinase fusions with low frequency (two occurrences) in our data, CUX1-ALK8 and FAM179A-ALK9 have been shown response to ALK TKIs. Meanwhile, it has been reported that genetic rearrangements can mediate drug resistance to TKI treatment10. FGFR3-TACC311,12 is the resistance mechanism against EGFR TKIs in NSCLC patients with EGFR L858R or 19del mutation. EML4-ALK13 and NCOA4-RET14 are mechanisms of resistance to EGFR TKIs in NSCLC patients with EGFR T790M. Meanwhile, KIF5B-RET15, CCDC6-RET16, EZR-ROS117, PHF20-NTRK118, and TRIM24-BRAF19 have been reported to function as resistance mechanisms to EGFR 19del upon EGFR TKIs treatment in NSCLC patients. Besides, HIP1-ALK20 and STRN-ALK21 have been reported to mediate resistance to ALK TKIs in NSCLC patients with other ALK rearrangement. Further studies should be conducted to deeply understand the function of kinase fusions in cancer therapy.
Fig. 2. Recurrent kinase fusions in lung cancer.
a Kinases involved in fusions in different lung cancer subtypes and the number of events in each lung cancer subtype is shown to the left of each bar. b Kinase fusions in different lung cancer subtypes and the number of events in each lung cancer subtype is shown on the left. The fusion in red is a novel kinase fusion that has not been previously reported in all cancer types. Fusions marked with * represent kinase fusions never reported in lung cancer. The ☆ showed the targetable kinase fusion and the • represent the fusion which was identified as resistance mechanism to EGFR TKI or fusion TKI. c Comparison of the prevalence of kinase fusions in ADC and SCC. The number of patients with each fusion and the total number of patients in ADC or SCC are shown on the top of each bar.
Seven types of recurrent ROS1 fusions were identified in our data, including the well described CD74-ROS1, EZR-ROS1, SDC4-ROS1, SLC34A2-ROS1, TPM3-ROS1, and MYH9-ROS122,23, as well as a novel recurrent fusion, SLC12A2-ROS1, which was identified as a novel fusion in inflammatory myofibroblastic tumors (Fig. 2b). This was the first time that SLC12A2-ROS1 was reported in lung cancer. For RET fusions, KIF5B-RET and CCDC6-RET, accompanied with other rarely-reported fusions, such as NCOA4-RET24, ANK3-RET, KIF13A-RET25, RASGEF1A-RET26, and TRIM24-RET27 were identified (Fig. 2b). Among which, RASGEF1A-RET26 was previously identified in breast cancer as a tumorigenic fusion sensitive to RET inhibitors, but not previously reported in lung cancer. Other recurrent fusions including BRAF, EGFR, ERBB2, and NTRK1 were also observed. Among them, EGFR-VSTM2A, CCT6A-EGFR, and ERBB2-IKZF3 have not previously been documented in lung cancer.
Importantly, only six recurrent fusions, including EML4-ALK, KIF5B-RET, EZR-ROS1, FGFR3-TACC3, SDC4-ROS1, and KIF5B-ALK were detected in ADC and SCC (Fig. 2c). Notably, FGFR3-TACC3 was the only fusion involving FGFR3 in this study and had a higher frequency in SCC patients (28.9%; 10/35) compared to ADC patients (1.3%; 12/906) (Fig. 2c).
Breakpoint patterns of known kinase fusions
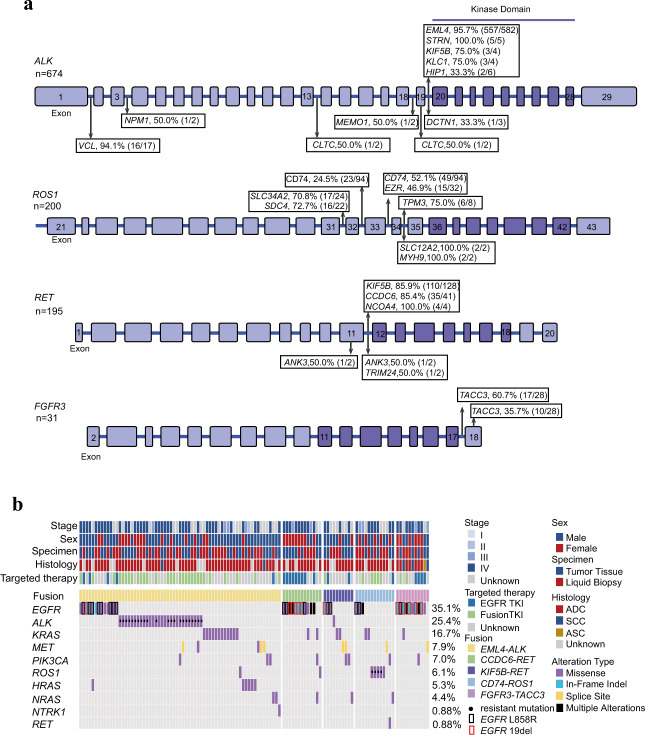
Although the kinases in our cohort all retained the integrity of the kinase domain, the breakpoints were variable when ligated with different partners. Such an observation may be caused by the structural and functional characteristics of each partner gene and may influence the tumorigenesis capacities of the fusions.
In STRN- (100.0%; 5/5), EML4- (95.7%; 557/582), KIF5B- (75.0%; 3/4), and KLC1-ALK (75.0%; 3/4) fusions, the breakpoints of ALK were almost exclusively in intron 19, which was consistent with previous studies28–31 (Fig. 3a). Interestingly, we identified a patient with a novel ALK breakpoint in intron 4 of a KIF5B-ALK fusion. Additionally, the ALK breakpoints in VCL-ALK fusions were primarily located in intron 1 (94.1%; 16/17), which was an uncommon breakpoint among ALK fusions and indicated a distinct rearrangement mechanism. Several novel breakpoints were also identified, including CLTC-ALK intron 13/exon 19 and MEMO1-ALK intron 18. The function of those fusion types should be further studied.
Fig. 3. The breakpoint locations for kinase genes and druggable driver mutations co-occurring with kinase fusions.
a The breakpoints for ALK with different partners; the breakpoints for ROS1 with different partners; the breakpoints for RET with different partners; the breakpoints for FGFR3 with different partners. The partners shown above the structure of each kinase belong to common partners. Some rare partners are shown under the kinase structure. The number of events involving each of the four kinases is shown to the left of each kinase structures. The percentage breakpoints with each partner, the number of events for each fusion in specific kinase breakpoints, and the total number of events for each fusion are shown. b The co-occurrence of kinase fusions and driver mutations is displayed in the hotspot plot. The incidences of driver gene mutations are shown to the left of the plot. Tissue/plasma samples were collected from 59 patients after developing resistance to EGFR TKI or fusion targeted treatment.
The most frequent fusion partner genes for ROS1 were CD74, EZR, SLC34A2, SDC4, and TPM3 (Supplementary Table 3). Although CD74 was the most common partner in lung cancer, only EZR-ROS1 and SDC4-ROS1 fusions were found in SCC (Supplementary Table 3). The majority of breakpoints located in common positions of ROS1 have been reported previously32,33 and we observed consistent results in our data. CD74 (52.1%; 49/94) and EZR (46.9%; 15/32) were primarily ligated to intron 33 of ROS1 (Fig. 3a). For most SDC4- (72.7%; 16/22) and SLC34A2-ROS1 (70.8%; 17/24) fusions, the ROS1 breakpoints were in intron 31, whereas TPM3-ROS1 (75.0%; 6/8) fusions mainly occurred in intron 34 of ROS1. We also identified several rare events with ROS1 breakpoints in intron 26 (SLC34A2-ROS1), intron 28 (CD74-ROS1 and SLC34A2-ROS1), and exon 28 (CD74-ROS1). ROS1 breakpoints involved in novel fusions with SLC12A2 and MYH9 were also in intron 34.
We also analyzed the most frequent fusion partner genes of RET, including KIF5B, CCDC6, and NCOA4, and found that they were primarily rearranged in intron 11 of RET at frequencies of 85.9% (110/128), 85.4% (35/41), and 100% (4/4), respectively (Fig. 3a). TACC3 was the main fusion partner of FGFR3 in both ADC (85.7%; 12/14) and SCC (100.0%; 10/10) (Supplementary Table 3), which frequently ligated to intron 17 (60.7%; 17/28) and exon 18 (35.7%; 10/28) of FGFR3 (Fig. 3a and Supplementary Table 3).
Analysis of mutations co-occurring with kinase fusions
The frequency of mutations co-occurring with CCDC6-RET (31.7%; 13/41) and FGFR3-TACC3 (39.3%; 11/28) was higher than those co-occurring with EML4-ALK (11.5%; 67/582), KIF5B-RET (7.8%; 10/128), and CD74-ROS1 (13.8%; 13/94) (Supplementary Fig. 2a). Strikingly, deep analysis of the concurrent mutations with CCDC6-RET and FGFR3-TACC3 showed significant enrichment in EGFR ex19del-mutant patients (p < 0.01), respectively (Supplementary Fig. 2b and Fig. 3b) but not in L858R mutant patients. Indeed, most patients with CCDC6-RET and FGFR3-TACC3 received prior EGFR TKI treatment; thus, suggesting that CCDC6-RET and FGFR3-TACC3 may function as resistance mechanisms to EGFR TKIs in lung cancer, and may also highlight the differences in acquired resistance to TKIs through gene fusions between EGFR L858R and ex19del-mutant variants.
In the EML4-ALK group, L858R more frequently co-occurred than other EGFR mutations (Fig. 3b). Additionally, the frequency of ALK mutations was 44.3% (31/70), including ALK G1202R, G1269A, and L1196M, which are well-described resistance mechanisms to ALK TKIs. The HRAS mutation was only identified in the EML4-ALK group. MET exon 14 skipping (77.8%; 7/9) occurred significantly more frequently than other types of MET alterations that co-occurred with kinase fusions.
Interestingly, an ADC patient harboring an EML4-ALK fusion also had a NTRK1 G595R mutation, which is considered to be an acquired resistant mutation to NTRK inhibitors (larotrectinib or entrectinib)34. However, whether this alteration is associated with ALK TKI resistance remains to be determined. We also found a patient carrying CD74-ROS1 and RET V804M alterations, however, the function of the concurrent RET V804M mutation requires further investigation.
RET fusions in lung cancer and the response to RET inhibitors
Since RET is a crucial therapeutic target in cancers, several multi-kinase inhibitors (MKIs) such as cabozantinib, vandetanib, alectinib, and apatinib have shown anti-RET activities. However, these MKIs have limited efficacy and patients with RET fusions showed poor responses with short progression free survival (PFS) times of generally less than 7.5 months27. In our cohort, targeted therapy information was available for 10 patients carrying different RET fusions (Fig. 4), among which nine patients were treated with MKIs and one was treated with the RET-selective inhibitor, pralsetinib. For the nine patients who received MKI therapy, five showed no response. Among the remaining four patients, three KIF5B-RET-positive cases received cabozantinib treatment and one achieved a PFS longer than 6 months. There were three CCDC6-RET-positive patients treated with apatinib in this study, but only one showed a response and achieved a PFS of 10 months. We also identified a patient with a novel AFF2-RET fusion who was treated with pralsetinib and maintained a response for 9 months. The clinical history of that AFF2-RET patient is described below.
Fig. 4. Progression Free Survival (PFS) of patients carrying RET fusions.
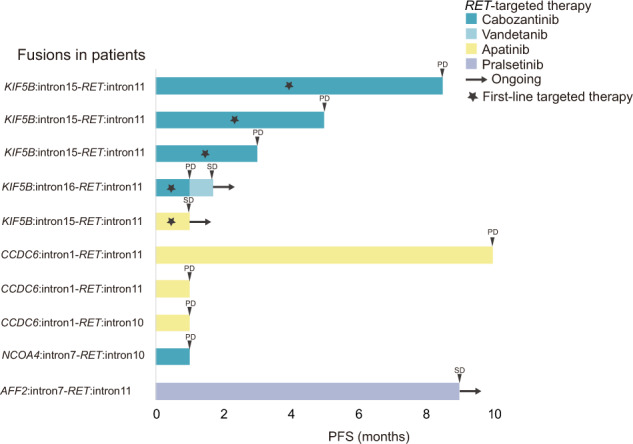
Swimmer plot showing treatment drugs and durations (months). Patients with different fusions are indicated.
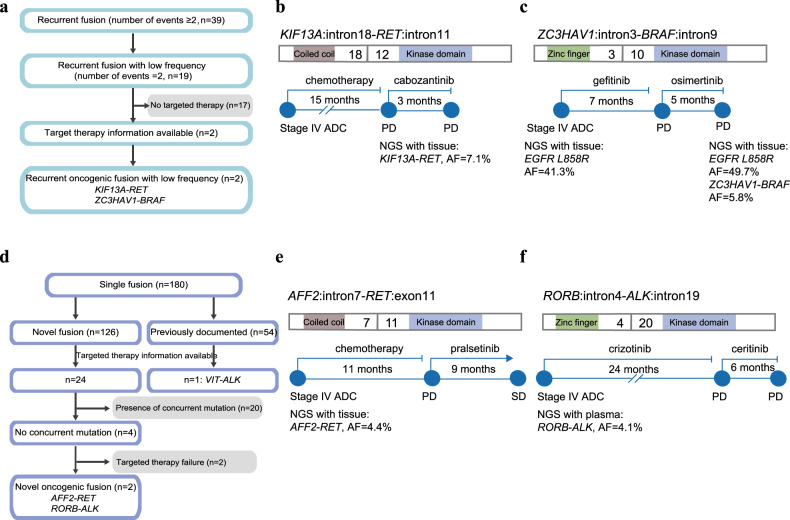
Novel fusions that demonstrated favorable clinical benefits of kinase inhibitors
Besides the well-described recurrent kinase fusions, we also identified 19 recurrent fusion types with low frequency (two occurrences) in our cohort (Fig. 2). To search for functional fusions, we focused only on cases with targeted therapy information available, which included two fusions, KIF13A-RET and ZC3HAV1-BRAF (Fig. 5a). The patient with the KIF13A-RET fusion was a 64-year-old male with stage IV ADC. After 15 months of chemotherapy, the patient’s tumor biopsy was subjected to NGS and revealed the potential driver fusion, KIF13A: exon18-RET: exon12. Subsequently, the patient was treated with cabozantinib, but failed to exhibit a response (Fig. 5b). The patient harboring the ZC3HAV1-BRAF fusion was a 56-year-old female with stage IV ADC. Her tissue and plasma biopsies were subjected NGS which identified the EGFR L858R mutation at mutation allele frequency (MAF) of 41.3% and 3.0%, respectively. The patient was administered gefitinib and achieved a PFS of 7 months. After the disease progressed, the patient was administered osimertinib treatment, but only responded for 5 months. The tumor progressed and an additional metastatic site was identified, and thus, the patient’s plasma and metastatic tissue biopsies were subjected to a second NGS test. That test identified a ZC3HAV1: exon3-BRAF: exon10 mutation at a MAF of 3.1% and 5.8%, respectively, thus, indicating the resistance function of the ZC3HAV1-BRAF fusion (Fig. 5c).
Fig. 5. Potentially druggable fusions in lung cancer patients.
a Pipeline of oncogenic fusions among all recurrent fusions. b The structure of KIF13A-RET and clinical information of the patient harboring that fusion. The domains are highlighted in different colors as: coil-coiled domain of KIF13A (brown) and RET tyrosine kinase domain (blue). c The structure of ZC3HAV1-BRAF and clinical information of the patient harboring that fusion. The domains are highlighted in different colors as: zinc finger domain of ZC3HAV1 (green) and BRAF tyrosine kinase domain (blue). d Pipeline of novel oncogenic fusions among single fusions. e The structure of AFF2-RET and clinical information of the patient harboring that fusion. The domains are highlighted in different colors as: coil-coiled domain of AFF2 (brown) and RET tyrosine kinase domain (blue). f The structure of RORB-ALK and clinical information of the patient harboring that fusion. The domains are highlighted in different colors as: zinc finger domain of RORB (green) and ALK tyrosine kinase domain (blue).
We also found 180 types of single kinase fusions, including 126 fusions that had not previously been reported. To scan for functional cases from novel kinase fusions, we evaluated the therapeutic history of each patient and focused only on fusions without concurrent driver mutations that benefited from fusion TKI treatments (Fig. 5d). We identified two novel fusion types, including AFF2-RET (AFF2: intron7-RET: exon11) and RORB-ROS1 (RORB: intron4-ALK: intron19). The patient harboring the AFF2-RET fusion was a male with stage IV ADC. His plasma biopsy was subjected to NGS, and the novel AFF2-RET fusion was identified at a MAF of 4.1%. After chemotherapy for 11 months, the patient received pralsetinib treatment and achieved a stable disease, which was maintained for 9 months. Additional investigations of the AFF domain identified a coil-coiled domain, which is capable of mediating dimerization (Fig. 5e). The patient carrying the RORB-ALK fusion was a 64-year-old male with stage IV ADC. His tumor biopsy was subjected to NGS, which revealed the potential driver fusion. Subsequently, the patient was treated with crizotinib and achieved a response for 24 months. After disease progression, the patient was treated with ceritinib and responded for 6 months. Structural analysis of RORB revealed a zinc finger domain that may lead to dimerization and activation of downstream ALK targets (Fig. 5f). These two novel kinase fusions both exhibited remarkable sensitivity to RET and ALK TKIs, respectively, which suggested that they were able to function as potentially druggable fusions.
Discussion
In this study, we used DNA-seq data from 17,442 Chinese lung cancer patients to identify potential driver fusions, and analyze the clinical and genomic features of patients with different kinase fusion types. Overall, 6.66% (1162/17,442) of patients contained kinase fusions (recurrent fusions account for 5.2%). Of the 219 fusion types observed, 39 were recurrent while 180 were observed in single cases. The most frequent kinases identified in patients were ALK, RET, ROS1, EGFR, and FGFR3.
An interesting observation was the identification of recurrent fusions that had not previously been reported in lung cancer, but were found in other cancer types, such as the VCL-ALK fusion in epithelioid fibrous histiocytoma35 and renal cell carcinoma36, the FN1-ALK fusion in ovarian cancer37 and gastrointestinal leiomyomas38, the NPM1-ALK fusion in anaplastic large cell lymphoma39, the VSTM2A-EGFR fusion in glioblastomas40, and the ERBB2-IKZF3 fusion in breast cancer41. Those results indicated that driver fusions in other cancers may also play crucial roles in lung cancer carcinogenesis. We also identified a novel recurrent fusion, CCT6A-EGFR, but the function of which remains to be determined.
Many driver mutations often co-occurred and played critical roles with kinase fusions during cancer progression. Thus, detailing the relationships between fusions and mutations may lead to the development of efficient treatment strategies. Therefore, we analyzed treatment-relevant driver alterations that co-occurred with the most frequent kinase fusions.
Further investigation of the breakpoints involved in different kinase fusions revealed preferences for the ligation sites of partners. Most of the fusions occurred in intron 11 and intron 17 of RET and FGFR3, respectively, but the breakpoints of ALK and ROS1 varied between different partners. Studies showed that translocations were not random, as the breakpoints in the fusion genes had both sequence and structure preferences42. It was reported that ALK fusion variants may affect clinical outcomes upon TKI treatment43.
By integrating mutation, copy number variation, and fusion data, we evaluated the genomic alterations co-occurring with each fusion, and focused mainly on alterations related to drug administration. Patients with CCDC6-RET and FGFR3-TACC3 had a higher prevalence of EGFR ex19del mutations (p < 0.01), which indicated that they may function as resistance mechanisms for EGFR TKIs. It has been reported that CCDC6-RET can mediate osimertinib resistance in NSCLC, and the combination of osimertinib and RET inhibitors leads to a rapid response in those patients44. In our cohort, nine patients with EGFR alterations were treated with EGFR TKIs, among which seven were treated with osimertinib. However, two patients with CCDC6-RET were only treated with gefitinib and ectinib, respectively, which indicated that the CCDC6-RET fusion functioned as a resistance alteration against first and third generation TKIs. The therapeutic approach to targeting those fusions should be further studied. FGFR3-TACC3 could confer resistance to EGFR TKIs and often had co-existing EGFR activating mutations in NSCLC11, which is consistent with our findings. We also revealed that the frequency of patients with METex 14 skipping was higher than that of other MET alterations co-occurring with kinase fusions. However, the underlying mechanisms leading to such observations should be studied in detail.
Since only 6% of cancer patients can benefit from the existing drugs targeting fusions, the identification of novel druggable fusions is crucial for expanding the therapeutic options for cancer patients. In this study, we focused only on fusions with intact kinase domains, without concurrent driver mutations, and studied the clinical history of each patient. We identified two novel fusions, including AFF2-RET and RORB-ALK with demonstrated responses to TKI treatment. We analyzed the domains of the two fusions and found that they both harbored domains that mediate protein dimerization and activate downstream signaling pathways. We also observed a patient with a novel AFF3-ALK fusion, without the dimerization domain (data not shown). As expected, that patient showed no response when treated with crizotinib.
This study has two main limitations. The first one is that the therapeutic information were incomplete or missing, which is also one of the weaknesses of real-world studies45. Although we used a system with error alert function to abstract medical record, errors might have occurred during information collection. The other limitation is that we did not validate the novel or concurrent fusions via other diagnostic approaches including reverse transcription polymerase chain reaction, or RNA sequencing which due to the insufficiency of tissue amount. A recent study46 showed that two non-canonical ALK fusions identified with DNA sequencing in a patient generated canonical EML4-ALK transcripts during mRNA maturation which was revealed via RNA sequencing, indicating a difference of fusion detected at DNA and RNA level. Therefore, other diagnostic approaches should be used for further validation of fusions observed with DNA sequencing.
In conclusion, we provided a comprehensive overview of the landscape of kinase fusions in a large Chinese lung cancer population, including lung ADC and SCC patients, and characterized the patterns of breakpoint locations and co-occurring mutations in patients with known kinase fusions. Furthermore, we reported novel kinase fusions and highlighted patients with recurrent kinase fusions with low frequency (two occurrences) that demonstrated favorable clinical outcomes on TKIs. Collectively, these findings not only advance our understanding of the spectrum of fusions involving kinases in lung cancer, but also have immediate implications for disease diagnosis and treatment.
Methods
Patients and samples
Using capture-based targeted NGS, this study retrospectively reviewed 17,442 Chinese NSCLC patients who underwent genetic testing at hospitals across China between June 2016 and July 2019, including The Second Affiliated Hospital of Zhejiang University School of Medicine, Fujian Medical University Cancer Hospital and Zhangzhou Zhengxing Hospital. Demographic and clinical data, including age, gender, histology type, pathological stage, metastasis sites, treatment regimens, and duration of TKI treatment were abstracted from the medical records provided by physicians. Genomic profiling was performed on formalin-fixed paraffin-embedded (FFPE) tumor and liquid biopsy specimens. All samples were sequenced in a Clinical Laboratory Improvement Amendments (CLIA)- and College of American Pathologists (CAP)-certified genomic testing facility (Nanjing Geneseeq Technology Inc., Nanjing, China). This study was approved by the ethical committee of each participating hospital and all patients provided written informed consent to participate.
Library preparation and sequencing
For targeted NGS, DNA extraction and sequencing libraries were prepared according to protocols described previously47. Briefly, genomic DNA was extracted from FFPE tumor and liquid biopsies using a QIAamp DNA FFPE Tissue Kit and a QIAamp Circulating Nucleic Acid kit (Qiagen), respectively. Library preparations were performed using the KAPA Hyper Prep Kit (KAPA Biosystems). Target enrichment was performed using the xGen lockdown probes targeting 425 cancer-related genes (Geneseeq Prime). The target-enriched libraries were quantified by qPCR using the KAPA Library Quantification kit (KAPA Biosystems) and sequenced on HiSeq NGS platforms (Illumina) to generate 2 × 150-bp reads following the manufacturer’s instructions.
Fusions detection and kinase fusion analysis
We used Delly fusion calling tool48 to identify the number of chimeric reads (sequencing paired ends mapped to different genes) and split reads (spanning a fusion breakpoint) from the targeted DNA-seq data. Fusions were filtered by removing fusions with (1) breakpoints involving intergenic loci; and (2) fusions with split reads <3 or paired reads <5. Then, kinase fusions matched to the Kinase Database (including 538 kinase genes; http://kinase.com/web/current/) were evaluated with the conservation of full catalytic kinase domains. All fusions were manually confirmed using the Integrative Genomics Viewer (IGV) and only kinase fusions retaining an intact kinase domain were included for analysis in this study. For patients with multiple tests, same fusion was counted only once regardless of whether it was detected in multiple samples or different sample sources. Samples’ therapeutic timelines, including treatment-naïve and post-TKI treatment, were considered unless this information was lacking.
Mutation calling
Somatic alterations concurrent with kinase fusions were analyzed as previously described49. In brief, we used Trimmomatic for FASTQ file quality control (below 15 or N bases were removed). Burrows–Wheeler Aligner (BWA v0.7.12) was used for mapping reads to the reference Human Genome (hg19). Local realignment around the indels and base quality score recalibration was performed with the Genome Analysis Toolkit (GATK 3.4.0). VarScan 2 was used for somatic mutations calling, with at least 0.2% MAF and with at least three supporting-reads. The Oncology Knowledge Base (oncoKB) was used to identify oncogenic alterations50. Information on targeted therapy-relevant hotspot alterations were also abstracted from the oncogenic alterations51.
Data analysis and statistics
The chi-squared test, Mann–Whitney U test, and Kruskal–Wallis H test were used for analyses of gender and age. A p-value < 0.05 was considered significant for all tests, unless otherwise indicated. Statistical analyses were performed in R (v.3.3.2) and Statistical Product and Service Solutions.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This research was financially by National Natural Science Foundation of China (Grant/Award Number: 81802684), Physician Elite in Training Scholarship of Zhejiang University (Grant/Award No. 171132002), Zhejiang Medical Science Foundation (Grant/Award No. 2017KY392) and Natural Science Foundation of Fujian Provincial Health Commission (Grant/Award No. 2017J01263).
Author contributions
Conceptualization, methodology, funding acquisition: Nong Lin. Data curation, writing-original draft preparation: Binghao Li, Hao Qu, Sisi Liu, and Ruting Guan. Visualization, investigation: Xuexin He and Dong Lin. Supervision: Youbin Xu, Xue Wu, and Yang Shao. Software, validation, formal analysis: Yong Wu, Qiuxiang Ou, and Hua Bao. Project administration, resources: Weibo Pan, Meng Liu, and Xiaobo Yan. Writing, reviewing and editing: Jing Zhang, Nong Lin, and Xin Huang.
Data availability
The data presented in the study are deposited in the Genome Sequence Archive for Human (GSA-Human) repository, accession number HRA001045.
Code availability
Chimeric reads were called with Delly tool (v0.7.8; available at https://github.com/dellytools/delly). Reads were mapped to the reference Human Genome with Burrows–Wheeler Aligner (BWA) (v0.7.12; available at https://github.com/lh3/bwa/). Local realignment around the indels and base quality score recalibration was applied with the Genome Analysis Toolkit (GATK) (v3.4.0; available at https://software.broadinstitute.org/gatk/). The code that supports the findings of this study are available from the corresponding authors on reasonable request.
Competing interests
S.L., R.G., Y.W., Q.O., H.B., X.W. and Y.S. are the employees of Nanjing Geneseeq Technology Inc., China. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Binghao Li, Hao Qu, Jing Zhang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-021-00221-z.
References
- 1.Lu T, et al. Trends in the incidence, treatment, and survival of patients with lung cancer in the last four decades. Cancer Manag. Res. 2019;11:943–953. doi: 10.2147/CMAR.S187317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medves S, Demoulin JB. Tyrosine kinase gene fusions in cancer: translating mechanisms into targeted therapies. J. Cell Mol. Med. 2012;16:237–248. doi: 10.1111/j.1582-4934.2011.01415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat. Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutchinson KE, et al. BRAF fusions define a distinct molecular subset of melanomas with potential sensitivity to MEK inhibition. Clin. Cancer Res. 2013;19:6696–6702. doi: 10.1158/1078-0432.CCR-13-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayliss R, Choi J, Fennell DA, Fry AM, Richards MW. Molecular mechanisms that underpin EML4-ALK driven cancers and their response to targeted drugs. Cell Mol. Life Sci. 2016;73:1209–1224. doi: 10.1007/s00018-015-2117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang DD, et al. HIP1-ALK, a novel ALK fusion variant that responds to crizotinib. J. Thorac. Oncol. 2014;9:285–294. doi: 10.1097/JTO.0000000000000087. [DOI] [PubMed] [Google Scholar]
- 7.Kelly LM, et al. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc. Natl Acad. Sci. USA. 2014;111:4233–4238. doi: 10.1073/pnas.1321937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang M, et al. CUX1-ALK, a novel ALK rearrangement that responds to crizotinib in non-small cell lung cancer. J. Thorac. Oncol. 2018;13:1792–1797. doi: 10.1016/j.jtho.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Yan J, Zhou X, Pan D. A case of one lung adenocarcinoma patient harboring a novel FAM179A-ALK (F1, A19) rearrangement responding to lorlatinib treatment. Lung Cancer. 2020;147:26–29. doi: 10.1016/j.lungcan.2020.06.026. [DOI] [PubMed] [Google Scholar]
- 10.Zhu VW, Klempner SJ, Ou SI. Receptor tyrosine kinase fusions as an actionable resistance mechanism to EGFR TKIs in EGFR-mutant non-small-cell lung cancer. Trends Cancer. 2019;5:677–692. doi: 10.1016/j.trecan.2019.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Ou SI, et al. Emergence of FGFR3-TACC3 fusions as a potential by-pass resistance mechanism to EGFR tyrosine kinase inhibitors in EGFR mutated NSCLC patients. Lung Cancer. 2017;111:61–64. doi: 10.1016/j.lungcan.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schrock AB, et al. Receptor tyrosine kinase fusions and BRAF kinase fusions are rare but actionable resistance mechanisms to EGFR tyrosine kinase inhibitors. J. Thorac. Oncol. 2018;13:1312–1323. doi: 10.1016/j.jtho.2018.05.027. [DOI] [PubMed] [Google Scholar]
- 13.Batra U, Sharma M, Amrith BP, Mehta A, Jain P. EML4-ALK fusion as a resistance mechanism to osimertinib and its successful management with osimertinib and alectinib: case report and review of the literature. Clin. Lung Cancer. 2020;21:e597–e600. doi: 10.1016/j.cllc.2020.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Xu H, et al. Characterization of acquired receptor tyrosine-kinase fusions as mechanisms of resistance to EGFR tyrosine-kinase inhibitors. Cancer Manag. Res. 2019;11:6343–6351. doi: 10.2147/CMAR.S197337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu YC, et al. The KIF5B-RET fusion gene mutation as a novel mechanism of acquired EGFR tyrosine kinase inhibitor resistance in lung adenocarcinoma. Clin. Lung Cancer. 2019;20:e73–e76. doi: 10.1016/j.cllc.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Klempner SJ, et al. Emergence of RET rearrangement co-existing with activated EGFR mutation in EGFR-mutated NSCLC patients who had progressed on first- or second-generation EGFR TKI. Lung Cancer. 2015;89:357–359. doi: 10.1016/j.lungcan.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 17.Niu X, et al. PS01.06: tumor heterogeneity in lesion specific response creates ROS1 fusion mediating resistance to gefitinib in EGFR 19 deletion lung adenocarcinoma. J. Thorac. Oncol. 2016;11:S273. doi: 10.1016/j.jtho.2016.09.042. [DOI] [Google Scholar]
- 18.Xia H, et al. Evidence of NTRK1 fusion as resistance mechanism to EGFR TKI in EGFR+ NSCLC: results from a large-scale survey of NTRK1 fusions in chinese patients with lung cancer. Clin. Lung Cancer. 2020;21:247–254. doi: 10.1016/j.cllc.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Sheikine Y. et al. BRAF in lung cancers: analysis of patient cases reveals recurrent BRAF mutations, fusions, kinase duplications, and concurrent alterations. JCO Precis. Oncol. 1–15 (2018). [DOI] [PMC free article] [PubMed]
- 20.Li M, Tang Q, Chen S, Wang Y. A novel HIP1-ALK fusion variant in lung adenocarcinoma showing resistance to Crizotinib. Lung Cancer. 2021;151:98–100. doi: 10.1016/j.lungcan.2020.11.014. [DOI] [PubMed] [Google Scholar]
- 21.Iida Y, et al. P1.03-009 a lung adenocarcinoma with a STRN-ALK rearrangement was poorly responsive to alectinib treatment. J. Thorac. Oncol. 2017;12:S1954. doi: 10.1016/j.jtho.2017.09.813. [DOI] [Google Scholar]
- 22.Davies KD, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin. Cancer Res. 2012;18:4570–4579. doi: 10.1158/1078-0432.CCR-12-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rozenblum AB, et al. Clinical impact of hybrid capture-based next-generation sequencing on changes in treatment decisions in lung cancer. J. Thorac. Oncol. 2017;12:258–268. doi: 10.1016/j.jtho.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, et al. RET fusion in advanced non-small-cell lung cancer and response to cabozantinib: a case report. Medicines. 2019;98:e14120. doi: 10.1097/MD.0000000000014120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, et al. Identification of a novel KIF13A-RET fusion in lung adenocarcinoma by next-generation sequencing. Lung Cancer. 2018;118:27–29. doi: 10.1016/j.lungcan.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Paratala BS, et al. RET rearrangements are actionable alterations in breast cancer. Nat. Commun. 2018;9:4821. doi: 10.1038/s41467-018-07341-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrara R, Auger N, Auclin E, Besse B. Clinical and translational implications of RET rearrangements in non-small cell lung cancer. J. Thorac. Oncol. 2018;13:27–45. doi: 10.1016/j.jtho.2017.10.021. [DOI] [PubMed] [Google Scholar]
- 28.Perot G, et al. Identification of a recurrent STRN/ALK fusion in thyroid carcinomas. PLoS ONE. 2014;9:e87170. doi: 10.1371/journal.pone.0087170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenbaum JN, et al. Genomic heterogeneity of ALK fusion breakpoints in non-small-cell lung cancer. Mod. Pathol. 2018;31:791–808. doi: 10.1038/modpathol.2017.181. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi K, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin. Cancer Res. 2009;15:3143–3149. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 31.Dietz, S. et al. Serial liquid biopsies for detection of treatment failure and profiling of resistance mechanisms in KLC1-ALK-rearranged lung cancer. Cold Spring Harb. Mol. Case Stud.5, a004630 (2019). [DOI] [PMC free article] [PubMed]
- 32.Bubendorf L, et al. Testing for ROS1 in non-small cell lung cancer: a review with recommendations. Virchows Arch. 2016;469:489–503. doi: 10.1007/s00428-016-2000-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park S, et al. Characteristics and outcome of ROS1-positive non-small cell lung cancer patients in routine clinical practice. J. Thorac. Oncol. 2018;13:1373–1382. doi: 10.1016/j.jtho.2018.05.026. [DOI] [PubMed] [Google Scholar]
- 34.Okamura, R. et al. Analysis of NTRK alterations in pan-cancer adult and pediatric malignancies: implications for NTRK-targeted therapeutics. JCO Precis. Oncol.2, 1–20 (2018). [DOI] [PMC free article] [PubMed]
- 35.Dickson BC, Swanson D, Charames GS, Fletcher CD, Hornick JL. Epithelioid fibrous histiocytoma: molecular characterization of ALK fusion partners in 23 cases. Mod. Pathol. 2018;31:753–762. doi: 10.1038/modpathol.2017.191. [DOI] [PubMed] [Google Scholar]
- 36.Smith NE, et al. VCL-ALK renal cell carcinoma in children with sickle-cell trait: the eighth sickle-cell nephropathy? Am. J. Surg. Pathol. 2014;38:858–863. doi: 10.1097/PAS.0000000000000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren H, et al. Identification of anaplastic lymphoma kinase as a potential therapeutic target in ovarian cancer. Cancer Res. 2012;72:3312–3323. doi: 10.1158/0008-5472.CAN-11-3931. [DOI] [PubMed] [Google Scholar]
- 38.Panagopoulos I, et al. Recurrent fusion of the genes FN1 and ALK in gastrointestinal leiomyomas. Mod. Pathol. 2016;29:1415–1423. doi: 10.1038/modpathol.2016.129. [DOI] [PubMed] [Google Scholar]
- 39.Lin JJ, Riely GJ, Shaw AT. Targeting ALK: precision medicine takes on drug resistance. Cancer Discov. 2017;7:137–155. doi: 10.1158/2159-8290.CD-16-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erdem-Eraslan L, et al. Identification of patients with recurrent glioblastoma who may benefit from combined bevacizumab and CCNU therapy: a report from the BELOB trial. Cancer Res. 2016;76:525–534. doi: 10.1158/0008-5472.CAN-15-0776. [DOI] [PubMed] [Google Scholar]
- 41.Gao Q, et al. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 2018;23:227–238. doi: 10.1016/j.celrep.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang D, et al. The structural characterization of tumor fusion genes and proteins. Comput. Math. Methods Med. 2015;2015:912742. doi: 10.1155/2015/912742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin JJ, et al. Impact of EML4-ALK variant on resistance mechanisms and clinical outcomes in ALK-positive lung cancer. J. Clin. Oncol. 2018;36:1199–1206. doi: 10.1200/JCO.2017.76.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piotrowska Z, et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov. 2018;8:1529–1539. doi: 10.1158/2159-8290.CD-18-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Camm AJ, Fox KAA. Strengths and weaknesses of ‘real-world’ studies involving non-vitamin K antagonist oral anticoagulants. Open Heart. 2018;5:e000788. doi: 10.1136/openhrt-2018-000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang J, et al. Complex ALK fusions are associated with better prognosis in advanced non-small cell lung cancer. Front. Oncol. 2020;10:596937–596937. doi: 10.3389/fonc.2020.596937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tong L, et al. Tumor-derived DNA from pleural effusion supernatant as a promising alternative to tumor tissue in genomic profiling of advanced lung cancer. Theranostics. 2019;9:5532. doi: 10.7150/thno.34070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin. Cancer Res. 2018;24:3097–3107. doi: 10.1158/1078-0432.CCR-17-2310. [DOI] [PubMed] [Google Scholar]
- 50.Chakravarty D, et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. 2017;1:1–16. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mayakonda A, Lin D-C, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in the study are deposited in the Genome Sequence Archive for Human (GSA-Human) repository, accession number HRA001045.
Chimeric reads were called with Delly tool (v0.7.8; available at https://github.com/dellytools/delly). Reads were mapped to the reference Human Genome with Burrows–Wheeler Aligner (BWA) (v0.7.12; available at https://github.com/lh3/bwa/). Local realignment around the indels and base quality score recalibration was applied with the Genome Analysis Toolkit (GATK) (v3.4.0; available at https://software.broadinstitute.org/gatk/). The code that supports the findings of this study are available from the corresponding authors on reasonable request.