Abstract
Recently a novel insertion element, IS1245, has been described and suggested for use as a probe in restriction fragment length polymorphism studies of Mycobacterium avium strains. An important issue in this context is the stability of the insertion element. We analyzed single colonies of M. avium cultures and found frequent small one- to two-band changes. However, following repeated in vitro passages over 1 year, similar one- to two-band changes were observed in the IS1245 patterns of only six M. avium strains investigated.
Mycobacteria belonging to the Mycobacterium avium complex (MAC) have long been known to be potentially pathogenic for humans, with pulmonary infections and lymphadenitis in small children as the most common clinical presentations (6). However, MAC is also capable of causing disseminated infection in severely immunocompromised individuals, and due to the human immunodeficiency virus (HIV) pandemic, an increase in these infections has been observed (5). As many as 25 to 40% of the HIV-infected patients contract MAC infections when their CD4 cell counts reach a level of 50 × 109 to 100 × 109/liter (8). This circumstance has created an increased interest in the epidemiology of MAC, with special attention being given to routes and sources of infection. Studies of these topics are dependent on reliable methods for strain differentiation. A considerable number of genetic tools have become available during recent years, e.g., pulsed-field gel electrophoresis (2, 7), PCR-based methods (10), and restriction fragment length polymorphism (RFLP) using different insertion elements (3, 12). Pulsed-field gel electrophoresis has been demonstrated to be a useful technique (13, 17, 18). However, RFLP has already been established in many mycobacteriology laboratories as the method of choice for the genotyping of Mycobacterium tuberculosis. A proposal for the standardization of RFLP of M. avium using the recently described insertion sequence IS1245 has been elaborated (16). Experiences in the studies of molecular epidemiology of M. tuberculosis show that such a standardization is of great value since it allows comparison of data obtained in different laboratories (4, 15).
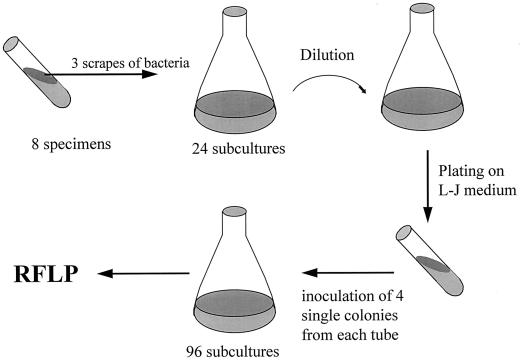
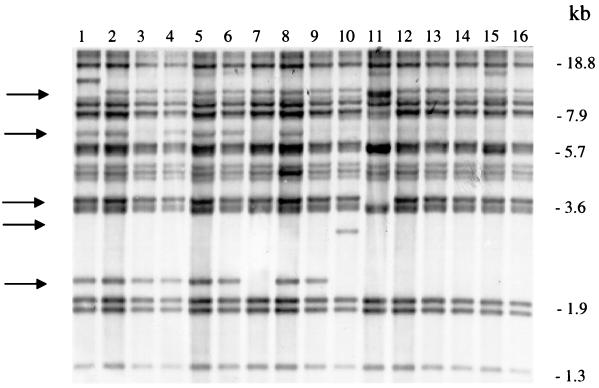
Knowledge of the stability of the insertion element in vivo and in vitro is crucial for interpretation of the results. With the original purpose to investigate the possible occurrence of polyclonal M. avium infections in HIV-infected patients, we undertook a study of three AIDS patients who all had disseminated M. avium infections. The study was based on the hypothesis that polyclonal colonization of the intestine might precede translocation of one or a very limited number of infectious organisms leading to monoclonal infection. Species identification was performed by DNA-RNA hybridization using Accu-Probe (Gen-Probe Inc., San Diego, Calif.). A total of eight cultures from the three patients were collected. All patients had positive cultures from stool and blood. One patient had one additional isolate cultured from a liver biopsy specimen, and another patient had M. avium cultured from sinus frontalis. From each specimen, three colonies were subcultured, and from the resulting 24 cultures purified single colonies were obtained. Four single colonies from each culture were further subcultured for RFLP analysis, for a total of 96 bacterial clones (Fig. 1). DNA for RFLP analyses was obtained from 87 isolates. RFLP analyses of these isolates were performed as previously described (14) with the only minor alterations that gels were run longer in order to obtain better separation of the hybridization bands. In brief, DNA was extracted and digested with PvuII. After electrophoresis on agarose gels, the digested DNA was transferred to nylon membranes (Hybond N+; Amersham) and probed with a chemiluminiscence-labelled 427-bp sequence of IS1245 generated by PCR as previously described (3). Colonies from various locations on the same patient in all cases exhibited almost identical IS1245 patterns. Yet differences affecting one to two bands were observed in 5 of 33 isolates from patient A, in 4 of 34 isolates from patient B, and in 11 of 20 isolates from patient C. An example of results from patient C is shown in Fig. 2. As is apparent from the autoradiogram, some hybridization bands appeared weaker than others. Similar weak bands have been observed in isolates analyzed in a previous study (1) and might be due to cross-hybridization with IS1311 as previously described (12).
FIG. 1.
Schematic presentation of the preparation of single-colony cultures from three patients.
FIG. 2.
Autoradiogram showing PvuII-digested IS1245 patterns of purified single colonies from patient C. Lanes: 1 to 7, RFLP patterns of single colonies from stool; 8 to 16, RFLP patterns of single colonies from blood. The arrows indicate positions where changes occur.
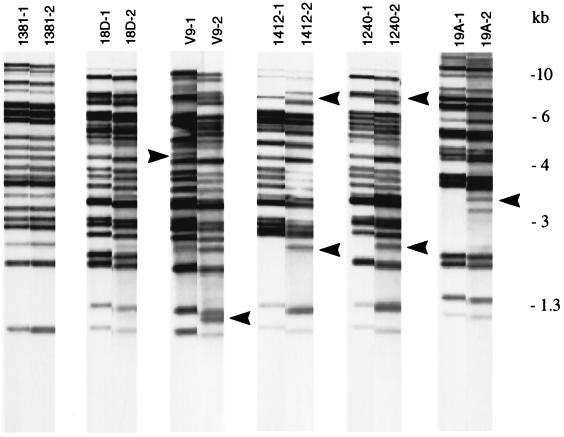
To further explore whether these small differences among the clones indicated a high degree of instability of IS1245, we decided to investigate the in vitro stability of the insertion element. A total of six M. avium cultures were collected, four of which had been analyzed by RFLP in another study (1) and two of which were single-colony isolates (Table 1). These six isolates were initially subcultured in Dubos Tween 80 medium. From November 1996 until November 1997, they were subcultured 33 times in liquid media at intervals of 1 to 2 weeks. The first and last subcultures from each isolate were analyzed by RFLP. As shown in Fig. 3, the isolates exhibited identical or almost identical IS1245 patterns following the subcultures. In four cases, differences in one or two bands could be observed. Considering the fact that human M. avium isolates cultured in Denmark almost exclusively exhibit IS1245 patterns with a high number of bands (1), one- to two-band differences were considered minor changes. Because the driving force of the observed instability of the IS1245 RFLP pattern is not fully understood, we do not know whether M. avium strains containing only a few IS1245 elements will exhibit the same degree of instability.
TABLE 1.
Sources of six M. avium isolates subcultured during 1 year
| Strain | Source |
|---|---|
| V9 | Peat |
| 18D | Single colony from blood (AIDS patient) |
| 19A | Single colony from stool (AIDS patient) |
| 1381 | Non-HIV patient with lymphadenitis |
| 1412 | Non-HIV patient with pulmonary infection |
| 1240 | Liver biopsy specimen from AIDS patient |
FIG. 3.
IS1245 patterns of six M. avium isolates before and after 33 subcultures. Arrowheads indicate changes in patterns.
Previous studies have demonstrated that the IS1245 pattern exhibited by isolates cultured from the same patient over time is quite stable (9, 11). The degree of changes in the IS1245 patterns of such serial isolates seems to be comparable to those observed in serial M. tuberculosis isolates cultured from the same patient (19). This indicates that the stability of the insertion element in vivo is good. In this study, an attempt to uncover possible polyclonal infections failed but demonstrated several small one- to two-band differences among single colonies from the same isolate. Similar observations were made by Pestel-Caron and Arbeit (9), who recently described the same biological variations within the same culture. It was further demonstrated that no changes in the IS1245 pattern could be observed following subcultures on solid media over several months. In this study, only one- to two-band variations were observed in four of six M. avium strains subcultured in liquid media 33 times over a period of 1 year. Since small changes, however, do seem to occur, the finding of indistinguishable IS1245 banding patterns among M. avium strains must consequently be a strong indicator that these strains are actually related. This study therefore concludes that the stability of IS1245 is sufficient to recommend its use as a tool for differentiation of M. avium strains.
REFERENCES
- 1.Bauer, J., A. B. Andersen, D. S. Askgaard, S. B. Giese, and B. Larsen. Submitted for publication.
- 2.Burki D R, Bernasconi C, Bodmer T, Telenti A. Evaluation of the relatedness of strains of Mycobacterium avium using pulsed-field gel electrophoresis. Eur J Clin Microbiol Infect Dis. 1995;14:212–217. doi: 10.1007/BF02310358. [DOI] [PubMed] [Google Scholar]
- 3.Guerrero C, Bernasconi C, Burki D, Bodmer T, Telenti A. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J Clin Microbiol. 1995;33:304–307. doi: 10.1128/jcm.33.2.304-307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hermans P W, Messadi F, Guebrexabher H, van Soolingen D, de Haas P E, Heersma H, de Neeling H, Ayoub A, Portaels F, Frommel D, et al. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia, and The Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J Infect Dis. 1995;171:1504–1513. doi: 10.1093/infdis/171.6.1504. [DOI] [PubMed] [Google Scholar]
- 5.Horsburgh C R., Jr Mycobacterium avium complex infection in the acquired immunodeficiency syndrome. N Engl J Med. 1991;324:1332–1338. doi: 10.1056/NEJM199105093241906. [DOI] [PubMed] [Google Scholar]
- 6.Inderlied C B, Kemper C A, Bermudez L E. The Mycobacterium avium complex. Clin Microbiol Rev. 1993;6:266–310. doi: 10.1128/cmr.6.3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazurek G H, Hartman S, Zhang Y, Brown B A, Hector J S, Murphy D, Wallace R J., Jr Large DNA restriction fragment polymorphism in the Mycobacterium avium-M. intracellulare complex: a potential epidemiologic tool. J Clin Microbiol. 1993;31:390–394. doi: 10.1128/jcm.31.2.390-394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nightingale S D, Byrd L T, Southern P M, Jockusch J D, Cal S X, Wynne B A. Incidence of Mycobacterium avium-intracellulare complex bacteremia in human immunodeficiency virus-positive patients. J Infect Dis. 1992;165:1082–1085. doi: 10.1093/infdis/165.6.1082. [DOI] [PubMed] [Google Scholar]
- 9.Pestel-Caron M, Arbeit R D. Characterization of IS1245 for strain typing of Mycobacterium avium. J Clin Microbiol. 1998;36:1859–1863. doi: 10.1128/jcm.36.7.1859-1863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Picardeau M, Vincent V. Typing of Mycobacterium avium isolates by PCR. J Clin Microbiol. 1996;34:389–392. doi: 10.1128/jcm.34.2.389-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritacco V, Kremer K, van der Laan T, Pijnenburg J E M, de Haas P E, van Soolingen D. Use of IS901 and IS1245 in RFLP typing of Mycobacterium avium complex: relatedness among serovar reference strains, human and animal isolates. Int J Tuberc Lung Dis. 1997;2:242–251. [PubMed] [Google Scholar]
- 12.Roiz M P, Palenque E, Guerrero C, Garcia M J. Use of restriction fragment length polymorphism as a genetic marker for typing Mycobacterium avium strains. J Clin Microbiol. 1995;33:1389–1391. doi: 10.1128/jcm.33.5.1389-1391.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slutsky A M, Arbeit R D, Barber T W, Rich J, von Reyn C F, Pieciak W, Barlow M A, Maslow J N. Polyclonal infections due to Mycobacterium avium complex in patients with AIDS detected by pulsed-field gel electrophoresis of sequential clinical isolates. J Clin Microbiol. 1994;32:1773–1778. doi: 10.1128/jcm.32.7.1773-1778.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Embden J D, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Embden J D, van Soolingen D, Heersma H, De Neeling A J, Jones M E, Steiert M, Grek V, Mooi F R, Verhoef J. Establishment of a European network for the surveillance of Mycobacterium tuberculosis, MRSA and penicillin-resistant pneumococci. J Antimicrob Chemother. 1996;38:905–907. doi: 10.1093/jac/38.5.905. [DOI] [PubMed] [Google Scholar]
- 16.van Soolingen D, Bauer J, Ritacco V, Cardoso Leão S, Pavlik I, Vincent V, Rastogi N, Gori A, Bodmer T, Garzelli C, Garcia M J. IS1245 restriction fragment length polymorphism typing of Mycobacterium avium isolates: proposal for standardization. J Clin Microbiol. 1998;36:3051–3054. doi: 10.1128/jcm.36.10.3051-3054.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Reyn C F, Jacobs N J, Arbeit R D, Maslow J N, Niemczyk S. Polyclonal Mycobacterium avium infections in patients with AIDS: variations in antimicrobial susceptibilities of different strains of M. avium isolated from the same patient. J Clin Microbiol. 1995;33:1008–1010. doi: 10.1128/jcm.33.4.1008-1010.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Reyn C F, Maslow J N, Barber T W, Falkinham III J O, Arbeit R D. Persistent colonisation of potable water as a source of Mycobacterium avium infection in AIDS. Lancet. 1994;343:1137–1141. doi: 10.1016/s0140-6736(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 19.Yeh R W, Ponce de Leon A, Agasino C B, Hahn J A, Daley C L, Hopewell P C, Small P M. Stability of Mycobacterium tuberculosis DNA genotype. J Infect Dis. 1998;177:1107–1111. doi: 10.1086/517406. [DOI] [PubMed] [Google Scholar]