Significance
Enzymes comprised of membrane-spanning acyltransferase-3 (AT-3) domains catalyze the O-acetylation of diverse extracytoplasmic glycans in all forms of life. In many cases, such as peptidoglycan, these glycans represent important cell wall components, and their O-acetylation confers resistance to lytic enzymes. The enzyme responsible for peptidoglycan O-acetylation in gram-positive bacteria, OatA, is a single bimodal protein of an AT-3 domain fused to an SGNH domain. The AT-3 domain adopts a different topology to that predicted in silico. Moreover, its utilization of a unique mechanism for the translocation of acetyl groups across the cytoplasmic membrane for their transfer to peptidoglycan involving catalytic Tyr and Ser residues may be broadly applicable to homologs involved in the modification of other important cell wall glycans.
Keywords: peptidoglycan, O-acetylation, O-acylransferase, acyltransferase 3 family, Staphylococcus aureus
Abstract
The O-acetylation of exopolysaccharides, including the essential bacterial cell wall polymer peptidoglycan, confers resistance to their lysis by exogenous hydrolases. Like the enzymes catalyzing the O-acetylation of exopolysaccharides in the Golgi of animals and fungi, peptidoglycan O-acetyltransferase A (OatA) is predicted to be an integral membrane protein comprised of a membrane-spanning acyltransferase-3 (AT-3) domain and an extracytoplasmic domain; for OatA, these domains are located in the N- and C-terminal regions of the enzyme, respectively. The recombinant C-terminal domain (OatAC) has been characterized as an SGNH acetyltransferase, but nothing was known about the function of the N-terminal AT-3 domain (OatAN) or its homologs associated with other acyltransferases. We report herein the experimental determination of the topology of Staphylococcus aureus OatAN, which differs markedly from that predicted in silico. We present the biochemical characterization of OatAN as part of recombinant OatA and demonstrate that acetyl-CoA serves as the substrate for OatAN. Using in situ and in vitro assays, we characterized 35 engineered OatA variants which identified a catalytic triad of Tyr-His-Glu residues. We trapped an acetyl group from acetyl-CoA on the catalytic Tyr residue that is located on an extracytoplasmic loop of OatAN. Further enzymatic characterization revealed that O-acetyl-Tyr represents the substrate for OatAC. We propose a model for OatA action involving the translocation of acetyl groups from acetyl-CoA across the cytoplasmic membrane by OatAN and their subsequent intramolecular transfer to OatAC for the O-acetylation of peptidoglycan via the concerted action of catalytic Tyr and Ser residues.
The O-acetylation of cell wall and extracellular polysaccharides is a common theme in nature for their protection from lysis and degradation. Examples include the sialic acid residues of glycoproteins that comprise the glycocalyx of eukaryotic cells (1), xylan of the secondary cell walls of plants (2), and the peptidoglycan (PG), secondary cell wall polysaccharides and biofilm components (e.g., cellulose, alginate) of bacterial cell walls or extracellular matrices (3, 4). PG is an essential component of bacterial cell walls, contributing to both cell shape and protection. Although over 50 chemotypes of PG have been identified (5), the glycan structure is conserved among bacteria. It is composed of repeating units of β-1,4–linked N-acetylglucosaminyl (GlcNAc) and N-acetylmuramoyl residues (MurNAc), and adjacent glycan strands are cross-linked together through peptides that stem from MurNAc residues.
PG O-acetylation occurs at the C-6 hydroxyl of MurNAc residues in both gram-positive and gram-negative bacteria (reviewed in ref. 6). Though rare, it can also occur at GlcNAc residues, such as in some lactobacilli (7). Initially described in 1958 (8), this modification is now known to occur in a large number of bacteria, particularly pathogens. For example, in a comprehensive study of the staphylococci, only pathogenic species (including Staphylococcus aureus) were found to produce O-acetylated PG (9). O-acetylation is a nonstoichiometric modification, and its extent can range from 20 to 70% depending on the organism, environmental conditions, and age of the culture (10–12). With Enterococcus faecalis, for example, PG O-acetylation levels increase by 10 to 40% as cultures enter the stationary phase and a further 10 to 16% as cells enter the viable but nonculturable state (13). It is not yet known with any bacterium if the O-acetyl moieties are dispersed throughout the PG sacculus or localized to particular regions (6).
The main pathobiological role of PG O-acetylation is the inhibition of lysozyme, an enzyme of the innate immunity system that hydrolyzes the glycosidic linkage between MurNAc and GlcNAc residues causing bacterial cells to lyse. The C-6 hydroxyl of MurNAc is important for the coordination of PG into the substrate-binding cleft of lysozyme (14) and so its O-acetylation sterically hinders this interaction (15, 16). With Listeria monocytogenes, in addition to providing resistance to lysozyme, PG O-acetylation is important for its growth in macrophages, conferring resistance to bacteriocins and β-lactam antibiotics and limiting the innate immune response (17). The ability of PG O-acetylation to modulate the immune system is of particular interest, as during S. aureus infection, the O-acetylation of PG limits T helper cell priming required to develop an effective protective response to systemic infection (18). Hence, PG O-acetylation is considered important for virulence in this (18–20) and numerous other pathogens (21), including L. monocytogenes (17), Streptococcus pneumoniae (22), E. faecalis (23), Neisseria meningitidis (24), Neisseria gonorrhoeae (25, 26), and Helicobacter pylori (27).
As with other cell wall and many glycocalyx components, the O-acetylation of PG is a postbiosynthetic modification. It occurs after the incorporation of GlcNAc-MurNAc-pentapeptide (Lipid II) precursors into the existing sacculus (28–31). Accordingly, acetyl groups must therefore be translocated across the cytoplasmic membrane from a cytoplasmic donor to the cell wall. Two distinct enzymatic systems have been identified in bacteria for this purpose, both of which are now thought to be the ancestors for homologous systems that evolved in eukaryotes (2). The system operating in gram-positive bacteria, which was first discovered in S. aureus (20), is analogous to those involved with the O-acetylation of exopolysaccharides in the Golgi of animals and fungi (2). In each of these cells, a single bimodal protein (named O-acetyltransferase A [OatA] in gram-positive bacteria) is postulated to catalyze both the translocation and transfer reactions. With OatA, its N-terminal domain (OatAN) is predicted to be a member of Acyl_transf_3 (AT-3) Acyltransferases (pfam PF01757; InterPro IPR002656) family of proteins and also as an acyltransferase_3/putative acetyl-CoA transporter (Transporter Classification Database number TC 9.B.97). As such, OatAN is thought to span the cytoplasmic membrane and function as an O-acyltransferase to shuttle acetyl groups from an unknown source for presentation to the C-terminal extracellular domain (OatAC), which subsequently transfers them onto MurNAc residues (4).
Our earlier structural and biochemical studies of OatAC from both S. aureus (32) and S. pneumoniae (33, 34) demonstrated that this domain functions like an SGNH hydrolase in which the C-6 hydroxyl group of muramoyl residues serves as the acceptor for PG O-acetylation instead of water as for a hydrolase. Nothing, however, is known mechanistically nor structurally about the cognate OatAN or other members of AT-3, and little insight has been gained from studies on any other system translocating acetyl groups across a membrane for their transfer to a glycan. The concept of the two-step coordinated reaction for the overall O-acetylation of extracytoplasmic glycans was first proposed by Higa et al. (35) in their study of a sialyl O-acetyltransferase embedded in the Golgi membrane of the rat liver. These early investigators used a chemical modification approach to identify the participation of an essential histidyl and lysyl residues for the first step translocation reaction. More recently, a site-directed mutagenesis of the gene encoding Salmonella Typhimurium O-antigen O-acetyltransferase A (OafA) confirmed the importance of an Arg-X-X-Arg motif (36) identified previously in this enzyme (37) and a Shigella flexneri homolog, Oac (38). The Arg and His residues in an Arg/Lys-X10-His motif were also found to be important for function in OafA, and they are proposed to interact with acetyl-CoA, assuming that this metabolite serves as the acetyl donor for the enzyme (36). Pearson et al. (36) also observed that, while this latter motif is present in all AT-3 homologs, the Arg-X-X-Arg motif together with a Gly-Gly-Phe-(Val/Ile/Leu)-Gly-Val-Asp-(Ile/Val/Leu) motif are found uniquely in AT-3 domains fused to a cognate SGNH domain. In addition to OatA and the O-antigen and O-acetyltransferases noted above, members of this subfamily of enzymes are present in other important bacterial pathogens. Examples include OatB and Lot3 that O-acetylate GlcNAc in Lactobacillus PG and N. meningitidis lipooligosaccharides, respectively. In the intervening years between these studies, little other knowledge has been gained.
Herein, we present a biochemical characterization of the AT-3 domain of OatA. We experimentally determined the topology of S. aureus OatAN (SaOatAN), which we find to differ significantly from that predicted in silico. Using two novel assays (one each in situ and in vitro) coupled with site-specific replacements of consensus amino acids, we identified catalytically essential Arg, His, and Tyr residues. Based on these data, combined with our identification of two acetyl intermediates, we propose a mechanism for the translocation of acetyl groups from cytoplasmic acetyl-CoA to an external loop of SaOatAN that serves as the acetyl donor for the O-acetylation of PG by OatAC.
Results
OatA Contributes to Lysozyme Resistance.
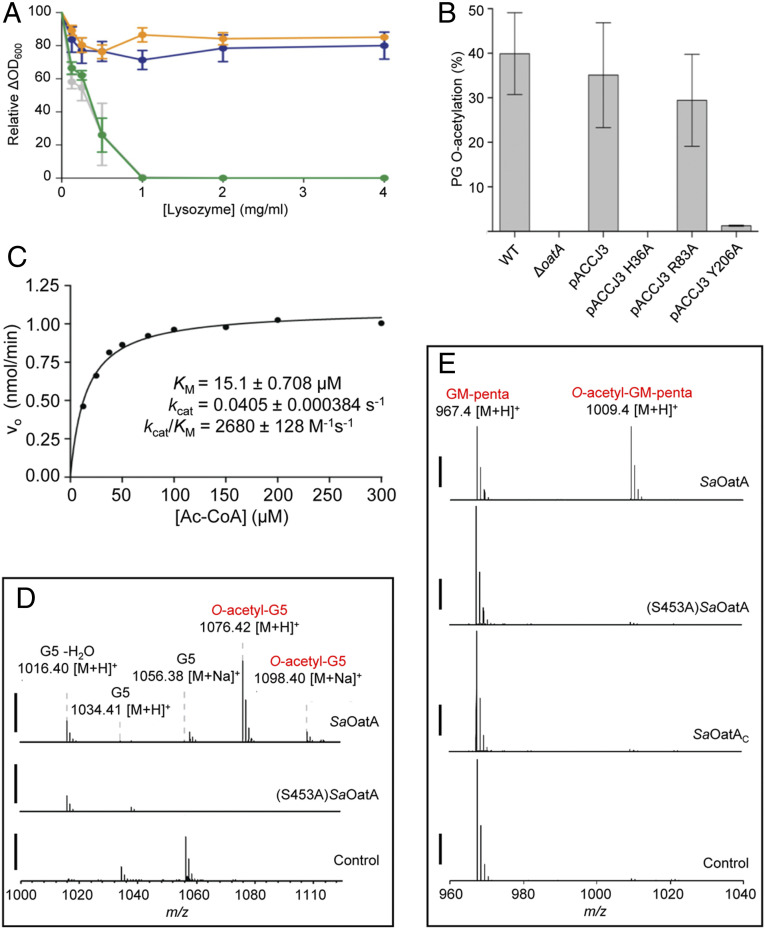
To assess the function of OatA in situ, we developed a phenotypic assay that harnessed lysozyme resistance as the read out. This utilized a mutant of S. aureus USA300 we engineered, S. aureus USA300 ∆oatA, that has a marker-less deletion of oatA. S. aureus USA300 and S. aureus USA300 ∆oatA were grown in liquid culture in the presence of increasing concentrations of lysozyme to determine minimal inhibitory concentration (MIC) values for lysozyme. Lysozyme only caused a lag in growth of S. aureus USA300 ∆oatA and did not result in complete cell death over an 18-h growth period (SI Appendix, Fig. S1A). It is known that the abolishment of both PG O-acetylation and wall teichoic acid (WTA) production results in increased lysozyme sensitivity compared to the lack of either alone (39). Tunicamycin is a selective and potent inhibitor of TarO, the enzyme that catalyzes the first committed step in WTA synthesis, without causing other significant growth defects in S. aureus at low concentrations in rich media (40). We therefore tested lysozyme sensitivity of the two S. aureus USA300 strains in the presence of 0.4 µg/mL tunicamycin. Under these conditions, an MIC of 1 mg/mL lysozyme was determined for S. aureus USA300 ∆oatA, with little effect on the wild-type strain at 4 mg/mL lysozyme (Fig. 1A and SI Appendix, Fig. S1B). Lysozyme resistance was restored to S. aureus USA300 ∆oatA by complementation in trans with His-tagged SaOatA on a constitutive expression plasmid (pACCJ3). Complementation with an empty vector (pALC2073) had no effect.
Fig. 1.
In situ and in vitro activity of SaOatA. (A) Effect of OatA production on lysozyme sensitivity in strains of S. aureus. Cultures of S. aureus USA300 (blue), S. aureus USA300 ∆oatA (green), S. aureus USA300 ∆oatA pACCJ3 (orange), and S. aureus USA300 pALC2073 (gray) were incubated for 12 h in the presence of 0.4 μg/mL tunicamycin and lysozyme at the concentrations indicated. Error bars denote SD (n = 3). (B) PG O-acetylation levels of S. aureus USA300 strains. Concentrations of muramic acid and base-labile acetyl groups in purified samples of PG were determined to provide the relative percentage of PG O-acetylation in each strain. Error bars denote SD (n = 3). (C) Kinetic analysis of SaOatA acting as an esterase with acetyl-CoA as substrate. The steady-state parameters of 5 µM SaOatA were determined in 50 mM sodium phosphate buffer, pH 7.0. “±” denote SD (n = 3); error bars are smaller than the height of the symbol. Activity of SaOatA as a PG O-acetyltransferase with (D) pentaacetyl-chitopentaose (G5) and (E) semisynthetic muroglycan oligomers as acceptors. Enzymes (5 µM) in 50 mM sodium phosphate buffer, pH 6.5 were incubated with 1 mM acetyl-CoA and either 1 mM pentaacetyl-chitopentaose or 10 µg/mL semisynthetic muroglycan oligomers (GM-penta) for 18 h prior to QTOF-MS analysis. Control, reaction mixture without added enzyme. The solid bars to the left of the spectrograms presented in D and E denote 2,500 ion counts.
Lysozyme Resistance Correlates with PG O-Acetylation.
To confirm the correlation between the lysozyme sensitivity observed in the functional complementation assay and OatA activity, we determined the PG O-acetylation levels in the two S. aureus USA300 strains. The PG O-acetylation level of S. aureus USA300 was ∼40%, and there was no detectable acetylation in S. aureus USA300 ∆oatA (Fig. 1B). In accordance with the functional complementation assay results, S. aureus USA300 ∆oatA complemented with pACCJ3 had ∼35% PG O-acetylation.
Both SaOatAC and Full-Length SaOatA Have Esterase Activity In Vitro.
The gene encoding full-length SaOatA was cloned with a His10-tag, expressed in Escherichia coli C43(DE3), and purified by immobilized metal ion affinity chromatography to apparent homogeneity as determined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis (SI Appendix, Fig. S2A). Yields were ∼3 mg protein per liter of culture. We also produced and purified SaOatAC (residues 445 to 601) as previously described (32). As an SGNH hydrolase domain, we recently demonstrated that SaOatAC functions as a weak esterase, with activity toward the pseudo substrates p-nitrophenyl acetate (pNP) and 4-methylumbelliferyl acetate (4MU-Ac), while the variant lacking a catalytic Ser residue was shown to be inactive (32). Consistent with this earlier study, purified SaOatAC in 5% dimethyl sulfoxide (DMSO) at pH 6.5 had KM and kcat/KM values of 112 µM and 42 M−1 ⋅ s−1, respectively, for 4MU-Ac as substrate. We found that full-length SaOatA was also active as an esterase on 4MU-Ac under the same conditions, but its specific activity was only 78% of that of SaOatAC (SI Appendix, Fig. S3). This apparent reduced level of activity of SaOatA likely results from its aggregation when prepared at concentrations required for in vitro studies; evidence for such aggregation was observed with SDS-PAGE analyses (SI Appendix, Fig. S2). To determine if SaOatAN alone can function as an esterase, we generated a recombinant variant of full-length SaOatA lacking a catalytically active SaOatAC by site-specific replacement of its catalytic nucleophile Ser453 with Ala (SI Appendix, Fig. S2B). (S453A)SaOatA was indeed active toward 4MU-Ac, but its specific activity was only 69% and 54% of that of full-length SaOatA and SaOatAC, respectively, suggesting that this substrate is not preferred by the AT-3 domain (SI Appendix, Fig. S3).
Only Full-Length SaOatA Uses Acetyl-CoA as a Substrate.
To test both full-length SaOatA and SaOatAC for thioesterase activity against acetyl-CoA, we adapted the chromogenic assay of Grassetti and Murray (41) that monitors the release of CoASH. With SaOatAC, we did not detect any release of CoASH from acetyl-CoA. This is in keeping with the earlier finding that SaOatAC is unable to O-acetylate pentaacetyl-chitopentaose (G5) when acetyl-CoA is supplied as the acetyl donor (34). However, full-length SaOatA was active toward acetyl-CoA (SI Appendix, Fig. S4A), indicating that the thioesterase active site is located within SaOatAN. Interestingly, (S453A)SaOatA was weakly active toward acetyl-CoA, retaining only 5.4% specific activity compared to SaOatA. This suggested that, whereas SaOatAC alone is not active on acetyl-CoA, both domains of SaOatA are required for efficient hydrolysis of this substrate.
We determined the steady-state parameters for full-length SaOatA acting as a thioesterase on acetyl-CoA as substrate (Fig. 1C). The determined KM value of 15 µM was 77- and 6-fold lower than that of SaOatAC for the pseudo substrates pNP-Ac and 4MU-Ac, respectively. Likewise, the overall catalytic efficiency of SaOatA, as reflected by kcat/KM, was much higher with acetyl-CoA as substrate compared to SaOatAC acting on these two pseudo substrates.
OatA Utilizes Acetyl-CoA for PG O-Acetylation.
The kinetic data presented above suggested that acetyl-CoA may be the natural substrate for the O-acetylation of PG by full-length SaOatA. To test this in vitro, we analyzed full-length SaOatA, SaOatAC, and (S453A)SaOatA independently as O-acetyltransferases using our previously established transferase assay (42) with acetyl-CoA as a donor and pentaacetyl-chitopentaose (G5) as a model acceptor. Reaction products were analyzed by liquid chromatography–mass spectrometry (LC-MS) and an acetylated product was only detected in reactions with wild-type, full-length SaOatA; neither SaOatAC nor (S453A)SaOatA produced acetylated G5 (Fig. 1D). Confirmation for the identification of O-acetylated G5 was obtained by tandem MS (MS/MS) analysis (SI Appendix, Fig. S4B). We also tested SaOatA’s ability to acetylate semisynthetically prepared oligomers of GlcNAc-MurNAc-pentapeptide (GM-penta) as a model acceptor for PG (34) using acetyl-CoA as donor. Reaction products were digested with mutanolysin for subsequent LC-MS/MS analysis as described previously (34). As observed with G5, only full-length SaOatA was capable of O-acetylating these muroglycans with acetyl-CoA as the donor (Fig. 1E). Combined, these data suggested that acetyltransfer to acceptor glycans requires OatAN to remove acetyl groups from acetyl-CoA and OatAC to transfer them onto the muroglycan acceptor substrate.
Topology of SaOatAN.
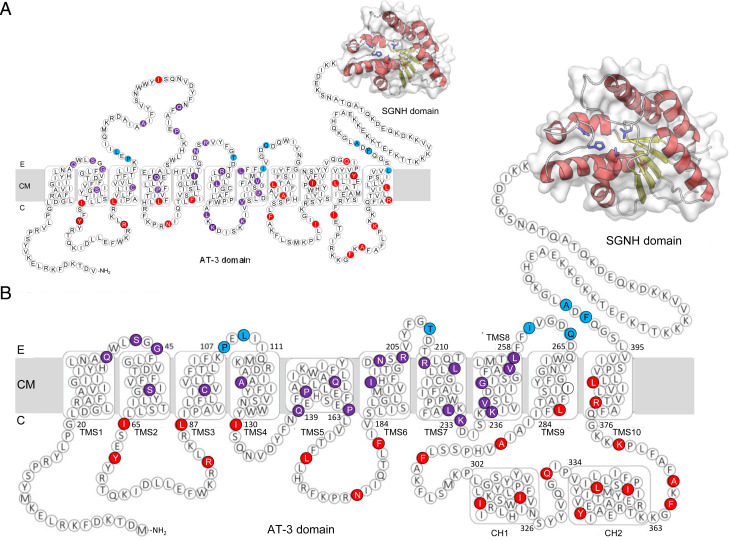
OatAN is a member of the Acyl_transf_3 (AT-3) Acyltransferases (Pfam PF01757; InterPro IPR002656), a superfamily that includes a wide variety of integral membrane acyltransferases from all forms of life that often acylate saccharides. It has also been identified as an acyltransferase_3/putative acetyl-CoA transporter (Transporter Classification Database no. TC 9.B.97). Using in silico tools, it is predicted to be an integral membrane domain involving 11 transmembrane segments (TMS), with a large periplasmic loop between TMS3 and TMS4, and a 20-residue linker from TMS11 to the extracytoplasmic C-terminal SGNH O-acetyltransferase domain, OatAC (Fig. 2A).
Fig. 2.
Topology maps of the AT-N domain of S. aureus OatA. (A) Consensus in silico prediction of SaOatAN using TopCons. The PhoA-LacZα fusion data are mapped onto the consensus in silico topology prediction. Each colored residue represents the terminal amino acid of truncation, and the color denotes the location of the truncation: red, cytoplasm; purple, transmembrane; blue, periplasm. (B) The adjusted topology model of SaOatAN to accommodate both the experimental PhoA-LacZα fusion data and in silico predictions of secondary structures. E, external; CM, cytoplasmic membrane; C, cytoplasm.
Initially, we attempted to apply the substituted cysteine accessibility method (SCAM) (43) for the experimental determination of S. aureus OatAN topology within the full-length form of the enzyme. This technique was chosen because it allows the topology to be determined on a nontruncated functional protein within its natural host. However, all attempts to label residues expected to be extracytoplasmic (Leu,42, Ile121, Val205, Gln264), including Gly403 within the putative linker sequence between OatAN and OatAC, failed. Based on these results, we hypothesized that the extracytoplasmic Cys residues were inaccessible to the methoxypolyethylene glycol maleimide (PEGmal)-labeling and 2-(trimethylammonium)ethyl methane thiosulfonate (MTSET)-blocking reagents due to interactions between SaOatAN and cognate SaOatAC and/or other membrane proteins.
Due to our lack of success with SCAM, we used the PhoA-LacZα method (44) to determine the topology of SaOatAN. This in situ method involves the generation and expression of chimeras composed of C-terminally truncated forms of the protein of interest with dual reporter enzymes, E. coli β-galactosidase LacZα, which is only active in the cytoplasm, and the E. coli alkaline phosphatase PhoA, which is only active in the bacterial periplasm. When localized to the periplasm, the PhoA-LacZα reporter will display high alkaline phosphatase activity but no β-galactosidase activity. Conversely, if the dual reporters are localized to the cytoplasm, only high β-galactosidase activity will be detected; detection of both activities indicates localization within the membrane. We generated both a random 3′ gene truncation library of oatA from S. aureus American Type Culture Collection 6538 fused to phoA-lacZα, and targeted truncations were cloned for regions of the protein lacking sufficient coverage in the random truncation library. Based on the data obtained for all 53 truncations, the normalized alkaline phosphatase:β-galactosidase activity ratio (NAR) cutoffs were set at <0.1 and >4.0 for cytoplasmic and extracellular locations, respectively; NAR values of 0.1 to 4.0 denoted transmembrane locations of the truncation. The results for all truncations are given in SI Appendix, Table S1, and they were mapped onto the predicted topology map for SaOatAN (Fig. 2A). Whereas these experimental data are consistent with the prediction for the first three transmembrane segments, the data deviate significantly for the rest of the domain.
To develop a topology model that more accurately reflects the experimental evidence, the PhoA-LacZα fusion tagging data were evaluated in context with in silico predictions (SI Appendix, Fig. S5A), including general rules for transmembrane proteins; ∼15 to 20 amino acids are needed for α-helical TMS to cross the membrane, and successive positive amino acids are not usually found in a TMS. For large regions of the protein found experimentally to be localized to the same compartment, helix prediction software was used to predict where helices would likely form. The overall result was a new topology model for SaOatAN with 10 TMS, four extracytoplasmic loops, and four cytoplasmic loops (Fig. 2B). Both the C-terminal and N-terminal ends of TMS5 were located in the cytoplasm, suggesting that TMS5 is a membrane-penetrating region, or a re-entrant helix, and not a transmembrane helix. The cytoplasmic loop between TMS8 and TMS9 was the longest, consisting of 92 residues, and it is predicted to form two cytoplasmic helices (CH1 and CH2). Neither CH1 nor CH2 are predicted to be amphipathic, but both are characterized as having alternating hydrophilic and hydrophobic ends (SI Appendix, Fig. S5B). Thus, the N-terminal end of CH1 and the C-terminal end of CH2 have low mean hydrophobicities (<H0> = 0.600 and 0.245, respectively), whereas the C-terminal and N-terminal ends of CH1 and CH2, respectively, are much more hydrophobic (<H0> = 1.023 and 1.204, respectively). This suggests that the C-terminal end of CHI may extend from the cytoplasm to associate with the membrane surface through a small hydrophobic patch, while the N terminus of CH2 associates with the membrane periphery, and its C-terminal end extends back into the cytoplasm.
Identification of Amino Acid Residues Essential for OatAN Function.
In situ studies.
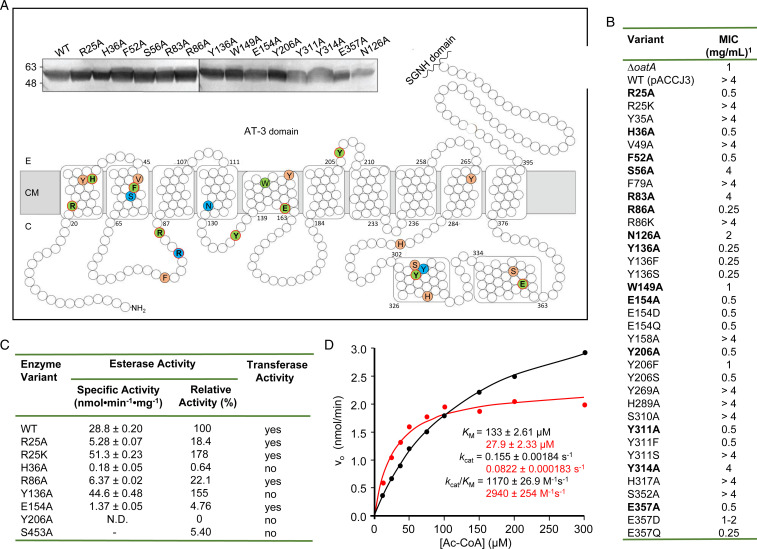
We applied the lysozyme sensitivity assay to identify amino acids essential within SaOatAN for the function of OatA in situ. A multiple sequence alignment was prepared to identify invariable or highly conserved residues among gram-positive OatA homologs (SI Appendix, Fig. S6), and then they were mapped onto our topology map (Fig. 3A). Of the 29 invariant and 49 highly conserved residues identified, several appear to be clustered in or near TMS1 and TMS2 as well as TMS5 and cytoplasmic helices 10 and 11. These include the Arg-X-X-Arg and Arg/Lys-X10-His motifs identified in earlier studies of other AT-3 domains that are associated with cognate SGNH domains like OatA (36–38). We replaced 23 targeted residues with Ala by site-directed mutagenesis of the oatA gene in pACCJ3, and the resulting plasmids were used to complement S. aureus USA300 ∆oatA. Both this latter strain transformed with empty vector and S. aureus USA300 ∆oatA pACCJ3 (viz. transformed with wild-type OatA) were used as controls. The growth of each complemented strain in the presence of 0.4 µg/mL tunicamycin and increasing concentrations of lysozyme was monitored to assess the effect of the respective amino acid replacements on lysozyme sensitivity as a measure of function. Whereas S. aureus USA300 ∆oatA complemented with wild-type oatA had full lysozyme resistance at the highest concentration tested (4 mg/mL), single amino acid substitutions caused significant lysozyme sensitivity for 10 of the conserved residues (Arg25, His36, Phe52, Arg86, Tyr136, Trp149, Glu154, Tyr206, Tyr311, and Glu357), while we found moderate lysozyme sensitivity for four others (Ser56, Arg83, Asn126, and Tyr314) (Fig. 3B).
Fig. 3.
Identification of conserved and functional residues in SaOatAN. (A) Invariable and highly conserved residues were identified by bioinformatic analysis of OatA homologs and mapped onto the topology map of SaOatAN. Selected residues were individually replaced with Ala, and the resultant SaOatA variants were assessed for their ability to provide lysozyme resistance in a ∆oatA background of S. aureus USA300. The amino acid color denotes the functional complementation assay result: completely lysozyme sensitive (green), partially lysozyme sensitive (blue), or completely lysozyme resistant (orange). E, external; CM, cytoplasmic membrane; C, cytoplasm. Residues identified as important for substrate-binding and the catalytic mechanism are outlined in red. (Top) The membranes from lysozyme-sensitive S. aureus USA300 ∆oatA-complemented strains (bolded variants listed in B) were isolated and probed for SaOatA by anti-His Western blotting to ensure proper expression and membrane localization. Molecular weight markers (kDa) are shown on Left. (B) In situ activity of S. aureus USA300 ΔoatA expressing single amino acid variants of SaOatA as monitored by their sensitivity to lysozyme. The MIC values of lysozyme were determined by culturing S. aureus USA300 ΔoatA expressing oatA in trans in the presence of 0.4 μg/mL tunicamycin and increasing concentrations of lysozyme (0 to 4 mg/mL). Each variant was tested in technical and biological triplicates. (C) Esterase and transferase activities of SaOatA variants with acetyl-CoA as acetyl donor. Reactions were conducted in 50 mM sodium phosphate buffer, pH 7.0, containing 5% (vol/vol) DMSO at 25 °C with 0.1 mM acetyl-CoA and 0.2 mM DTDP (5% ethanol replaced DMSO for reactions with S453A). Transferase assays included 1 mM pentaacetyl-chitopentaose (G5) as an acceptor, and products were detected by LC-MS as described in the legend to Fig. 1E. “±” denotes SD; N.D. = no detectable activity. (D) Determination of the steady-state parameters of (Y136A)SaOatA (black) and (R25A)SaOatA (red) acting as esterases on acetyl-CoA. The reaction conditions for these determinations are described in the legend to Fig. 1C.
To ensure that the 14 functionally impaired SaOatA variants were expressed and localized correctly, the membranes from each S. aureus USA300 ∆oatA-complemented strain were isolated and probed for SaOatA presence by anti-His Western analysis. Each SaOatA variant was found in the membrane in approximately equal amounts, suggesting that the observed lack of function was not due to insufficient expression or improper membrane insertion of the OatA variants (Fig. 3 A, Inset). Confirmation of the correlation between lysozyme sensitivity and OatA function was again obtained by analyzing the PG from representative mutants for O-acetylation. The PG purified from the mutants producing single amino acid variants (H36A)SaOatA and (Y206A)SaOatA and displaying full lysozyme sensitivity had 0 and 1.3% O-acetylation, respectively, while the strain producing (R83A)SaOatA and displaying only partial lysozyme sensitivity had ∼29% PG O-acetylation (Fig. 1B).
We investigated the importance of charge or functional groups associated with some of these residues on activity by replacing them with alternate amino acids. The replacement of Arg25 and Arg86 individually with Lys had no effect on lysozyme resistance, suggesting that the positive charge of Lys can compensate for that of Arg at these positions (Fig. 3B). We found that neither Asp nor Gln could compensate for the functional role of Glu154, but partial activity was retained with the replacement of Glu357 with Asp. As with Glu154, we found both Tyr136 and Tyr206 to be essential residues; their replacement with either Phe or Ser resulted in mutant strains sensitive to lysozyme. However, the mutant producing (Y311S)SaOatA retained wild-type levels of lysozyme resistance.
In vitro studies.
We subcloned and expressed a selection of the mutated oatA genes in E. coli, and the recombinant SaOatA variants were purified to apparent homogeneity in the same manner as for SaOatA and (S453A)SaOatA (SI Appendix, Fig. S2). We tested each for esterase and transferase capacity by incubating them with acetyl-CoA alone (esterase) and together with pentaacetyl-chitopentaose (G5) as a pseudoacceptor (transferase). For the latter, we assayed for G5 O-acetylation by LC-MS/MS. The (R25A), (R86A), and (E154A)SaOatA variants retained 18.4%, 22.1%, and 4.76% esterase activity, respectively (Fig. 3C), with low amounts of O-acetylated G5 detected by LC-MS (SI Appendix, Fig. S7). (H36A)SaOatA retained only 0.64% apparent esterase activity and no transferase activity was detected. The (Y206A)SaOatA variant was devoid of both esterase and transferase activity. Interestingly, (Y136A)SaOatA displayed 155% esterase activity toward acetyl-CoA; however, no acetylated G5 was detected by LC-MS in the transferase assay, suggesting that this residue does not contribute directly to the catalytic mechanism of SaOatAN. (R25K)SaOatA also displayed increased esterase activity compared to wild-type (178%); however, this variant was functional as an O-acetyltransferase.
We determined the steady-state kinetic parameters for the (Y136A) and (R25K)SaOatA variants as esterases (Fig. 3D). The KM of (R25K)SaOatA for acetyl-CoA was almost double that of wild-type OatA. However, the Vmax was also double, such that the overall efficiency of the enzyme, as reflected by kcat/KM, was only slightly increased compared to wild-type. Due to limitations of the assay, we could not saturate (Y136A)SaOatA with acetyl-CoA, but extrapolation of the data suggests a significantly increased KM of 133 µM and decreased kcat/KM of 1,170 M−1 ⋅ s−1.
Trapping O-Acetyl-Enzyme Intermediates.
Previously, we demonstrated the accumulation of an acetyl intermediate within SaOatAC involving its catalytic nucleophile Ser453 by a real-time analysis of its reaction with pNP-Ac as an acetyl donor (32). Recognizing that SaOatA appears to have two catalytic centers, we postulated that a residue within SaOatAN may become acetylated in the translocation/transferase mechanism and that this residue may serve as the acetyl donor for SaOatAC. Thus, we attempted to repeat the trapping experiment using full-length SaOatA in the presence of acetyl-CoA. However, being a large integral membrane protein, we experienced solubility issues during the chromatography step of the LC-MS analysis. We found that we could (barely) detect native, untreated SaOatA with a deconvoluted mass of 70,871.6 Da, the expected mass for the unbound protein, but only when a low concentration (4.3 mg/mL; 60 μM) sample was analyzed. However, when injected in the presence of acetyl-CoA, a suppression of the already low-intensity multiply charged SaOatA ions was observed and precluded spectral deconvolution to monoisotopic mass species. This included the loss of the monoisotopic mass peak at 70,871.6 that corresponded to unmodified SaOatA, suggesting that acetylated species were nonetheless produced. We suspected that, unlike the situation with OatAC with its single active center, full-length OatA possesses at least two which may give rise to a mixed population of ionic species with different extents of acetylation, compounded by the potential of additional sodium adducts.
We encountered similar obstacles with the matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) MS analysis of SaOatA and its possible adducts. Despite the amount of protein applied to the chips, we detected very weak signals (SI Appendix, Fig. S8). Moreover, as with the MALDI-TOF MS analyses of other membrane proteins (e.g., ref. 45), the signals were very broad, and their center of mass was shifted over 4,000 Da from the expected mass of the protein. This shift has been attributed to the presence of protein-bound lipids and/or detergents. However, despite the breadth of the signals, we observed a difference between the average m/z ratios of SaOatA in the absence and presence of acetyl-CoA of 124.9 Da, the approximate mass of three acetyl groups (expected mass, 126.03 Da).
Despite our failure to detect modified SaOatA directly by intact protein LC-MS but encouraged by the MALDI-TOF MS data, we attempted to identify site(s) of acetylation within a protease digest of full-length SaOatA. LC–quadrupole-TOF MS/MS analysis of the peptides from a control digestion of untreated SaOatA provided 56% coverage of the native SaOatA sequence; not surprisingly, there was minimal coverage of the N-terminal membrane-spanning helices (46–48). Searching for peptides modified by acetyl groups (viz. +42.01 Da) using the Peaks XPro software, we detected the nonspecific acetylation of His557 and Tyr581, albeit with low confidence and high mass error. These two residues are surface exposed on separate faces of SaOatAC, and their side-chain O atoms are located 24.3 and 18.8 Å from the catalytic Ser453 side-chain oxygen, respectively; presumably, they became nonspecifically acetylated in the E. coli expression host. A similar treatment and analysis of a protease digest of SaOatA preincubated with acetyl-CoA yielded a slightly better coverage of 60%, and again, we observed nonspecific acetylation of Tyr581 together with another surface-exposed residue Tyr490, also located in SaOatAC and 28.4 Å from the active site. The only unique acetylated peptide species, which could be detected in multiple scans of both datasets with high confidence and low mass error and confirmed by subsequent MS/MS fragmentation analysis, mapped to catalytic Ser453 of SaOatAC (SI Appendix, Fig. S9). This was expected given this acetyl intermediate had been identified previously in our earlier study with isolated SaOatAC (32). Nonetheless, this confirmed that while the isolated domain alone is unable to utilize acetyl-CoA as an acetyl donor, acetyl groups from this natural metabolite are provided directly to it through SaOatAN for PG O-acetylation. Interestingly, we also observed the Ser453-Lys464 peptide with a moderate score of acetylation in the digest of the untreated control preparation of SaOatA. Presumably, this acetylation occurred by the enzyme’s acquisition of acetyl from acetyl-CoA pools in the cytoplasm of the E. coli expression host.
Recognizing that the specific activity of (S453A)SaOatA as a thioesterase is significantly lower than that of the wild-type enzyme, we postulated that we may be able to trap any acetyl-product of the N-terminal acyltransferase domain within this enzyme variant. LC-MS/MS analysis of a protease digest of (S453A)SaOatA pretreated with acetyl-CoA provided 65% coverage of the protein. As with the other analyses, we detected low levels of nonspecific acetylations of Tyr574 and Tyr581 and additionally His557, which all mapped to our structure of OatAC as surface-exposed residues distant from the catalytic center. However, we also observed with high confidence and in multiple scans the unique acetylated peptides Val205–Lys232 and Val205-Lys233 containing the essential Tyr residue, Tyr206. The precursor ion was reliably matched to the OatA sequence, and the mass shift of 42.01 Da of Tyr206 was confirmed by MS/MS fragmentation with high confidence and low mass error, indicating its acetylation (SI Appendix, Fig. S9). This finding is consistent with essential Tyr206 serving as the catalytic nucleophile for the acquisition of acetyl groups from acetyl-CoA.
O-Acetyl-Tyr Is a Substrate for OatAC.
The data presented above indicated the presence of two active sites within SaOatA, involving Tyr206 and Ser453 as the catalytic nucleophiles within SaOatAN and SaOatAC, respectively. Being located on a small extracellular loop linking transmembrane segments, we postulated that Tyr206 also functions to directly transfer acetyl groups between the two catalytic domains and serves as the acetyl donor for SaOatAC. To test this, we prepared as potential substrates synthetic peptides based on the Tyr206 extracellular loop that were O-acetylated at the Tyr hydroxyl group. Given the poor solubility of these acetylated peptides, we needed to conduct the assays in the maximal permissible DMSO concentration. To determine this, we tested the activity of SaOatAC toward 4MU-Ac in increasing DMSO concentrations up to 60% (SI Appendix, Fig. 10A). To our surprise, we found that esterase activity peaked at 40% DMSO. It would appear that DMSO helps to stabilize the activity of the SaOatAC in the same manner as has been observed with other PG metabolizing enzymes, such as the high MW PBPs and SEDS proteins, which are active in vitro only in the presence of 20 to 30% DMSO (e.g., ref. 49). In 35% DMSO, the KM of SaOatAC for 4MU-Ac was similar to that in 5% DMSO, but its kcat was significantly higher, resulting in an overall efficiency of the enzyme that was sixfold greater under these conditions (SI Appendix, Fig. S10 B and C).
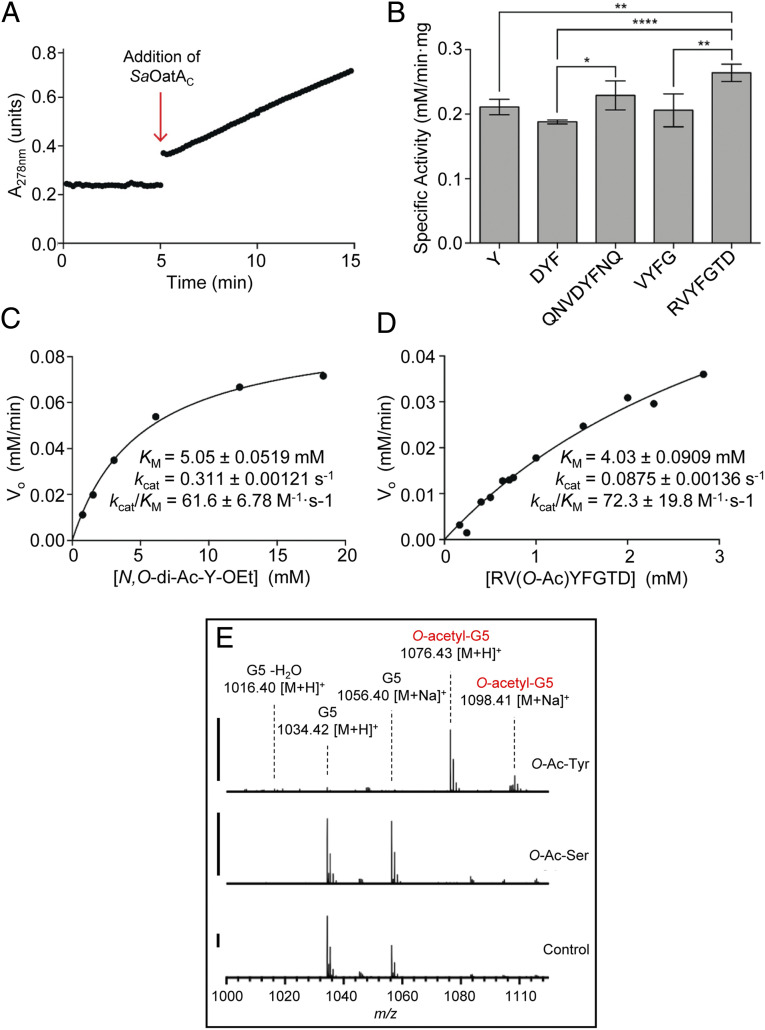
We prepared N-acetyl-Val-(O-acetyl)-Tyr-Phe-amide (VYF), N-acetyl-Val-(O-acetyl)-Tyr-Phe-Gly-amide (VYFG), and N-acetyl-Arg-Val-(O-acetyl)-Tyr-Phe-Thr-Asp-amide (RVYFGTD) as representative short and long peptides, respectively, of the Tyr206 extracytoplasmic loop. Unfortunately, the extremely low solubility of VYF prevented its use for activity assays. N-acetyl-Asp-(O-acetyl)-Tyr-Phe-amide (DYF) and N-acetyl-Gln-Asn-Val-Asp-(O-acetyl)-Tyr-Phe-Asn-Gln-amide (QNVDYFNQ) were also prepared as representatives of a cytoplasmic loop in SaOatA (residues 130 to 139). To determine activity, we developed a spectrophotometric assay that monitors the resulting increase in absorbance at 278 nm with the removal of the acetyl group from the Tyr residue of the donor peptide (Fig. 4A); the acetylation of Tyr residues suppresses absorbance at 278 nm (50). Unfortunately, substrate availability and solubility, even in 35% DMSO, limited the scope of analyses, and it was only possible to obtain a specific activity with some peptides. For this reason, the O-acetylated peptides were tested in 44% DMSO to maximize substrate solubility while maintaining enzyme activity.
Fig. 4.
Esterase and O-acetyltransferase activity of OatAC with O-acetyl-Tyr peptides as acetyl donor. (A) Representative assay for determination of esterase activity. The absorbance of acetylated peptide in 50 mM sodium phosphate pH 6.5 and DMSO was monitored at 278 nm for spontaneous hydrolysis of the acetyl group. SaOatAC (5 µM) was then added to the reaction (indicated by red arrow), and the rate of esterase activity was monitored. (B) Determination of the specific activity of 5 µM SaOatAC for each O-acetylated peptide at 1 mM in 50 mM sodium phosphate pH 6.5 containing 35% (vol/vol) DMSO. Error bars denote SD (n = 3). *P ≤ 0.05; **P ≤ 0.01; ****P ≤ 0.0001. (C and D) The steady-state parameters were determined for 5 µM SaOatAC in 50 mM sodium phosphate pH 6.5 and 44% (vol/vol) DMSO using (C) 0.76 to 18.4 mM O-Ac-Tyr-ethyl ester or (D) 0.24 to 2.8 mM Arg-Val-(O-Ac)-Tyr-Phe-Gly-Thr-Asp-amide as substrate. (E) LC-MS analysis of reaction products following 18 h incubation of 5 µM SaOatAC in phosphate buffer containing 4% DMSO with 1 mM G5 as a glycan acceptor and 1 mM of either O-acetyl-Tyr-ethyl ester or O-acetyl-Ser as an acetyl donor. The solid bars to the left of the spectrograms denote 5,000 ion counts.
O-acetyl-Tyr-ethyl ester and each of the peptides containing O-acetyl-Tyr served as substrates for SaOatAC acting as an esterase (Fig. 4B). Whereas the range of determined specific activities for each of these peptides was small, that for the peptide RVYFGTD was the highest. This peptide alone is predicted to form a loop, which would likely be stabilized by a salt bridge between Arg204 and Asp210, a feature consistent with its position in our topology map of SaOatAN. SaOatAC was less active toward the internal tetrapeptide of this loop, VYFG, and both DYF and QNVDYFNQ, peptides that comprise in an intracellular loop linking TMS4 to TMS5. We determined the Michaelis–Menten parameters of SaOatAC for N,O-di-acetyl-Tyr-ethyl ester and RVYFGTD (Fig. 4 C and D), and whereas the KM values were relatively high, the overall catalytic efficiency of the enzyme for these O-acetylated peptides was nearly double that for the simple pseudo substrate 4MU-Ac (SI Appendix, Fig. S10).
Finally, we found that O-acetyl-Tyr-ethyl ester, but not O-acetyl-Ser, may function as a donor substrate for the O-acetylation of G5 by SaOatAC. The enzyme was incubated with 1 mM of each potential donor substrate and 1 mM G5 as an acceptor for 2 h, and reaction products were analyzed by LC-MS. O-Acetylated G5 was detected only after a reaction with O-acetyl-Tyr-ethyl ester (Fig. 4E) as a donor.
Discussion
Membrane proteins are notoriously difficult to study due to their poor solubility and low stability in vitro. As such, structural and biochemical information for many membrane-bound enzymes is very limited. This applies to the wide variety of acetyltransferases responsible for the O-acetylation of extracellular glycans, including OatA and all other AT-3 family proteins. The extracytoplasmic C-terminal SGNH hydrolase domain of OatA has been well characterized as a PG O-acetyltransferase (32–34), but nothing was known previously about the cognate N-terminal domain beyond homology-based predictions. In this study, we experimentally determined the topology and function of SaOatAN and provide molecular insights of the acetyl translocation process of any membrane-bound enzyme catalyzing extracytoplasmic O-acetylations of glycans.
The topology map of SaOatAN generated by PhoA-LacZα fusion mapping enabled a two-dimensional model to be generated (Fig. 2B), which differed significantly from that determined using the same method for another AT-3 family protein, Oac, an O-acetyltransferase responsible for the O-acetylation of O-antigen in S. flexneri (38). However, only 16 truncations of Oac were assayed, and certain regions of the protein were not mapped at all (38), which may account for discrepancies between the Oac and SaOatAN experimental models. Indeed, our study of 53 truncations of SaOatAN revealed several unanticipated structural features. Whereas each approach has its limitations, experimental mapping methods have their advantage over in silico predictions which have rigid criteria. Predictions assume each TMS crosses the membrane, and they do not account for certain structural features commonly found in integral membrane proteins, such as peripheral membrane, re-entrant, or partial helices. Each of these elements appears to be present in SaOatAN including the re-entrant helix TMS5. Our interpretation that TMS5 is a re-entrant helix is based on finding both Ile130 and Leu168 to be located in the cytoplasm. As a typical transmembrane helix has an average length of 25 residues (51), it is unlikely the intervening 38 amino acid residues would form two transmembrane helices. Furthermore, helix prediction software predicts one helix in this region encompassing residues Leu145 to Arg171. We set the bounds of TMS5 by Gln139 and Pro163 because both were located to the membrane, but it is not possible to ascertain whether TMS5 is one long or two short helices based solely on the data obtained. While not being predicted, re-entrant helices are observed in transmembrane proteins with solved structures. For example, E. coli undecaprenyl pyrophosphate phosphatase possesses two short adjacent re-entrant helices (52, 53), and the ClC chloride channel from the same bacterium is characterized by six re-entrant regions (54).
The region between TMS9 and TMS10 involving two predicted α-helices was located to the cytoplasm by 11 truncations (Fig. 2B). Although these helices are not predicted to be amphipathic, both CH1 and CH2 have large hydrophobic dipole moments over their lengths, suggesting peripheral association with the membrane through their C- and N-terminal ends, respectively (SI Appendix, Fig. 5B). Peripheral membrane helices are also a common occurrence in membrane proteins. A recently discovered example is with Streptococcus thermophilus DltB, a member of the membrane-bound O-acyltransferase (MBOAT) family of proteins involved in the d-alanylation of WTAs. The structural model for this protein solved by X-ray crystallography (55) presents several short peripheral membrane helices that contain important functional residues, features that are not predicted in silico. These examples demonstrate the limitations of in silico tools and highlight the value of experimental approaches to determining the structure and topology of membrane proteins.
Our ability to assay SaOatA in vitro permitted us to identify acetyl-CoA as its preferred acetyl donor for PG O-acetylation (Fig. 1). The kinetic parameters we established for this reaction are consistent with acetyl-CoA being its natural substrate. This is supported by our identification of three Arg residues on the cytoplasmic face of SaOatAN that likely function to coordinate the phosphoryl groups of coenzyme A for its productive binding. These three Arg residues comprise the Arg-X-X-Arg and Arg/Lys-X10-His motifs identified earlier in the Oac homologs by others (36–38). The results of our in situ assay (Fig. 3B) coupled with the in vitro kinetic analyses of some of the recombinant SaOatA variants confirmed the importance of Arg86 and, to a lesser extent, Arg83 of the Arg-X-X-Arg motif and both Arg25 and His36 of the Arg/Lys-X10-His motif. Concerning the latter motif, the (R25K)SaOatA variant was generally more active in vitro as a thioesterase, indicating that the retention of positive-charge character is important for function. However, its KM for acetyl-CoA was approximately twofold higher than that for the wild-type enzyme, strongly supporting the notion that Arg25 and its counterparts in other AT-3 acetyltransferases contributes to acetyl-CoA–binding (36–38). On the other hand, the Arg residues of the Arg-X-X-Arg motif have been proposed to be critical for the assembly of S. flexneri Oac (38), but we found no differences in the in situ expression of the (R83A) and (R86A)SaOatA variants (Fig. 3 A, Inset) suggesting otherwise. Unlike the earlier models for AT-3 topology in which the Arg-X-X-Arg motif is predicted to comprise TMS3, we located both Arg83 and Arg86 on a cytoplasmic loop between TMS2 and TMS3. This positioning is consistent with the rule that positively charged amino acids are usually found in cytoplasmic loops of transmembrane proteins. Moreover, comprising a loop would provide flexibility for the clustering of Arg83 and Arg86 near the cytoplasmic face of TMS1, the location of Arg25, and thereby form an appropriate binding site to accommodate and stabilize the three phosphoryl groups of coenzyme A (Fig. 5A). This hypothesis is supported by structural evidence obtained in the examination of acetyl-CoA interactions with other acetyltransferases, such as N. meningitidis polysialic acid O-acetyltransferase OatWY (56) and E. coli galactoside acetyltransferase (57), in which a set of electrostatic interactions involving three Arg/His/Lys residues directly stabilizes these phosphate groups.
Fig. 5.
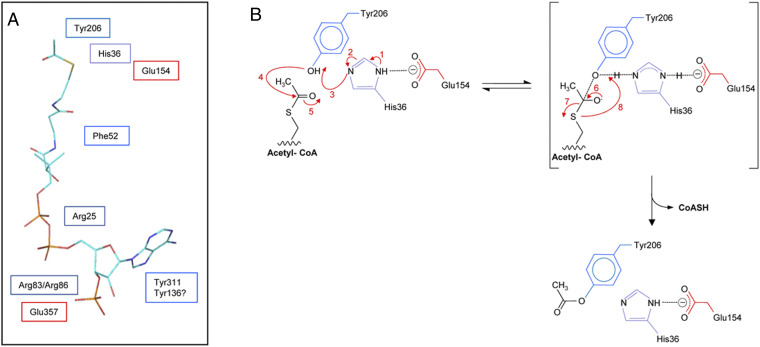
Proposed binding site of acetyl-CoA and mechanism of action of SaOatAN. (A) Proposed positioning of residues identified as essential for SaOatAN activity relative to acetyl-CoA. The structural conformation of acetyl-CoA presented was adopted from its complex with galactoside acetyltransferase as determined by X-ray crystallography (57) (Protein Data Bank Acc. No. 1KRR). (B) Postulated mechanism of SaOatAN. Aided by its interaction with Glu154 (reaction 1, 2; red labeling), His36 acts as a base to abstract the proton from the hydroxyl group of Tyr206 (reaction 3), rendering it nucleophilic. An attack on the carbonyl carbon of the thioester of acetyl-CoA by the nucleophilic Oη of Tyr206 (reaction 4) leads to a putative tetrahedral oxyanion intermediate (reaction 5), which collapses (reaction 6) to release CoASH (reaction 7, 8) and the formation of the O-acetyl-Tyr206 product.
Other typical features of binding sites for acetyl-CoA in acetyltransferases include a stacking interaction between an aromatic amino acid residue (often Tyr) and the adenine ring of coenzyme A and the stabilization of the phosphopantothenyl arm of CoA by both polar and nonpolar interactions with the side chains of residue(s) as observed with, for example, N. meningitidis OatWY (56). Based on their observed locations, importance for function, and flexibility (or lack thereof) for replacement, we postulate that invariant residues Phe52, Tyr311, Glu357, and possibly Tyr136 contribute to these functions (Fig. 5A). Instead of comprising integral membrane segments as initially predicted, Tyr311 and Glu357 are located within the cytoplasm. Tyr311 could be replaced with Ser, but not Ala nor Phe, for the retention of in situ activity, suggesting the requirement for hydrogen bonding capacity. Similarly, Glu357 could be replaced with Asp, but neither Gln nor Ala, indicating the need for anionic character at its position. These characteristics are consistent with these two residues contributing to and stabilizing the binding site for coenzyme A at the cytoplasmic face of the membrane. Phe52, on the other hand, was predicted and observed to be located within the middle of TMS2. This is an appropriate position for hydrophobic interactions with the phosphopantothenyl arm of CoA if the acetylated coenzyme extends up from the cytoplasm within a channel of the protein formed by TMS1, TMS2, and others to present the acetyl group toward the extracytoplasmic face of the protein (Fig. 5A). Such an orientation of the acetyl donor would be expected for the translocation of the acetyl group to Tyr206 on the extracellular surface of the protein to serve as substrate for SaOatAC and eventual transfer to PG. The importance of invariant Phe52 is underscored by it comprising a conserved sequence motif, VxxFFx(I/V/L)SG(F/WY), that was first identified in several O-antigen O-acetyltransferases (38, 58).
The functional role of Tyr136 is less clear. This invariant residue was initially predicted to comprise a large extracytoplasmic loop, but our topology mapping located it within a short cytoplasmic loop linking TMS4 to the re-entrant helix TMS5. This residue could not be replaced with either Ala, Ser, or Phe without the impairment of in situ activity, and the KM of the (Y136A)SaOatA variant for acetyl-CoA was an order of magnitude higher than that of the wild-type enzyme, while their respective kcat values were similar. These features are consistent with a substrate-binding role for Tyr136, and it is conceivable that, like Arg83 and Arg86, its location on a cytoplasmic loop provides its opportunity to comprise the acetyl-CoA–binding site at the cytoplasmic face of the protein (Fig. 5A). On the other hand, while lacking transferase activity, (Y136A)SaOatA is more active as an esterase, indicating that Tyr136 in some way contributes to the appropriate positioning of Tyr206 for the effective transfer of acetyl groups between SaOatAN and SaOatAC. With our understanding of typical integral membrane proteins, these two direct functions involving both cytoplasmic and extracellular events would seem to be mutually exclusive. However, a number of the SaOatAN transmembrane helices are predicted to be either re-entrant or oblique (SI Appendix, Fig. S5A), while others were observed to be cytoplasmic. These features are consistent with the overall conformation of the enzyme to be similar to that of the MBOAT family member, d-alanyl carrier protein (DltB). DltB contains a ring of 11 peripheral transmembrane helices, which form an intracellular concave surface and an extracellular structural funnel that extends into the middle of the lipid bilayer of the cytoplasmic membrane and surrounds a central thin structural core (55). The active site located within the central core catalyzes the translocation of d-alanyl residues across the cytoplasmic membrane of gram-positive bacteria for their addition to teichoic acids. It is conceivable that this unique structure may represent a new paradigm for other families of membrane-associated acyltransferases, such as the AT-3 proteins. If so, then it may be possible for a residue such as Tyr136 in SaOatA to serve the two roles of contributing to acetyl-CoA–binding and structurally aligning the catalytic center. Of course, the delineation of Tyr136’s role in the function of SaOatAN, and those of the other invariant residues discussed above, awaits structural information at the atomic level.
We identified invariant His36 within TMS1 as being essential for SaOatA activity both in vitro and in situ. Its equivalent has been recognized as being important in many other membrane-bound acetyltransferases, including AT-3 proteins (36). Being 10 residues apart, His36 and Arg25 would be positioned on the same face of TMS1 and separated by a distance of ∼16.5 Å, the approximate length of the phosphopantothenyl arm of coenzyme A (Fig. 5A). Also recognizing this, Pearson et al. (32) postulated that homologous residues Arg14 and His25 in S. Typhimurium OafA provide a potential interaction site for acetyl-CoA. Our finding that (H36A)SaOatA retains only 0.6% residual activity as an esterase compared to wild-type SaOatA and lacks the ability to catalyze acetyl transfer to glycan acceptors suggests an important catalytic role for the residue in the enzyme’s mechanism of action. Invariant Glu154 of the re-entrant TMS5 was also determined to be important for catalytic activity. Its replacement with Ala, Asp, or Gln generated weakly active forms of SaOatA, also suggesting a mechanistic role for this acidic residue. If Tyr136 does interact with the adenine ring of coenzyme A, TMS5 would have to be associated with TMS1 and TMS2 and thereby position Glu154 in close proximity to His36.
Considering all of the findings discussed above, we hypothesize that Tyr206, His36, and Glu154 of SaOatAN form a catalytic triad for the translocation of acetyl groups from the cytoplasm to the extracellular surface of SaOatA (Fig. 5B and SI Appendix, Fig. S11), which would function in a manner analogous to the catalytic Ser, His, Asp catalytic triad of OatAC (32–34). Thus, we propose that Glu154 forms a salt bridge with His36, enabling His36 to function as a base to deprotonate Tyr206. Rendered nucleophilic, Tyr206 would attack the thioester carbonyl carbon of acetyl-CoA generating a tetrahedral transition state, which then collapses into a covalently bound acetyl-enzyme intermediate. In this study, Tyr206 was identified as the site of acetylation (SI Appendix, Fig. S9), and kinetic studies with model peptides support the notion that O-acetyl-Tyr206 serves as the acetyl donor for SaOatAC. It is theoretically possible that His36 functions as the nucleophile to directly attack the carbonyl of the acetyl-CoA thioester to form an initial covalent adduct and that the acetyl group then migrates to Tyr206. Indeed, an unrelated membrane acetyltransferase, lysosomal heparan sulfate acetyl-CoA:α-glucosaminide N-acetyltransferase, was experimentally shown to form an acetyl intermediate on a His residue (59, 60). Our analysis of SaOatA incubated with acetyl-CoA by MALDI-TOF MS did suggest the possibility of three acetyl adducts. However, the MALDI-TOF MS data were not conclusive, and the kinetic data obtained with the various SaOatA variants does not support this mechanism. Typically, the replacement of the catalytic nucleophile with Ala leads to a complete loss of enzymatic activity as observed with (Y206A)SaOatA and previously with OatAC (32–34), while small amounts of residual activity remain with the replacement of residues providing assistance as detected with the (H36A) and (E154A)SaOatA variants (0.6% and 4.8% residual activity, respectively). Moreover, the replacement of Tyr206 with Ala resulted in a complete abolishment of both esterase and transferase activity; if His36 were the catalytic nucleophile, (Y206A)SaOatA would still be able to function as an esterase.
With Tyr206, His36, and Glu154 of SaOatAN (SI Appendix, Fig. S6) together with Ser453, His578, and Asp575 of SaOatAC (32) being invariant among all OatA homologs, we propose that the mechanism presented in Fig. 5B (and SI Appendix, Fig. S11) applies to all OatA enzymes. Moreover, as noted above, a catalytically important His residue homologous to His36 has been identified in the first TMS of all AT-3 proteins. Our alignment of all characterized AT-3 domains fused to SGHN domains for both translocation and the transfer of acetyl groups to extracytoplasmic acceptors indicated that both Glu154 and Tyr206 are also invariant in this subfamily of proteins (SI Appendix, Fig. S12). This would suggest that, in addition to all other OatAs, this same mechanism of action applies to other AT-3/SGNH enzymes. The identification and/or assignment of function of the corresponding catalytic nucleophile in the AT-3 domain of the other members of this subfamily of enzymes may have been missed by others given the likely incorrect prediction of topology of the sequence harboring it. Regardless, identification of the catalytic nucleophile of enzymes is key for their further characterization as potential targets for antibacterial development. Indeed, while this subfamily of AT-3 proteins includes hundreds of members, those beyond the Oat enzymes of gram-positive bacteria that have been studied function to modify cell wall glycans in important gram-negative pathogens, such as N. gonorrhoeae, N. meningitidis, Haemophilus influenzae, and species of Salmonella.
The location of Tyr206 on an external loop would provide some flexibility for the residue to first function as the nucleophile at the active site of SaOatAN and then as the acetyl donor for the acetyltransferase reaction at the active site of SaOatAC. Presumably, the two SaOatA domains fold over each other to sandwich a PG acceptor glycan strand between them. This would be consistent with our inability to apply the SCAM method for topology determination as discussed above. Such an association would also explain why the active site of isolated OatAC appeared to be unusually surface exposed (32–34).
Also, with acetyl-Tyr206 serving as the acetyl donor for OatAC, its binding “site” for this substrate would be, in effect, the entire contact surface area between it and OatAN. As such, OatAC would be less dependent on discrete binding site/subsites for the productive binding of its substrate O-acetyl-Tyr as typical of most other enzymes, including OatAN for its donor substrate acetyl-CoA. The lack of a discrete binding site would account for the poor binding parameters and virtual lack of specificity observed with SaOatAC for the model peptides containing O-acetyl-Tyr that we tested as substrates in vitro.
Methicillin-resistant S. aureus continues to be a major threat, costing the lives of over 10,000 people per year in the United States alone. Hence, there is continued interest in developing new antibiotics for its effective treatment. OatA represents a potential target for antibiotic development, and using this enzyme as a model, this study presents a biochemical characterization of an AT-3 domain. Our findings will help form the basis for further studies of this acetyltransferase and other AT-3 family enzymes with a large array of roles. In particular, our elucidation of the interaction between the AT-3 and SGNH domains of OatA will inform the search and development of inhibitors as potential antibiotic leads. Indeed, the lack of a discrete binding site for the first half of the acetyltransferase reaction catalyzed by OatAC has significant ramifications for this search. With the knowledge gleaned from this study, investigators may be better served if in vitro screening assays were designed to target the acceptor half of the reaction. Better still, the application of the in situ assay we developed based on lysozyme sensitivity may prove to be a more effective approach.
Materials and Methods
Bacterial Strains and Growth Conditions.
Cultures of bacteria (SI Appendix, Table S2) were grown at 37 °C with aeration in Luria-Bertani broth (Difco) or Tryptic Soy broth (TSB) (Difco) as indicated. Media were supplemented with appropriate antibiotics (E. coli: 100 μg/mL ampicillin, 50 μg/mL kanamycin; S. aureus: 10 μg/mL chloramphenicol) when required.
Cloning, Engineering, and Production of S. aureus OatA.
General protocols.
Plasmids generated and/or used in this study are listed in SI Appendix, Table S3. Custom DNA oligonucleotide primers (SI Appendix, Table S4) were obtained from Integrated DNA Technologies. The protocols for the deletion of oatA in S. aureus USA300 (S. aureus USA300 ΔoatA) using the method of Bae and Schneewind (61), the cloning of the gene encoding full-length S. aureus OatA (residues 1 to 603) (SaOatA), and the generation of SaOatA variants possessing site-specific amino acid replacements by site-directed mutagenesis are described in SI Appendix, Supplementary Methods.
Functional Complementation (In Situ) Assay of OatA Variants in S. aureus.
S. aureus USA300 ΔoatA was transformed with pACCJ3 carrying the desired mutation. MIC assays were performed with lysozyme in the absence and presence of tunicamycin (0.4 μg/mL) in 200 μL volumes in a 96-well microtitre plate. A 20 h overnight culture of S. aureus USA300 ΔoatA pACCJ3 was diluted 1/10,000 in sterile TSB. For experiments including tunicamycin, 0.4 μg/mL of the antibiotic (in DMSO) was added such that the final concentration of DMSO was constant at 1% (vol/vol). Filter-sterilized hen egg-white lysozyme (Bio Basic, Inc.) dissolved in water was added at concentrations ranging from 0.25 to 4 mg/mL to each well. Each plate was covered with a Breathe-Easy sealing membrane (Millipore Sigma) and incubated in a Synergy plate reader at 37 °C with double-orbital shaking for 20 h, monitoring optical density at 600 nm every 20 min.
Overproduction and Purification of Full-Length SaOatA and Its Variants.
The overproduction of full-length SaOatA and its variants possessing site-specific single amino acid replacements was conducted using E. coli C43(DE3) transformed with pACCJ2 or pACCJ2 carrying the desired mutation. The recombinant enzymes were isolated and purified to apparent homogeneity, as determined by SDS-PAGE analysis, by immobilized metal affinity chromatography using fresh resin each time to prevent cross-contamination. Further details are presented in SI Appendix, Supplementary Methods. The production and purification of SaOatAC was performed as described in Jones et al. (32).
Determination of Catalytic Activity and Kinetic Parameters.
As an esterase.
Routine determinations of SaOatA and SaOatAC as an esterase were conducted spectrophotometrically as described previously (32) using 4MU-Ac as substrate in 50 mM sodium phosphate buffer pH 7 containing 5% (vol/vol) DMSO at 25 °C. Unless otherwise stated, all reactions were performed in triplicate, and errors are expressed as SD. Measurements of thioesterase activity toward acetyl-CoA were determined using a modified aldrithiol assay (41) as described in SI Appendix, Supplementary Methods.
As a transferase.
The MS-based transferase assay previously developed (42) was modified as described in SI Appendix, Supplementary Methods to detect the ability of SaOatA to acetylate pseudoacceptors and muroglycans. The muroglycans enzymatically prepared as acceptor substrate had a degree of polymerization (of Lipid II) of 4 to 10 GM-pentapeptides (poly-GM5).
Topology Mapping of SaOatAN within SaOatA.
SCAM.
The protocol of Bogdanov et al. (43) was used in an attempt to determine the topology of SaOatAN as described in SI Appendix, Supplementary Methods.
PhoA-LacZα truncation fusion method.
Details for the cloning and engineering of the gene encoding full-length SaOatA (residues 1 to 603) to produce libraries of random exonuclease III–generated truncation fusions to phoA-lacZα following the method of Alexeyev and Winkler (44) and their transformation into E. coli DH5-α as an α-complementing host strain are presented in SI Appendix, Supplementary Methods. Cells were plated on dual indicator plates containing 1.5% (wt/vol) agar, 1% (wt/vol) tryptone, 0.5% (wt/vol) yeast extract, 0.5% (wt/vol) NaCl, 80 mM K2HPO4, 80 mg/mL 5-bromo-4-chloro-3-indolyl phosphate disodium salt (Sigma-Aldrich), 100 mg/mL 6-chloro-3-indolyl-β-d-galactopyranoside (Sigma-Aldrich), 1 mM isopropyl β-D-1-thiogalactopyranoside, and 100 μg/mL ampicillin (45). The β-galactosidase and alkaline phosphatase assays were performed as described by Manoil (62) with adaptation to a 96-well plate format (see SI Appendix for adaptation methodology and data manipulation).
Identifying In Situ Location of Full-Length SaOatA by Western Blot.
The protocol used to detect and locate full-length SaOatA and its variants in transformed strains of S. aureus USA300 ΔoatA by western immunoblotting (63) is described in SI Appendix, Supplementary Methods.
Trapping and Identification of Acetyl-SaOatAN Intermediate(s).
Details for the preparation of samples of SaOatA treated with acetyl-CoA and their MS analyses are described in SI Appendix, Supplementary Methods. Briefly, for MALDI-TOF MS, samples were concentrated to 35 μM, washed with 10 mM ammonium acetate, and then mixed with equal volumes of sinapinic acid matrix prior to direct analysis. For LC-MS/MS analyses, reduced and alkylated SaOatA samples were digested with a trypsin/Lys-C mixture followed by MS-grade endoproteinase GluC. The resulting digests were injected directly for LC-MS/MS analysis. To identify sites of any acetylation, the MS/MS data were analyzed using Peaks XPro (Bioinformatics Solutions, Inc.) against the sequences of wild-type SaOatA or (S453A)SaOatA variant according to the protein digested. Variable peptide modifications considered included Asn/Glu deamidation (−0.98 Da), Met oxidation (+15.99 Da), and acetylation of Tyr, His, and/or Ser (+42.01 Da).
Assay of O-Acetylated Tyr-Containing Peptides as Substrate.
The synthesis of the O-acetylated peptides, the determination of their concentration, and use as substrates for SaOatAC are described in SI Appendix, Supplementary Methods. Briefly, the specific activity of 5 μM SaOatAC acting as an esterase toward O-acetyl-Tyr–containing peptides was determined spectrophotometrically by monitoring the increase in 278 nm absorbance of peptides (50) in 50 mM sodium phosphate pH 6.5 containing 44% (vol/vol) DMSO.
Other Analytical Techniques.
All other analytical techniques used in this study are described in SI Appendix, Supplementary Methods.
Supplementary Material
Acknowledgments
We thank Dr. David Heinrichs, Western University, for his gift of S. aureus strains USA300 and RN4220 and plasmids pKOR1 and pALC2073. We also thank Drs. Dyanne Brewer and Armen Charchoglyan of the Mass Spectrometry Facility (Advanced Analysis Centre, University of Guelph) and Valerie Goodfellow (Mass Spectrometry Facility, University of Waterloo) for expert technical assistance and advice and Maria A. Eng, Bryan J. Fraser, Catherine Jany, and Laura Thompson for technical assistance with some of the enzyme assays. These studies were supported in part by operating grants from the Canadian Institutes of Health Research to A.J.C. (TGC 114045) and the Canadian Glycomics Network (https://canadianglycomics.ca/). C.S.J. and A.C.A. were supported in part by graduate scholarships from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103602118/-/DCSupplemental.
Data Availability
The raw unprocessed files for SaOatA tryptic peptide LC-MS, corresponding Peaks XPro search results, and annotated MS2 spectra for supporting peptides have been deposited to Figshare (46–48). All other study data are included in the main text and/or SI Appendix.
References
- 1.Park S. S., Post-glycosylation modification of sialic acid and its role in virus pathogenesis. Vaccines (Basel) 7, 171 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pauly M., Ramírez V., New insights into wall polysaccharide O-acetylation. Front. Plant Sci. 9, 1210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sukhithasri V., Nisha N., Biswas L., Anil Kumar V., Biswas R., Innate immune recognition of microbial cell wall components and microbial strategies to evade such recognitions. Microbiol. Res. 168, 396–406 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Moynihan P. J., Clarke A. J., O-acetylated peptidoglycan: Controlling the activity of bacterial autolysins and lytic enzymes of innate immune systems. Int. J. Biochem. Cell Biol. 43, 1655–1659 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Schleifer K. H., Kandler O., Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moynihan P. J., Sychantha D., Clarke A. J., Chemical biology of peptidoglycan acetylation and deacetylation. Bioorg. Chem. 54, 44–50 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Bernard E., et al., Characterization of O-acetylation of N-acetylglucosamine: A novel structural variation of bacterial peptidoglycan. J. Biol. Chem. 286, 23950–23958 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brumfitt W., Wardlaw A. C., Park J. T., Development of lysozyme-resistance in Micrococcus lysodiekticus and its association with an increased O-acetyl content of the cell wall. Nature 181, 1783–1784 (1958). [DOI] [PubMed] [Google Scholar]
- 9.Bera A., Biswas R., Herbert S., Götz F., The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect. Immun. 74, 4598–4604 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke A. J., Extent of peptidoglycan O acetylation in the tribe Proteeae. J. Bacteriol. 175, 4550–4553 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johannsen L., Labischinski H., Reinicke B., Giesbrecht P., Changes in the chemical structure of walls of Staphylococcus aureus grown in the presence of chloramphenicol. FEMS Microbiol. Lett. 16, 313–316 (1983). [Google Scholar]
- 12.Swim S. C., Gfell M. A., Wilde C. E. III, Rosenthal R. S., Strain distribution in extents of lysozyme resistance and O-acetylation of gonococcal peptidoglycan determined by high-performance liquid chromatography. Infect. Immun. 42, 446–452 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfeffer J. M., Strating H., Weadge J. T., Clarke A. J., Peptidoglycan O acetylation and autolysin profile of Enterococcus faecalis in the viable but nonculturable state. J. Bacteriol. 188, 902–908 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips D. C., The hen egg-white lysozyme molecule. Proc. Natl. Acad. Sci. U.S.A. 57, 483–495 (1967). [Google Scholar]
- 15.Brumfitt W., The mechanism of development of resistance to lysozyme by some gram-positive bacteria and its results. Br. J. Exp. Pathol. 40, 441–451 (1959). [PMC free article] [PubMed] [Google Scholar]
- 16.Pushkaran A. C., et al., Understanding the structure-function relationship of lysozyme resistance in Staphylococcus aureus by peptidoglycan O-acetylation using molecular docking, dynamics, and lysis assay. J. Chem. Inf. Model. 55, 760–770 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Aubry C., et al., OatA, a peptidoglycan O-acetyltransferase involved in Listeria monocytogenes immune escape, is critical for virulence. J. Infect. Dis. 204, 731–740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez M., et al., O-acetylation of peptidoglycan limits helper T cell priming and permits Staphylococcus aureus reinfection. Cell Host Microbe 22, 543–551.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baranwal G., et al., Impact of cell wall peptidoglycan O-acetylation on the pathogenesis of Staphylococcus aureus in septic arthritis. Int. J. Med. Microbiol. 307, 388–397 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Bera A., Herbert S., Jakob A., Vollmer W., Götz F., Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55, 778–787 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Brott A. S., Clarke A. J., Peptidoglycan O-acetylation as a virulence factor: Its effect on lysozyme in the innate immune system. Antibiotics (Basel) 8, 94 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crisóstomo M. I., et al., Attenuation of penicillin resistance in a peptidoglycan O-acetyl transferase mutant of Streptococcus pneumoniae. Mol. Microbiol. 61, 1497–1509 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Hébert L., et al., Enterococcus faecalis constitutes an unusual bacterial model in lysozyme resistance. Infect. Immun. 75, 5390–5398 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veyrier F. J., et al., De-O-acetylation of peptidoglycan regulates glycan chain extension and affects in vivo survival of Neisseria meningitidis. Mol. Microbiol. 87, 1100–1112 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Fleming T. J., Wallsmith D. E., Rosenthal R. S., Arthropathic properties of gonococcal peptidoglycan fragments: Implications for the pathogenesis of disseminated gonococcal disease. Infect. Immun. 52, 600–608 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blundell J. K., Smith G. J., Perkins H. R., The peptidoglycan of Neisseria gonorrhoeae: O-acetyl groups and lysozyme sensitivity. FEMS Microbiol. Lett. 9, 259–261 (1980). [Google Scholar]
- 27.Wang G., Lo L. F., Forsberg L. S., Maier R. J., Helicobacter pylori peptidoglycan modifications confer lysozyme resistance and contribute to survival in the host. MBio 3, e00409–e00412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gmeiner J., Kroll H.-P., Murein biosynthesis and O-acetylation of N-acetylmuramic acid during the cell-division cycle of Proteus mirabilis. Eur. J. Biochem. 117, 171–177 (1981). [DOI] [PubMed] [Google Scholar]
- 29.Gmeiner J., Sarnow E., Murein biosynthesis in synchronized cells of Proteus mirabilis. Quantitative analysis of O-acetylated murein subunits and of chain terminators incorporated into the sacculus during the cell cycle. Eur. J. Biochem. 163, 389–395 (1987). [DOI] [PubMed] [Google Scholar]
- 30.Lear A. L., Perkins H. R., O-acetylation of peptidoglycan in Neisseria gonorrhoeae. Investigation of lipid-linked intermediates and glycan chains newly incorporated into the cell wall. J. Gen. Microbiol. 132, 2413–2420 (1986). [DOI] [PubMed] [Google Scholar]
- 31.Snowden M. A., Perkins H. R., Wyke A. W., Hayes M. V., Ward J. B., Cross-linking and O-acetylation of newly synthesized peptidoglycan in Staphylococcus aureus H. J. Gen. Microbiol. 135, 3015–3022 (1989). [DOI] [PubMed] [Google Scholar]
- 32.Jones C. S., Sychantha D., Howell P. L., Clarke A. J., Structural basis for the O-acetyltransferase function of the extracytoplasmic domain of OatA from Staphylococcus aureus. J. Biol. Chem. 295, 8204–8213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sychantha D., Clarke A. J., Peptidoglycan modification by the catalytic domain of Streptococcus pneumoniae OatA follows a ping-pong bi-bi mechanism of action. Biochemistry 57, 2394–2401 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Sychantha D., et al., In vitro characterization of the antivirulence target of gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA). PLoS Pathog. 13, e1006667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higa H. H., Butor C., Diaz S., Varki A., O-acetylation and de-O-acetylation of sialic acids. O-acetylation of sialic acids in the rat liver Golgi apparatus involves an acetyl intermediate and essential histidine and lysine residues–A transmembrane reaction? J. Biol. Chem. 264, 19427–19434 (1989). [PubMed] [Google Scholar]
- 36.Pearson C. R., et al., Acetylation of surface carbohydrates in bacterial pathogens requires coordinated action of a two-domain membrane-bound acyltransferase. MBio 11, e01364–e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kintz E., et al., A BTP1 prophage gene present in invasive non-typhoidal Salmonella determines composition and length of the O-antigen of the lipopolysaccharide. Mol. Microbiol. 96, 263–275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thanweer F., Verma N. K., Identification of critical residues of the serotype modifying O-acetyltransferase of Shigella flexneri. BMC Biochem. 13, 13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bera A., et al., Influence of wall teichoic acid on lysozyme resistance in Staphylococcus aureus. J. Bacteriol. 189, 280–283 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell J., et al., Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 6, 106–116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grassetti D. R., Murray J. F. Jr., Determination of sulfhydryl groups with 2,2′- or 4,4′-dithiodipyridine. Arch. Biochem. Biophys. 119, 41–49 (1967). [DOI] [PubMed] [Google Scholar]
- 42.Moynihan P. J., Clarke A. J., Assay for peptidoglycan O-acetyltransferase: A potential new antibacterial target. Anal. Biochem. 439, 73–79 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Bogdanov M., Zhang W., Xie J., Dowhan W., Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM(TM)): Application to lipid-specific membrane protein topogenesis. Methods 36, 148–171 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexeyev M. F., Winkler H. H., Membrane topology of the Rickettsia prowazekii ATP/ADP translocase revealed by novel dual pho-lac reporters. J. Mol. Biol. 285, 1503–1513 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Bechara C., et al., MALDI-TOF mass spectrometry analysis of amphipol-trapped membrane proteins. Anal. Chem. 84, 6128–6135 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Jones C. S., Anderson A., Clarke A. J., Full-length SaOatA digest raw MS data. Figshare. 10.6084/m9.figshare.13309427. Deposited 30 November 2020. [DOI] [Google Scholar]
- 47.Jones C. S., Anderson A., Clarke A. J., Full-length SaOatA peptide library search results. Figshare. 10.6084/m9.figshare.13309463. Deposited 30 November 2020. [DOI] [Google Scholar]
- 48.Jones C. S., Anderson A., Clarke A. J., Supporting MS2 spectra for identified acetyl-peptides. Figshare. 10.6084/m9.figshare.13309505. Deposited 30 November 2020. [DOI] [Google Scholar]
- 49.Sjodt M., et al., Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex. Nat. Microbiol. 5, 813–820 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riordan J. F., Vallee B. L., [42] o-acetyltyrosine. Methods Enzymol. 25, 500–506 (1972). [DOI] [PubMed] [Google Scholar]
- 51.Saidijam M., Azizpour S., Patching S. G., Comprehensive analysis of the numbers, lengths and amino acid compositions of transmembrane helices in prokaryotic, eukaryotic and viral integral membrane proteins of high-resolution structure. J. Biomol. Struct. Dyn. 36, 443–464 (2018). [DOI] [PubMed] [Google Scholar]
- 52.El Ghachi M., et al., Crystal structure of undecaprenyl-pyrophosphate phosphatase and its role in peptidoglycan biosynthesis. Nat. Commun. 9, 1078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Workman S. D., Worrall L. J., Strynadka N. C. J., Crystal structure of an intramembranal phosphatase central to bacterial cell-wall peptidoglycan biosynthesis and lipid recycling. Nat. Commun. 9, 1159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dutzler R., Campbell E. B., MacKinnon R., Gating the selectivity filter in ClC chloride channels. Science 300, 108–112 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Ma D., et al., Crystal structure of a membrane-bound O-acyltransferase. Nature 562, 286–290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee H. J., et al., Structural and kinetic characterizations of the polysialic acid O-acetyltransferase OatWY from Neisseria meningitidis. J. Biol. Chem. 284, 24501–24511 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X. G., Olsen L. R., Roderick S. L., Structure of the lac operon galactoside acetyltransferase. Structure 10, 581–588 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Lück P. C., et al., A point mutation in the active site of Legionella pneumophila O-acetyltransferase results in modified lipopolysaccharide but does not influence virulence. Int. J. Med. Microbiol. 291, 345–352 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Bame K. J., Rome L. H., Acetyl-coenzyme A:α-glucosaminide N-acetyltransferase. Evidence for an active site histidine residue. J. Biol. Chem. 261, 10127–10132 (1986). [PubMed] [Google Scholar]
- 60.Durand S., Feldhammer M., Bonneil E., Thibault P., Pshezhetsky A. V., Analysis of the biogenesis of heparan sulfate acetyl-CoA:α-glucosaminide N-acetyltransferase provides insights into the mechanism underlying its complete deficiency in mucopolysaccharidosis IIIC. J. Biol. Chem. 285, 31233–31242 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bae T., Schneewind O., Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Manoil C., Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 34, 61–75 (1991). [DOI] [PubMed] [Google Scholar]
- 63.Moynihan P. J., Clarke A. J., O-acetylation of peptidoglycan in gram-negative bacteria: Identification and characterization of peptidoglycan O-acetyltransferase in Neisseria gonorrhoeae. J. Biol. Chem. 285, 13264–13273 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw unprocessed files for SaOatA tryptic peptide LC-MS, corresponding Peaks XPro search results, and annotated MS2 spectra for supporting peptides have been deposited to Figshare (46–48). All other study data are included in the main text and/or SI Appendix.