Significance
In floral meristem (FM), the transcription factor (TF) WUS and the secreted peptide CLV3 form a feedback loop to maintain robust stem cell activities. We found that the TF KNU is a temporally regulated repressor of WUS. However, how the spatial expression patterns of CLV3 and WUS are fully extinguished during floral organogenesis is unknown. Here, we show that KNU has position-specific multifunctions that achieve termination of robust FM. Besides repressing WUS in the organizing center, in the central zone, KNU directly represses CLV3 and inhibits WUS from sustaining CLV3. Furthermore, KNU represses CLV1 and disrupts WUS–WUS and WUS–HAM1 interactions. Thus, KNU plays an essential role to promote FM determinacy for the proper formation of floral reproductive organs.
Keywords: floral meristem, CLV3, WUS
Abstract
Floral organs are properly developed on the basis of timed floral meristem (FM) termination in Arabidopsis. In this process, two known regulatory pathways are involved. The WUSCHEL (WUS)-CLAVATA3 (CLV3) feedback loop is vital for the spatial establishment and maintenance of the FM, while AGAMOUS (AG)-WUS transcriptional cascades temporally repress FM. At stage 6 of flower development, a C2H2-type zinc finger repressor that is a target of AG, KNUCKLES (KNU), directly represses the stem cell identity gene WUS in the organizing center for FM termination. However, how the robust FM activity is fully quenched within a limited time frame to secure carpel development is not fully understood. Here, we demonstrate that KNU directly binds to the CLV1 locus and the cis-regulatory element on CLV3 promoter and represses their expression during FM determinacy control. Furthermore, KNU physically interacts with WUS, and this interaction inhibits WUS from sustaining CLV3 in the central zone. The KNU–WUS interaction also interrupts the formation of WUS homodimers and WUS–HAIRYMERISTEM 1 heterodimers, both of which are required for FM maintenance. Overall, our findings describe a regulatory framework in which KNU plays a position-specific multifunctional role for the tightly controlled FM determinacy.
Plant aerial tissues are mainly formed by the shoot apical meristem (SAM) that harbors a stem cell population at its apex defined as the central zone (CZ). Within the CZ, stem cells slowly divide, and the daughter cells are displaced into the lateral flanks of the CZ, forming the peripheral zone (PZ) (1). Cells in the PZ will later develop into lateral organs (i.e., leaves and flowers). Unlike SAM, which is active throughout the entire life of plants, floral meristem (FM) activity is arrested by precisely coordinated developmental programs that secure the accurate formation of floral reproductive organs (2, 3).
The WUSCHEL (WUS) gene functions to establish and maintain the SAM and FM, and it is expressed in the organizing center (OC), located beneath the CZ containing the stem cells (4). The null mutant wus-1 prematurely abolishes the SAM and causes random phyllotaxy, producing a few carpel-less flowers with only one to two stamens (4). The WUS protein has a homeodomain (HD) for DNA binding, two homodimerization domains (HOD), an acidic region, a WUS-box, and an EAR-like motif (5). Through movement from the OC to the overlaying stem cells via plasmodesmata, WUS activates the expression of the stem cell marker gene CLAVATA3 (CLV3) in the CZ (6, 7).
CLV3 encodes a polypeptide that can diffuse from stem cells to the OC (8, 9). CLV3 can be perceived by the receptor complexes, which may be composed of CLV1, CLV2, CORYNE (CRN), BARELY ANY MERISTEMS (BAMs), RECEPTOR-LIKE PROTEIN KINASE 2 (RPK2) or CLAVATA3 INSENSITIVE RECEPTOR KINASES (CIKs) (10), thereby restricting WUS transcription through signaling cascades (1, 2). A recent study showed that various redundant compensation mechanisms may exist for the canonical CLV ligand-receptor signaling system for stem cell activity control among different plant species (11). In Arabidopsis, the CLV3-WUS feedback loop plays the essential role of maintaining stem cell homeostasis in the SAM and FM (1).
In the SAM and FM, HAIRYMERISTEM (HAM) family proteins are involved in the control of stem cell homeostasis through collaboration with WUS (12). The direct interactions between HAM proteins and WUS are required for stem cell maintenance, and the spatial expression of CLV3 is confined by both HAM proteins and WUS (13). Furthermore, a recent study has shown that the HD transcription factors (TFs) WUS and SHOOT MERISTEMLESS (STM) form heterodimers and bind to the CLV3 promoter to enhance stem cell activity (14).
Upon flower development, WUS activates the C-class gene AGAMOUS (AG) together with LEAFY in stage 3 floral buds (15, 16). AG may directly repress WUS by recruiting the Polycomb Repressive Complex 1 (PRC1) factor TERMINAL FLOWER 2 (TFL2) (17) via a chromatin looping mechanism (18). However, overexpression of AG has subtle effects on carpel development in 35S:AG flowers (19), indicating that additional factors may function together with AG for effective FM termination (2).
At floral stage 6, AG directly promotes the activity of a C2H2-type zinc finger protein KNUCKLES (KNU) for direct WUS repression (20–22). KNU initially associates with a histone deacetylase complex for transcriptional repression of WUS (23), and WUS is later further silenced by a KNU-recruited PRC2 complex that deposits trimethylation of lysine 27 of histone H3 (H3K27me3) on WUS chromatin for stable epigenetic silencing (22).
For homeostatic maintenance of stem cell population, there is a reported compensatory mechanism dependent on CLV3 activity for WUS recovery. Even if CLV3 messenger RNA (mRNA) fluctuates from 33 to 320% of the wild-type level, meristem activity can still be regularly maintained (24). Similarly, reduction of WUS mRNA levels in plants with overexpression of ARABIDOPSIS RESPONSE REGULATOR 7 (ARR7) may not lead to noticeable meristem defects (25). These results demonstrate the robustness of the WUS-CLV3 loop that confers the developmental plasticity of meristem tissue upon environmental perturbation (1). We recently found that KNU could repress both WUS and CLV genes (22), hinting at a mechanism for the effective termination of the robust floral stem cells and differentiation control. Therefore, how KNU functions to interrupt the CLV-WUS regulatory loop for timely termination of the FM requires further investigation.
In this study, we present a regulatory framework mediated by KNU for FM determinacy control. KNU plays an essential role in floral stem cells via direct repression of both CLV1 and CLV3 as well as silencing of CLV3 through H3K27me3-mediated epigenetic mechanisms. In addition, KNU physically interacts with WUS, thereby inhibiting WUS from sustaining CLV3. Furthermore, KNU may disrupt the homodimer formation of WUS and heterodimer formation of HAM1–WUS, both of which are required for meristem maintenance (5, 12). Thus, our work provides a tightly controlled mechanism for FM determinacy in which KNU plays a pivotal role via its multiple functions.
Results
Spatial and Temporal Expression of KNU and CLV3.
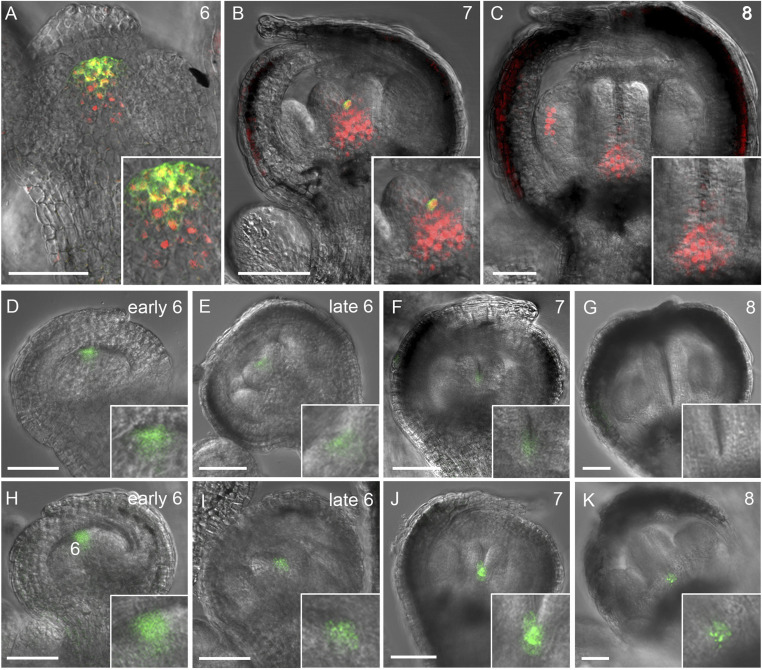
We have previously shown that KNU may repress CLV3 expression in stage 6 floral buds (22). To further analyze this, we first created a line doubly transgenic for pKNU:KNU-VENUS (21) and pCLV3:GFP-ER (26, 27). In a stage 6 floral bud, KNU activity can be detected in the stem cells, where CLV3 is prominently expressed (Fig. 1A and SI Appendix, Fig. S1 A–D). The transient overlap of KNU and CLV3 expression can still be observed in a stage 7 floral bud (Fig. 1B and SI Appendix, Fig. S1 E–H), in which CLV3 activity converges in a few cells at the basal center of two carpel primordia (Fig. 1B and SI Appendix, Fig. S1 H and I). In stage 8 floral buds, CLV3 activity becomes undetectable and stem cell activity ceases, and KNU is actively expressed in the basal center of two carpel primordia and stamen primordia (Fig. 1C and SI Appendix, Fig. S1 J–M). These results indicate that KNU may repress CLV3 cell autonomously, similar to KNU repression of WUS (22).
Fig. 1.
KNU and CLV3 expression patterns. (A–C) Confocal observation of doubly transgenic pKNU:KNU-VENUS (red) and pCLV3:GFP-ER (green) flowers in stage 6 (A), stage 7 (B), and stage 8 (C). (D–K) CLV3 activity in wild-type (D–G) and knu-2 (H–K) floral buds in early stage 6 (D and H), late stage 6 (E and I), stage 7 (F and J), and stage 8 (G and K) (Scale bars, 50 µm). The insets in A–K are the close-up views.
WUS expression is no longer detectable beyond floral stage 6 (SI Appendix, Fig. S1 N–P), while stem cell activity indicated by CLV3 expression is maintained up to late stage 7 (Fig. 1 D–G and SI Appendix, Fig. S1I). In the pCLV3:GFP-ER line, CLV3 activity can be weakly detected in both early and late stage 7 buds (Fig. 1F and SI Appendix, Fig. S1I), demonstrating the prolonged stem cell activity even without WUS after floral stage 6 (SI Appendix, Fig. S1 O and P). By contrast, in flower buds of the knu-2 null mutant, CLV3 activity is strongly expressed in stages 6 to 8 compared to the wild type (Fig. 1 D–K). Consistently, we noticed the prolonged expression of WUS-GFP (green fluorescent protein) in stage 6 to 8 floral buds of knu-2 pWUS:WUS-linker-GFP, in which C terminus of WUS and GFP is spaced by a 30-amino-acid glycine–serine linker and results in a robust activity of the fusion protein (7) (SI Appendix, Fig. S1 Q–S). All these results indicate that KNU activity may be required for the repression of CLV3 to terminate the prolonged floral stem cell activity beyond floral stage 6.
KNU Directly Represses CLV3.
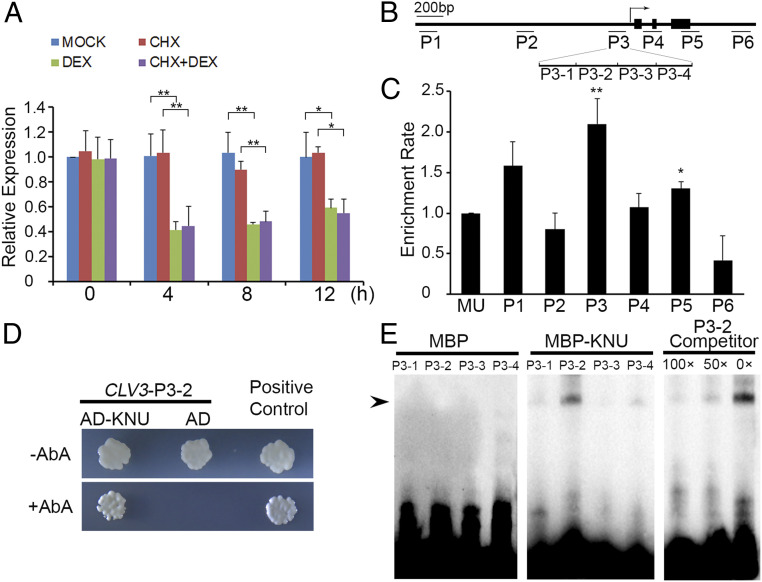
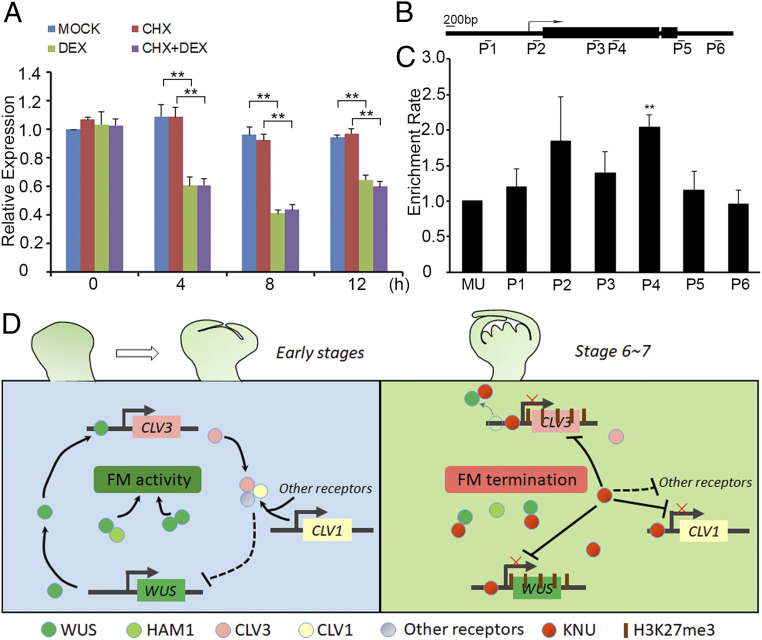
To examine the timing of CLV3 repression by KNU, we used ap1 cal 35S:KNU-GR-myc, a transgenic line enriched for meristematic tissues that produces a fusion protein between KNU and the steroid-binding domain of the rat glucocorticoid receptor (GR) tagged with myc, thereby conferring inducible KNU activity upon dexamethasone (DEX) treatment. We also generated knu-2 35S:KNU-GR-myc, in which the knu-2 phenotype can be rescued by 3 times DEX treatments (SI Appendix, Fig. S2C and Table S1). In ap1 cal 35S:KNU-GR-myc, we observed an ∼60% decrease of the CLV3 transcript level at 4 h relative to the 0-h time point after a single (DEX) treatment (Fig. 2A). There were slight increases of CLV3 mRNA at 8 and 12 h, potentially caused by WUS recovery, although WUS was also repressed by KNU within 4 h (22). Furthermore, we treated ap1 cal 35S:KNU-GR-myc plants with the protein synthesis inhibitor cycloheximide (CHX) as well as CHX combined with DEX. The results showed that CLV3 repression was irrelevant to protein synthesis inhibition (Fig. 2A), hinting at direct repression of CLV3 by KNU.
Fig. 2.
KNU directly represses CLV3. (A) CLV3 expression levels in ap1 cal 35S:KNU-GR-myc after single DEX treatment, CHX treatment, and DEX + CHX treatment. CLV3 transcript levels were quantified by qPCR. Tip41-like gene (At4g34270) served as the internal control. The error bars represent SD of three biological replicates. The asterisks indicate significant differences between samples treated with different chemicals (*P < 0.05 and **P < 0.01, Student’s t test). (B) Schematic diagram of CLV3 locus and primer sets P1 to P6 used for ChIP assays. (C) ChIP assay using ap1 cal 35S:KNU-GR-myc inflorescences. Nuclear proteins were immunoprecipitated with anti–c-Myc agarose beads, and the enriched DNA was used for qPCR assays. The y-axis shows relative enrichment compared with no antibody (negative control). Mu-like transposon (MU) served as a negative control locus, and the values of MU were calibrated to 1. The error bars represent SD of three biological replicates. The asterisks indicate significant differences between MU and different primer sets on CLV3 (*P < 0.05 and **P < 0.01, Student’s t test). (D) Yeast one-hybrid assays show that KNU interacts with the P3 region of CLV3. AbA, Aureobasidin A. (E) EMSAs confirm that KNU binds to the P3-2 fragment. The black arrow indicates the DNA–protein complex. Nonlabeled oligonucleotides were used as competitors. MBP was used as a negative control.
To test whether KNU directly binds to CLV3, we performed chromatin immunoprecipitation (ChIP) assays in ap1 cal 35S:KNU-GR-myc after DEX treatment and noticed that KNU was enriched on the CLV3 proximal promoter in the region from −256 to −62 base pair (bp) upstream of the ATG start codon (primer set P3, SI Appendix, Table S2), with a peak of 2.1-fold enrichment (Fig. 2 B and C and SI Appendix, Fig. S3 A and B). This binding was also confirmed by yeast one-hybrid assays (Fig. 2D) and electrophoretic mobility shift assays (EMSAs) (Fig. 2E). Four fragments within P3 (P3-1 to P3-4) were biotin labeled and incubated with maltose binding protein (MBP)-tagged KNU protein. P3-2 produced a clearly shifted band, which could be significantly weakened when unlabeled competitor probes were added (Fig. 2E). In addition, we synthesized unlabeled competitor probes for P3-2 in five mutated forms (named as M1 to M5) and noticed that the binding was almost unaffected by M4, in which the sequence of AACTATGATA (−174 to −165 bp) was mutated to CCTGGCTGCG (SI Appendix, Fig. S3 C and D). Furthermore, we tested 10 competitor probes (M4-1 to M4-10), each of which had a single-nucleotide change and noticed that the binding was slightly weakened by five probes (M4-1 to M4-4 and M4-6) (SI Appendix, Fig. S3 C and D), indicating that the AACTNT sequence (−174 to −169 bp) is the putative core for KNU binding.
To investigate the binding strength between KNU and P3-2 double-stranded DNA (dsDNA), we expressed and purified the recombinant MBP-tagged KNU (MBP–KNU) in Escherichia coli (SI Appendix, Fig. S3E). We then examined the binding affinity between MBP–KNU and P3-2 dsDNA by size-exclusion chromatography and noticed a clear shift of the peak position expected for protein–DNA complex formation (SI Appendix, Fig. S3F). The results demonstrate that KNU has a strong binding capacity to P3-2 dsDNA to form a stable protein–DNA complex.
As CLV3 chromatin is also modified by PRC2-mediated H3K27me3 repressive mark (28), we checked H3K27me3 levels on the CLV3 locus by using D0 and D4 inflorescence samples (corresponding to synchronized flower buds of stages 1 to 2 and stages 6 to 7) of ap1 cal 35S:AP1-GR after single DEX treatment (22). We noticed that the H3K27me3 level on the CLV3 locus was significantly higher at day 4 compared to day 0 (SI Appendix, Fig. S4 A and B). In addition, in the knu-2 ap1 cal 35S:AP1-GR plants, we only detected a basal level of H3K27me3 repressive mark on the CLV3 locus at D0 and D4 after DEX treatment (SI Appendix, Fig. S4C). These results suggested that the H3K27me3 deposition on CLV3 is also mediated by KNU. As KNU directly interacts with FIE (for FERTILIZATION-INDEPENDENT ENDOSPERM), a core protein of PRC2 which catalyzes H3K27me3, hence KNU recruits PRC2 complex on WUS for H3K27me3-mediated silencing of WUS locus (22). It is possible that KNU may work in the same way for CLV3 silencing.
Effects of KNU Expression in Floral Stem Cells.
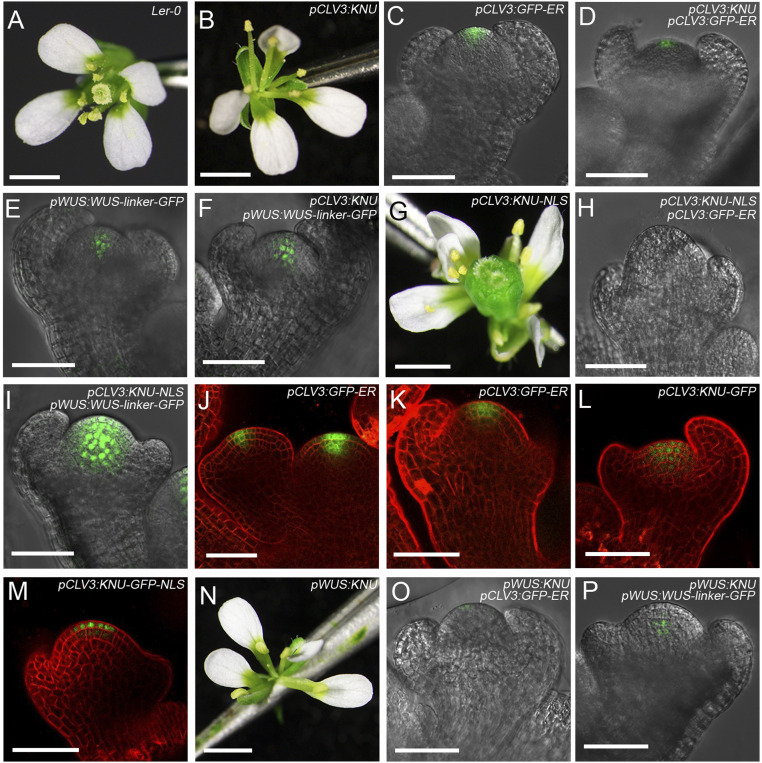
To investigate the role of KNU in floral stem cells in stage 6 (Fig. 1A), we created the line pCLV3:KNU, in which KNU CDS is encompassed by 3.1-kb promoter upstream of CLV3 start codon and 2.2-kb sequence downstream of CLV3 stop codon, both of which fragments contain reported cis-regulatory sites for WUS binding (6). Strikingly, 23 of 113 (20.4%) T1 transgenic plants showed adventitious growth of shoots (SI Appendix, Fig. S5 A–C and Table S1; categorized as a moderate phenotype), generating flowers with reduced stamen numbers and filamentous-like carpels compared to the wild type (Fig. 3 A and B and SI Appendix, Fig. S5D and Table S1). Moreover, 5 of 113 (4.4%) T1 transgenic plants showed severely arrested SAM, resembling the wus-1 mutant (SI Appendix, Fig. S5 A, E, and F and Table S1; categorized as a wus-like phenotype).
Fig. 3.
KNU represses both CLV3 and WUS in the FM. (A) Wild-type flower. (B) pCLV3:KNU flower. (C and D) Expression patterns of CLV3 in stage 3 flower buds of pCLV3:GFP-ER (C) and pCLV3:KNU pCLV3:GFP-ER (D). (E and F) Expression of WUS in stage 3 flower buds of pWUS:WUS-linker-GFP (E) and pCLV3:KNU pWUS:WUS-linker-GFP (F). (G) pCLV3:KNU-NLS flower. (H and I) Expression of CLV3 (H) and WUS (I) in stage 3 flower buds of pCLV3:KNU-NLS pCLV3:GFP-ER and pCLV3:KNU-NLS pWUS:WUS-linker-GFP. (J and K) GFP signal in the SAM (J) and stage 3 flower buds (K) of pCLV3:GFP-ER. (L and M) GFP signals in stage 3 flower buds of pCLV3:KNU-GFP (L) and pCLV3:KNU-GFP-NLS (M). (N) pWUS:KNU flower. (O and P) Expression of CLV3 (O) and WUS (P) in stage 3 flower buds from pWUS:KNU pCLV3:GFP-ER and pWUS:KNU pWUS:WUS-linker-GFP (Scale bars, 1 mm in A, B, G, and N; 50 μm in C–F, H–M, O, and P).
To observe CLV3 and WUS expression, we generated lines of pCLV3:KNU pCLV3:GFP-ER and pCLV3:KNU pWUS:WUS-linker-GFP. Compared to wild-type flowers, CLV3 expression was noticeably weaker in a stage 3 flower bud of pCLV3:KNU pCLV3:GFP-ER plants than in pCLV3:GFP-ER (Fig. 3 C and D and SI Appendix, Fig. S5G). In contrast, WUS expression was slightly higher in pCLV3:KNU pWUS:WUS-linker-GFP than in pWUS:WUS-linker-GFP (Fig. 3 E and F and SI Appendix, Fig. S5G), possibly due to weakened CLV3 activity. Hence, repression of CLV3 in floral stem cells may be an essential function of KNU for FM determinacy.
To test whether stem cell activity is further repressed through enhancing KNU activity by forcing localization of KNU in the nucleus, we generated the line pCLV3:KNU-NLS in which a nuclear localization tag (29) was fused to the C terminus of KNU. Unexpectedly, 133 of 228 (58.3%) pCLV3:KNU-NLS T1 plants produced flowers with more floral organs (3 to 4 carpels and 6 to 7 stamens) compared to the wild type (categorized as a phenotype of enhanced FM; SI Appendix, Fig. S5 A and D and Table S1), and 74 of the 133 plants displayed a clv3-like fasciated inflorescence meristem (SI Appendix, Fig. S5 J and K) and clv3-like flowers (Fig. 3 A and G) with enlarged gynoecia composed of multiple fused carpels (categorized as a clv3-like phenotype) (SI Appendix, Fig. S5 L and M).
We then generated lines of pCLV3:KNU-NLS pCLV3:GFP-ER and pCLV3:KNU-NLS pWUS:WUS-linker-GFP. For pCLV3:KNU-NLS pCLV3:GFP-ER plants, which showed the clv3-like phenotype, the GFP signal under the control of CLV3 promoter was rarely detected in a stage 3 flower bud (Fig. 3H), whereas the WUS-GFP signal became prominently expressed in a larger domain in pCLV3:KNU-NLS pWUS:WUS-linker-GFP than in the wild-type background stage 3 floral bud (Fig. 3 E and I). Both CLV3 and WUS expression were verified by qPCR assays (SI Appendix, Fig. S5G). As endogenous KNU activity is not detectable before floral stage 6 in wild-type background (SI Appendix, Fig. S5H and ref. 20), in a stage 3 floral bud of pCLV3:KNU-NLS, CLV3 promoter–controlled KNU expression may have no effect on the silenced endogenous KNU locus. By qPCR, the endogenous KNU expression level in early stage buds (no later than stage 7) of pCLV3:KNU-NLS is also indistinguishable from wild-type flowers (SI Appendix, Fig. S5I). Thus, the overproliferated floral stem cells in pCLV3:KNU-NLS are due to the strong derepression of WUS, which might be the effect of highly suppressed CLV3 by KNU-NLS. Furthermore, we introduced a pCLV3:KNU-NLS transgene into the weak mutant wus-7 background, and wus-7 pCLV3:KNU-NLS plants and flowers resembled wus-7 (SI Appendix, Fig. S6 A–F), indicating that KNU’s function in floral stem cells is through affecting the CLV3-WUS regulatory pathway.
CLV3 promoter activity has been detected from the L1 to L3 layers in the SAM (30) (Fig. 3J and SI Appendix, Fig. S7 A–F). Here, we also noticed that in pCLV3:GFP-ER flower buds, the CLV3-GFP signal was observed within L1 to L3 of the FM (Fig. 3 J and K and SI Appendix, Fig. S7 D–I). To monitor KNU expression driven by the CLV3 promoter in the FM, we created the line pCLV3:KNU-GFP, and the GFP signal was similarly detected from L1 to L3 layers in the FM of pCLV3:KNU-GFP (Fig. 3L and SI Appendix, Fig. S7 J–L). Like pCLV3:KNU, 18 of 91 (19.8%) of pCLV3:KNU-GFP T1 plants produced flowers with reduced numbers of stamens and carpels (categorized as a moderate phenotype, SI Appendix, Fig. S7M). Also, we generated the line pCLV3:KNU-GFP-NLS (SI Appendix, Fig. S7 N–Q) with enhanced KNU activity in the nucleus, and 43 of 73 (58.9%) of the T1 plants produced flowers with increased floral organ numbers (categorized as enhanced FM, SI Appendix, Fig. S7Q), and the GFP signal was only noticeable in the L1 and L2 layers of the FM in pCLV3:KNU-GFP-NLS (Fig. 3M and SI Appendix, Fig. S7 N–P). For pCLV3:KNU-GFP-NLS plants that produce flowers with variably increased carpel numbers (SI Appendix, Fig. S8 A–C), we noticed a reduced GFP signal in floral buds showing increased FM size that may correspond to flowers with more carpels (SI Appendix, Fig. S8 D and E). Through ChIP assays, we noticed that KNU binding to the CLV3 promoter (peaked at primer sets P1 and P3) was significantly higher in inflorescences of pCLV3:KNU-GFP-NLS than in pCLV3:KNU-GFP (SI Appendix, Fig. S8G). In contrast, the RNA polymerase II (Pol II) level on the CLV3 proximal promoter (primer set P3) was clearly reduced in pCLV3:KNU-GFP-NLS compared to pCLV3:KNU-GFP (SI Appendix, Fig. S8H). All these data suggest that CLV3 promoter activity can be efficiently suppressed by KNU.
To examine the function of KNU in the OC, we also generated pWUS:KNU plants. In T1 plants of pWUS:KNU, 14 of 138 (10.2%) produced flowers with reduced numbers of stamens and carpels (Fig. 3N and SI Appendix, Fig. S9 A–C and Table S1; categorized as a moderate phenotype). Besides, 58 of 138 (42.0%) T1 plants showed a wus-like phenotype, and flowers were rarely produced (SI Appendix, Fig. S9 A and D and Table S1). We also generated lines of pWUS:KNU pCLV3:GFP-ER and pWUS:KNU pWUS:WUS-linker-GFP. In pWUS:KNU floral buds, both CLV3 and WUS expression levels were noticeably reduced compared to the wild type (Fig. 3 C, E, O, and P and SI Appendix, Fig. S9E). These results agree with our previous finding that KNU can directly repress WUS (22).
Since CLV3 and WUS are repressed by KNU in the CZ and OC, respectively, we next crossed pCLV3:KNU (moderate line) with pWUS:KNU (moderate line) to generate pCLV3:KNU pWUS:KNU. Compared to pWUS:KNU (moderate line), 19 of 43 (44.2%) F1 plants of pCLV3:KNU pWUS:KNU showed the stronger phenotype with adventitious growth of stems (SI Appendix, Fig. S9F) that bore few flowers with 1 to 2 stamens but no carpels (SI Appendix, Fig. S9 C and G), reminiscent of wus-1. In addition, 24 of 43 (55.8%) F1 plants resembled wus-1 plants, even without flowering (SI Appendix, Fig. S9H). Thus, the enhanced meristem defects in pCLV3:KNU pWUS:KNU indicate that KNU represses CLV3 and WUS simultaneously in the FM. Also, to reduce KNU activity in the CZ and OC, we generated artificial microRNAs (31) of KNU driven by CLV3 and WUS promoters, respectively. Flowers of both pCLV3:amiR-KNU and pWUS:amiR-KNU plants normally generate three to four carpels compared to the wild type (SI Appendix, Fig. S10 A–D), indicating that compromised KNU expression in either CZ or OC leads to enhanced FM activity. Unlike in knu mutant flowers, reiterated ectopic stamens and carpels inside the primary gynoecium were not observed in either pCLV3:amiR-KNU or pWUS:amiR-KNU flowers (SI Appendix, Fig. S10 E–H), both of which only showed a weak indeterminate FM. Altogether, these results suggest that KNU functions in both the CZ and OC for effective control of FM determinacy.
KNU Functions in Different Stem Cell Layers.
Different stem cell layers are clonally distinct in the SAM and FM, and the epidermal layer cells can generate a mobile signal miR394 that confers the stem cell competence by repressing LEAF CURLING RESPONSIVENESS (LCR) in subtending cells and thus enables CLV3 activation by WUS (32). Previous studies also showed that GIR1 (for GLABRA2-Interacting Repressor 1) is specifically expressed in the L1 layer (33). ARABIDOPSIS THALIANA MERISTEM LAYER 1 (AtML1) is specifically expressed in the epidermal layer of meristems (34), and MEI2 C-TERMINAL RRM ONLY LIKE 1 (MCT1) is expressed in the region including the L1 and L2 layers (33). To test KNU activity in different stem cell layers, we generated a chimeric KNU protein by fusing eGFP (enhanced green fluorescent protein) protein coding sequences with the C terminus of the KNU CDS. The fusion construct was expressed from the native promoters of GIR1, AtML1, and MCT1, respectively, to generate the lines pGIR1:KNU-eGFP, pAtML1:KNU-eGFP, and pMCT1:KNU-eGFP.
In floral buds of the above transgenic lines, GFP signals could be specifically detected in the L1 layer in pGIR1:KNU-eGFP (Fig. 4A), the epidermal layer in pAtML1:KNU-eGFP (Fig. 4B), and the L1 to L2 layers in pMCT1:KNU-eGFP (Fig. 4C). For pGIR1:KNU, we obtained 101 T1 plants, and 23 (22.8%) had flowers with decreased stamen numbers (categorized as a mild phenotype; Fig. 4D and SI Appendix, Fig. S11 A and B and Table S1), suggesting that FM activity is slightly weakened when KNU is specifically expressed in L1 of the FM alone. For pAtML1:KNU, 31 of 87 (35.6%) T1 plants showed reduced stamen numbers (SI Appendix, Fig. S11 A–C, categorized as a mild phenotype; SI Appendix, Table S1), and 3 of 87 plants produced flowers without carpels (Fig. 4E). For pMCT1:KNU, 22 of 122 (18.0%) T1 plants produced flowers with a reduced number of stamens (SI Appendix, Fig. S11 A, B, and D, categorized as a mild phenotype; SI Appendix, Table S1), and 19 of 122 (15.6%) T1 plants produced flowers with filamentous-like carpels (Fig. 4F, categorized as a moderate phenotype; SI Appendix, Table S1). In flower buds of pGIR1:KNU, pAtML1:KNU, and pMCT1:KNU with reduced floral organ numbers, we detected via qPCR reduced CLV3 expression but generally only slightly enhanced WUS expression (SI Appendix, Fig. S11E), as in pCLV3:KNU flowers (SI Appendix, Fig. S5G). These results suggest that KNU activity in each stem cell layer may contribute to FM determinacy. There seems to be a dosage effect in KNU-expressing cells, as fewer stamens and carpels are generally observed in pCLV3:KNU and pMCT1:KNU than in pGIR1:KNU and pAtML1:KNU (SI Appendix, Fig. S11B), hinting at a potential role of KNU in regulating stamen and carpel numbers during flower development.
Fig. 4.
Effects of specific expression of KNU in different stem cell layers. (A–C) GFP signal in stage 3 floral buds of pGIR1:KNU-eGFP (A), pAtML1:KNU-eGFP (B), and pMCT1:KNU-eGFP (C). (D–F) Flowers of pGIR1:KNU (D), pAtML1:KNU (E), and pMCT1:KNU (F) (Scale bars, 50 μm in A–C; 1 mm in D–F).
In pGIR1:KNU, pAtML1:KNU and pMCT1:KNU, KNU expression domains all overlap with the L1 layer that generates miR394 (32). To test whether KNU affects miR394 expression for FM regulation, we used ap1 cal 35S:KNU-GR-myc inflorescences with a single DEX treatment. However, qPCR assays did not detect noticeable changes in MIR394B mRNA levels at 4 or 8 h compared to 0 h (SI Appendix, Fig. S11F), suggesting that KNU activity in L1 contributes to FM determinacy independent of miR394 signaling.
KNU Physically Interacts with WUS.
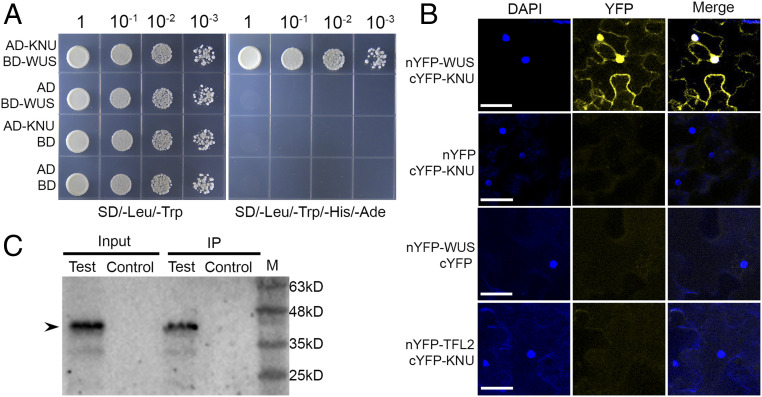
Because both KNU and WUS are expressed in floral stem cells (SI Appendix, Fig. S12 A–D) and have opposite effects on CLV3 expression (Fig. 2) (6, 7), we tested the possibility that KNU can physically interact with WUS using a yeast two-hybrid assay (Y2H). Using the KNU full-length coding sequence as a prey indicated an interaction between KNU and the full-length WUS protein (Fig. 5A), and no interaction was detected between KNU and CLV3 by Y2H (SI Appendix, Fig. S12E).
Fig. 5.
KNU physically interacts with WUS. (A) Y2H using full-length KNU and WUS. Transformed yeast cells were grown on media lacking leucine and tryptophan (SD/−Leu/−Trp) and lacking leucine, tryptophan, histidine, and adenine (SD/−Leu/−Trp/−His/−Ade). AD or BD refers to empty only. (B) Bimolecular fluorescent complementation analysis in tobacco leaves. Merge refers to merged images for yellow fluorescent protein (YFP) and DAPI fluorescence. WUS and KNU were fused to nYFP and cYFP to generate nYFP–WUS and KNU–cYFP, respectively. No interaction between TFL2 and KNU was detected, and this served as a negative control (Scale bars, 50 μm). (C) Co-IP assay. Nuclear extracts were incubated with anti–c-Myc agarose beads. The co-IPed KNU fusion protein (arrowhead) was detected by anti-GFP antibody. IP represents immunoprecipitation. Test and control represent samples from stage 6 flower buds of ap1 cal 35S:AP1-GR pKNU:KNU-VENUS pWUS:WUS-myc and ap1 cal 35S:AP1-GR, respectively. M represents protein marker.
To verify the Y2H result, we performed bimolecular fluorescent complementation analysis in tobacco (Nicotiana tabacum) leaves and noticed an in vivo interaction between KNU and WUS in the nucleus (Fig. 5B). In contrast, TFL2 as a negative control did not show an in vivo interaction with KNU (Fig. 5B). Furthermore, we carried out a coimmunoprecipitation (co-IP) analysis of nuclear extracts from stage 6 flowers of ap1 cal 35S:AP1-GR pKNU:KNU-VENUS pWUS:WUS-myc, and the co-IP results confirmed the in vivo interaction between KNU and WUS (Fig. 5C and SI Appendix, Fig. S12F). All these results indicated that KNU could physically interact with WUS.
Next, we tested which domains of KNU and WUS were required for their interaction using Y2H assays. The KNU protein has the C2H2 domain and a C-terminal EAR-like motif (35) (SI Appendix, Fig. S13A). Deleting the N-terminal fragment of KNU including the C2H2 domain (amino acids 1 to 100) abolished the interaction of KNU with WUS. In contrast, deleting the C-terminal domain of KNU (amino acids 101 to 161) harboring the EAR-like motif had no effect on the KNU–WUS interaction. In addition, the truncated KNU protein without the C2H2 domain (amino acids 40 to 60) failed to interact with WUS (SI Appendix, Fig. S13 A and B).
The WUS protein consists of the HD domain, the HOD domain, the HBD domain, the acidic region, a WUS-box, and an EAR-like motif (5) (SI Appendix, Fig. S13A). The Y2H results showed that truncation of the WUS C-terminal fragment (amino acids 237 to 292) consists of a partial acidic region; the WUS box and the EAR-like motif do not affect the KNU–WUS interaction (SI Appendix, Fig. S13B). Lack of the region consisting of HD, HOD, and HBD (amino acids 1 to 236) abolished WUS binding with KNU (SI Appendix, Fig. S13B). In addition, three fragments, the N-terminal region containing HD and HOD1 domain (amino acids 1 to 133), the HOD2 domain (amino acids 134 to 208), and the HBD domain (amino acids 203 to 236) could interact with KNU independently (SI Appendix, Fig. S13B).
Together, these results indicate that the C2H2 domain of the KNU protein and the HD, HOD, and HBD domains of the WUS protein all contribute to the KNU–WUS interaction.
Effects of KNU–WUS Interaction in the FM.
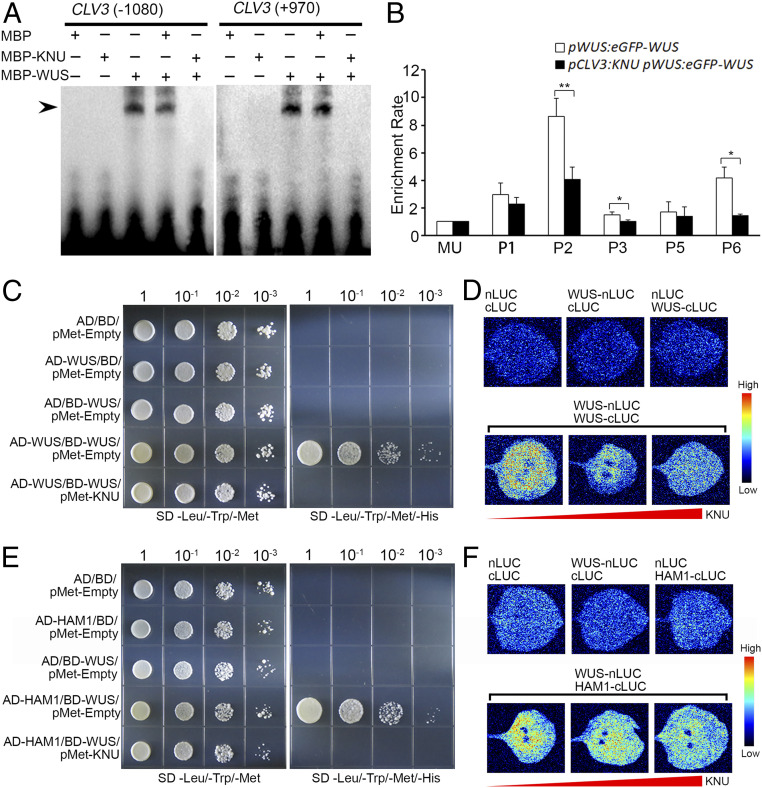
WUS directly binds to the CLV3 locus to activate CLV3 expression in stem cells (6, 30). We wondered whether the KNU–WUS interaction could affect this activation. Two fragments, a 25-bp fragment (−1,090 to −1,066 bp upstream of the ATG start codon) and a 28-bp fragment (+920 bp to +947 bp downstream of the ATG start codon), from the reported WUS-binding sites on CLV3 were used as probes for EMSA experiments (6, 30). We noticed that the binding of WUS to the two fragments of CLV3 was inhibited by the presence of KNU (Fig. 6A).
Fig. 6.
Effects of KNU–WUS interaction. (A) EMSA results showing that KNU inhibits the binding of WUS to CLV3. The black arrow indicates a DNA–protein complex. (B) ChIP assay using early flowers from pWUS:eGFP-WUS and pCLV3:KNU pWUS:eGFP-WUS. Nuclear proteins were immunoprecipitated with anti-GFP antibody, and the enriched DNA was used for qPCR assays. The y-axis shows relative enrichment using no antibody control. MU served as a negative control locus, and the values of MU were calibrated to 1. The error bars represent SD of three biological replicates. The asterisks indicate significant differences between two samples at certain primer sets on CLV3 (*P < 0.05 and **P < 0.01, Student’s t test). (C and D) WUS–WUS interaction is disrupted by KNU in Y3H (C) and BiLC assays (D). (E and F) WUS–HAM1 interaction is disrupted by KNU in Y3H (E) and BiLC assays (F). For Y3H assays, transformed yeast cells were grown on nonselective medium lacking leucine, tryptophan, and methionine (SD/−Leu/−Trp/−Met) and lacking leucine, tryptophan, methionine, and histidine (SD/−Leu/−Trp/−Met/−His) supplemented with 10 mM 3-AT for (C) or 75 mM 3-AT for (E). For BiLC assays, nLUC and cLUC refer to the N-terminal and C-terminal of luciferase, respectively. WUS-nLUC indicates WUS-nLUC fusion; WUS-cLUC indicates WUS-cLUC fusion, and HAM1-cLUC indicates HAM1-cLUC fusion. The color column on the right presents the range of luminescence intensity.
To confirm the EMSA results, we generated the line pCLV3:KNU pWUS:eGFP-WUS and performed ChIP assays by using pCLV3:KNU pWUS:eGFP-WUS and pWUS:eGFP-WUS (6) inflorescences. The ChIP results showed that WUS was enriched on P2 and P6 fragments harboring the aforementioned two reported WUS binding sites in inflorescences of pWUS:eGFP-WUS. In contrast, the enrichment levels were significantly reduced on both P2 and P6 in pCLV3:KNU pWUS:eGFP-WUS (Fig. 6B), showing that the presence of KNU in floral stem cells inhibits the binding of WUS on CLV3.
Studies have shown that CLV3 is activated by a low concentration of WUS that may appear as a monomer and is repressed by high concentrations of WUS that tend to form homodimers (30). The homodimerization of WUS is critical for the maintenance of meristem activity (5, 7). Both the WUS N-terminal containing the HD domain (amino acids 1 to 133) and the HOD2 domain (amino acids 134 to 208) mediate WUS homodimerization (5) and also contribute to the KNU–WUS interaction (SI Appendix, Fig. S13B). Thus, we were curious as to whether WUS homodimer formation could be affected by the presence of KNU. To test this, we carried out yeast three-hybrid assays (Y3H); the results showed that WUS–WUS interaction is disrupted by KNU (Fig. 6C). The formation of WUS homodimers was also interfered with and titrated by KNU in bimolecular luciferase complementation (BiLC) assays in tobacco leaves (Fig. 6D).
Previous studies have shown that the HBD domain (amino acid 203 to 236) of WUS mediates the WUS–HAM interaction that is required for meristem maintenance (12). In our Y2H assay, we noticed that the HBD domain of WUS also contributed to the KNU–WUS interaction (SI Appendix, Fig. S13B). Thus, we wished to know whether KNU could affect WUS–HAM interaction. Our Y3H and BiLC results both indicated that the WUS–HAM1 interaction was disrupted by KNU (Fig. 6 E and F), suggesting that KNU may prevent heterodimer formation by WUS and HAM1. In addition, it was recently shown that the WUS–STM interaction is required for the reinforcement of CLV3 expression in the SAM (14). Thus, we tested whether the WUS–STM interaction could be affected by KNU through Y3H assays. However, both the Y3H and BiLC results showed that KNU did not interfere with the WUS–STM interaction (SI Appendix, Fig. S14). This could be due to that the acidic domain and the adjacent upstream interdomain region of WUS (amino acids 209 to 249) are required for WUS–STM interaction (14), while the region including HD, HOD, and HBD domains (amino acids 1 to 236) of WUS are required for WUS–KNU interaction (SI Appendix, Fig. S13B). It seems that WUS–STM interaction at the acid domain of WUS (amino acids 229 to 249) (14) may not be affected by KNU.
KNU Represses CLV1 and Other CLV Signaling Components.
We previously showed that CLV1 transcripts could also be repressed by KNU (22). To test whether this repression is direct, we used the ap1 cal 35S:KNU-GR-myc plants treated with DEX, CHX, and CHX combined with DEX. We observed an ∼60% decrease of CLV1 transcript level by qPCR at 8 h relative to the 0-h time point after a single (DEX) treatment, and this repression was independent of protein synthesis (Fig. 7A). Furthermore, ChIP assays showed that KNU could directly bind to the CLV1 locus on both the proximal promoter (primer set P2, −167 to −43 bp upstream of the ATG start codon) and the first exon of CLV1 (primer set P4, +1,824 to +1,986 bp downstream of the ATG start codon) (Fig. 7 B and C). The peak binding on P4 was verified by EMSA experiments showing that KNU binding on CLV1 is sequence specific (SI Appendix, Fig. S15A).
Fig. 7.
CLV1 is directly repressed by KNU and the regulatory framework mediated by KNU for FM determinacy. (A–C) KNU directly represses CLV1. (A) CLV1 expression in ap1 cal 35S:KNU-GR-myc after a single DEX treatment, CHX treatment, and DEX + CHX treatment. CLV1 transcript levels were quantified by qPCR. The Tip41-like served as the internal control. The error bars represent SD of three biological replicates. The asterisks indicate significant differences between samples treated with different chemicals (**P < 0.01, Student’s t test). (B) The Schematic diagram of CLV1 locus and primer sets P1 to P6 used for ChIP assays. (C) ChIP assay using ap1 cal 35S:KNU-GR-myc inflorescences. Nuclear proteins were immunoprecipitated with anti–c-Myc agarose beads, and the enriched DNA was used for qPCR assays. The y-axis shows relative enrichment compared with no antibody (negative control). MU served as a negative control locus, and the values of MU were calibrated to 1. The error bars represent SD of three biological replicates. The asterisks indicate significant differences between MU and different primer sets on CLV1 (**P < 0.01, Student’s t test). (D) Model for FM determinacy mediated by KNU in a comprehensive manner. Floral stem cell homeostasis is maintained by the CLV-WUS feedback loop at early stages (before stage 6) in the FM. At early stages, both WUS–WUS homodimers and WUS–HAM1 heterodimers are essential for the activity of the FM. At floral stages 6 and 7, KNU promotes the control of FM activity in multiple ways. First, KNU directly binds to CLV3 promoter and mediates the deposition of repressive mark H3K27me3; meanwhile, KNU inhibits WUS binding to CLV3 by KNU–WUS interaction. Second, both WUS–WUS and WUS–HAM1 interactions are interrupted by KNU. Third, KNU represses CLV1 and other CLV-like receptors as well as several CLEs.
To test the effect of CLV1 repression by KNU, we generated the lines pCLV1:KNU and pCLV1:KNU-eGFP. A clear GFP signal was observed in the FM of early floral buds (SI Appendix, Fig. S15B), and the GFP signal distribution in early FM of pCLV1:KNU-eGFP was similar to a reported expression pattern of the yellow fluorescent protein Ypet driven by the CLV1 promoter (36). For pCLV1:KNU, we noticed that 14 of 91 (15.4%) T1 plants produced flowers with three to four carpels (SI Appendix, Fig. S15 C–E and Table S1), resembling the clv1 mutant (SI Appendix, Fig. S15 F and G; categorized as a clv1-like phenotype). In these plants, we detected reduced CLV1 but significantly increased CLV3 and WUS transcripts by qPCR (SI Appendix, Fig. S15H). In contrast, 19 of 91 (20.9%) pCLV1:KNU T1 plants showed adventitious growth of shoots bearing flowers with filamentous-like carpels (SI Appendix, Fig. S15 I and J; categorized as a moderate phenotype, SI Appendix, Table S1), in which CLV1, CLV3, and WUS were all significantly repressed (SI Appendix, Fig. S15H). In addition, 12 of 91 (13.2%) of pCLV1:KNU T1 plants appeared indistinguishable from the wus-1 mutant (SI Appendix, Fig. S15K and Table S1).
There is compensatory BAM1/2/3 genes expression in SAM and FM when CLV1 activity is compromised (36). So we examined BAM1/2/3 expression by qPCR in pCLV1:KNU and found that BAM1 and BAM3 are slightly up-regulated, and BAM2 remains unchanged in both pCLV1:KNU (moderate) and pCLV1:KNU (clv1-like) flower buds (SI Appendix, Fig. S16A). To test if KNU directly regulates BAM1/2/3, we searched for KNU putative binding element of “AACTNT” on BAM1/2/3 loci and designed primers accordingly for ChIP assays. However, we couldn’t detect obvious enrichment of KNU on BAM1/2/3 (SI Appendix, Fig. S16 B–G), suggesting that KNU may not directly regulate these three genes. There are other reported compensation mechanisms if the function of the ligand–receptor pair of CLV1–CLV3 is compromised (11, 36, 37), so we tested whether KNU may affect the expression of the reported compensatory CLV-like signaling components [i.e., receptor genes of CLV2, CRN, RPK2, BAMs, CIKs, and ligand genes of CLEs (10, 11, 36, 38–40)] by using ap1 cal 35S:KNU-GR-myc inflorescences with a single DEX treatment. At 4 h, we noticed reduced mRNA levels of CLV2, CRN, BAM2, CIK2, and CIK4 as well as several CLEs by qPCR assays (SI Appendix, Fig. S16 H and I). These results suggest that KNU may potentially repress multiple compensatory CLV-like signaling components for effective control of the robust FM activities.
Discussion
The control of FM determinacy requires multiple factors that switch FM activity from a dynamic balance toward timed termination in a programmed manner. Previous studies have shown that the CLV3-WUS feedback loop is robustly maintained in both SAM and FM. Temporal fluctuations of CLV3 concentration do not effectively influence meristem function (24), and strong repression of WUS by overexpression of Type-A ARRs may not lead to SAM defects (25). These findings show the robustness of stem cell niches. Hence, timed FM determinacy control requires a precise regulatory network to arrest floral stem cell activity for proper carpel development. In FM determinacy control, AG can repress WUS from floral stage 3 onward in a mild but direct manner via recruiting TFL2 to the WUS locus (17). From late floral stage 3, the zinc finger protein SUPERMAN (SUP), which defines the boundary between whorls 3 and 4, regulates FM determinacy by directly repressing auxin biosynthesis genes YUC1/4 through interaction with the PRC2 factor CLF and PRC1 factor TFL2 (41). From floral stage 6, CRABS CLAW (CRC), another direct target of AG, is also involved in FM determinacy control by fine-tuning auxin homeostasis and indirectly inhibiting WUS in an auxin-dependent manner (42, 43).
We have shown that in floral stage 6, KNU as a direct target of AG (20, 21) mediates the direct repression and silencing of WUS (22). However, even WUS is terminated in floral stage 6; CLV3 expression can still be detected in both early and late stage 7 floral buds (Fig. 1F and SI Appendix, Fig. S1I), suggesting that the repression of CLV3 occurs later than for WUS. This could be due to the fact that WUS is repressed by multiple pathways and factors including AG, SUP, CRC, and KNU in a programmed manner (2). Another possibility is that floral stem cells can be temporarily maintained from floral stage 6 independently of WUS. A recent study showed that CLV3 is initiated by several WOX genes during embryonic initiation of shoot meristem stem cells, while WUS is dispensable for this process (44). Thus, it is possible that these WOX factors may have functional redundancy to maintain CLV3 in flower development, even if WUS becomes absent from floral stage 6. Therefore, it hints at additional repression mechanisms for floral stem cells. Similarly, during plant senescence, loss of CLV3 expression is also observed later than that of WUS in the SAM (45).
Our EMSA results show that the C2H2-type zinc finger TF KNU directly binds to an AACTNT motif of the CLV3 promoter (Fig. 2 and SI Appendix, Fig. S3 C and D). Di19 (Drought-induced 19), another C2H2-type zinc finger TF, was shown to bind the TACA(A/G)T element (46). Several other abiotic-stress–related C2H2 zinc finger TFs such as AZF1 (Arabidopsis Zinc Finger protein 1), AZF2, AZF3, and ZAT10 (Zinc Finger of Arabidopsis 10), specifically bind to the repeat sequences of A(G/C)T in their target promoters. The DNA sequences of A(AG/CT)CNAC, TGCTANNATTG, and TACAAT motifs are also putative binding elements of C2H2 TFs in plants (47). In one study, CLV3 can also be directly repressed by a TF FHY3 (FAR-RED ELONGATED HYPOCOTYL3) to promote FM determinacy (48). The binding sites on the CLV3 promoter of FHY3 (48) and KNU are overlapped, hinting at potential cross-talk between the two repression mechanisms for CLV3.
We showed that specific expression of KNU in stem cell layers resulted in reduced floral organ numbers (Fig. 4 A–F and SI Appendix, Fig. S11 A and B), which indicate that KNU functions in all stem cell layers for FM determinacy. In addition, our results imply that the upper and lower layers of the FM may be responsible for stamen and carpel formation, respectively (Fig. 4 A–F and SI Appendix, Fig. S11 A and B), agreeing with a previous report that the stamen primordia and the gynoecium primordia mainly originate from L2 cells and L3 cells, respectively (49). Besides, both pWUS:KNU and pCLV3:KNU plants show flowers with filamentous-like carpels and reduced stamen numbers, and both pWUS:amiR-KNU and pCLV3:amiR-KNU produce flowers with three to four carpels. Hence, the repressor activity of KNU in both CZ and OC contributes to the robust control of FM determinacy.
In our study, we noticed a threshold-dependent effect of KNU for CLV3 repression. In pGIR1:KNU, pAtML1:KNU, pMCT1:KNU, and pCLV3:KNU plants that produced flowers with reduced floral organ numbers (Figs. 3B and 4 D–F), we observed decreased CLV3 expression but generally slightly increased WUS expression in floral buds (Fig. 3 C–F and SI Appendix, Fig. S11E). These could be due to WUS–WUS interaction required for FM maintenance (5, 12) being disrupted by KNU in stem cell layers. For pCLV3:KNU-NLS plants that produced flowers with more floral organs due to higher KNU protein levels in the nucleus, we noticed that CLV3 expression was barely detectable, while the derepressed WUS expression domain was greatly expanded in the FM (Fig. 3 H and I). Thus, it seems that the change in the relative ratio of CLV3 and WUS levels can trigger a shift of stem cells from differentiation to proliferation, hinting at an unknown mechanism for control of stem cell homeostasis. Meanwhile, we found that KNU could repress CLV3 promoter activity. In pCLV3:GFP-ER plants, the GFP signal was observed from L1 to L3 of FM (Fig. 3K and SI Appendix, Fig. S7 D–I). In contrast, in pCLV3:KNU-GFP-NLS flowers, the GFP signal was only detectable in L1 to L2 (Fig. 3M and SI Appendix, Fig. S7 N–P). These results suggest that the CLV3 promoter activity is further repressed when KNU protein concentration becomes higher in the nucleus. For pCLV3:KNU plants, the clv3-like phenotype was not observed, but inflorescences and flowers resembling the weak wus mutant were produced. This phenotype is in contrast to pCLV3:HECATE1(HEC1) plants that produce clv3-like inflorescences due to simultaneous repression of WUS and CLV3 by the TF HEC1, which could uncouple stem cell fate from CLV3 expression and stimulate cell proliferation independent of WUS (50).
Our Y3H and BiLC results show that KNU can disrupt WUS–WUS interaction and WUS–HAM1 interaction (Fig. 6 C–F), both of which are required for FM maintenance (5, 12). Similarly, a previous study showed that protein interactions among CUP SHAPED COTYLEDON (CUC) TFs such as formation of CUC2–CUC2 homodimers and CUC2–CUC3 heterodimers can be disrupted by the TF TEOSINTE BRANCHED1/CYCLOIDEA/PCF 4 (TCP4) in preventing the formation of serrations in leaflets of Arabidopsis (51). Thus, KNU activities in disruption of the WUS–WUS interaction and the WUS–HAM1 interaction suggest another tier of regulation for FM determinacy. This may account for the phenotypic differences between pCLV3:KNU and pCLV3:KNU-NLS flowers. For pCLV3:KNU, KNU is expressed in L1 to L3 (Fig. 3L), partially overlapping with OC, where KNU may disrupt WUS–WUS and WUS–HAM1 interactions that are required for FM maintenance (5, 12). Therefore, pCLV3:KNU produces flowers with reduced floral organ numbers (Fig. 3B). In contrast, for pCLV3:KNU-NLS, KNU is mainly expressed in L1 (Fig. 3M), where KNU may have little effect on WUS–HAM1 interaction. Thus, in pCLV3:KNU-NLS, KNU may mainly function to repress CLV3 intensively (Fig. 3H and SI Appendix, Fig. S5G), thereby leading to the greatly up-regulated WUS expression level and expanded WUS expression domain which result in clv3-like flowers (Fig. 3 G and I).
In pCLV1:KNU-eGFP floral buds, KNU-eGFP signal can be observed in both stem cells and OC (SI Appendix, Fig. S15B), thereby the transgene activity may also repress CLV3 and WUS. In agreement with this, in pCLV1:KNU (moderate) flowers, we detected obvious repression of CLV1, CLV3, and WUS (SI Appendix, Fig. S15H), and these lead to reduced FM activity and filamentous-like carpels (SI Appendix, Fig. S15J), while in pCLV1:KNU (clv1-like) flowers, reduced CLV1 expression but obviously increased CLV3 and WUS expression were observed (SI Appendix, Fig. S15H). These unlikely could be due to the suppression of CLV1 alone by KNU, but a complex feedback regulation may exist, which is worth further investigation.
Altogether, KNU plays a comprehensive role in terminating floral stem cell activity via multiple modes (Fig. 7D), including repressing and silencing both WUS and CLV3, repressing CLV1 and other CLV-like signaling components, inhibiting WUS from sustaining CLV3 expression, and preventing the stem cell maintenance by disrupting WUS–WUS and WUS–HAM1 interactions. All these functions of KNU contribute to controlling FM determinacy, thereby guaranteeing the proper formation of floral reproductive organs.
Materials and Methods
All plants were grown in soil and maintained in a greenhouse at 22 °C under continuous light. Standard molecular biology and genetic methods were used for vector construction and for crossing and plant transformation. Confocal imaging, quantitative real-time PCR, EMSA experiments and ChIP assays were performed as previously described (22). Plant phenotypic statistics are listed in SI Appendix, Table S1. Primers used in this study are listed in SI Appendix, Table S2. Detailed results of statistical analyses are available in SI Appendix, Table S3. All the details are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jan U. Lohmann for the pWUS:WUS-linker-GFP seeds, G. Venugopala Reddy for the pWUS:eGFP-WUS and pCLV3:GFP-ER seeds, Limin Pi for the wus-7 seeds, and Jiawei Wang for BiLC vectors. This work was supported by the National Natural Science Foundation of China (32070200 and 31670308 to B.S.), the Fundamental Research Funds for the Central Universities (0208/14380167 to B.S.), and by Grant-in-Aid for Scientific Research A (Grant No. 20H00470 to T.I.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102826118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Han H., Liu X., Zhou Y., Transcriptional circuits in control of shoot stem cell homeostasis. Curr. Opin. Plant Biol. 53, 50–56 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Shang E., Ito T., Sun B., Control of floral stem cell activity in Arabidopsis. Plant Signal. Behav. 14, 1659706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Y., Yamaguchi N., Gan E.-S., Ito T., When to stop: An update on molecular mechanisms of floral meristem termination. J. Exp. Bot. 70, 1711–1718 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Mayer K. F., et al., Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez K., et al., DNA-dependent homodimerization, sub-cellular partitioning, and protein destabilization control WUSCHEL levels and spatial patterning. Proc. Natl. Acad. Sci. U.S.A. 113, E6307–E6315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yadav R. K., et al., WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 25, 2025–2030 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daum G., Medzihradszky A., Suzaki T., Lohmann J. U., A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 14619–14624 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo T., et al., A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845–848 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y., A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5, 578–580 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Hu C., et al., A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants 4, 205–211 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez-Leal D., et al., Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat. Genet. 51, 786–792 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y., et al., Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517, 377–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y., et al., HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers. Science 361, 502–506 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su Y. H., et al., Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U.S.A. 117, 22561–22571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenhard M., Bohnert A., Jürgens G., Laux T., Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105, 805–814 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Lohmann J. U., et al., A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Liu X., et al., AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23, 3654–3670 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo L., et al., A chromatin loop represses WUSCHEL expression in Arabidopsis. Plant J. 94, 1083–1097 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Mizukami Y., Ma H., Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell 9, 393–408 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun B., Xu Y., Ng K. H., Ito T., A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 23, 1791–1804 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun B., et al., Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 343, 1248559 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Sun B., et al., Integration of transcriptional repression and polycomb-mediated silencing of WUSCHEL in floral meristems. Plant Cell 31, 1488–1505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bollier N., et al., At-MINI ZINC FINGER2 and Sl-INHIBITOR of MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in Arabidopsis and tomato. Plant Cell 30, 83–100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller R., Borghi L., Kwiatkowska D., Laufs P., Simon R., Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell 18, 1188–1198 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leibfried A., et al., WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438, 1172–1175 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Reddy G. V., Meyerowitz E. M., Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science 310, 663–667 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Yadav R. K., Tavakkoli M., Reddy G. V., WUSCHEL mediates stem cell homeostasis by regulating stem cell number and patterns of cell division and differentiation of stem cell progenitors. Development 137, 3581–3589 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Zhang X., et al., Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5, e129 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rihs H.-P., Jans D. A., Fan H., Peters R., The rate of nuclear cytoplasmic protein transport is determined by the casein kinase II site flanking the nuclear localization sequence of the SV40 T-antigen. EMBO J. 10, 633–639 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perales M., et al., Threshold-dependent transcriptional discrimination underlies stem cell homeostasis. Proc. Natl. Acad. Sci. U.S.A. 113, E6298–E6306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D., Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18, 1121–1133 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knauer S., et al., A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 24, 125–132 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Yadav R. K., Girke T., Pasala S., Xie M., Reddy G. V., Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. U.S.A. 106, 4941–4946 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu P., Porat R., Nadeau J. A., O’Neill S. D., Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8, 2155–2168 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Payne T., Johnson S. D., Koltunow A. M., KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131, 3737–3749 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Nimchuk Z. L., Zhou Y., Tarr P. T., Peterson B. A., Meyerowitz E. M., Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142, 1043–1049 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diss G., Ascencio D., DeLuna A., Landry C. R., Molecular mechanisms of paralogous compensation and the robustness of cellular networks. J. Exp. Zoolog. B Mol. Dev. Evol. 322, 488–499 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Kayes J. M., Clark S. E., CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125, 3843–3851 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Müller R., Bleckmann A., Simon R., The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20, 934–946 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinoshita A., et al., RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137, 3911–3920 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Xu Y., et al., SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. EMBO J. 37, e97499 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaguchi N., Huang J., Xu Y., Tanoi K., Ito T., Fine-tuning of auxin homeostasis governs the transition from floral stem cell maintenance to gynoecium formation. Nat. Commun. 8, 1125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi N., et al., Chromatin-mediated feed-forward auxin biosynthesis in floral meristem determinacy. Nat. Commun. 9, 5290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z., Tucker E., Hermann M., Laux T., A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev. Cell 40, 264–277.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Wang Y., et al., Morphological and physiological framework underlying plant longevity in Arabidopsis thaliana. Front Plant Sci 11, 600726 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu W. X., et al., Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol. Plant 6, 1487–1502 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Han G., et al., C2H2 zinc finger proteins: Master regulators of abiotic stress responses in plants. Front Plant Sci 11, 115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li D., et al., FAR-RED ELONGATED HYPOCOTYL3 activates SEPALLATA2 but inhibits CLAVATA3 to regulate meristem determinacy and maintenance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, 9375–9380 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jenik P. D., Irish V. F., Regulation of cell proliferation patterns by homeotic genes during Arabidopsis floral development. Development 127, 1267–1276 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Schuster C., et al., A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev. Cell 28, 438–449 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Rubio-Somoza I., et al., Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr. Biol. 24, 2714–2719 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.