Significance
Accumulation of DNA variants is considered a cause of organ dysfunction during natural aging. Here, we directly assessed the frequency of DNA damage of the murine myocardium during natural aging and in hearts from four mouse models of premature aging. In contrast to the prevailing dogma, our results provide no evidence for an enrichment in variants in the naturally aging heart until 2 y of age in Hutchinson–Gilford Progeria Syndrome hearts or in hearts with reduced telomere lengths. Accumulation of randomly distributed variants was, however, evident in Ercc1 knockout mice with deficient DNA repair machinery and in the intestine, liver, and lung of naturally aging mice. These results launch avenues to dissect the fundamental biological processes underlying cardiac aging.
Keywords: genomic instability, aging, DNA repair, oxidative stress, RNA-seq
Abstract
Genomic instability, the unresolved accumulation of DNA variants, is hypothesized as one of the contributors to the natural aging process. We assessed the frequency of unresolved DNA damage reaching the transcriptome of the murine myocardium during the course of natural aging and in hearts from four distinct mouse models of premature aging with established aging-related cardiac dysfunctions. RNA sequencing and variant calling based on total RNA sequencing was compared between hearts from naturally aging mice, mice with cardiomyocyte-specific deficiency of Ercc1, a component of the DNA repair machinery, mice with reduced mitochondrial antioxidant capacity, Tert-deficient mice with reduced telomere length, and a mouse model of human Hutchinson–Gilford progeria syndrome (HGPS). Our results demonstrate that no enrichment in variants is evident in the naturally aging murine hearts until 2 y of age from the HGPS mouse model or mice with reduced telomere lengths. In contrast, a dramatic accumulation of variants was evident in Ercc1 cardiomyocyte-specific knockout mice with deficient DNA repair machinery, in mice with reduced mitochondrial antioxidant capacity, and in the intestine, liver, and lung of naturally aging mice. Our data demonstrate that genomic instability does not evidently contribute to naturally aging of the mouse heart in contrast to other organs and support the contention that the endogenous DNA repair machinery is remarkably active to maintain genomic integrity in cardiac cells throughout life.
As Cicero stated “Senectus autem aetatis est peractio tanquam fabulae” (old age the crown of life, our play's last act) in this famous aphorism from his philosophical work De senectute, age is one of the most familiar phenomena that every person will unmistakably face. Yet, aging remains a poorly understood aspect of human biology; almost every significant discovery in the history of cellular and molecular biology has led to the development of a new theory of aging, and when Medvedev in the ’90s tried to give a comprehensive overview on the subject, he had to face a list of more than 300 different and sometimes opposite theories (1). Nowadays, aging is regarded as a complex process resulting from the synergistic contribution of several causes, such as epigenetic, proteotoxic, and oxidative stresses, telomere shortening, and unresolved DNA damages (2–5). The growing number of alterations of the molecular structures inside the cells gradually hamper key functions and eventually provoke effects at the organ and organism level, the manifestation of which is aging (6).
DNA is susceptible to environmental and intracellular assaults, including ultraviolet (UV) radiations, tobacco smoke, oxidative damage, and errors introduced by DNA replication. These factors can cause oxidation or deamination of specific bases or the introduction of single- and double-strand breaks followed by genome rearrangements. To ensure genomic stability, an intricate machinery of repair, damage tolerance, and checkpoint pathways has evolved that can ultimately determine the survival of the cell or its replicative senescence and death (5).
Cardiovascular diseases (CVD), such as heart failure, atherosclerosis, and myocardial infarction, are commonly considered as aged-related pathological conditions: in fact, both the prevalence and incidence of these illnesses are higher among the elderly (7). With increasing age, the cardiovascular system gradually undergoes a series of functional, structural, and cellular changes that contribute to CVD occurrence, including alterations in the left ventricular (LV) systolic and diastolic function, increased incidence of sinus dysfunctions, myocardial hypertrophy, arterial stiffness, and fibrosis (8–10).
It is generally accepted that age-associated CVDs develop due to the accumulation of the aforementioned cardiovascular risk factors in combination with the genetic make-up of each individual (8–10). On the contrary, very little is known about the molecular pathways underlying these pathologies. Studies have been performed to assess genomic instability during natural cardiac aging as a possible contributor to the development of CVDs. By using transgenic mice, it has been shown that the mutation spectrum in the old heart is characterized by large genome rearrangements rather than point mutations (11, 12).
Here, we addressed whether genomic or transcriptomic instability contributes to myocardial natural aging compared to four models of premature senescence: mice with cardiomyocyte-specific knock-out (KO) of excision repair cross-complementation group 1 (Ercc1), a component of the DNA repair machinery; mice manifesting reduced mitochondrial antioxidant capacity due to apoptosis inducing factor mitochondria associated 1 (Aifm1) haploinsufficiency, a telomerase reverse transcriptase (Tert)-deficient mouse model with reduced telomere length; and finally, a mouse model of human Hutchinson-Gilford progeria syndrome (HGPS). In particular, we focused on the impact that DNA damage accumulation can have on the transcriptomic stability. Surprisingly, via total RNA- and DNA-sequencing (RNA-seq and DNA-seq, respectively) and variant calling analysis, we demonstrated a general transcriptomic stability of hearts of natural and accelerated aging mice reflecting a comparable genomic stability. Our results question the actual decrease in functionality of the DNA repair machinery traditionally considered one of the causes of senescence.
Results
Variant Calling in Natural and Accelerated Aging.
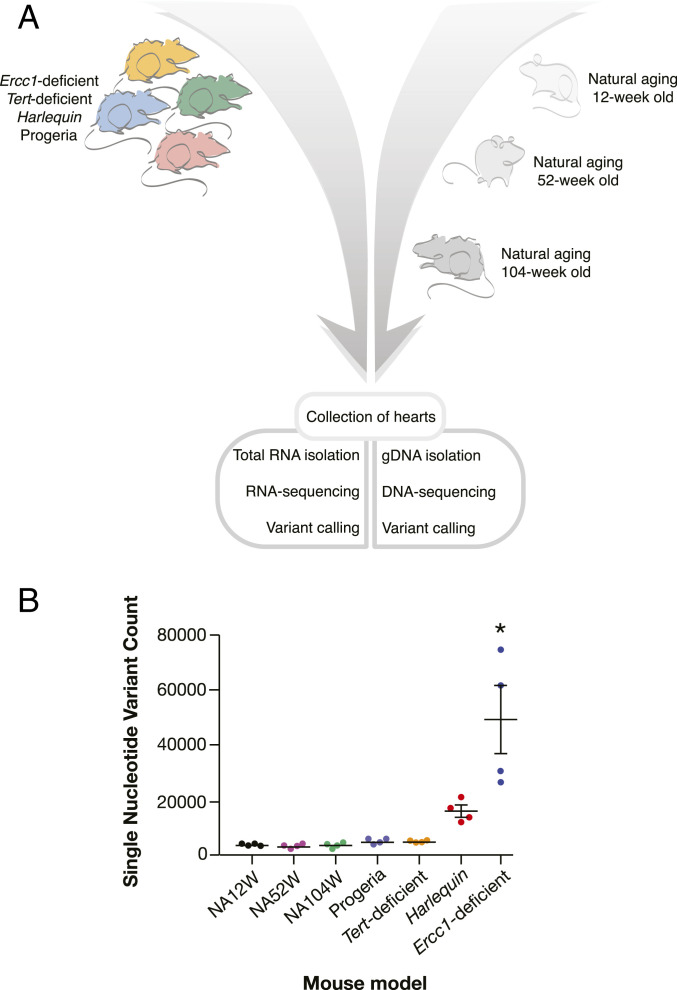
To address whether genomic instability occurs in the progressively aging myocardium, we performed variant calling in RNA-seq data from cardiac tissue of naturally aging mice of 12, 52, and 104 wk of age. We also included hearts from four different mouse models of accelerated aging: mice with cardiomyocyte specific KO of Ercc1, Tert KO mice, mice harboring the Hq mutation characterized by haploinsufficiency of Aifm1, and LmnaG609G/G609G mice geno- and phenocopying the point mutation found in HGPS patients (Fig. 1A). The (prematurely) aged models differentially manifested spontaneous cardiac phenotypes, ranging from no symptoms to severe heart failure (SI Appendix, Fig. 1).
Fig. 1.
Experimental design and variant calling results summary. (A) Timeline of heart tissue collected for subsequent sequencing and variant calling. (B) SNP counts per each of the considered aging mouse models (n = 4 for each model). NA12W, natural aging 12-wk old; NA52W, natural aging 52-wk old; and NA104W, natural aging 104-wk old. Data are represented as mean ± SEM * indicates P < 0.0001 versus NA12W.
When we compared the three distinct points in naturally aging mice against the reference genome GRCm38, remarkably few transcriptomic variants were observed, and the number of variants also did not accumulate in the heart over time, averaging to 3,138 [95% CI (2821;3456)], 2,905 [95% CI (2114;3697)], and 3,220 [95% CI (2071;4368)] at 12, 52, and 104 wk, respectively (SI Appendix, Table 1). Between the different natural aging models, the majority of transcripts expressed was detected at all time points (88 to 92%). Next, we compared the naturally aged mice of 12 wk of age with the four distinct models of premature aging, since this time point in natural aging was the closest to the maximal life span of the other models (Fig. 1A). The accelerated aging models fell into two categories. In the first category, the Hq and Ercc1-deficient models accumulated a significant higher number of single-nucleotide variants (SNPs). Although only the Ercc1 model showed clear statistical significance, both models reached counts of no less than five- and 15-fold more variants compared to any time point of the naturally aged hearts, respectively (mean 15,050 ± 2,106 and mean 48,366 ± 11,920 variants; SI Appendix, Table 1). We anticipate that increasing the sample size would show a strong statistical significance for both models. On the other hand, the LmnaG609G/G609G and Tert knockout (G3 with short telomeres) models did not accumulate significantly more SNPs compared to natural aging (mean 4,187 ± 262 and 4,167 ± 115 for LmnaG609G/G609G and Tert knockout mice, respectively). Taken together, the dispersion plot representation of total number of variants in each model essentially demonstrates the absence of genomic instability in all naturally aged models, the LmnaG609G/G609G and the Tert knockout mice. In contrast, in Hq mice with excess mitochondrial reactive oxygen species accumulation and in Ercc1-deficient mice with deficiency in the DNA repair machinery, an increasing number of variants accumulated in their transcriptomes (Fig. 1B).
Variants Accumulate across the Genome.
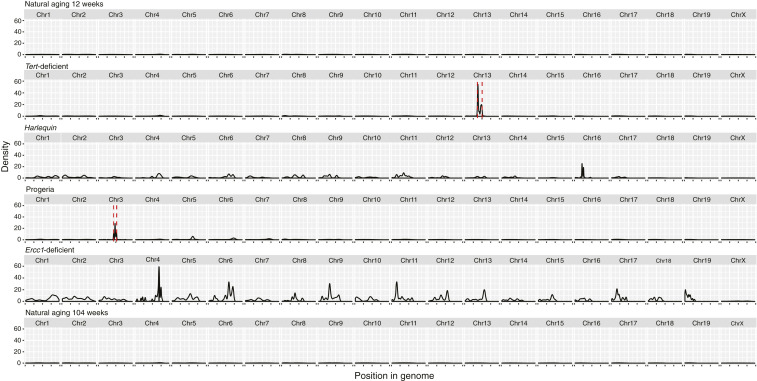
Next, variant distribution was visualized in a density plot across the mouse genome (Fig. 2). The graphical representation of the data demonstrates that the entire genome accumulated mutations roughly evenly distributed in the Hq and in the Ercc1-deficient mice while essentially no variants were observed in all other models.
Fig. 2.
Variant distribution across the transcriptome. Density plots of variants across the transcriptome for each mouse model. The red-dashed lines include the regions affected by the model construction of Tert-deficient (27.3 Mb around Tert gene) and Progeria (18.1 Mb around Lmna gene) and subsequently excluded from further analysis.
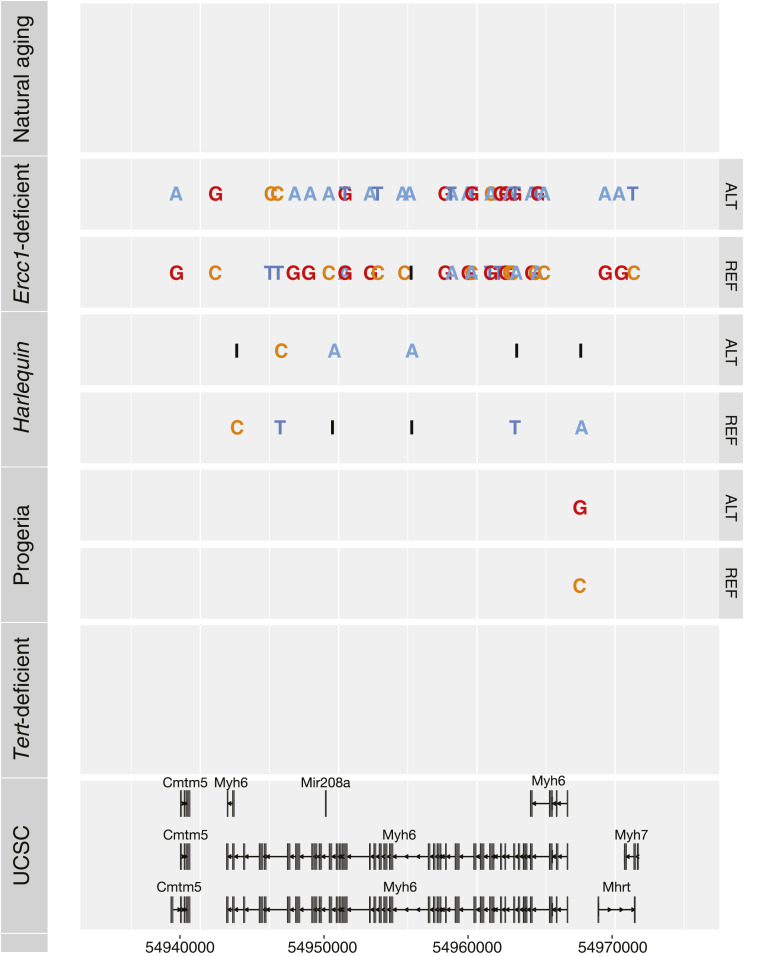
The myocardium contains multiple cell types, such as endothelial cells, fibroblasts, and inflammatory cells in addition to cardiomyocytes. The genetic material derived from cardiomyocytes is proportionally lower than the nonmyocytes fraction and decreases with age (13–15); thus, to specifically assess the genomic stability of cardiomyocytes, we focused on Myh6, a canonical cardiomyocyte-specific gene. The Myh6 gene in natural aging, LmnaG609G/G609G, and Tert KO mice do not show accumulation of mutations compared to the reference genome. A roughly evenly distributed number of variants throughout the genomic locus was observed in the Hq model and Ercc1-deficient mice, even affecting intragenic genes such as miR-208a and the long-noncoding RNA Mhrt (Fig. 3). Conclusively, these results confirm the previously described discordant genomic instability patterns in the distinct mouse models. Interestingly, this trend is confirmed when analyzing genetic loci that are expressed specifically in cardiomyocytes.
Fig. 3.
Variant distribution in the Myh6 locus. Variants accumulating in the Myh6 locus for each model. UCSC, University of California Santa Cruz.
Variant Distribution Pattern in Ercc1-Deficient and Wild-Type Mice Is Similar at the Whole-Genome Level.
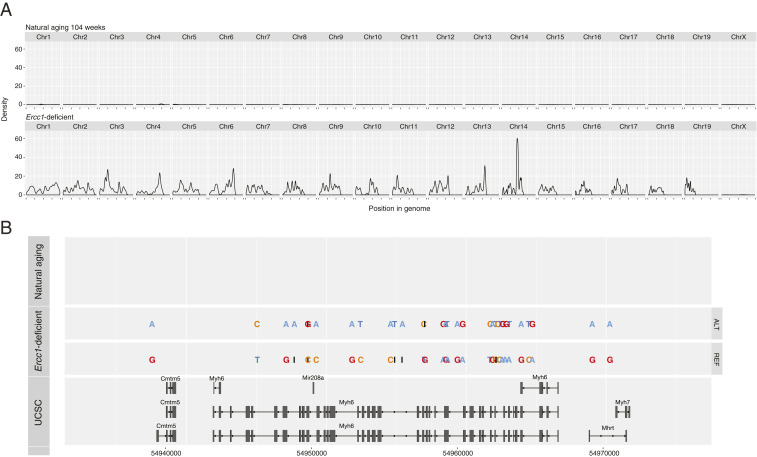
To validate the results obtained from the RNA-seq analysis and to exclude possible bias of the SNPs calling coming from expressed RNA transcripts only, we performed whole-genome sequencing in a heart of naturally aged mice of 104 wk of age and from mice with cardiomyocyte specific Ercc1-deficiency. The naturally aged heart displayed 18,274 variants, the Ercc1-deficient heart 1,492,451. The distribution of the variants across the mouse genome is visualized in a density plot (Fig. 4A). The data confirm that genetic variants are distributed across the genome of Ercc1-deficient mice, while essentially no variants are observed in the naturally aged model. These results are in accordance with the results obtained with RNA-seq, indicating that the differences in mutation rates at the transcriptome level between the aging models reflect differences in the DNA repair machinery efficiency. Variant accumulation was independent of transcriptional or cell cycle activation status (SI Appendix, Figs. 1 and 2). Finally, we also zoomed in to the locus of the cardiomyocyte-specific gene Myh6 (Fig. 4B). While in natural aging mice, no accumulation of variants was observed in Myh6 or any of the intragenic genes such as miR-208a and the long-noncoding RNA Mhrt compared to the reference genome, variants were roughly evenly distributed in Ercc1-deficient mice.
Fig. 4.
Variant distribution across the genome and in the Myh6 locus. (A) Density plot of the variants across the genome of NA104W and Ercc1-deficient mice (n = 1). (B) Visualization of individual DNA variants accumulating in the Myh6 locus of NA104W and Ercc1-deficient mice. The gene structure of the Myh6 locus from the University of California Santa Cruz (UCSC) genome browser is provided in the bottom.
Finally, whole-genome sequencing and variant calling was performed on small intestine, lung, and liver from naturally aged mice of 12 wk and 104 wk of age. Small intestine, lung, and liver are highly proliferative tissues and characterized by spontaneous tumor growth in senescent mice, since the accumulation of unresolved mutations is one of the leading causes of neoplasia. As described previously for liver, lung, and small intestine (16, 17), we do observe a significant accumulation of mutational burden in small intestine, lung, and liver with age (SI Appendix, Table 2) as well as differences for the mutational burden between these organs, proving the sensitivity and specificity of our next-generation sequencing (NGS) approach. Taken together, whole-genome sequencing confirmed the reliability of the variant calling technique and reinforced the conclusion that discordant genomic instability patterns are evident in distinct aging mouse models and among different organs.
Discussion
Here, we investigated the impact of transcriptomic accumulation variants in the aging myocardium. Our main results demonstrate that no enrichment in variants is evident in the naturally aging murine hearts nor in hearts from the LmnaG609G/G609G or the G3 Tert KO models. In contrast, a substantial accumulation of alterations were found in Ercc1 cardiomyocyte-specific knockout mice, as can be expected by the depletion of one of the components of the DNA repair machinery. A similar trend was observed in hearts from the Hq model, characterized by higher oxidative stress sensitivity; however, possibly due to the sample size, this tendency did not reach statistical significance. Whole-genome sequencing results support the striking difference observed between naturally aged and Ercc1 cardiomyocyte-specific knockout mice. Importantly, genomic instability was not correlated with the severity of cardiac phenotypes in the models, since LmnaG609G/G609G mice show no significant accumulation of genomic variants, while Ercc1-deficient mice did show a high mutational burden, even though both models suffer from spontaneous development of heart failure. While we do not observe an accumulation of mutations in the naturally aging heart, we do observe such phenomena in liver, spleen, and intestinal tissues as described previously (16, 17). This clearly supports the notion that different tissues are accumulating mutations at divergent rates and, in fact, that our observation of the absence of such a phenomenon in myocardial tissue is a valid observation.
The first and unexpected outcome of our findings is the conservation of transcriptome integrity in the naturally aged myocardium until 2 y of age, which contradicts previous studies supporting age-related genomic instability as a contributor to cellular loss and overall degeneration of the heart occurring with senescence (11, 12). Past studies used LacZ-transgenic reporter mice to evaluate pattern and frequency of DNA mutations in a particular locus; the integration of a LacZ reporter, though, can change the epigenetic genomic landscape and sensitizes the transgenic genomic region to distorted accumulation of mutations. In contrast, our approach based on total RNA-seq can detect whether DNA variants accumulate anywhere in the coding and noncoding transcriptome. These studies also describe the mutation spectrum in the old murine heart as characterized by large genome rearrangements rather than point mutations. In contrast, we did not detect such large rearrangements in our transcriptome and genome data, suggesting that most genomic deletions do not contribute to transcriptome or genome alterations to a large extent. Notwithstanding the possibility that DNA mutations might still occur and possibly increase with age in genomic regions unrelated to transcriptional control, our data strongly suggest that the DNA repair machinery remains remarkably functional during the physiological stages of cardiac aging.
The adult murine myocardium is composed of ∼30% myocytes, 40% endothelial cells, 15% hematopoietic-derived cells, and 15% fibroblasts (18); the vast majority of cardiomyocytes is postmitotic, binuclear, and withdraw from the cell cycle soon after birth, excluding a very small fraction (<1%) which conserves proliferating capacity as mononuclear cells. With aging, the number of myocytes decreases, and interstitial fibrosis doubles as a reparative mechanism (13–15). Although it could be argued that cardiomyocyte nuclei constitute maximally 50% of myocardial DNA content and genomic instability could be a phenomenon that occurs primarily in nonmyocytes, the transcriptional output of heart muscle cells accounts for the most abundant cardiac RNA transcripts of genes encoding prototypical sarcomere components and ion channels. Therefore, we additionally focused on Mhy6 transcripts encoding the exclusively heart muscle–expressed cardiac alpha-myosin heavy chain and could demonstrate the same pattern of genomic instability—negligible in physiologically aging myocardium and hearts from Tert KO and LmnaG609G/G609G mice and abundant in hearts lacking a component of the DNA repair machinery (Ercc1 knockouts) or exposed to excessive oxidative stress (Hq haploinsufficient mutants). Finally, the prematurely aged Ercc1 knockout mouse model was generated by deleting a floxed allele selectively in heart muscle cells upon activation of Cre recombinase under control of the myosin heavy chain promoter, which is inactive in nonmyocytes postnatally. Accordingly, our data suggest that even in adult postmitotic cardiomyocytes, the DNA repair machinery seems fully operative and able to maintain genomic and transcriptomic stability. Our findings are consistent with the notion that heart muscle cells continuously renew themselves from within and in response to environmental stimuli (such as metabolic and/or hemodynamic stresses) by protein turnover, meaning that if not actively proliferating, this cell type is transcriptionally very active to synthetize and renew large amounts of new protein products. A well-functioning DNA repair machinery becomes therefore essential to avoid permanent alterations of the primary sequence of the genome and chromosomal rearrangements, which would most likely have consequences at the transcriptome and proteome level. In fact, any metabolically active cell, even when not replicating, will require an efficient repair system to preserve the integrity of the DNA, especially of the most-transcribed loci, since transcription itself has the potential to introduce mutations by stimulating loss of heterozygosity and by generating diverse types of rearrangements, such as deletions, duplications, inversions, and translocations (19).
As generally accepted and extensively reported previously (4, 5), our results agree with the postulated premise that excessive oxidative stress can directly cause cardiac DNA damage. Indeed, in the transcriptome of Hq mice, characterized by reduced efficiency of mitochondrial antioxidant activity, an accumulation of variants was detected, albeit not statistically significant with the current number of biological replicates, suggesting that high levels of reactive oxygen species (ROS) can affect genome and transcriptome integrity. Therefore, we were able to provide a proof of the effect of oxidative stress on DNA integrity in the context of a model showing a considerable shortening of lifespan and severe cardiovascular and skeletal muscle pathologies (20), thus correlating genomic instability with oxidative stress, aging, and CVD. Of note, however, our observation that the transcriptome remains stable in naturally aging hearts, in which ROS accumulation is not as dramatic as the one observed in the Hq model, questions whether gradually increasing oxidative stress and concomitant genomic instability plays a direct causative role during physiological cardiac aging. A second and more provocative explanation is that the mechanisms underlying natural myocardial aging in mice and humans are fundamentally too distinct to allow for a direct comparison and the use of the mouse as an experimental model for research in this field. While for humans over 65 y, the most common causes of mortality are CVD, senescent laboratory rodents usually die from neoplasia, with very rare occurrence of cardiovascular abnormalities (21, 22).
In conclusion, our results demonstrate that genomic and transcriptomic instability do not evidently contribute to the natural aging of murine hearts nor to the phenotype of the hearts from a mouse model that faithfully mimics the human HGPS genetic disorder. Only in prematurely aged mouse models with heart-restricted deletion of the DNA repair machinery or severely impaired mitochondrial antioxidant capacity was genomic instability abundant. Our data are in support of a very-active DNA repair machinery to maintain genomic integrity in cardiac cells.
Methods
Mouse Models.
As models for natural aging, male and female wild-type B6129F1 mice of 12-, 52-, and 104-wk-old were included to represent the young (12 wk), adult (52 wk), and old (104 wk) phases of the murine lifespan. Wild-type B6129F1 mice show normal development. In 104-wk-old mice, signs of fibrosis, LV wall thinning, ventricular dilation, and reduced contractility are evident (23).
As a first model for accelerated aging, female mice harboring a null mutation in the Tert gene on a C57BL/6J genetic background and crossbred to the third generation (G3) to achieve critical telomere shortening were used (24, 25). Tert is responsible for a de novo addition of telomeric repeats on chromosomal ends. In mice and humans, telomerase is silenced after birth, leading to progressive telomere shortening throughout the lifespan (26–28). Age-related shortening of telomeres triggers the DNA damage response and the induction of cellular senescence or apoptosis. G3 Tert mice with shortened telomeres secondary to telomerase deficiency only show spontaneous development of mild fibrosis and no other signs of cardiac dysfunction (SI Appendix, Fig. 3).
As a second model for accelerated aging, male mice between 17 and 20 wk of age carrying the Hq mutation on a B6CBACa-Aw-J/A (B6CBA) background were used that are hemizygous for a proviral insertion in the first intron of Aifm1 resulting in an 80% reduction in the expression of this gene (29). Aifm1 is localized at the mitochondrial intermembrane space level with a prosurvival role as free radical scavenger. Hq mice show a wide spectrum of pathological conditions, including neuronal loss, progressive retinal degeneration, ataxia, and reduced lifespan to less than 6 mo. Hq animals display no discernible spontaneous cardiovascular abnormalities. The introduction of cardiac injury by ischemia/reperfusion, however, results in a more severe outcome in Hq mice, with increased cardiomyocyte apoptotic and necrotic death and accelerated progression toward maladaptive LV remodeling (30).
A third progeroid model is the LmnaG609G/G609G knock-in mouse model (31). We used 16-wk-old male mice on a C57BL/6J genetic background carrying a point mutation (c.1827C > T; p.G609G) in the Lmna gene that mimics the human genetic disorder HGPS (32). This mutation results in ubiquitous production of progerin, a mutant form of protein lamin A. Progerin lacks the proteolytic cleavage site required for normal lamin A maturation and stays permanently anchored to the inner part of the nuclear membrane, making it stiffer, fragile, and more sensitive to mechanical stress (33, 34). LmnaG609G/G609G mice display most of the clinical manifestations of HGPS patients, including postnatal growth impairment, lipodystrophy, and reduced survival. Compared with age-matched wild-type mice, 16-wk-old homozygous LmnaG609G/G609G mice present fibrosis and electrocardiographic alterations (lower heart rate, prolonged PQ and QT intervals, and T-wave flattening), preserved systolic function indicated by normal values for both LV and right ventricular ejection fraction (EF), diastolic dysfunction indicated by reduced mitral valve E/A ratio, and increased isovolumetric relaxation time (IVRT) (35). Taken together, these results suggest that LmnaG609G/G609G mice develop heart failure with preserved ejection fraction.
Finally, male and female Ercc1 cardiomyocyte-specific knockout mice of 16 wk of age were used as fourth model of accelerated aging. Mice harboring floxed alleles in Ercc1 were generated in a 129P2/OlaHsd background as described previously (36) and crossbred with mice harboring Cre recombinase under control of the murine Myh6 promoter in a C57BL/6J background (37), resulting in cardiomyocyte-specific Ercc1 gene deletion. Ercc1 is a component of the nucleotide excision repair pathway required for removing UV-induced DNA damages and interstrand crosslinks (38). Mice harboring a homozygous Ercc1 null mutation exhibit impaired growth and several features of premature aging and a markedly reduced lifespan to a maximal age of 8 to 10 wk (39, 40). Cardiomyocyte-specific Ercc1-deficient mice spontaneously develop all characteristics of systolic heart failure with extensive fibrosis, severe reduction of contractile function, and induction of fetal genes, and they succumb to heart failure within a lifespan of 23 wk (SI Appendix, Fig. 1).
All protocols were performed according to institutional guidelines and approved by local Animal Care and Use Committees. Hq mice were housed on a 12-hr light:12-hr dark cycle in a temperature-controlled environment with ad libitum access to water and chow at Innoser Netherlands BV, a commercial mouse breeding company with a quarterly animal health monitoring system that complies with the Federation of European Laboratory Animal Science Associations (FELASA) guidelines and recommendations. All other models were housed at local Institutes in Madrid, Hannover, and Rotterdam on a 12-hr light:12-hr dark cycle in a temperature-controlled environment with ad libitum access to water and chow with animal health monitoring systems that comply with FELASA guidelines and recommendations. Randomization of subjects to experimental groups or data analysis was based on a single sequence of random assignments. Animal caretakers blinded investigators to animal group allocation before NGS sequencing and/or when assessing the outcome.
Transthoracic Echocardiography.
Noninvasive, echocardiographic parameters were measured using a RMV707B (15 to 45 MHz) scan-head interfaced with a Vevo-770 high frequency ultrasound system (VisualSonics). Long-axis electrocardiogram-triggered cine loops of the LV contraction cycle were obtained in B-mode to assess end-diastolic/systolic volume. Short-axis recordings of the LV contraction cycle were taken in M-mode to assess wall thickness of the anterior/posterior wall at the midpapillary level. From B-mode recordings, LV length from basis to apex, LV internal diameter in systole (LVIDs) and diastole (LVIDd) were determined. From M-mode recordings, LV posterior wall thickness in systole (LV PWs) and diastole (LV PWd) were determined. LV mass was calculated with the following formula: (0,8*(1.04*(((LVIDd + LV PWd + IVSd)^3)-((LVIDd)^3))+0,6), and fractional shortening was calculated with the following formula: (LVIDd-LVIDs)/LVIDd*100). EF was calculated as ((SV/Vd)*100) with Vs, systolic volume (3,1416*(LVIDs^3)/6), Vd, diastolic volume (3,1416*(LVIDd^3)/6), and SV, stroke volume (Vd-Vs) (41).
Histological Analysis.
Hearts were arrested in diastole, fixed with 4% paraformaldehyde/phosphate-buffered saline solution, embedded in paraffin, and sectioned at 4 μm. Paraffin sections were stained with Sirius Red for the detection of fibrillar collagen or fluorescein isothiocyanate-labeled antibody against wheat-germ-agglutinin to visualize and quantify the myocyte cross-sectional area (1:100; Sigma-Aldrich). Cell surface areas and fibrotic areas were determined using ImageJ imaging software (https://rsb.info.nih.gov/ij/).
RNA Isolation.
Total RNA was extracted from myocardial tissue of mice euthanized at the timepoints reported above using the Direct-zol RNA MiniPrep method (Thermo Fisher), TriFast (Peqlab), or the RNeasy (Fibrous Tissue) Mini Kit (QIAGEN) including DNase digestion following the manufacturer’s protocol.
qPCR.
Total RNA (1 μg) was reverse-transcribed using hexameric random primers. The housekeeping gene ribosomal protein L7 (Rpl7), Tbp, or Gapdh were used for normalization. Fold changes were determined using the 2-ΔΔCT method. Real-time PCR primer sequences used in the study are: mouse Nppa, 5′-TCTTCCTCGTCTTGGCCTTT-3′ and 3′-CCAGGTGGTCTAGCAGGTTC-5′; mouse Nppb, 5′-TGGGAGGTCACTCCTATCCT-3′ and 3′-GGCCATTTCCTCCGACTTT-5′ or 5′-CTGAAGGTGCTGTCCCAGAT-3′ and 5′-GTTCTTTTGTGAGGCCTTGG-3′; mouse Acta1, 5′-CCGGGAGAAGATGACTCAAA-3′ and 3′-GTAGTACGGCCGGAAGCATA-5′; mouse Myh7, 5′-CGGACCTTGGAAGACCAGAT-3′ and 3′-GACAGCTCCCCATTCTCTGT-5′ or 5′-TCTCCTGCTGTTTCCTTACTTGCT-3′ and 5′-CAGGCCTGTAGAAGAGCTGTACTC-3′.; mouse Rpl7, 5′-GAAGCTCATCTATGAGAAGGC-3′ and 3′-AAGACGAAGGAGCTGCAGAAC-5′; mouse Tbp, 5′-TGGAATTGTACCGCAGCTTCA-3′ and 5′-CTGCAGCAAATCGCTTGGGA-3′; and mouse Gapdh, 5′-TTCACCACCATGGAGAAGGC-3′ and 5′-GGCATGGACTGTGGTCATGA-3′.
DNA Isolation.
Total DNA was extracted from heart tissues of naturally aged mice of 104 wk of age and of mice with cardiomyocyte-specific Ercc1 gene deletion. DNA was extracted using the Quick-DNA Miniprep Plus Kit (Zymo Research) following the manufacturer’s protocol.
RNA-seq.
Quality control of total RNA was performed using the RNA 6000 Pico Kit (Agilent Bioanalyzer) yielding RNA integrity number values of 6.3 and higher. Removal of ribosomal RNA (rRNA) was carried out using the NEBNext rRNA Depletion Kit Human/Mouse/Rat (NEB) followed by strand-specific complementary DNA NGS library preparation (NEBNext Ultra II Directional RNA Library Prep Kit for Illumina, NEB). The size of the resulting library was controlled by use of a D1000 ScreenTape (Agilent 2200 TapeStation) and quantified using the NEBNext Library Quant Kit for Illumina (NEB). Equimolar pooled libraries were sequenced in a paired end mode (75 cycles) on the NextSEq. 500 System (Illumina) using version 2 chemistry yielding in an average QScore distribution of 84% ≥ Q30 score.
DNA-seq.
A total of 500 ng of the genomic DNA was sheared on the S220 instrument (Covaris) followed by NGS library preparation using the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB). The size of the resulting library was controlled by use of a D1000 ScreenTape (Agilent 2200 TapeStation) and quantified using the NEBNext Library Quant Kit for Illumina (NEB). Equimolar pooled libraries were sequenced in a paired-end mode (80 cycles) on the NextSEq. 500 System (Illumina) using version 2 chemistry yielding in an average QScore distribution of 85% ≥ Q30 score. To rule out that we may have missed an accumulation of mutational burden in myocardial tissue (type II error), we sequenced genomic DNA derived from the spleen, liver, and small intestine, in which such an accumulation is well-established through the literature (16, 17), in order to prove that our NGS approach is sensitive enough to pick up the signals.
RNA-seq Variant Calling.
After quality control with FastQC (Andrews, S., 2014, FastQC A Quality Control tool for High Throughput Sequence Data; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), raw sequencing data were read trimmed using trimmomatic (42) and aligned to the mouse reference genome GRCm38 with hisat2 version 2.1.0 (43). After marking duplicate reads, SNPs of the RNA reads were called separately using GATK (44) version 3.7 according to the best practices for RNAsEq. (45, 46) The most crucial step, in which the variant calling for RNA-seq differs from the DNA-seq, is SplitNCigarReads in which the reads are split into their respective exon segments. After calling the variants through Haplotypecaller, they were filtered by Fisher strand values (30) and Quality by depth (2.0). The resulting variants were called on the basis of the reference genome GRCm38; however, the mice sequenced for the models of Hq, Ercc1, and natural aging were hybrid mice of different strains. To adjust for the influence of different strains in the hybrid mice, we removed strain-specific variants from the set of called variants. The strain specific variants for the strains 129S1, 129P2/OlaHsd, and CBA were provided by the Mouse Genomes Project (47). Additionally, we observed a peak of variants in the region around the affected genes in the Tert and Progeria models. Since they were likely introduced by recombination during model construction, they were removed from further analysis. For the Tert model, this is a region of 27.3 Mb surrounding the Tert gene and for the Progeria model it is an 18.1 Mb region around the Lmna gene.
Whole-Genome Variant Calling.
After quality control with FastQC, the raw sequencing data were read-trimmed using trimmomatic and aligned to the mouse reference genome GRCm38 with Burrows–Wheeler Aligner Maximal Exact Match version 0.7.12 (48). In both samples, an average genome-wide coverage > 9 was reached, which we deem sufficient for the validation of the RNA-seq detection approach. After marking duplicate reads and base recalibration, SNPs were called using GATK version 3.7 Haplotypecaller. The resulting SNPs were filtered for Fisher strand values (30) and Quality by depth (2.0). Since the Ercc1-deficient and natural aging models were hybrid mice, the variants were also filtered for strain-specific variants. This resulted in 24076 variants for natural aging and 1687460 for Ercc1-deficient. A technical summary of mapped reads and alignment rates for all sequencing analyses is provided in SI Appendix, Table 3.
Statistical Analysis.
The results are presented as mean ± SD (SD), SEM (SEM), and lower and higher 95% CIs. Statistical approaches for bioinformatics analyses are described above. Normal distribution of the samples was tested using International Business Machines SPSS Statistics software applying the Shapiro–Wilk test. Sample size was determined by a power calculation based upon an echocardiographic effect size. All other statistical analyses were performed using Prism software (GraphPad Software Inc.) and consisted of ANOVA followed by a Bonferroni’s multiple comparison test when group differences were detected at the 5% significance level. Differences were considered significant when P < 0.05.
Supplementary Material
Acknowledgments
F.D.M. is supported by HS-BAFTA and Kootstra fellowships of Maastricht University and a CVON-ARENA-PRIME fellowship. L.M. is supported by the fund Innovative Medical Research of the University of Münster Medical School (RÜ121510). M.R.H. is supported by a Juan de la Cierva contract from the Spanish Ministerio de Ciencia, Innovación y Universidades (IJC2019-040798-I). R.M.N. is the beneficiary of a predoctoral contract from the Spanish Ministerio de Educación, Cultura y Deporte (FPU16/05027). V.A. is supported by the Spanish Ministerio de Ciencia e Innovación (PID2019-108489RB-I00) and the Instituto de Salud Carlos III (ISCIII) (AC16/00091) as member of the ERA-CVD JCT2016 EXPERT Network (European Union's Horizon 2020 Framework Programme), with cofunding from the European Regional Development Fund (“Una manera de hacer Europa”). The Centro Nacional de Investigaciones Cardiovasculares Carlos III (CNIC) is supported by the Ministry for Research, Science and Innovation (MICIN), the ISCIII, the Pro-CNIC Foundation, and is a Severo Ochoa Center of Excellence. C.B. was supported by the Deutsche Forschungsgemeinschaft (DFG) (BA5631/2-1). T.T. was supported by the European Research Council (ERC) Consolidator Grant LONGHEART, by ERA-CVD JCT2016 EXPERT, and the DFG (TH903/22-1). D.J.D., M.S., and L.J.D.W. acknowledge support from the Dutch CardioVascular Initiative: the Netherlands Heart Foundation, Dutch Federation of University Medical Centers, ZonMW, and the Royal Netherlands Academy of Sciences (CVON-ARENA-PRIME, CVON-RACE-V, CVON-PREDICT-2). B.S. acknowledges funding by the Netherlands Heart Foundation (Dr. Dekker 2014T105 and CVON-SHE-PREDICTS-HF) and a VIDI Award 917.14.363 from the Dutch Research Council (NWO). A.S.A. was funded by Association Française contres les Myopathies (AFM 18802). F.D.M., T.T., and L.J.D.W. are supported by ERA-CVD JCT2016 EXPERT. M.S. is funded by the DFG (RTG2220, Project 281125614) and Marie Skłodowska-Curie Grant Agreement 81371. L.J.D.W. was further supported by ERC Consolidator Grant 311549 CALMIRS, a VICI Award 918-156-47 from NWO and Marie Skłodowska-Curie Grant Agreements 813716 and 765274.
Footnotes
Competing interest statement: T.T. is the founder and holds shares of Cardior Pharmaceuticals GmbH. L.J.D.W. is the cofounder and stockholder of Mirabilis Therapeutics BV. All other authors have no conflict of interest to declare.
This article is a PNAS Direct Submission. G.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022974118/-/DCSupplemental.
Data Availability
Anonymized sequencing data have been deposited in Gene Expression Omnibus (GSE124087). All other study data are included in the article and/or SI Appendix.
References
- 1.Medvedev Z. A., An attempt at a rational classification of theories of ageing. Biol. Rev. Camb. Philos. Soc. 65, 375–398 (1990). [DOI] [PubMed] [Google Scholar]
- 2.Childs B. G., Durik M., Baker D. J., van Deursen J. M., Cellular senescence in aging and age-related disease: From mechanisms to therapy. Nat. Med. 21, 1424–1435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cournil A., Kirkwood T. B., If you would live long, choose your parents well. Trends Genet. 17, 233–235 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Hasty P., The impact of DNA damage, genetic mutation and cellular responses on cancer prevention, longevity and aging: Observations in humans and mice. Mech. Ageing Dev. 126, 71–77 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Hoeijmakers J. H., DNA damage, aging, and cancer. N. Engl. J. Med. 361, 1475–1485 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Liochev S. I., Which is the most significant cause of aging? Antioxidants 4, 793–810 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.North B. J., Sinclair D. A., The intersection between aging and cardiovascular disease. Circ. Res. 110, 1097–1108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamczyk M. R., del Campo L., Andrés V., Aging in the cardiovascular system: Lessons from Hutchinson-Gilford progeria syndrome. Annu. Rev. Physiol. 80, 27–48 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Lakatta E. G., Levy D., Arterial and cardiac aging: Major shareholders in cardiovascular disease enterprises: Part I: Aging arteries: A “set up” for vascular disease. Circulation 107, 139–146 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Steenman M., Lande G., Cardiac aging and heart disease in humans. Biophys. Rev. 9, 131–137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busuttil R. A., Dollé M., Campisi J., Vijga J., Genomic instability, aging, and cellular senescence. Ann. N. Y. Acad. Sci. 1019, 245–255 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Dollé M. E., Snyder W. K., Gossen J. A., Lohman P. H., Vijg J., Distinct spectra of somatic mutations accumulated with age in mouse heart and small intestine. Proc. Natl. Acad. Sci. U.S.A. 97, 8403–8408 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee I., Fuseler J. W., Price R. L., Borg T. K., Baudino T. A., Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am. J. Physiol. Heart Circ. Physiol. 293, H1883–H1891 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Lin J., et al., Age-related cardiac muscle sarcopenia: Combining experimental and mathematical modeling to identify mechanisms. Exp. Gerontol. 43, 296–306 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yutzey K. E., Cardiomyocyte proliferation: Teaching an old dogma new tricks. Circ. Res. 120, 627–629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dollé M. E., et al., Increased genomic instability is not a prerequisite for shortened lifespan in DNA repair deficient mice. Mutat. Res. 596, 22–35 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Haines D. C., Chattopadhyay S., Ward J. M., Pathology of aging B6;129 mice. Toxicol. Pathol. 29, 653–661 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Pinto A. R., et al., Revisiting cardiac cellular composition. Circ. Res. 118, 400–409 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim N., Jinks-Robertson S., Transcription as a source of genome instability. Nat. Rev. Genet. 13, 204–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armand A. S., et al., Apoptosis-inducing factor regulates skeletal muscle progenitor cell number and muscle phenotype. PLoS One 6, e27283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snyder J. M., Ward J. M., Treuting P. M., Cause-of-death analysis in rodent aging studies. Vet. Pathol. 53, 233–243 (2016). [DOI] [PubMed] [Google Scholar]
- 22.S. National Center for Health , “Health, United States” in Health, United States, 2016: With Chartbook on Long-term Trends in Health (National Center for Health Statistics [US], Hyattsville, MD, 2017), pp. 10–18. [PubMed] [Google Scholar]
- 23.van Almen G. C., et al., MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell 10, 769–779 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y., Kha H., Ungrin M., Robinson M. O., Harrington L., Preferential maintenance of critically short telomeres in mammalian cells heterozygous for mTert. Proc. Natl. Acad. Sci. U.S.A. 99, 3597–3602 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y., et al., The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr. Biol. 10, 1459–1462 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Canela A., Vera E., Klatt P., Blasco M. A., High-throughput telomere length quantification by FISH and its application to human population studies. Proc. Natl. Acad. Sci. U.S.A. 104, 5300–5305 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Meyer T., et al., Telomere length as cardiovascular aging biomarker: JACC review topic of the week. J. Am. Coll. Cardiol. 72, 805–813 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Vera E., Bernardes de Jesus B., Foronda M., Flores J. M., Blasco M. A., The rate of increase of short telomeres predicts longevity in mammals. Cell Rep. 2, 732–737 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Klein J. A., et al., The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature 419, 367–374 (2002). [DOI] [PubMed] [Google Scholar]
- 30.van Empel V. P., et al., Downregulation of apoptosis-inducing factor in harlequin mutant mice sensitizes the myocardium to oxidative stress-related cell death and pressure overload-induced decompensation. Circ. Res. 96, e92–e101 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Osorio F. G., et al., Splicing-directed therapy in a new mouse model of human accelerated aging. Sci. Transl. Med. 3, 106ra107 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Hamczyk M. R., et al., Vascular smooth muscle-specific progerin expression accelerates atherosclerosis and death in a mouse model of Hutchinson-Gilford progeria syndrome. Circulation 138, 266–282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahl K. N., et al., Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U.S.A. 103, 10271–10276 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noda A., et al., Progerin, the protein responsible for the Hutchinson-Gilford progeria syndrome, increases the unrepaired DNA damages following exposure to ionizing radiation. Genes Environ. 37, 13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fanjul V., et al., Identification of common cardiometabolic alterations and deregulated pathways in mouse and pig models of aging. Aging Cell 19, e13203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doig J., et al., Mice with skin-specific DNA repair gene (Ercc1) inactivation are hypersensitive to ultraviolet irradiation-induced skin cancer and show more rapid actinic progression. Oncogene 25, 6229–6238 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Agah R., et al., Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J. Clin. Invest. 100, 169–179 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryu H., et al., ERCC1 expression status predicts the response and survival of patients with metastatic or recurrent cervical cancer treated via platinum-based chemotherapy. Medicine (Baltimore) 96, e9402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McWhir J., Selfridge J., Harrison D. J., Squires S., Melton D. W., Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat. Genet. 5, 217–224 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Weeda G., et al., Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr. Biol. 7, 427–439 (1997). [DOI] [PubMed] [Google Scholar]
- 41.da Costa Martins P. A., et al., MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nat. Cell Biol. 12, 1220–1227 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenna A., et al., The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DePristo M. A., et al., A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van der Auwera G. A., et al., From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 43, 11.10.1–11.10.33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keane T. M., et al., Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized sequencing data have been deposited in Gene Expression Omnibus (GSE124087). All other study data are included in the article and/or SI Appendix.