Significance
Bifunctional enzymes are widely distributed throughout bacteria and are involved in modulating bacterial phenotypes; however, regulatory mechanisms that control the activities of the opposing output domains have remained elusive. Studies on DcpG demonstrate that binding of ligands to the sensor globin domain differentially affect GGDEF and EAL domain activities and highlight a role for protein conformational changes in modulating enzymatic activity. Unusual sensor globin domain characteristics, including heme midpoint potentials, are likely important for the unique regulatory properties of DcpG. As Paenibacillus dendritiformis responds to changes in the gaseous environment by modulating biofilm formation, DcpG is likely important in modulating physiological responses to changes in O2 and NO levels, identifying a role for heme sensor signaling in the bacterium.
Keywords: bifunctional enzyme, cyclic di-GMP, heme sensor
Abstract
Cyclic dimeric guanosine monophosphate (c-di-GMP) serves as a second messenger that modulates bacterial cellular processes, including biofilm formation. While proteins containing both c-di-GMP synthesizing (GGDEF) and c-di-GMP hydrolyzing (EAL) domains are widely predicted in bacterial genomes, it is poorly understood how domains with opposing enzymatic activity are regulated within a single polypeptide. Herein, we report the characterization of a globin-coupled sensor protein (GCS) from Paenibacillus dendritiformis (DcpG) with bifunctional c-di-GMP enzymatic activity. DcpG contains a regulatory sensor globin domain linked to diguanylate cyclase (GGDEF) and phosphodiesterase (EAL) domains that are differentially regulated by gas binding to the heme; GGDEF domain activity is activated by the Fe(II)-NO state of the globin domain, while EAL domain activity is activated by the Fe(II)-O2 state. The in vitro activity of DcpG is mimicked in vivo by the biofilm formation of P. dendritiformis in response to gaseous environment, with nitric oxide conditions leading to the greatest amount of biofilm formation. The ability of DcpG to differentially control GGDEF and EAL domain activity in response to ligand binding is likely due to the unusual properties of the globin domain, including rapid ligand dissociation rates and high midpoint potentials. Using structural information from small-angle X-ray scattering and negative stain electron microscopy studies, we developed a structural model of DcpG, providing information about the regulatory mechanism. These studies provide information about full-length GCS protein architecture and insight into the mechanism by which a single regulatory domain can selectively control output domains with opposing enzymatic activities.
Many pathways in bacteria use signaling proteins to sense and respond to the environment by converting extracellular cues into intracellular signals (1). A subset of these signaling proteins, termed bifunctional enzymes contain two output domains with opposing enzymatic activities, such as the synthesis and degradation of a metabolite or signaling molecule. Bifunctional enzymes have been shown to play important roles in the balance of intracellular signals (2–5), but, despite their near ubiquity in bacteria, the signaling mechanisms that regulate the opposing enzymatic activities are poorly understood and the proteins are often referred to as enzymatic conundrums (6–10). A famous example of bifunctional enzymes is the RelA/SpoT family of proteins, which both synthesize and hydrolyze (p)ppGpp a bacterial alarmone signal (11–13).
Within bacteria, bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) (5, 14) is a key intracellular secondary messenger used to control numerous phenotypes, including biofilm formation and motility, in response to changes in the environment (5, 14). Typically, high concentrations of c-di-GMP promote sessility while low concentrations promote motility, and c-di-GMP-related processes have been related to infection and host colonization (14, 15). C-di-GMP levels are regulated by the opposing activities of GGDEF and EAL enzyme domains. The GGDEF domain, named after its conserved active site motif (Gly-Gly-Asp-Glu-Phe), possesses diguanylate cyclase (DGC) activity that synthesizes c-di-GMP from two molecules of guanosine triphosphate (GTP) (16, 17); the EAL domain, named after its conserved active site motif (Glu-Ala-Leu), possesses c-di-GMP–specific phosphodiesterase (PDE) activity that hydrolyzes c-di-GMP into linear pGpG (16, 18). Bifunctional enzymes that contain both GGDEF and EAL domains have been identified in genomes of diverse bacteria and can regulate c-di-GMP concentration (6–10). While sensor domains linked to bifunctional GGDEF-EAL domains have been demonstrated to affect GGDEF-EAL enzyme activities (6, 8–10), a model for understanding how the opposing enzymatic activities are regulated within a single polypeptide chain has yet to be described.
A challenge in dissecting the mechanism of regulation of bifunctional enzymes is the need for signals that differentially regulate the two output domains. To address this issue, we have searched for bifunctional GGDEF-EAL enzymes that contain a sensor globin regulatory domain. Proteins with sensor globin domains linked to one or more output domains are termed globin-coupled sensor (GCS) proteins. Ligand (O2/NO/CO) binding to the sensor globin heme has been shown to regulated output domain activity in other members of the GCS family. GCS proteins are predicted based on sequence similarity throughout the bacterial kingdom, as well as in archaea and some lower eukaryotes (19, 20), and our searches identified a GCS that contained GGDEF and EAL output domains within Paenibacillus dendritiformis, which we have named DcpG, for Diguanylate Cyclase Phosphodiesterase with sensor Globin.
While neither EAL-containing nor bifunctional GGDEF/EAL GCS proteins have been previously described, characterization of GGDEF-containing GCS proteins demonstrated that their diguanylate cyclase activity is activated when the heme is in the Fe(II)-O2 state, as compared to Fe(II), Fe(II)-NO, and Fe(II)-CO states (21–24). These data suggested that a GCS containing GGDEF and EAL domains could provide a system whereby different gaseous ligands could be used to gain insight into the activation/inhibition of the two output domains. In addition, P. dendritiformis generates unique swarming patterns in response to its environment, but the chemical signals that alter behavior of the bacterium are poorly understood (25, 26). Given the links between c-di-GMP levels and motility, understanding the effects of different heme ligation states on DcpG activity may provide new information about how the gaseous environment affects P. dendritiformis behavior (27–31).
In this manuscript, we biochemically and electrochemically characterize DcpG and demonstrate that DcpG GGDEF and EAL domains are differentially regulated by heme ligation state of the sensor globin domain, likely due to the unusual heme characteristics that the sensor globin exhibits. To further probe the signaling mechanism, small-angle X-ray scattering (SAXS) and negative stain electron microscopy (EM) were used to characterize the conformation of DcpG in the Fe(II)-O2 state. Finally, the effects of O2, NO, and anaerobic conditions on biofilm formation were probed. These results provide insights regarding differential regulation of GGDEF-EAL enzymatic activities within a single polypeptide by binding of multiple ligands to a sensory domain, as well as full-length structural information on a GCS.
Results and Discussion
Sequence Analysis of DcpG.
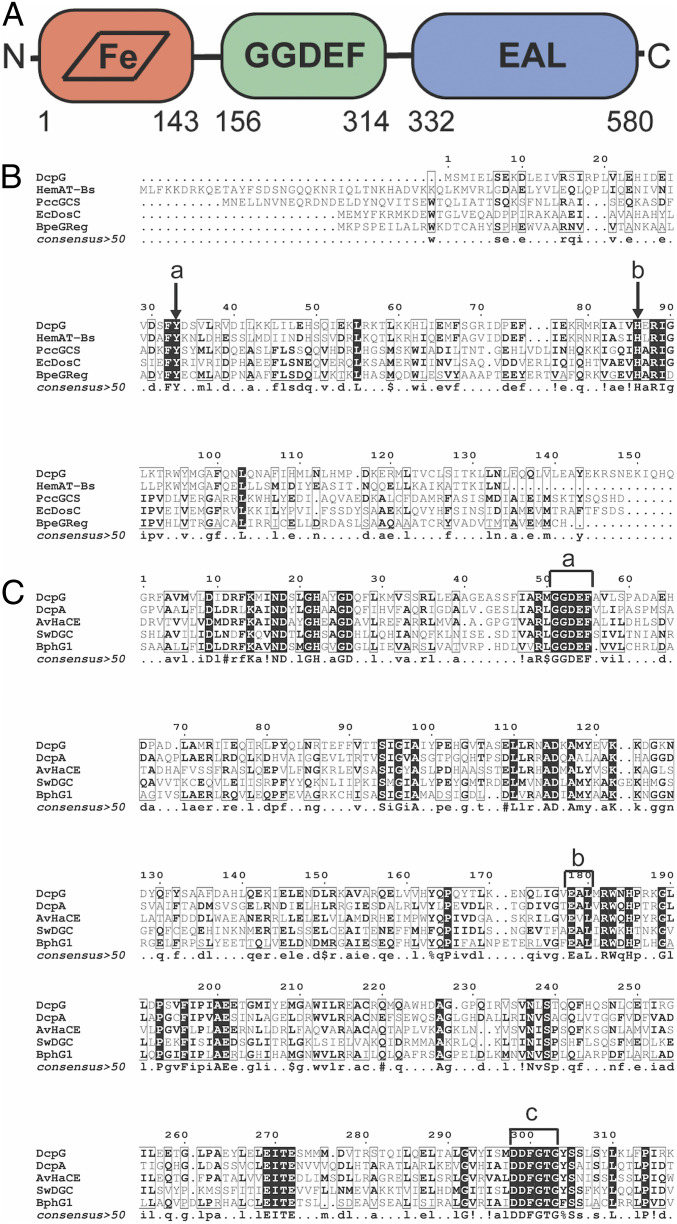
Analysis of the DcpG amino acid (AA) sequence identified an N-terminal sensor globin domain (AA 1 to 143), a short linker (AA 144 to 155), a GGDEF domain (AA 156 to 314), and a C-terminal EAL domain (AA 332 to 580) (Fig. 1A) (32). The heme-bound sensor globin domain of GCS proteins is responsible for binding diatomic ligands that control output domain activity (33, 34). Therefore, we aligned the globin domain of DcpG with the sensor globin domains of previously characterized GCS proteins to provide insight into the heme pocket sensor (Fig. 1B). DcpG globin has a sequence identity of 35% with HemAt-Bs globin domain, a GCS from Bacillus subtilis that contains a methyl accepting protein chemotaxis protein output domain that is involved in aerotactic signaling (19). When compared to GCS proteins containing a diguanylate cyclase (GGDEF) domain, DcpG globin has 13, 10, and 11% sequence identity with the GCS proteins from Escherichia coli (EcDosC), Pectobacterium carotovorum (PccGCS), and Bordetella pertussis (BpeGReg) globin domains, respectively (23, 28, 29). The alignment of DcpG, HemAt-Bs, EcDosC, PccGCS, and BpeGReg globin domains reveals that DcpG contains the residues previously identified in heme and ligand binding, including the absolutely conserved proximal histidine (H86) that ligates the heme and a highly conserved tyrosine residue (Y33) that is located on the distal side of the heme and has been previously shown to stabilize bound O2 (Fig. 1A) (19, 21, 23, 24, 28, 29, 35, 36). Although not conserved, GCS proteins often have a second hydrogen bond donor to stabilize bound O2; based on sequence alignments, DcpG contains a distal threonine (T58) in the heme pocket, which is analogous to HemAt-Bs.
Fig. 1.
Domain arrangement of DcpG and sequence alignments with GCS globin domains and active GGDEF-EAL proteins. (A) Monomer domain arrangement of DcpG globin, GGDEF, and EAL domains. (B) Globin domain alignments with DcpG (H3SIC7), HemAt-Bs (O07621), EcDosC (P0AA89), PccGCS (C6D9C2), and BpeGreg (Q7VTL8). (a) Conserved distal tyrosine; (b) conserved proximal histidine. (C) GGDEF-EAL domain alignments with DcpG (H3SIC7), DcpA (P9WM13), AvHaCE (B9JYX6), BphG1 (Q3IUZ1), and SwDGC (B1KIH5). (a) GGDEF active site; (b) EAL active site; and (c) DDFGTG sequence essential for PDE activity.
The alignment of DcpG GGDEF-EAL domains with previously characterized GGDEF-EAL–containing proteins from Mycobacterium smegmatis (DcpA), Agrobacterium vitis (AvHaCE), Rhodobacter sphaeroides (BphG1), and Shewanella woodyi (SwDGC) (Fig. 1C) and sequence analysis confirms that DcpG contains the conserved GGDEF and EAL active-site AA motifs for DGC and PDE enzymes, respectively (6, 10, 30, 37, 38). In addition, the DcpG EAL domain contains a DDFGTG motif (Asp-Asp-Phe-Gly-Thr-Gly) that has been shown to be essential for PDE activity (18). Since DcpG contains the conserved motifs that are found in active GGDEF-EAL proteins, we hypothesized that DcpG is a bifunctional enzyme that possesses both c-di-GMP diguanylate cyclase and phosphodiesterase activities. Taken together, the domain organization predicted that the binding of diatomic ligands (O2, CO, NO) to the sensor globin domain should control activity of the c-di-GMP enzymatic domains, providing a unique opportunity to probe how domains with opposing enzymatic activities can be controlled within a single protein.
Diatomic Gas Binding to DcpG.
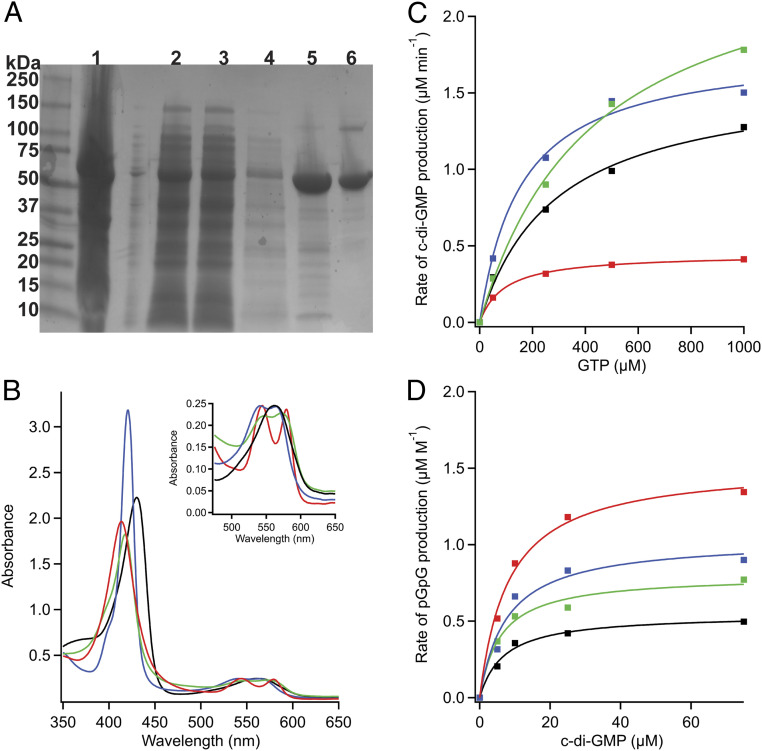
To identify which gaseous ligands can bind to DcpG (34), the full-length protein was heterologously expressed and purified. Purified DcpG migrated to positions of 67 and 134 kDa when analyzed by gel electrophoresis [the 67 kDa band corresponds to the monomeric form of the protein and the 134 kDa band to the dimeric form (Fig. 2A)], and dynamic light scattering experiments found that DcpG is present in solution as a monodisperse dimer (SI Appendix, Fig. S12 and Table S1). To assess which ligands bind to the DcpG globin domain, ultraviolet-visible absorption spectroscopy was used to determine ligand-dependent shifts in the spectra of the heme cofactor. DcpG purifies in the Fe(II)-O2 state with a λmax of the Soret band centered at 414 nm, consistent with a six-coordinate heme; the Fe(II)-O2 complex can also be generated by reducing DcpG and then exposing the protein to O2 (23, 34). DcpG is readily reduced by sodium dithionite, resulting in a red shift to 430 nm (five-coordinate complex). Unligated ferrous DcpG can then readily bind both NO and CO, resulting in blue shifts to 417 nm (six-coordinate complex) and 421 nm (six-coordinate complex), respectively (Fig. 2B). The α/β bands also show the expected increased splitting upon the addition of these diatomic gases (Fig. 2 B, Inset). Oxidation of DcpG to form the Fe(III) state of the heme is very slow, regardless of whether the protein is oxidized chemically or electrochemically. Furthermore, DcpG Fe(II)-O2 does not exhibit autooxidation of the heme, demonstrating that the Fe(II)-O2 complex is very stable (SI Appendix, Fig. S1). Taken together, the ligand-dependent absorption spectra for DcpG are similar to previously characterized GCS proteins (19, 23, 29, 39), indicating that DcpG likely binds and/or senses O2, NO, and/or CO in vivo.
Fig. 2.
Biochemical characterization of DcpG. (A) SDS-PAGE. Lanes: 1, cell pellet; 2, supernatant; 3, flow through; 4, wash; 5, nickel column; 6, S200 gel filtration column. DcpG is recalcitrant to complete unfolding, yielding mainly monomer with a small dimer band on SDS-PAGE gels. (B) UV-visible spectra of DcpG. Fe(II), black; Fe(II)-O2, red; Fe(II)-CO, blue; Fe(II)-NO, green. The Soret band of DcpG is sensitive to the identity of the axial ligand. (Inset) The α/β bands exhibit increased splitting as the diatomic gases bind to the heme. (C) Representative Michaelis–Menten curves for DcpG Fe(II) (black), Fe(II)-O2 (red), Fe(II)-NO (green), and Fe(II)-CO (blue) DGC activity. (D) Representative Michaelis–Menten curves for DcpG Fe(II) (black), Fe(II)-O2 (red), Fe(II)-NO (green), and Fe(II)-CO (blue) PDE activity.
DGC and PDE Activities of DcpG.
To determine the effect of different ligands on activity of the two output domains, enzymatic activities of DcpG GGDEF and EAL domains were investigated with the sensor globin domain heme in different ligation/oxidation states [Fe(II), Fe(III), Fe(II)-O2, Fe(II)-CO, and Fe(II)-NO]. Separate monitoring of the GGDEF and EAL domain activities was achieved through use of different substrates; GTP was used as the substrate for the GGDEF domain DGC activity to measure the rate of c-di-GMP production, while c-di-GMP was used as the substrate for the EAL domain PDE activity to measure the rate of pGpG production (Fig. 2 C and D).
DcpG DGC activity was quantified over a range of GTP and protein concentrations, and the reaction was coupled with EcDosP (23, 28), a c-di-GMP PDE enzyme, to ensure rapid c-di-GMP hydrolysis and prevent potential product inhibition. Although DcpG does not contain the conserved RxxD inhibitory site (I-site) upstream of the GGDEF active site that is necessary for noncompetitive product inhibition of DGC activity (EASS in DcpG) (40, 41), product inhibition was observed during DGC assays in the absence of EcDosP (SI Appendix, Fig. S1) (42). These data suggest that either DcpG can bind c-di-GMP within the GGDEF active site and inhibit DGC activity or that there is some c-di-GMP binding at the noncanonical I-site (40).
As listed in Table 1, DcpG Fe(II)-O2 ligation state resulted in a kcat of 0.45 min−1 and a KM of 80.9 µM, DcpG Fe(II) unligated yielded a kcat of 1.21 min−1 and a KM of 284.4 µM, DcpG Fe(III) resulted in a kcat of 0.22 min−1 and a KM of 88.6 µM, DcpG Fe(II)-CO state exhibited a kcat of 1.32 min−1 and a KM of 236.6 µM, and DcpG Fe(II)-NO state resulted in a kcat of 2.02 min−1 and a KM of 443.7 µM. It is unusual for a GCS containing a GDDEF to have the highest kcat for Fe(II)-NO; other GGDEF-containing GCS proteins such as HemDGC from Desulfotalea psychrophila, BpeGReg from B. pertussis, EcDosC from E. coli, and PccGCS from P. carotovorum exhibit the highest diguanylate cyclase activity in the Fe(II)-O2 state (23, 24, 28, 29).
Table 1.
Kinetic parameters for DcpG ligand-dependent DGC activity at 25 °C (mean ± SD of at least three independent experiments)
| Ligation state | kcat (min−1) | KM (µM) | kcat/KM (M−1/min−1) |
| Fe(II) | 1.21 ± 0.07 | 284.4 ± 123.3 | 4,243 |
| Fe(II)-O2 | 0.45 ± 0.04 | 80.9 ± 33.5 | 5,562 |
| Fe(II)-NO | 2.02 ± 0.36 | 443.7 ± 186.4 | 4,574 |
| Fe(II)-CO | 1.32 ± 0.09 | 236.6 ± 71.9 | 5,569 |
| Fe(III) | 0.22 ± 0.05 | 88.6 ± 35.0 | 2,483 |
To probe DcpG PDE activity, a dual enzyme activity assay was developed to monitor the conversion of c-di-GMP to pGpG. A 5′-nucleotidase was included in the reaction mixtures to cleave the 5′-phosphorlytic site of the pGpG product and release phosphate, which is detected. As the 5′-nucleotidase can only hydrolyze pGpG and not c-di-GMP due to its cyclic structure (SI Appendix, Fig. S2), we could monitor the production of pGpG through quantification of phosphate release. Using this assay, the kcat and the KM for DcpG PDE activity were measured for each ligation state (Table 2). DcpG Fe(II)-O2 ligation state yielded the highest activity, with a kcat of 0.62 ± 0.07 min−1 and a KM of 7.82 ± 6.14 µM. The Fe(II), Fe(II)-CO, and Fe(II)-CO states all exhibited the same low-level PDE activity. The kcat and KM could not be measured for DcpG Fe(III) PDE activity because the KM for c-di-GMP was greater than 100 µM, and the high c-di-GMP levels resulted in artifacts in the kinetics. The combination of the low enzymatic activity, high heme midpoint potentials (discussed in DcpG Exhibits Two High Midpoint Potentials), and the slow kinetics of oxidation, whether through chemical or electrochemical means, make it unlikely that DcpG serves as a redox sensor in vivo. These data suggest that the DcpG EAL domain responds to O2, similar to EcDosP, a monofunctional PDE with a heme-bound PAS domain in which binding of O2 enhances its PDE activity (28, 43).
Table 2.
Kinetic parameters for DcpG ligand-dependent PDE activity at 25 °C (mean ± SD of at least three independent experiments)
| Ligation state | kcat (min−1) | KM (µM) | kcat /KM (M−1/min−1) |
| Fe(II) | 0.22 ± 0.01 | 8.35 ± 4.78 | 25,746 |
| Fe(II)-O2 | 0.54 ± 0.08 | 11.94 ± 4.65 | 44,848 |
| Fe(II)-NO | 0.27 ± 0.04 | 6.66 ± 2.09 | 40,943 |
| Fe(II)-CO | 0.17 ± 0.01 | 7.82 ± 2.96 | 21,987 |
Our data demonstrates that the Fe(II)-O2 ligation state results in low DGC activity but the highest PDE activity, demonstrating that a single sensor domain can exhibit opposite effects on two output domains, activating one while inhibiting the other. As GTP concentrations in dividing bacteria are typically in the low mM range (44), the differences in cyclase KM in the various ligation states likely will not contribute to physiological function during log phase growth. However, it is possible that GTP levels in P. dendritiformis may decrease during stationary phase and/or within biofilms, which could affect activity of the DGC domain. In addition, DcpG may interact with other proteins within the cell, further regulating DGC and PDE activity. For example, when growing aerobically, intracellular conditions and/or interactions within P. dendritiformis could result in DcpG exhibiting undetectable DGC activity and high PDE activity, in contrast to the low, but measurable, DGC activity measured in vitro. In addition, changes in c-di-GMP levels, which typically range from submicromolar to low micromolar, could further regulate DcpG PDE activity (5, 16, 45–47). The potential effects of the cellular milieu could prohibit DcpG from participating in a futile cycle of GTP to c-di-GMP to pGpG within the cell.
The comparison of GGDEF and EAL domains finds that DcpG Fe(II), Fe(II)-NO, and Fe(II)-CO states exhibit the same low level of PDE activity; however, DGC activity increases in the following order of ligation states, Fe(II)-O2 << Fe(II) < Fe(II)-CO < Fe(II)-NO. This is surprising because previously characterized GGDEF-containing GCS proteins are activated by the Fe(II)-O2 state (23, 24, 28, 29). Our results also differ from previously characterized GGDEF-EAL proteins that are associated with H-NOX (Heme Nitric oxide/OXygen binding) domains (48), in which the sensor H-NOX domain and the two output domains are within two separate polypeptide chains (H-NOX and GGDEF-EAL). The H-NOX associated GGDEF-EAL protein from Legionella pneumophila, lpg1057, exhibits DGC activity that is inhibited by the Fe(II)-NO state (7), as does the activity of the H-NOX associated GGDEF-EAL protein in S. woodyii, SwDGC (30). Therefore, DcpG is the first GCS protein and GGDEF-EAL bifunctional enzyme with DGC activity that is activated by the Fe(II)-NO state of the heme. This is analogous to the mammalian H-NOX–containing protein, soluble guanylate cyclase (sGC), which converts GTP to 3′,5′-cGMP and is also activated by the Fe(II)-NO state (49, 50).
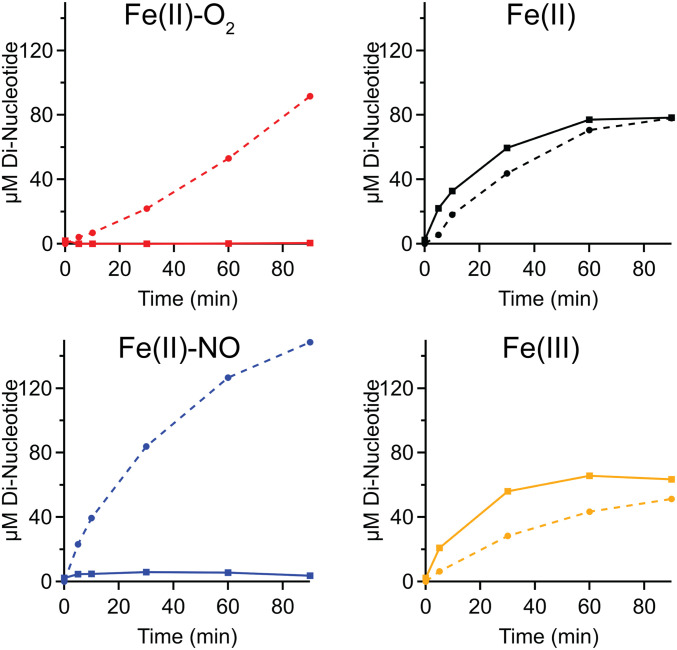
To probe pseudocellular conditions where c-di-GMP synthesized by the DcpG DGC domain is likely hydrolyzed by the PDE domain, rather than a separate PDE, c-di-GMP and pGpG produced by DcpG were monitored in the absence of an additional c-di-GMP PDE (EcDosP; Fig. 3). As expected from the single domain enzyme kinetics, the high affinity of the EAL domain for c-di-GMP results in rapid hydrolysis of c-di-GMP produced by the GGDEF domain. Even DcpG Fe(II)-NO, which exhibits the highest DGC activity and low PDE activity, did not result in appreciable buildup of c-di-GMP. It also is possible that a burst of c-di-GMP was rapidly produced but not observed due to the temporal limitations of the assay. The rapid turnover of GTP to c-di-GMP to pGpG by DcpG suggests that downstream signaling partners may either make protein–protein interactions, allowing direct hand off of c-di-GMP, or are colocalized to allow for rapid c-di-GMP binding. Either scenario could allow c-di-GMP produced by DcpG DGC domain to be sensed by downstream proteins prior to hydrolysis by the EAL domain and would be consistent with recent studies that have highlighted the importance of localized c-di-GMP signaling in a number of bacteria (51–56). Another possibility is that that cellular conditions/binding partners may further regulate DcpG activity and lead to inhibition of DGC activity under aerobic conditions and inhibition of PDE activity in the presence of NO.
Fig. 3.
Tandem DcpG DGC and PDE kinetics in various ligation states. Solid lines represent c-di-GMP levels, and dashed lines represent pGpG levels.
DcpG is a heme protein for which O2 and NO binding to the heme result in the opposite regulatory effects on an output domain. The findings that a single ligand (O2) can serve as both agonist and antagonist for the two domains within DcpG and the fact that O2 and NO exhibit opposing effects on DGC suggests that DcpG can serve as a unique system to probe signaling within a bifunctional protein.
Effects of Gaseous Conditions on P. dendritiformis Biofilm Formation.
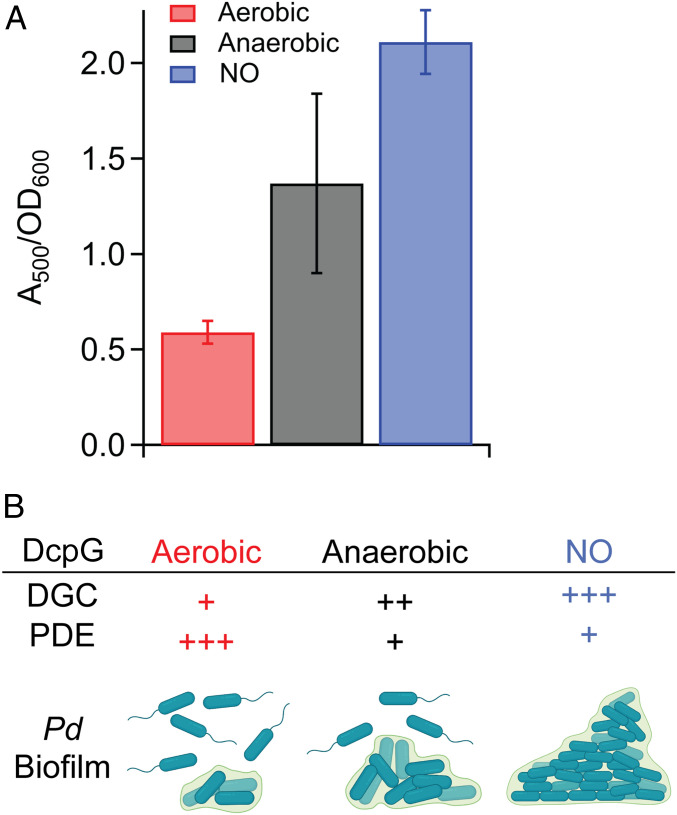
To gain insight into a potential physiological role for DcpG as an O2/NO sensor, the impacts of O2 and NO on P. dendritiformis biofilm formation were investigated. Growth of P. dendritiformis cells in aerobic conditions resulted in low biofilm formation; anaerobic growth resulted in 2.3-fold more biofilm formation, and growth in the presence of NO resulted in ∼4-fold more biofilm formation, as compared to aerobic growth (Fig. 4A and SI Appendix, Fig. S3). The results of the biofilm studies are consistent with enzymatic characterization of purified DcpG; DcpG Fe(II)-O2 exhibits low DGC activity and high PDE activity, which should result in low levels of c-di-GMP and minimal biofilm formation. In contrast, DcpG Fe(II) exhibits moderate DGC activity and low PDE activity, resulting in medium levels of c-di-GMP and moderate biofilm formation, and DcpG Fe(II)-NO exhibits high DGC activity and low PDE activity, which should result in high levels of c-di-GMP (Fig. 4A). Given that NO is an important signaling messenger in both eukaryotes and prokaryotes (57, 58), but can also be toxic to organisms (59, 60), DcpG sensing of NO may result in increased biofilm formation to protect the organism from the toxic effects. Taken together, these data demonstrate that P. dendritiformis alters biofilm formation in response to its gaseous environment and that DcpG likely serves as an O2/NO sensor to regulate biofilm formation (Fig. 4B).
Fig. 4.
Gaseous environment alters P. dendritiformis biofilm formation. (A) Biofilm formation quantified by Congo Red staining of P. dendritiformis grown in the presence/absence of O2 and NO. A500/A600 is a measurement of the amount of Congo Red bound to the cell pellet relative to the cell density. Differences between all measurements are significant (P < 0.003). (B) Comparison of the relative in vitro DGC and PDE activities of DcpG in various ligation states [Fe(II)-O2, Fe(II), and Fe(II)-NO] and P. dendritiformis biofilm formation when grown under aerobic, anaerobic, and anaerobic +NO conditions.
O2 and NO Dissociation Kinetics of DcpG Wild Type (WT).
The surprising finding that heme ligands differentially regulate DcpG output domain activities leads us to investigate the characteristics of the globin domain. To provide more insight into how the DcpG sensor globin domain responds to O2/NO binding, the O2 and NO dissociation rates were measured using stopped-flow UV-visible spectroscopy. The dissociation rate of O2 from DcpG was measured and fit to a double exponential function (SI Appendix, Fig. S4 and Table S2), resulting in rate constants of k1 = 12.07 s−1 (17.8%) and k2 = 87.30 s−1 (82.2%), which are relatively fast for a sensor globin. (Table 3). The observed biphasic O2 dissociation is typical of GCS proteins and has been attributed to multiple conformations of the heme pocket (35, 36, 61). Rapid O2 dissociation rates also have been observed for the GCS protein HemAt-Bs (Table 3) and were proposed to be due to an apolar distal pocket (61). However, other characterized GCS proteins (EcDosC from E. coli and BpeGReg from B. pertussis) have similar apolar distal pockets but exhibit nearly 100-fold slower O2 dissociation rates (Table 3), suggesting that the regulation of ligand affinity is more complicated (28, 62, 63).
Table 3.
O2 and NO dissociation kinetics of DcpG and other heme sensor proteins
| Protein | k1 (s−1) | k2 (s−1) | % k 1 | % k 2 | Ref. | |
| O2 dis. | DcpG | 12.07 | 87.30 | 17.8 | 82.2 | TW |
| PccGCS | 0.56 | 3.87 | 56.0 | 44.0 | (23) | |
| BpeGReg | 0.82 | 6.30 | 39.3 | 60.7 | (23) | |
| EcDosC | 13 | — | — | — | (21) | |
| HemAt-Bs | 87 | 1,900 | NR | NR | (61) | |
| NO dis. | DcpG | 0.16 | 0.012 | 48.2 | 50.8 | TW |
| sGC | 0.0038 | 0.0012 | 35 | 65 | (65) |
TW, this work. dis., dissociation; NR, not reported.
A complementary experiment also was conducted, in which the rate of NO dissociation from DcpG was measured (Table 3), to gain further insight into DcpG ligand affinity. The NO dissociation data best fit a double exponential function (SI Appendix, Fig. S5), yielding rate constants of k1 = 0.16 s−1 (48.2%) and k2 = 0.012 s−1 (50.8%). The NO dissociation rates have not been characterized for other GCS proteins, but based on the “sliding-scale” rule proposed by Olson and coworkers (64), which postulates that affinity for all ferrous ligands of histidyl ligated heme proteins are inherently linked and any mutations will result in increases/decreases (or slides) of all ligands, the rapid rates of O2 dissociation from DcpG should be mirrored in fast dissociation rates of other ferrous heme ligands, such as NO. In comparison to soluble guanylate cyclase (sGC), the mammalian NO receptor that has picomolar affinity for NO, DcpG NO dissociation rates were approximately an order of magnitude faster than sGC, indicating that DcpG has a weaker affinity for NO (Table 3) and suggesting that the sensor globin heme exhibits unusual ligand binding properties, compared to other globins (65–67).
DcpG Exhibits Two High Midpoint Potentials.
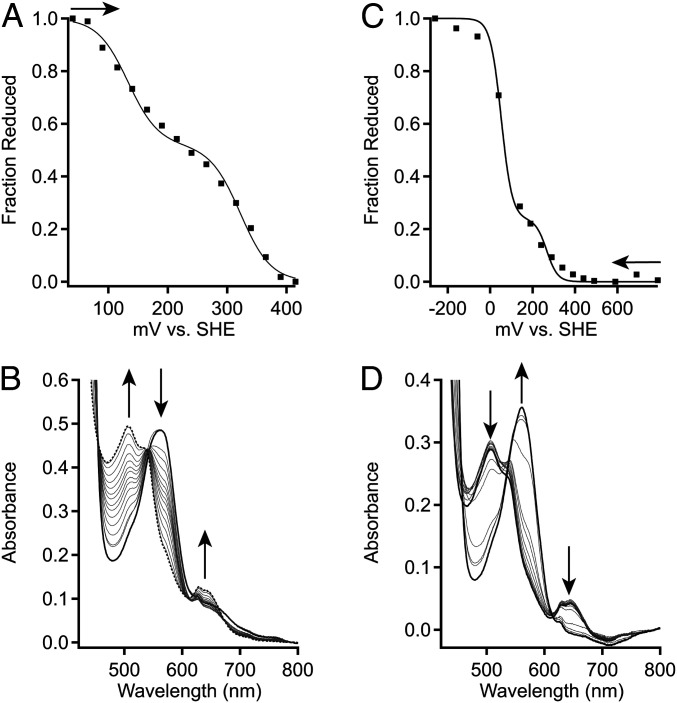
To further understand the unusually fast O2 and NO dissociation rates exhibited by DcpG, we hypothesized that the observed rapid O2 dissociation kinetics and lack of measurable rate of autooxidation (SI Appendix, Fig. S5) for DcpG might be due to a high heme midpoint potential, as previous work has demonstrated that heme midpoint potential modulates O2 affinity of heme proteins (68). To investigate this possibility, coupled spectro-electrochemical titrations of DcpG were performed. As DcpG was found to exhibit a slow transition from five-coordinate to six-coordinate heme in the Fe(III) state (SI Appendix, Fig. S7), a variety of buffers and pHs were tested, identifying 100 mM potassium phosphate pH 7.0 as the optimal condition for relatively rapid DcpG Fe(III) equilibration (SI Appendix, Figs. S8 and S9). Using the optimized buffer conditions, both reductive and oxidative titrations (following either chemical oxidation or reduction) were performed on DcpG. The oxidative titration yielded two midpoint potentials of 151 mV (48%) and 358 mV (52%) versus standard hydrogen electrode (SHE), while reductive titration yielded midpoint potentials of 81 mV (88%) and 339 mV (13%) versus SHE. (Table 4 and Fig. 5 and SI Appendix, Fig. S10). The two midpoint potentials may be the result of nonequivalent hemes within the DcpG homodimer, the difference in the lower potential depending on type of titration may be due to slow changes in conformation and/or H2O/−OH binding in the Fe(III) state.
Table 4.
Midpoint potentials of DcpG and PccGCS
| Protein | Titration | pH | Midpoint 1 | Midpoint 2 | % 1 | % 2 |
| DcpG | Reductive | 7.0 | 81 ± 6 | 339 ± 36 | 13 | 87 |
| DcpG | Oxidative | 7.0 | 151 ± 36 | 358 ± 72 | 40 | 60 |
| PccGCS | Ox/red | 7.0 | −7 ± 21 | 246 ± 64 | 88 | 12 |
Values are reported as an average of two or three experiments ± range or ± SD, respectively.
Fig. 5.
Electrochemical characterization of DcpG. (A) Representative oxidative titration of DcpG Fe(II) in 100 mM potassium phosphate 50 mM KCl pH 7.0. Midpoint values were determined by plotting the fraction of reduced heme against the potential. The fraction of reduced heme was determined by tracking the spectral changes at 571 and 641 nm. The representative data (black-filled squares) were fit using SI Appendix, Eq. S1, which describes a species with two redox active components. (B) Absorption spectra of DcpG from optically transparent thin layer electrochemical cell (OTTLE) cell oxidative titration. UV-visible spectra were recorded for each set potential (Top). (C) Representative reductive titration of DcpG Fe(III) in 100 mM potassium phosphate 50 mM KCl pH 7.0. Midpoint values were determined by plotting the fraction of reduced heme against the potential. The fraction of reduced heme was determined by tracking the spectral changes at 571 and 641 nm. The representative data (black-filled squares) were fit using SI Appendix, Eq. S1, which describes a species with two redox active components. (D) Absorption spectra of DcpG from OTTLE cell reductive titration. Arrows show direction of titration [oxidative (A)] or reductive (C) or change in peak intensities (B and D).
To determine if nonequivalent hemes are found in other GCS proteins, a coupled spectro-electrochemical titration was performed on PccGCS, the GCS from the plant pathogen P. carotovorum (23). PccGCS binds O2 more tightly than DcpG but also displays biphasic O2 dissociation kinetics and forms oligomers (tetramer, dimer) in solution. The oxidative and reductive titration of PccGCS was reversible and yielded two midpoint potentials of −7.0 mV (81%) and 245.8 mV (18%) versus SHE (Table 4 and SI Appendix, Fig. S11). The lower midpoint potential of PccGCS, as compared to DcpG, could be due to differences in heme-edge residues, which can alter heme electronics (69); PccGCS has a tryptophan residue at the heme edge, as compared to histidine in DcpG. In addition, the lower midpoint potential value for PccGCS likely contributes to its slower O2 dissociation rates compared to DcpG, as lower midpoint potentials have previously been linked to tighter O2 affinity (69, 70).
As observed for PccGCS, proteins with a globin fold typically have much lower midpoint potentials (Table 5); for example, the midpoint potentials of equine skeletal myoglobin and human hemoglobin are +47 mV and 175 mV versus SHE, respectively (71–73). Other gas sensors with alternative heme domain folds exhibit lower midpoint potentials as well; the PAS heme-domain protein BjFixLH exhibits a midpoint potential of +68 mV versus SHE (21, 74) and heme-bound H-NOX domains such as sGC (the mammalian NO sensor) and Tt H-NOX have midpoint potentials of +187 mV and +167 mV versus SHE, respectively (68, 75). DcpG midpoint potentials fall outside of the typical range of any of the previously characterized five-coordinate heme gas sensors and are closest to that of a six-coordinate cytochrome c (for example, cytochrome c6 from Chlamydomonas reinhardtii midpoint = +370 mV versus SHE) (76).
Table 5.
Midpoint potentials values for five-coordinate histidyl ligated heme proteins
As P. dendritiformis, which natively encodes DcpG, is a facultative anaerobic bacterium (26), DcpG may have evolved to have a high midpoint potential to limit O2 binding and thereby allow responsiveness to other gases and/or to alter the O2 concentration at which the organism changes c-di-GMP–dependent phenotypes (26). The high midpoint potentials also should allow DcpG to serve as an O2/NO sensor rather than a redox sensor, as it is unlikely that DcpG can be oxidized in vivo (77, 78).
DcpG Structural Analysis.
Given that DcpG regulates DGC and PDE activity based on the ligation state of the heme-iron and that the heme-iron has unusual properties, we hypothesized that structural and/or conformational changes in the globin domain caused by ligand binding could be a part of the signal transduction mechanism to regulate both enzymatic domains. The globin domain of DcpG consists of an alpha-helical structure that is common among other globin sensor proteins, and we hypothesized that the alpha-helices may rearrange based on the ligation state of the heme-iron. However, circular dichroism spectroscopy of DcpG in various ligation states (SI Appendix, Fig. S12 and Table S4) did not result in a significant change in the helical content (79) regardless of ligation state, suggesting that there is minimal (if any) change in globin domain secondary structure in response to ligand binding. Dynamic light scattering demonstrated that the Fe(II)-O2, Fe(II), and Fe(II)-NO states of DcpG are dimeric in solution but did identify small changes in hydrodynamic diameter between DcpG Fe(II) unligated and ligand-bound states (SI Appendix, Fig. S13). To further probe potential differences, chemical cross-linking analysis (SI Appendix, Figs. S14 and S15 and Table S6) identified cross-links between the globin and DGC domains in multiple ligation states. DcpG Fe(II) unligated yielded cross-links between the DGC domain and residues on two globin helices (αF and αG), whereas DcpG Fe(II)-NO yielded one globin-DGC cross-link and DcpG Fe(II)-O2 only cross-links within the globin domain. These results suggest that DcpG undergoes conformational changes in response to heme ligation state. Therefore, to gain further insights into the structure of DcpG and link catalysis with conformational changes, SAXS and negative stain EM data were obtained for DcpG in the Fe(II)-O2 ligation state (Fig. 6).
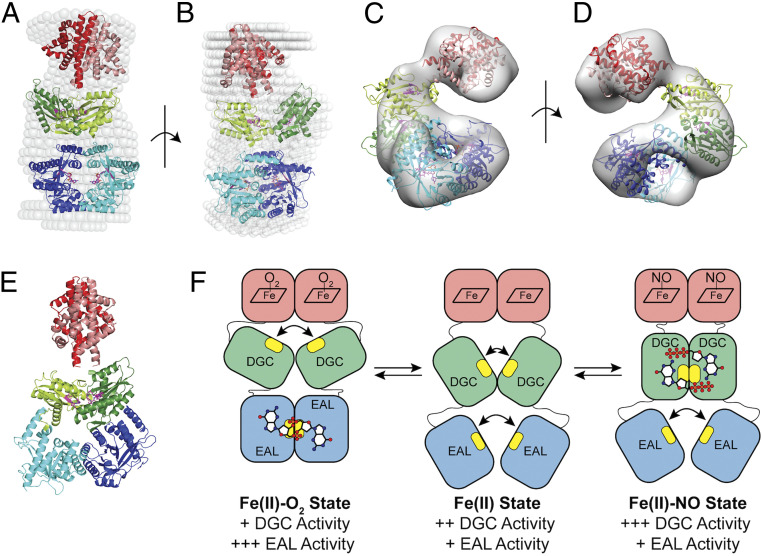
Fig. 6.
SAXS and negative stain models of DcpG Fe(II)-O2. (A and B) Two perpendicular views of an overlay of the SAXS solution envelope (white spheres) on the DcpG EAL active dimer model. Monomer 1 is shown in darker colors, while monomer 2 is depicted in lighter shades. The EAL domains (blue) form a ring stabilizing the active site residues inside. The GGDEF domains (green) are in an inactive conformation with their active sites ∼17 Å apart; globin domains are shown in red. (C and D) Two views (180° rotation) of DcpG negative stain EM density and model (ribbon colors are the same as in the SAXS model; σ level = 0.600). (E) A model of DcpG that allow dimerization and activity of the GGDEF domains. The active sites, shown in magenta, are ∼7 Å apart, as seen in other diguanylate cyclase active dimer structures. (PDB: 4URG, 6EIB, and 3I5B) (83–85). The EAL dimer is in a nonproductive form also seen in an earlier inactive dimer structure (PDB: 4RNI) (81). (F) A cartoon model of the DcpG conformational states. Active sites are highlighted in yellow, with GTP and c-di-GMP depicted as ball/stick models. Relative in vitro activity of the enzymatic domains is shown below.
DcpG WT SAXS and negative stain EM structure—PDE high-activity state.
Analysis of the SAXS data found that DcpG wild type (WT) is a dimer with a height of 143 Å and a width of 82 Å (Fig. 6 and SI Appendix, Fig. S16) and has an envelope structure obeying an approximate twofold symmetry, suggesting similar domain organization in the two monomers. Based on the SAXS envelope, two structural models fit the observed data with similar fitting coefficients: 1) an extended structure wherein dimerization occurs only at the globin domains, with the GGDEF and EAL domains within each monomer associating and resulting in the EAL active site facing the GGDEF active site within each DcpG monomer (SI Appendix, Fig. S16); and 2) a linear dimer wherein the globin, GGDEF, and EAL domains are arranged such that the globin and EAL domains of both monomers are in close contact with each other, while the GGDEF domains are physically apart (Fig. 6 A and B and SI Appendix, Fig. S16).
As the resolution limits of SAXS do not allow for the two models to be distinguished, we utilized Gradient Fixation (GraFix) (80) and negative stain EM to gain further insight into the structure of DcpG. The GraFix approach separates complexes in a glycerol gradient that contains cross-linking reagents, resulting in stabilization of protein complexes and conformations. Using this method, we were able to identify and image fractions that contained dimeric DcpG (SI Appendix, Fig. S17). The DcpG model generated from the negative stain EM data (Fig. 6 C and D) maintains the dimeric EAL domains observed in the linear dimer SAXS model (Fig. 6 A and B), supporting a structure of DcpG that contains a dimeric globin, GGDEF domains that are physically separated, and dimeric EAL domains. Previous studies have established that dimerization of EAL domains at the active sites results in increased phosphodiesterase activity (81), and, in the case of the EAL domain from YahA, substrate binding resulted in 100-fold increase in dimerization affinity (82). This model represents a high PDE activity state, as expected based on enzyme kinetic assays (Table 2), and the two EAL domains form a closed ring at the dimer interfaces with the two active sites facing each other. In contrast, the GGDEF active site loops in the two DGC domains separated by ∼17 Å, resulting in low DGC activity (Table 1) and suggesting that DGC inactivation may be a result of a physical separation of the monomers.
Although the EAL domain dimer is maintained both the SAXS and EM models, the globin domains in the negative stain EM model are bent relative to the axis of the GGDEF/EAL domains, as compared to the SAXS model (SI Appendix, Fig. S17). The GGDEF domains also are rotated relative to each other, although the distance between GGDEF active sites is maintained, minimizing the formation of active cyclase dimer. The differences in the envelopes obtained by SAXS and negative stain EM support the flexibility of DcpG, especially in the linker regions, and suggest that this flexibility is important for transmitting the ligand binding signal and/or regulation of enzymatic activity.
Model of DcpG fully extended state—DGC high-activity state.
Previous studies have established that a dimer of GGDEF domains is essential for DGC activity as each monomer binds one GTP substrate molecule, and cyclization occurs across the dimer interface (83). Using previously published high-resolution structures of GGDEF domains in an active dimer conformation (Protein Data Bank [PDB] ID: 4URG, 6EIB, and 3I5B) (83–85), maintaining the globin dimer interface that is observed in sensor globin structures (35, 63), and using the SAXS model of DcpG extended monomer, we constructed a model of dimeric DcpG in an active-DGC state (Fig. 6E). The model of the active-DGC state suggests that the domains may rotate relative to each other to allow for DGC versus PDE activation/inhibition.
Based on the structure and ligand-dependent changes in enzyme activity and cross-linking mass spectrometry, we propose a model of signal transduction within DcpG (Fig. 6F). The short linker regions between the domains are likely flexible and should allow the domains to change orientation relative to each other. In the Fe(II)-O2 state (low DGC activity, high PDE activity), the protein adopts the observed structure, with the DGC domain active sites physically separated and the EAL domains forming an active dimer (Fig. 6 A–D). Upon O2 dissociation, the EAL domain physically separates, resulting in low PDE activity, while the GGDEF domains are closer together and/or able to interact more frequently, resulting in increased DGC activity. Finally, in the Fe(II)-NO state, the GGDEF domains interact (as modeled in Fig. 6E), resulting in high DGC activity, while the EAL domains are physically separated and exhibit low activity. Taken together, the SAXS and negative stain EM-derived models for DcpG WT provides full-length structural information on a GCS protein. In addition, the structural information has yielded insights in the regulation of two domains with opposing enzymatic activities within a single protein.
Conclusions
In this manuscript, we have reported a biochemical and structural characterization of DcpG, a bifunctional globin-coupled sensor protein with GGDEF and EAL output domains. DcpG provided a unique opportunity to interrogate the mechanism by which one sensor domain can differentially control activity of two opposing output domains within a single polypeptide chain, likely due to the unusual characteristics of the sensor globin heme. The SAXS- and negative stain EM-derived models of DcpG have yielded structural insight into a full-length globin-coupled sensor protein and evidence of a mechanism by which conformational changes regulated output domain activity. These studies identify a multigas sensor in which two output domains (GGDEF and EAL) are differentially regulated in response to the ligation state of the heme. In addition, DcpG likely serves as a physiological O2/NO sensor within P. dendritiformis, allowing the bacterium to control biofilm formation in response to the gaseous environment.
Materials and Methods
Detailed materials and methods can be found in SI Appendix. These methods include conditions for protein expression and purification, heme complex formation, heme ligand dissociation kinetics, enzyme kinetic assays, and biofilm formation. In addition, full details on SAXS and negative stain EM data collection and model generation are included.
Supplementary Material
Acknowledgments
This work was supported by NSF Grants CHE1352040 (E.E.W.) and CHE2003350 (E.E.W.) and Frasch Foundation Grant 824-H17 (E.E.W.). D.C.P. is funded by an NSF Graduate Research Fellowship (1745025). We thank members of the E.E.W. laboratory for assistance and helpful suggestions. We sincerely appreciate the Bollinger-Krebs lab for the use of their anaerobic stopped flow to determine DcpG NO dissociation rates. This work is based on research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the NSF and NIH/National Institute of General Medical Sciences (NIGMS) via NSF Award DMR-1829070, and the MacCHESS resource is supported by NIH/NIGMS Award GM-124166. We sincerely appreciate the generous assistance from Drs. Qing Qiu Huang and Richard Gillilan at CHESS.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100657118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Hengge R., Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Rider M. H., et al., 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase: Head-to-head with a bifunctional enzyme that controls glycolysis. Biochem. J. 381, 561–579 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z., Goronzy J. J., Weyand C. M., The glycolytic enzyme PFKFB3/phosphofructokinase regulates autophagy. Autophagy 10, 382–383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J.-W., Park Y.-H., Seok Y.-J., Rsd balances (p)ppGpp level by stimulating the hydrolase activity of SpoT during carbon source downshift in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 115, E6845–E6854 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Römling U., Galperin M. Y., Gomelsky M., Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bharati B. K., et al., A full-length bifunctional protein involved in c-di-GMP turnover is required for long-term survival under nutrient starvation in Mycobacterium smegmatis. Microbiology (Reading) 158, 1415–1427 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Carlson H. K., Vance R. E., Marletta M. A., H-NOX regulation of c-di-GMP metabolism and biofilm formation in Legionella pneumophila. Mol. Microbiol. 77, 930–942 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira R. B. R., Antunes L. C. M., Greenberg E. P., McCarter L. L., Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J. Bacteriol. 190, 851–860. (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levet-Paulo M., et al., The atypical two-component sensor kinase Lpl0330 from Legionella pneumophila controls the bifunctional diguanylate cyclase-phosphodiesterase Lpl0329 to modulate bis-(3′-5′)-cyclic dimeric GMP synthesis. J. Biol. Chem. 286, 31136–31144. (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarutina M., Ryjenkov D. A., Gomelsky M., An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J. Biol. Chem. 281, 34751–34758 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Seyfzadeh M., Keener J., Nomura M., spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 90, 11004–11008 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vinella D., Albrecht C., Cashel M., D’Ari R., Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol. Microbiol. 56, 958–970 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Xiao H., et al., Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266, 5980–5990 (1991). [PubMed] [Google Scholar]
- 14.Ryan R. P., Fouhy Y., Lucey J. F., Dow J. M., Cyclic di-GMP signaling in bacteria: Recent advances and new puzzles. J. Bacteriol. 188, 8327–8334 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenal U., Reinders A., Lori C., Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 15, 271–284 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Simm R., Morr M., Kader A., Nimtz M., Römling U., GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53, 1123–1134 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Ross P., et al., Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325, 279–281 (1987). [DOI] [PubMed] [Google Scholar]
- 18.Schmidt A. J., Ryjenkov D. A., Gomelsky M., The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: Enzymatically active and inactive EAL domains. J. Bacteriol. 187, 4774–4781 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou S., et al., A globin-coupled oxygen sensor from the facultatively alkaliphilic Bacillus halodurans C-125. Extremophiles 5, 351–354 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Vinogradov S. N., Tinajero-Trejo M., Poole R. K., Hoogewijs D., Bacterial and archaeal globins—A revised perspective. Biochim. Biophys. Acta 1834, 1789–1800 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Kitanishi K., et al., Important roles of Tyr43 at the putative heme distal side in the oxygen recognition and stability of the Fe(II)-O2 complex of YddV, a globin-coupled heme-based oxygen sensor diguanylate cyclase. Biochemistry 49, 10381–10393 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Wu C., et al., Oxygen promotes biofilm formation of Shewanella putrefaciens CN32 through a diguanylate cyclase and an adhesin. Sci. Rep. 3, 1945–1951 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burns J. L., Deer D. D., Weinert E. E., Oligomeric state affects oxygen dissociation and diguanylate cyclase activity of globin coupled sensors. Mol. Biosyst. 10, 2823–2826 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Sawai H., et al., Molecular oxygen regulates the enzymatic activity of a heme-containing diguanylate cyclase (HemDGC) for the synthesis of cyclic di-GMP. Biochim. Biophys. Acta 1804, 166–172 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Be’er A., Strain S. K., Hernández R. A., Ben-Jacob E., Florin E.-L., Periodic reversals in Paenibacillus dendritiformis swarming. J. Bacteriol. 195, 2709–2717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tcherpakov M., Ben-Jacob E., Gutnick D. L., Paenibacillus dendritiformis sp. nov., proposal for a new pattern-forming species and its localization within a phylogenetic cluster. Int. J. Syst. Bacteriol. 49, 239–246 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Schmidt A., et al., Oxygen-dependent regulation of c-di-GMP synthesis by SadC controls alginate production in Pseudomonas aeruginosa. Environ. Microbiol. 18, 3390–3402 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Tuckerman J. R., et al., An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 48, 9764–9774 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Wan X., et al., Globins synthesize the second messenger bis-(3′-5′)-cyclic diguanosine monophosphate in bacteria. J. Mol. Biol. 388, 262–270 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu N., et al., Nitric oxide regulation of cyclic di-GMP synthesis and hydrolysis in Shewanella woodyi. Biochemistry 51, 2087–2099 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Burns J. L., et al., Oxygen-dependent globin coupled sensor signaling modulates motility and virulence of the plant pathogen Pectobacterium carotovorum. ACS Chem. Biol. 12, 2070–2077 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 33.Martínková M., Kitanishi K., Shimizu T., Heme-based globin-coupled oxygen sensors: Linking oxygen binding to functional regulation of diguanylate cyclase, histidine kinase, and methyl-accepting chemotaxis. J. Biol. Chem. 288, 27702–27711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker J. A., Rivera S., Weinert E. E., Mechanism and role of globin-coupled sensor signalling. Adv. Microb. Physiol. 71, 133–169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W., Phillips G. N. Jr, Structure of the oxygen sensor in Bacillus subtilis: Signal transduction of chemotaxis by control of symmetry. Structure 11, 1097–1110 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Ohta T., Yoshimura H., Yoshioka S., Aono S., Kitagawa T., Oxygen-sensing mechanism of HemAT from Bacillus subtilis: A resonance Raman spectroscopic study. J. Am. Chem. Soc. 126, 15000–15001 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Nesbitt N. M., Arora D. P., Johnson R. A., Boon E. M., Modification of a bi-functional diguanylate cyclase-phosphodiesterase to efficiently produce cyclic diguanylate monophosphate. Biotechnol. Rep. (Amst.) 7, 30–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu N., Pak T., Boon E. M., Characterization of a diguanylate cyclase from Shewanella woodyi with cyclase and phosphodiesterase activities. Mol. Biosyst. 6, 1561–1564 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Thijs L., et al., Characterization of a globin-coupled oxygen sensor with a gene-regulating function. J. Biol. Chem. 282, 37325–37340 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Yang C.-Y., et al., The structure and inhibition of a GGDEF diguanylate cyclase complexed with (c-di-GMP)(2) at the active site. Acta Crystallogr. D Biol. Crystallogr. 67, 997–1008 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Chan C., et al., Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. U.S.A. 101, 17084–17089 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christen B., et al., Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 281, 32015–32024 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Ishitsuka Y., et al., Arg97 at the heme-distal side of the isolated heme-bound PAS domain of a heme-based oxygen sensor from Escherichia coli (Ec DOS) plays critical roles in autoxidation and binding to gases, particularly O2. Biochemistry 47, 8874–8884 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Bennett B. D., et al., Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paul R., et al., Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18, 715–727 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinhouse H., et al., c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 416, 207–211 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Reinders A., et al., Expression and genetic activation of cyclic Di-GMP-specific phosphodiesterases in Escherichia coli. J. Bacteriol. 198, 448–462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plate L., Marletta M. A., Nitric oxide-sensing H-NOX proteins govern bacterial communal behavior. Trends Biochem. Sci. 38, 566–575 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ignarro L. J., Buga G. M., Wood K. S., Byrns R. E., Chaudhuri G., Endothelium-derived relaxing factor produced and released from artery and vein is nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 84, 9265–9269 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Derbyshire E. R., Marletta M. A., Structure and regulation of soluble guanylate cyclase. Annu. Rev. Biochem. 81, 533–559 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Sudarsan N., et al., Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321, 411–413 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Römling U., Amikam D., Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9, 218–228 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Tuckerman J. R., Gonzalez G., Gilles-Gonzalez M.-A., Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J. Mol. Biol. 407, 633–639 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Petters T., et al., The orphan histidine protein kinase SgmT is a c-di-GMP receptor and regulates composition of the extracellular matrix together with the orphan DNA binding response regulator DigR in Myxococcus xanthus. Mol. Microbiol. 84, 147–165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee V. T., et al., A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65, 1474–1484 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicastro G. G., et al., c-di-GMP-related phenotypes are modulated by the interaction between a diguanylate cyclase and a polar hub protein. Sci. Rep. 10, 3077 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martínez-Ruiz A., Lamas S., Two decades of new concepts in nitric oxide signaling: From the discovery of a gas messenger to the mediation of nonenzymatic posttranslational modifications. IUBMB Life 61, 91–98 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Spiro S., Regulators of bacterial responses to nitric oxide. FEMS Microbiol. Rev. 31, 193–211 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Justino M. C., Vicente J. B., Teixeira M., Saraiva L. M., New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J. Biol. Chem. 280, 2636–2643 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Pacher P., Beckman J. S., Liaudet L., Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 87, 315–424 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang W., Olson J. S., Phillips G. N. Jr, Biophysical and kinetic characterization of HemAT, an aerotaxis receptor from Bacillus subtilis. Biophys. J. 88, 2801–2814 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tarnawski M., Barends T. R., Schlichting I., Structural analysis of an oxygen-regulated diguanylate cyclase. Acta Crystallogr. D Biol. Crystallogr. 71, 2158–2177 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Rivera S., et al., Structural insights into oxygen-dependent signal transduction within globin coupled sensors. Inorg. Chem. 57, 14386–14395 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Tsai A.-L., Berka V., Martin E., Olson J. S., A “sliding scale rule” for selectivity among NO, CO, and O2 by heme protein sensors. Biochemistry 51, 172–186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winger J. A., Derbyshire E. R., Marletta M. A., Dissociation of nitric oxide from soluble guanylate cyclase and heme-nitric oxide/oxygen binding domain constructs. J. Biol. Chem. 282, 897–907 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Wood K. C., et al., Picomolar nitric oxide signals from central neurons recorded using ultrasensitive detector cells. J. Biol. Chem. 286, 43172–43181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Batchelor A. M., et al., Exquisite sensitivity to subsecond, picomolar nitric oxide transients conferred on cells by guanylyl cyclase-coupled receptors. Proc. Natl. Acad. Sci. U.S.A. 107, 22060–22065 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Olea C., Boon E. M., Pellicena P., Kuriyan J., Marletta M. A., Probing the function of heme distortion in the H-NOX family. ACS Chem. Biol. 3, 703–710 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weinert E. E., Phillips-Piro C. M., Marletta M. A., Porphyrin π-stacking in a heme protein scaffold tunes gas ligand affinity. J. Inorg. Biochem. 127, 7–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhagi-Damodaran A., et al., Insights into how heme reduction potentials modulate enzymatic activities of a myoglobin-based functional oxidase. Angew. Chem. Int. Ed. Engl. 56, 6622–6626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Faulkner K. M., Bonaventura C., Crumbliss A. L., A spectroelectrochemical method for differentiation of steric and electronic effects in hemoglobins and myoglobins. J. Biol. Chem. 270, 13604–13612 (1995). [DOI] [PubMed] [Google Scholar]
- 72.Addison A. W., Burman S., Ligand-dependent redox chemistry of Glycera dibranchiata hemoglobin. Biochim. Biophys. Acta 828, 362–368 (1985). [Google Scholar]
- 73.Antonini E., et al., Studies on the oxidation-reduction potentials of heme proteins. I. Human hemoglobin. J. Biol. Chem. 239, 907–912 (1964). [PubMed] [Google Scholar]
- 74.Balland V., et al., Functional implications of the propionate 7-arginine 220 interaction in the FixLH oxygen sensor from Bradyrhizobium japonicum. Biochemistry 45, 2072–2084 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Makino R., et al., Oxygen binding and redox properties of the heme in soluble guanylate cyclase: Implications for the mechanism of ligand discrimination. J. Biol. Chem. 286, 15678–15687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gorman D. S., Levine R. P., Photosynthetic electron transport chain of Chlamydomonas reinhardi. V. Purification and properties of Cytochrome 553 and Ferredoxin. Plant Physiol. 41, 1643–1647 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oktyabrskii O. N., Smirnova G. V., Redox potential changes in bacterial cultures under stress conditions. Microbiology 81, 131–142 (2012). [Google Scholar]
- 78.Shapiro H. M., “Flow cytometry of bacterial membrane potential and permeability” in New Antibiotic Targets, Champney W. Scott, Ed. (Humana Press, 2008), pp. 175–186. [DOI] [PubMed] [Google Scholar]
- 79.Micsonai A., et al., Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 112, E3095–E3103. (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kastner B., et al., GraFix: Sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Phippen C. W., et al., Formation and dimerization of the phosphodiesterase active site of the Pseudomonas aeruginosa MorA, a bi-functional c-di-GMP regulator. FEBS Lett. 588, 4631–4636 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Sundriyal A., et al., Inherent regulation of EAL domain-catalyzed hydrolysis of second messenger cyclic di-GMP. J. Biol. Chem. 289, 6978–6990 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deepthi A., Liew C. W., Liang Z.-X., Swaminathan K., Lescar J., Structure of a diguanylate cyclase from Thermotoga maritima: Insights into activation, feedback inhibition and thermostability. PLoS One 9, e110912 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chouhan O. P., Roske Y., Heinemann U., Biswas S., Structure of the active GGEEF domain of a diguanylate cyclase from Vibrio cholerae. Biochem. Biophys. Res. Commun. 523, 287–292 (2020). [DOI] [PubMed] [Google Scholar]
- 85.De N., Navarro M. V., Raghavan R. V., Sondermann H., Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J. Mol. Biol. 393, 619–633 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Andersen J. F., et al., Kinetics and equilibria in ligand binding by nitrophorins 1-4: Evidence for stabilization of a nitric oxide-ferriheme complex through a ligand-induced conformational trap. Biochemistry 39, 10118–10131 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.