Significance
Drugs that inhibit specific kinases now represent one of the main classes of targeted therapies. A majority of drug development efforts in this space focus on a given kinase in isolation as the target. However, our work demonstrates that in the context of the RAF/MEK/ERK pathway, the relevant target for compounds developed as allosteric MEK inhibitors (MEKi) is not free MEK but RAF/MEK complexes. Because signaling through this pathway is essential in normal tissues, indiscriminate blockade of all MEK activity is not tolerated. Our findings imply that selective inhibition of specific RAF/MEK complexes is an accessible route for development of new MEKi, and they provide a biophysical foundation for better understanding and applying current clinical agents.
Keywords: BRAF, MEK, allosteric kinase inhibitor, MEK inhibitor, X-ray crystallography
Abstract
The RAF/MEK/ERK pathway is central to the control of cell physiology, and its dysregulation is associated with many cancers. Accordingly, the proteins constituting this pathway, including MEK1/2 (MEK), have been subject to intense drug discovery and development efforts. Allosteric MEK inhibitors (MEKi) exert complex effects on RAF/MEK/ERK pathway signaling and are employed clinically in combination with BRAF inhibitors in malignant melanoma. Although mechanisms and structures of MEKi bound to MEK have been described for many of these compounds, recent studies suggest that RAF/MEK complexes, rather than free MEK, should be evaluated as the target of MEKi. Here, we describe structural and biochemical studies of eight structurally diverse, clinical-stage MEKi to better understand their mechanism of action on BRAF/MEK complexes. We find that all of these agents bind in the MEK allosteric site in BRAF/MEK complexes, in which they stabilize the MEK activation loop in a conformation that is resistant to BRAF-mediated dual phosphorylation required for full activation of MEK. We also show that allosteric MEK inhibitors act most potently on BRAF/MEK complexes rather than on free active MEK, further supporting the notion that a BRAF/MEK complex is the physiologically relevant pharmacologic target for this class of compounds. Our findings provide a conceptual and structural framework for rational development of RAF-selective MEK inhibitors as an avenue to more effective and better-tolerated agents targeting this pathway.
The RAS/RAF/MEK/ERK pathway mediates response to signaling from receptor tyrosine kinases (RTKs) and controls many aspects of cell physiology, including proliferation and differentiation (1–3). Activating, somatic mutations in components of this pathway are the cause of diverse cancers. RTKs (e.g., EGFR), RAS, and BRAF are frequently mutated in cancer, and though rare, oncogenic mutations in MEK have also been described (4, 5). Therefore, individual components of this pathway have been subjected to intense drug discovery and development efforts, which have resulted in a number of clinical candidates and approved drugs, revolutionizing cancer care (6–9). As the central kinase in this pathway and the only known substrate of RAF, MEK is an important cancer drug target (5). However, MEKi have found limited clinical application to date. Trametinib, cobimetinib, and binimetinib are approved for treatment of malignant melanoma driven by BRAF V600E but only in combination with BRAF inhibitors (10). MEKi selumetinib has recently been approved for treatment of certain pediatric patients with neurofibromatosis type 1 who have inoperable plexiform neurofibromas (11).
In humans, there are three RAF proteins (ARAF, BRAF, and CRAF), two MEK proteins (MEK1 and MEK2), and two ERK proteins (ERK1 and ERK2); for simplicity, we refer to them collectively as RAF, MEK, and ERK. Signaling through this three-tiered kinase cascade is initiated by ligand-induced activation of RTKs on the cell surface, which results in GTP-loading of RAS. GTP-bound RAS binds and activates RAF, and RAF in turn activates MEK by phosphorylating it on two serine residues in its activation loop (A-loop), S218 and S222. MEK then phosphorylates ERK, its sole substrate. This description implies a relay-like handoff of phosphorylation marks from upstream toward downstream factors. In accord with this linear or “stepwise” view of the pathway, inhibitors targeting these kinases are widely thought of as “blockers” of a given phosphorylation step (i.e., MEK inhibitors inhibit phosphorylation of ERK). However, this simple model is at odds with the complex pharmacology of MEK inhibitors. Certain MEKi have long been known to differ in their efficacy depending on the mechanism of pathway activation. Cobimetinib and PD0325901 are much more potent inhibitors of proliferation of cell lines driven by the oncogenic BRAFV600E mutant than they are of those driven by oncogenic KRAS mutants, while other MEK inhibitors (GDC-0623 and CH5126766) are potent in both contexts (12, 13). How do MEK inhibitors “distinguish” between MEK activated by these differing upstream mutations?
Drug action on RAF/MEK complexes, rather than inhibition of free MEK, would provide a potential resolution of this paradox. A given agent might then be expected to differ in potency on a BRAFV600E/MEK complex as compared with a CRAF/MEK complex. Because CRAF is thought to be the dominant RAF isoform activated by oncogenic KRAS (12, 14–16), lack of potency on CRAF/MEK complexes could explain the lack of efficacy of a particular MEKi in the setting of oncogenic KRAS. Almost all clinical stage MEKi are allosteric (5); they bind a pocket in the inactive conformation of the kinase that is formed in part by the regulatory C-helix, which is displaced outward, and by an α-helix in the N-terminal portion of the kinase A-loop (17). The A-loop helix encompasses the S218 and S222 activating phosphorylation sites, and some MEKi have long been known to interfere with its phosphorylation by RAF (12, 13, 18–21). Allosteric MEKi are also known to affect the stability of RAF/MEK complexes, with certain inhibitors reported to either stabilize or destabilize the complex (12, 13). Furthermore, recent structural and biochemical studies show that MEK and RAF are maintained in a mutually autoinhibited complex in the quiescent state (22–25). This quiescent complex also contains a 14–3-3 dimer—a key regulatory subunit—and the integrated RAF/MEK/14–3-3 complex, rather than RAF alone, functions as the RAS-activated switch that initiates signaling through the pathway (24). Collectively, these observations suggest that allosteric MEKi may act on the RAF/MEK complex to inhibit MEK signaling or, at a minimum, that their mechanism of action can be impacted by RAF. Indeed, one MEKi, CH5126766, has been described as a dual RAF/MEK inhibitor that inhibits MEK phosphorylation and also allows drug-bound MEK to act in a dominant negative manner by binding to RAF, blocking its activity (12, 20).
Enzyme inhibitors can exhibit idiosyncratic mechanisms of action beyond simple enzyme inhibition, including trapping of enzyme/substrate complexes or by binding primarily to the substrate rather than the enzyme. For example, PARP inhibitors exert their cytotoxic effect in part by trapping PARP–DNA complexes (26). Topoisomerase I inhibitors such as topotecan stack between base pairs in substrate DNA to trap a covalent topoisomerase I DNA adduct (27). Despite the allosteric binding site of MEK inhibitors and their known effects on phosphorylation of MEK itself, MEK inhibitors are widely regarded as agents that inhibit the ability of MEK to phosphorylate ERK. Furthermore, most structural studies of MEK inhibitors have employed MEK alone (12, 13, 17, 28–32). To better understand their mechanism of action on BRAF/MEK complexes, we undertook comparative structural and biochemical studies of eight structurally diverse, clinical-stage MEK inhibitors (SI Appendix, Fig. S1). We describe crystal structures of seven MEKi bound to the BRAF/MEK complex as well as an inhibitor-free structure of the complex. We compare features of these structures to each other and to existing structural information on MEK inhibitor complexes. We further dissect MEKi mechanism of action through a range of in vitro activity and binding assays. Collectively, our studies indicate that all the allosteric MEKi studied act on BRAF/MEK complexes to stabilize the MEK activation segment in a native or near-native conformation that is resistant to dual-phosphorylation by BRAF.
Results
Crystal Structures of Allosteric MEKi Bound to the BRAF/MEK1 Kinase Module.
We and others have recently described crystal structures of a mutually autoinhibited BRAF/MEK1 kinase domain complex with MEKi GDC-0623 and without a MEK inhibitor (24, 25). Although these crystal structures contain only the kinase domain of BRAF, they reveal the same inactive conformations of both MEK1 and BRAF kinase domains observed in the cryogenic electron microscopy (cryo-EM) structure of the complete BRAF/MEK1/14–3-3 complex in the autoinhibited state (24). We now report structures with seven additional MEKi (trametinib, selumetinib, cobimetinib, binimetinib, pimasertib, CH5126766, and PD0325901; refer to SI Appendix, Fig. S1 for chemical structures of the inhibitors) as well as an additional structure of inhibitor-free BRAF/MEK complex with an ATP analog crystallized in the same conditions as the majority of our MEKi complexes (Materials and Methods and SI Appendix, Table S1). All eight structures superimpose closely with each other and with the previously reported structures (24, 25) (RMSD ∼0.2 to 0.45 Å for the MEK portion of the structures). However, we do notice a small change in the conformation of the BRAF and MEK activation segment in a subset of the structures that we attribute to differing crystallization conditions and pH (6.5 versus 8.5) rather than effects of inhibitor binding (SI Appendix, Fig. S2).
As previously described (24, 25), the BRAF and MEK kinase domains bind with their active sites juxtaposed, and both kinases exhibit characteristic inactive conformations (Fig. 1A). MEK1 and BRAF interact extensively in this complex, with contacts between their N-lobes, activation segments, and αG helices. In the BRAF kinase domain, an inhibitory turn in the A-loop enforces an outward, inactive position of the αC-helix (Fig. 1B). Interactions among the phosphate binding loop, inhibitory turn, and bound ATP analog Adenosine 5′(β,γ-imido)triphosphate (AMPPNP) further stabilize this inactive state, which exhibits a very “closed” relative orientation of the kinase N- and C-lobes. In MEK1, the C-helix also adopts an outward, inactive position where it is stabilized by the A-loop helix (Fig. 1B). The overall MEK1 conformation, including the N-terminal helix which aids in maintaining the inhibited state, is closely similar to that observed for the intact MEK kinase domain alone (28). It is also quite similar to that in a prior structure of a BRAF/MEK complex determined with BRAF in an active, dimeric configuration, although the N-terminal helix was deleted in the latter structure (22).
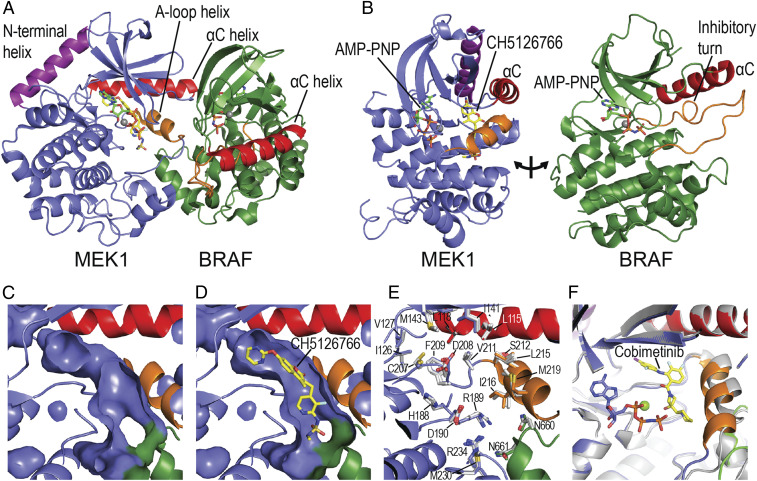
Fig. 1.
Overall structure of the BRAF/MEK complex and allosteric inhibitor site. (A) Ribbon diagram showing the overall structure of the mutually autoinhibited BRAF/MEK complex, with AMP-PNP bound in both kinase active sites and CH5126766 in the MEK allosteric site. (B) An approximate “open-book” view of the complex, showing the relationship of the ATP site and allosteric site in MEK and of the A-loop helix (orange) and C-helix (red) in forming the allosteric site. In both MEK and BRAF, the C-helix is in an outward, inactive position, stabilized by the A-loop helix in MEK and the inhibitory turn in BRAF. (C and D) Surface representations of the allosteric site in the inhibitor-free crystal structure (C) and the CH5126766 complex (D). (E) Most inhibitors have little effect on the conformation of the preformed allosteric pocket. The inhibitor-free structure is shown as a ribbon diagram, with sidechains of residues lining the allosteric pocket shown in stick form and colored as in A above. Side chains of three representative inhibitor bound structures (CH5126766, trametinib, and binimetinib) are superimposed and shown with carbon atoms colored white. (F) In the cobimetinib complex, the A-loop helix (orange) is rotated inward compared with its position in the absence of inhibitor, narrowing the allosteric pocket by ∼2 Å toward its C terminus. The cobimetinib complex is colored as in A, and the inhibitor-free structure is shown as a white ribbon.
Comparison of the BRAF/MEK1 structures with and without bound MEKi shows that the compounds bind in an elongated, preformed pocket that runs the length of the MEK A-loop helix (Fig. 1 C and D). Although the binding pocket is largely formed by MEK1, it is closed on one end by BRAF residues BRAFN660, BRAFN661, and BRAFR662, which lie at the N terminus of its αG helix. With the exception of cobimetinib, the bound inhibitors do not appreciably alter the conformation of binding pocket (Fig. 1 E and F).
All the MEKi studied here insert an aromatic group, typically a 4-bromo-2-fluorophenyl or 4-iodo-2-fluorophenyl group, into a lipophilic site at the back of the inhibitor-binding pocket (Fig. 2 and SI Appendix, Fig. S3). MEKi invariably position a polar or hydrogen bond–accepting group to interact with the mainchain amide of MEKS212 in the MEK activation segment (Fig. 2). This interaction is crucial for MEK inhibition and has been proposed to contribute to stabilization of the helical conformation of the activation segment, rendering it resistant to phosphorylation by RAF (13). A subset of MEK inhibitors contain a polar arm that forms hydrogen bonds with phosphate oxygens of the bound nucleotide. Cobimetinib, trametinib, and CH5126766 lack this feature and instead make additional hydrophobic interactions along the activation segment helix (Fig. 2).
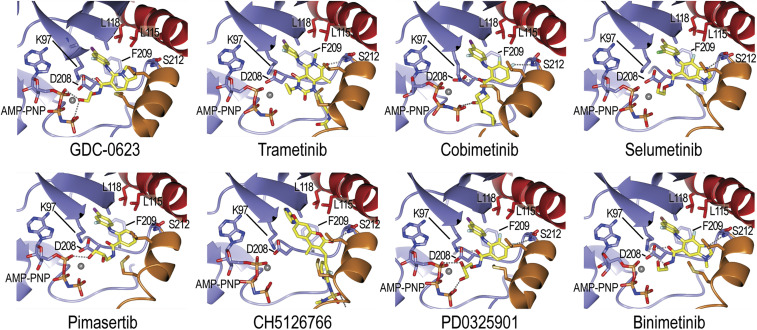
Fig. 2.
Details of MEK allosteric inhibitor binding. Inhibitors are shown in stick form with carbon atoms colored yellow. Hydrogen bonds are shown as dashed lines. Structural elements are colored as in Fig. 1 (A-loop helix orange, αC helix red), and selected side chains that form the allosteric binding pocket are labeled. Note that all inhibitors form a hydrogen bond with the mainchain amide of MEKS212 in the A-loop helix. Refer also to SI Appendix, Fig. S3 for corresponding electron density maps.
We also compared our results to available crystal structures of MEKi bound to MEK alone (12, 28, 33, 34). MEK conformations vary considerably in these prior structures. Without stabilizing interactions with RAF, the MEK C-helix usually assumes a somewhat more outward orientation, and the activation segment conformation is quite variable (SI Appendix, Fig. S4). Despite this variability, all MEKi adopt similar binding poses in both MEK alone and the BRAF/MEK complex (SI Appendix, Fig. S4). In the BRAF/MEK complexes, most of the MEKi do not directly contact BRAF, but CH5126766 and trametinib are exceptions. CH5126766 is in van der Waals contact with BRAFN660 and BRAFN661, and trametinib contacts BRAFR662.
The structure of trametinib bound to MEK1 in complexes with KSR1 and KSR2 has recently been described (35). Kinase suppressor of RAS (KSR) proteins are regulators of RAS/RAF/MEK/ERK pathway signaling that have a pseudokinase domain that, like the RAF kinase domain, can bind to MEK (36, 37). A comparison of these structures with the trametinib complex described here reveals a similar overall binding mode, despite the altered conformation of MEK1 when bound to KSR1/2 versus BRAF (SI Appendix, Fig. S5). As with the free MEK1 structures, the MEK1 C-helix and activation segment helix assume a more outward position in the KSR complex, likely because KSR does not make N-lobe contacts analogous to those of BRAF.
Collectively, our structural analysis reveals that the allosteric MEKi studied here bind in a pocket formed primarily by MEK but with minor direct contributions by BRAF. In addition, BRAF shapes the MEK portion of the pocket by altering the position of the MEK C-helix and A-loop helix. Despite the fact that inhibitors engage similarly regardless of whether MEK is free or bound to BRAF, these observations show clearly that the BRAF/MEK complex represents a structurally distinct target as compared with free MEK. Likewise, the allosteric pocket in the BRAF/MEK complex differs from that in the KSR/MEK complexes (35).
MEKi Vary Widely in Potency against Active, Phosphorylated MEK.
To examine further the mechanism of allosteric MEKi, we compared the potency of these inhibitors in two assays. The first was a cascade assay in which BRAF was used to phosphorylate unphosphorylated MEK (uMEK), and the inhibition was measured by monitoring levels of ERK phosphorylation. The second was a direct assay in which we measured inhibition of ERK phosphorylation by phosphorylated MEK (pMEK). The pMEK was preactivated via phosphorylation by BRAF and subsequently repurified to remove BRAF. For both formats, we used a sensitive radiometric assay and low concentrations of MEK (0.5 nM for both uMEK and pMEK) to allow reasonable discrimination in the activity of these very potent inhibitors.
As expected, all inhibitors were potent in the cascade assay format with uMEK, with low nanomolar or subnanomolar half-maximal inhibitory concentration (IC50) values, comparable to previously reported values for these agents (Table 1 and SI Appendix, Fig. S6). However, potencies for the same inhibitor differed markedly in the direct inhibition assay with pMEK, displaying a wide range of differences between their effect on pMEK vs. uMEK. Trametinib was the most potent (IC50 = 11 nM) and exhibited a shift of ∼25-fold from its IC50 in the cascade assay (0.42 nM). Conversely, CH5126766 inhibited pMEK only very weakly (IC50 >2 μM), a shift of more than 200-fold from its potency in the cascade assay (6.6 nM). Three structurally similar compounds exhibited the narrowest differences in potency on uMEK versus pMEK; GDC-0623, selumetinib, and binimetinib exhibited shifts in potency of seven-, three-, and fivefold, respectively. PD0325901, cobimetinib, and pimasertib exhibited intermediate potencies on pMEK, with shifts of 40- to 80-fold relative to their activity in the cascade assay with uMEK. Overall, these experiments show that potency on the BRAF/MEK complex is separable from potency on free pMEK.
Table 1.
MEK inhibitor potency (IC50) against BRAF/uMEK versus pMEK
| Inhibitor | BRAF/uMEK nM | Literature (ref.) nM | pMEK nM | pMEK/uMEK ratio |
| Selumetinib | 9.4 | 14 (45) | 35 | 3.7 |
| Binimetinib | 20.8 | 12 (46) | 104 | 5.0 |
| GDC-0623 | 4.8 | 0.13 (13) | 34 | 7.1 |
| Cobimetinib | 1.4 | 0.92 (47) | 57 | 40.7 |
| Pimasertib | 2.7 | 5–11 (48) | 180 | 66.7 |
| PD0325901 | 1.6 | 0.33 (49) | 130 | 81.3 |
| Trametinib | 0.42 | 0.92 (50) | 11.2 | 26.7 |
| CH5126766 | 6.6 | 19 (20)* | >2,000 | >200 |
In the referenced study, potency of CH5126766 was measured in an assay for inhibition of MEK phosphorylation by BRAF.
MEK Inhibitors Suppress Dual S218/S222 Phosphorylation and Differentially Modulate Single-Site Phosphorylation as Well as BRAF/MEK Complex Affinity.
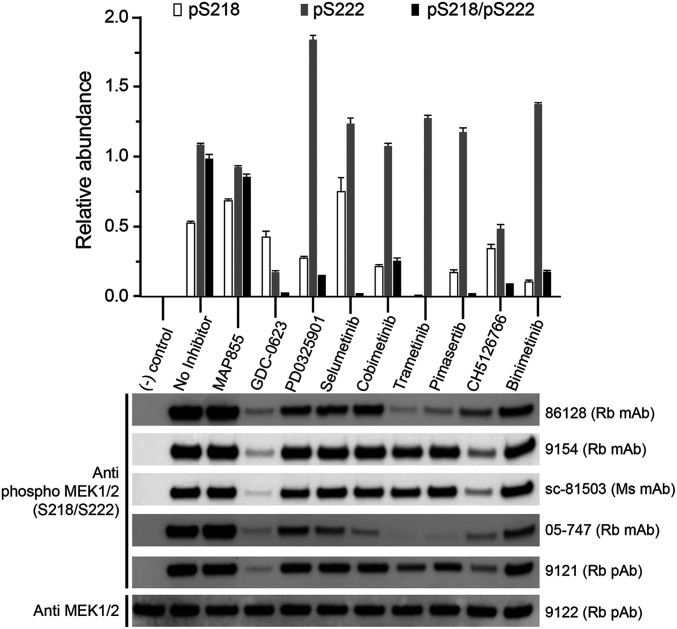
Several allosteric MEKi are known to affect phosphorylation of MEK by RAF, and a subset have been shown to alter the affinity of MEK for RAF (12, 13, 18–20). Either or both of these effects could explain their greater potency in the cascade assay with uMEK, but to our knowledge, the effects of MEK inhibitors on these properties have not been systematically investigated across diverse MEK inhibitor chemotypes. We examined the effect of the eight allosteric MEK inhibitors and one ATP-competitive MEK inhibitor (MAP855) on MEK phosphorylation by BRAF in an in vitro kinase assay with purified uMEK (kinase dead mutant) and a purified full-length active BRAF/14–3-3 complex. We used a saturating inhibitor concentration (10 μM), which was five times the uMEK concentration (2 μM). We measured phosphorylation in two ways: Western blotting with a panel of five anti-pMEK antibodies and a quantitative mass spectrometry assay calibrated with synthetic peptides. Strikingly, all the allosteric inhibitors markedly decreased dual pS218/pS222 phosphorylation of the MEK activation segment, with GDC-0623, selumetinib, trametinib, and pimasertib showing the levels less than 5% of that in the absence of inhibitor (Fig. 3, Upper). Cobimetinib, PD0325901, CH5126766, and binimetinib also diminished dual phosphorylation, albeit to a somewhat lesser extent. Effects on single-site phosphorylation were far more variable. GDC-0623 and CH5126766 were unique in decreasing single-site phosphorylation at both pS218 and pS222. The other six allosteric inhibitors all showed increased single-site phosphorylation on MEKS222. In all cases, the extent of single-site phosphorylation on MEKS222 was less than the sum of MEKS222 and MEKS218/S222 phosphorylation in the absence of inhibitor. Thus, the apparent increase in single-site MEKS222 phosphorylation likely stems from a decrease in MEKS218 phosphorylation, leading to less doubly phosphorylated MEKS218/S222 rather than from an increase in MEKS222 phosphorylation. Single-site phosphorylation of MEKS218 was also quite variable; with trametinib, it was inhibited almost entirely, while with selumetinib, it was modestly increased relative to the inhibitor free reaction (Fig. 3, Upper). The ATP-competitive inhibitor MAP855 had little effect on MEK phosphorylation in this assay.
Fig. 3.
Allosteric MEK inhibitors suppress dual phosphorylation of the MEK A-loop. Kinase-dead MEK1 was treated with active, full-length BRAF in the presence or absence of the indicated inhibitors at a concentration of 10 μM, and reaction mixtures were analyzed by quantitative mass spectrometry (Upper) and Western blotting (Lower). The control lane was left untreated; MAP855 is an ATP-competitive inhibitor. In the upper panel, the abundance of each phosphorylated species is normalized to that of the doubly-phosphorylated pS218/pS222 peptide in the sample with no inhibitor. Western blotting was carried out with a panel of five phospho-specific anti-MEK pS218/pS222 antibodies (9121, 9154, and 86128 are from Cell Signaling Technologies; sc-81503 from Santa Cruz Biotechnology; and 05–747 from Millipore-Sigma) and with an Anti-MEK1/2 antibody (9122, Cell Signaling Technologies). Antibody type is indicated as follows: Rb, rabbit; Ms, mouse; mAb, monoclonal antibody; and pAb, polyclonal antibody.
With antibody detection, we noticed dramatic variability among commercially available anti-pMEK antibodies (Fig. 3, Lower). All five antibodies cleanly discriminate between uMEK and MEK that was phosphorylated in the absence of inhibitor. However, detection varied widely and sometimes in a manner that was not obviously correlated with actual phosphorylation state as measured by mass spectrometry, likely due to complex effects of single-site phosphorylation. As an aside, we note that accumulation of reactivity with anti-pMEK antibodies is observed in cell-based experiments with PD0325901 and certain other MEKi and has been attributed to a block of feedback inhibition of the pathway. This effect is not observed with CH5126766, GDC-0623, and, to some extent, trametinib (12, 13), and these MEKi have been termed “feedback busters” (5). We observe the same pattern in this in vitro experiment in which feedback does not occur; rather, it is due to the direct action of MEK inhibitors on the BRAF/MEK complex.
We also measured the effect of allosteric inhibitors on the affinity of MEK for BRAF using biolayer interferometry (BLI). In these experiments, the His6-tagged BRAF kinase domain was immobilized on the sensor tip, and the affinity of uMEK or pMEK was measured in the presence or absence of each of the inhibitors at a concentration of 2 μM. Consistent with prior studies, we observed high affinity binding of uMEK to BRAF (20 nM) and much lower affinity interaction with pMEK (330 nM) (12, 13). The association/dissociation traces for uMEK binding to BRAF reveal a relatively slow on, slow off kinetic behavior (SI Appendix, Fig. S7A). The pMEK traces, in contrast, show a more rapid association phase and a much faster rate of dissociation (SI Appendix, Fig. S7B). Introducing a point mutation in the MEK αG helix (MEKL314E) that disrupts interaction with BRAF (13) resulted in a loss of binding, confirming the specificity of the assay (SI Appendix, Fig. S7C). As shown in Table 2, inhibitors modulated the affinity of uMEK for BRAF, with effects ranging from modestly increased affinity (∼3 nM in the presence of PD0325901) to severalfold-decreased affinity (96 nM in the presence of cobimetinib). With pMEK, all of the inhibitors tested increased affinity for BRAF, typically only modestly (1.5- to fivefold). CH5126766 was a notable exception; in the presence of this compound we measured an affinity of ∼10 nM for pMEK to BRAF, similar to the affinity of uMEK (Table 2 and SI Appendix, Fig. S7D). Taken together, our results show that MEK inhibitors exert distinct effects on BRAF-mediated uMEK phosphorylation. While all MEKi tested suppress dual phosphorylation of MEK, they vary in their ability to inhibit single-site phosphorylation and to modulate BRAF/MEK complex affinity.
Table 2.
Effect of MEK inhibitors on affinity of uMEK and pMEK for BRAF kinase*
| Inhibitor | uMEK Kd (nM) | pMEK Kd (nM) |
| No MEKi | 20 ± 1.9 | 330 ± 7.0 |
| Selumetinib | 48 ± 5.9 | 110 ± 12 |
| Binimetinib | 20 ± 2.3 | 220 ± 11 |
| GDC0623 | 13 ± 2.0 | 190 ± 20 |
| Cobimetinib | 96 ± 26 | 190 ± 18 |
| Pimasertib | 77 ± 9.6 | 65 ± 9.4 |
| PD0325901 | 3.3 ± 0.5 | 64 ± 5.2 |
| Trametinib | 68 ± 16 | 120 ± 6.6 |
| CH5126766 | 26 ± 5.5 | 10 ± 0.6 |
Measured by BLI, with immobilized BRAF kinase.
Discussion
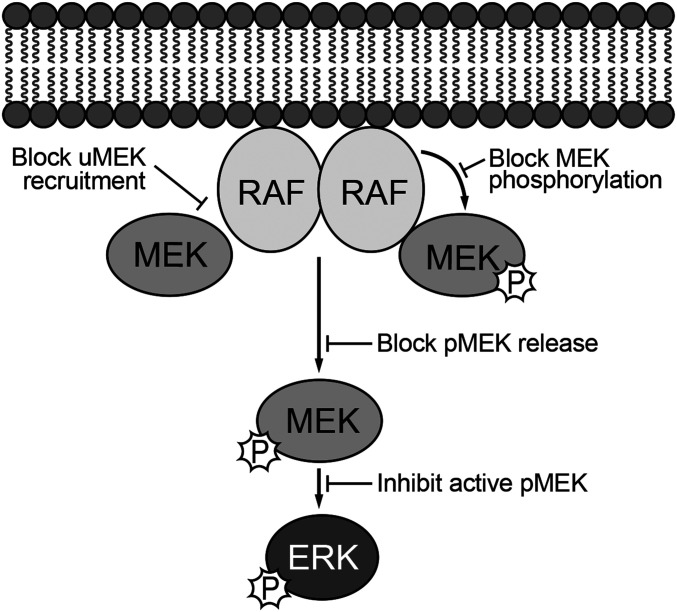
In principle, allosteric MEKi could act via one or more of at least four distinct mechanisms to block activation of RAF/MEK/ERK signaling pathway: 1) by binding to uMEK and inducing a conformation that blocks its recruitment to RAF; 2) by allowing binding of uMEK to RAF but stabilizing or inducing a phosphorylation-resistant conformation of MEK; 3) by preventing release of pMEK from RAF, which would likely preclude ERK phosphorylation; or 4) inhibiting free pMEK to block its ability to phosphorylate ERK (Fig. 4). Our findings show that as a class, allosteric MEK inhibitors act most potently on BRAF/MEK complexes to block MEK activation, primarily by preventing dual-phosphorylation of MEKS218/S222 by BRAF. Cocrystal structures with the BRAF/MEK complex show that they do so by filling the large preexisting pocket behind the MEK A-loop, where they establish favorable interactions that apparently stabilize it in a phosphorylation-resistant conformation.
Fig. 4.
Paradigm for understanding the mechanisms of action of allosteric MEK inhibitors. Allosteric inhibitors could in principle act on free uMEK to interfere with its recruitment to RAF or on RAF-MEK complexes to block or alter phosphorylation of MEK by RAF or its release from RAF once phosphorylated. Allosteric MEK inhibitors could also inhibit free, active pMEK. We find that all inhibitors studied here act most potently on BRAF/MEK1 complexes in which they inhibit the dual phosphorylation of MEK that is required for its activation. Inhibition of active pMEK was also observed but with variable and sometimes dramatic loss of potency.
While the results reported here suggest that allosteric MEKi do not act primarily by preventing binding of uMEK to RAF, we do observe that these agents modulate affinity of uMEK for BRAF. Thus, it is possible that this effect may contribute in a cellular context where MEK concentrations are more limiting. Treatment of cells with trametinib and certain other MEK inhibitors is reported to decrease coimmunoprecipitation of MEK with RAFs, while others, including CH5126766 and GDC-0623 appear to stabilize RAF/MEK complexes (12, 13, 20). However, differential phosphorylation of MEK is also at play in these cell-based experiments and will lead to differences in complex stability as a secondary effect. With respect to preventing release of pMEK from RAF, all inhibitors tested at least modestly increased the affinity of pMEK for BRAF at the high inhibitor concentrations tested (2 μM), and one did so dramatically (CH5126766). However, the contribution of this mechanism is likely to be minor because these inhibitors also prevent MEK phosphorylation. In terms of direct pMEK inhibition, we do observe that MEKi inhibit free pMEK, albeit with dramatically decreased potency in most cases. Although no crystal structure of pMEK is currently available, based on what is known for other kinases, phosphorylation is expected to “close” the allosteric inhibitor binding pocket, thus antagonizing allosteric inhibitor binding, in agreement with the loss of potency we observe here.
Because they are most potent as inhibitors of MEK activation, allosteric MEK inhibitors are in effect RAF inhibitors that happen to bind primarily to the substrate in the enzyme/substrate complex, rather than to the enzyme. Because they inhibit phosphorylation of MEK—which normally promotes its release from RAF—MEKi can trap RAF/MEK complexes. This effect is pronounced with CH5126766 and GDC-0623 (12, 13), the two MEKi that suppress both single-site and dual phosphorylation of MEKS218/S222 (Fig. 3). While most of these drugs also inhibit pMEK, albeit at higher concentrations, potency on pMEK is clearly separable from potency on the RAF/MEK complex. The chemotype exemplified by selumetinib exhibits rather narrow selectivity, while that of CH5126766 exceeds two orders of magnitude (Table 1).
Given these observations, one can imagine rational development of RAF-selective allosteric MEK inhibitors—for example, an agent that would be potent at inhibiting BRAF/MEK but able to spare CRAF/MEK and/or ARAF/MEK. Isoform selectivity or even mutant isoform selectivity, together with a lack of potency on active pMEK, could lead to more effective and better-tolerated therapeutics for particular indications in which this pathway is activated. Direct assessment of the potency of existing MEKi against ARAF/MEK, BRAF/MEK, and CRAF/MEK complexes is an important area for further study. Although they were not developed to be RAF selective, some current MEKi may exhibit a degree of isoform selectivity. Cobimetinib, for example, was observed to be a more-potent inhibitor in cascade assays with BRAFV600E than with CRAF (13). Furthermore, the lack of efficacy of PD0325901 in KRAS-mutant cell lines as compared with BRAFV600E-driven cell lines has been attributed to activation of CRAF by oncogenic KRAS (12), an observation that is consistent with more-potent inhibition of BRAFV600E/MEK complexes by this agent. These examples and the results we describe here support the notion that each isoform-specific complex represents a unique target for drug development. More generally, in the context of targeting kinase-mediated signaling pathways, considering kinase complexes as the target, rather than the kinase in isolation, may open opportunities for drug discovery and development, in particular with allosteric inhibitors.
Materials and Methods
Insect Cell Expression and Purification of the BRAF/MEK Kinase Complex.
For insect cell expression of the BRAF kinase domain in complex with MEK1, two recombinant baculovirus species were employed. The first was prepared using baculoviral transfer vector pFastBac Dual and encoded the BRAF kinase domain (BRAF residues 445 through 723, fused to an N-terminal His6 tag and a C-terminal chitin-binding domain) and human chaperone CDC37. The second baculovirus encoded full-length human MEK1SASA (S218A/S222A). For protein production, liter-scale suspension cultures of Sf9 cells were coinfected with both viruses. Cells were harvested 60 to 66 h postinfection and resuspended in lysis buffer (50 mM Tris pH 8.0, 250 mM NaCl, 5% glycerol and 20 mM imidazole, and 1 mM Tris (2-carboxyethyl)-phosphine [TCEP]) with protease inhibitor mixture (Roche). Resuspended cells were disrupted by sonication on wet ice, and the lysate was clarified by ultracentrifugation at 40,000 rpm for 2 h. Clarified lysate was batch-bound to nickel-nitrilotriacetic acid (Ni-NTA) agarose beads and washed extensively with binding buffer before elution with elution buffer (50 mM Tris pH 8.0, 250 mM NaCl, 250 mM imidazole, and 1 mM TCEP). The elution fractions were pooled and treated with 1:1,000 molar ratio of tobacco etch virus nuclear-inclusion-a endopeptidase (TEV protease) and 100 mM β-mercaptoethanesulfonic acid (MESNA) overnight to cleave N-terminal and C-terminal tags and further purified by size-exclusion chromatography (Superdex 200 10/300, GE Healthcare) in storage buffer (50 mM Hepes pH 7.5, 150 mM NaCl, and 1 mM TCEP). The fractions were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and fractions corresponding to the BRAF/MEK kinase domain complex were pooled, concentrated to 8 mg/mL, and flash-frozen. The final yield of the BRAF/MEK protein complex was ∼10 mg/6L preparation.
Expression and Purification of the BRAF Kinase Domain for Biophysical and Kinase Assays.
We employed the same BRAF baculoviral expression approach described above to obtain the isolated kinase domain but without coinfection with the MEK-expressing virus. After initial batch-mode Ni-NTA purification, the C-terminal chitin-binding domain was cleaved by overnight incubation of the elution fractions with 100 mM MESNA. The N-terminal His6 tag was retained for use as the capture “handle” for BLI experiments. In preparation for ion exchange chromatography (IEX), the Ni-NTA eluate was carefully diluted 1:3 into IEX buffer A (50 mM Hepes pH 7.5, 1 mM TCEP) and applied to a 5/50 Mono S cation exchange column. After ∼50 column volumes (CV) washing with 5% IEX buffer A, the BRAF kinase was eluted along a 5 to 50% IEX buffer B (50 mM Hepes pH 7.5, 1 M NaCl, 1 mM TCEP) gradient for 20 CVs. Fractions were analyzed by SDS-PAGE, relevant fractions were pooled and concentrated to 16 µM, and 10 µL aliquots were flash-frozen in liquid nitrogen for storage at −80 °C. The final yield from 6L Sf9 culture was 0.8 mg.
Expression and Purification of MEK1.
MEK kinase (residues 62 through 393) was engineered with an N-terminal, TEV-cleavable His6 tag for purification purposes and expressed in Escherichia coli strain BL21 DE3. Cells were grown at 37 °C to an optical density of 0.5 to 0.7, and expression was induced by addition of 0.5 mM IPTG and overnight incubation at 18 °C. Cells were pelleted by centrifugation and resuspended in roughly 20 mL per L of culture of Bind/Wash buffer (50 mM Hepes pH 7.5, 250 mM sodium chloride, 30 mM imidazole, and 1 mM TCEP) with 1 mM phenylmethylsulfonyl fluoride. Cells were disrupted by sonication, and lysate was clarified by centrifugation for 2 h at 17,000 rpm. After centrifugation, the cleared lysate was filtered (0.8-μm syringe filter) and applied to a pre-equilibrated 5 mL HisTrap column. The column was washed with 50 to 80 mL Bind/Wash buffer on an AKTATM Explorer FPLC (fast protein liquid chromatography) until a relatively flat ultraviolet absorbence (UV280) reading was reached, and MEK162-393 was eluted using a gradient from 0 to 50% Elution buffer (50 mM Hepes pH 7.5, 250 mM sodium chloride, 250 mM imidazole, and 1 mM TCEP) and collected in 10-mL fractions. Fractions were pooled, and the His6 tag was then cleaved by addition of TEV protease and incubation overnight at 4 °C. The next day, the MEK/TEV mixture was concentrated using a 30,000 molecular weight cut-off (MWCO) Amicon spin concentrator prerinsed with Bind/wash buffer to no more than 15 mg/mL. Concentrated protein was further purified over a GE Superdex 10/300 S75 column pre-equilibrated in size-exclusion chromatography (SEC) Buffer (50 mM Hepes pH 7.5, 250 mM sodium chloride, and 1 mM TCEP). Elution fractions were analyzed by SDS-PAGE, and relevant fractions were pooled and concentrated to 20 to 25 mg/mL and aliquoted and frozen. The yield of MEK62-393 was ∼10 mg per liter of culture.
BRAF/MEK1/MEKi Kinase Domain Crystallization and Structure Determination.
For crystallization, aliquots of the BRAF/MEK kinase complex were incubated with 5 mM MgCl2 and 2 mM AMPPNP in storage buffer at 4 °C overnight. Rod-shaped crystals suitable for structure determination were obtained by vapor diffusion in hanging drops with a reservoir solution containing 22% PEG3350, 0.1 M Tris pH 8.5, and 0.2 M Lithium Sulfate at room temperature. To obtain complexes with binimetinib, trametinib, cobimetinib, pimasertib, selumetinib, and CH5126766, 2-µL drops containing crystals of the AMPPNP-bound BRAF/MEK complex were spiked with 1 µL of the reservoir solution containing 0.2 to 0.5 mM of the inhibitor of interest for 20 to 30 min prior to harvesting. For the PD0325901 complex, the BRAF/MEK kinase complex was incubated with 5 mM MgCl2, 2 mM AMPPNP, and 0.2 mM PD0325901 in storage buffer at 4 °C overnight, and rod-shaped crystals suitable for structure determination were obtained by vapor diffusion in hanging drops with a reservoir solution containing 22% PEG3350, 0.1M Bis-Tris pH 6.5, and 0.2 M Ammonium Sulfate at room temperature. For all structures, crystals were harvested and flash-frozen in liquid nitrogen using additional 20 to 22% glycerol as a cryoprotectant. X-ray diffraction data were collected at 100 K using NE-CAT beamline ID-24-C at the Advanced Photon Source, Argonne National Laboratory, at a wavelength of 0.9786 Å. Data were integrated and merged using XDS (38) and scaled using Aimless in the CCP4 suite (39) or by the Xia2 suite using the Dials mode (40). Molecular replacement of the AMPPNP-only and all other inhibitor-bound structures was performed using the GDC0623 structure as a search model in PHENIX.PHASER (41, 42). Inhibitors were placed into positive density in an initial Fo-Fc map and included in subsequent rounds of refinement using PHENIX.REFINE (43). Successive manual refinement was performed using COOT (44). The data collection and refinement statistics are presented in SI Appendix, Table S1.
BRAF/uMEK Cascade and pMEK Activity Assays.
Radiometric inhibition assays were performed in Kinase Buffer (100 mM Hepes, pH 7.5, 150 mM sodium chloride, 5 mM magnesium chloride, and 1 mM TCEP). A total 1 µCi of γ-32P ATP (Perkin-Elmer) was diluted 1:1,200 into a 1-mM ATP stock solution, which was further diluted 10-fold for a final ATP concentration of 100 µM. MEK inhibitors were titrated from a starting concentration of 3 µM using eight threefold serial dilutions. Kinase-dead ERK2 was used as a substrate at a final concentration of 1 µM. For the coupled enzyme assays, BRAF445-723 was used at a final concentration of 4 nM, and uMEK62-393 was used at a final concentration of 500 pM. For inhibition assays of pMEK, 500 pM of pMEK was used. The reaction was started by addition of the ATP stock at room temperature (25 °C) and stopped by the 1:1 addition of 0.1 M EDTA after 1 h. Samples were resolved by SDS-PAGE, and the gels were dried for 4 h before exposure to a detection plate overnight. Counts were measured using a phosphorimager, and relative activity was determined by densitometry using ImageJ software. IC50 values were determined using the GraphPad Prism software.
BLI Binding Experiments.
Experiments measuring the binding affinity of MEK1 for the BRAF kinase were performed on an Octet Red 384 Biolayer interferometer (Molecular Devices/ForteBio) at the Center for Macromolecular Interactions at Harvard Medical School. Ni-NTA–coated biosensors (ForteBio/Molecular Devices) were prewetted in Octet Soaking buffer (50 mM Hepes pH 7.5, 150 mM sodium chloride, 5 mM magnesium chloride, 1 mM TCEP, and 0.1% wt/vol bovine serum albumin) for 10 min before the start of the experiment. The preloading baseline was determined by dipping the biosensors in Octet Assay buffer 1 (50 mM Hepes pH 7.5, 150 mM sodium chloride, 5 mM magnesium chloride, 1 mM TCEP, 0.1% wt/vol BSA, and 0.02% Tween-20) for 60 s. His6-tagged BRAF kinase was diluted in Octet Assay buffer 1 to a concentration of 20 µg/mL and loaded onto the biosensors over a period of 120 s. The postloading baseline was determined by dipping the biosensors in Octet Assay buffer 2 (50 mM Hepes pH 7.5, 150 mM sodium chloride, 5 mM magnesium chloride, 1 mM TCEP, 0.1% wt/vol BSA, 0.02% Tween-20, and either 0.5 mM ATP or 0.5 mM AMPPNP) for 120 s. Association measurements were determined by dipping the biosensors into seven threefold serial dilutions of MEK1 (starting concentration 1 µM) and one no-MEK1 well for 300 s followed by a 900-s dissociation phase in the same Octet Assay buffer 2 wells. Dimethyl sulfoxide (DMSO, 0.5% vol/vol) was added to all buffers, and a fixed concentration of 2-µM inhibitor was used. Double subtraction was performed using the no-MEK1 condition as a sample blank and a full dataset collected without loading BRAF onto the sensors. Kinetic and steady-state fits were determined using the Octet Data Analysis HT 11.0 Suite. We report the steady-state fits in Table 2.
MEK Phosphorylation Assay.
Unphosphorylated full-length MEK1 (D190N, kinase dead) was prepared by in vitro dephosphorylation with lambda phosphatase, and complete dephosphorylation was confirmed by mass spectrometry and Western blotting. For the phosphorylation assay, uMEK (KD) at a concentration of 2 μM was incubated with 5 nM active, full-length BRAF/14–3-3 complex prepared in insect cells as previously described (16). Reaction was carried out in presence of absence of each inhibitor at a concentration of 10 μM for 1 h and quenched by addition of EDTA (100 mM final concentration). Western blotting was performed with a panel of five phospho-specific anti-MEK pS218/pS222 antibodies (9121, 9154, and 86128 from Cell Signaling Technologies; sc-81503 from Santa Cruz Biotechnology; and 05–747 from Millipore-Sigma) and with an Anti-MEK1/2 antibody (9122, Cell Signaling Technologies).
To quantify MEK1 S218/S222 phosphorylation by mass spectrometry, standard stable isotope labeled peptides synthesized by New England Peptide were spiked into kinase assays at 2.5-pmol peptides to 0.5-µg MEK1. Standard peptides included the tryptic MEK1 A-loop phosphorylation sites (LCCAMDFGVSGQLIDSMANSFVGTR) in unphosphorylated, singly, and doubly phosphorylated states as well as two proteotypic peptides (YPIPPPDAK and LEAFLTQK). Samples were denatured in 8 M urea and subjected to reduction and carbamidomethylation with TCEP and iodoacetamide. Samples were diluted to 2 M urea and digested with trypsin (1:50 ratio of trypsin:protein) overnight at 32 °C. Digestions were brought to 1% formic acid (FA), and peptides were dried by vacuum centrifugation and desalted over a C18 column. Peptides were resuspended in 5% acetonitrile (MeCN)/1% FA and analyzed on an Ultimate 3000 RSLCnano system coupled to an Orbitrap Eclipse mass spectrometer (Thermo Fisher Scientific). Peptides were separated across a 75-min gradient of 5 to 43% MeCN in 1% FA over a 50-cm C18 column (ES803A, Thermo Fisher Scientific) and electrosprayed (2.15 kV, 300 °C) into the mass spectrometer with an EasySpray ion source (Thermo Fisher Scientific). Precursor ion scans (300 to 2,000 m/z) were obtained in the orbitrap at 120,000 resolution in profile (RF lens % = 30, Max IT = 100 ms). Fragment ion scans with a 0.7 m/z isolation window, HCD (higher-energy C-trap dissociation) at 30% NCE (normalized collision energy), and 30,000 resolution were prioritized for a precursor inclusion list including m/z values for native and synthetic peptides containing the A-loop sites at z = 3+ and proteotypic peptides at z = 2+, which were previously determined to be the most abundant charge states. Peptide sequence and phosphorylation site localization were confirmed from fragment ion spectra and retention time, and precursor abundances were quantified from extracted ion chromatograms. Ratios of native-to-standard peptides were normalized to those of proteotypic peptides for comparison across treatments. Experiments were conducted in triplicate.
Supplementary Material
Acknowledgments
We thank M. Kostic for helpful discussions and critical reading and editing of the manuscript and K. Arnett at the Center for Macromolecular Interactions in Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School for guidance in the BLI studies. This work was supported by Grants P50CA165962, R35CA242461, and P01CA154303 from the NIH and by the Pediatric Low-Grade Astrocytoma fund of the Pediatric Brain Tumor Foundation.
Footnotes
Competing interest statement: M.J.E. has received sponsored research support from Novartis Institutes for Biomedical Research and Takeda Pharmaceuticals. M.J.E. is a consultant for Novartis Institutes for Biomedical Research and H3 Biomedicines.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107207118/-/DCSupplemental.
Data Availability
Atomic coordinates and structure factors for the BRAF/MEK crystal structures reported here have been deposited in the Protein Data Bank and are available at https://www.rcsb.org/ with the following accession codes: 6V2W (AMPPNP), 7M0T (selumetinib complex), 7M0U (binimetinib complex), 7M0V (cobimetinib complex), 7M0W (pimasertib complex), 7M0X (PD0325901 complex), 7M0Y (trametinib complex), and 7M0Z (CH5126766 complex).
References
- 1.Lavoie H., Therrien M., Regulation of RAF protein kinases in ERK signalling. Nat. Rev. Mol. Cell Biol. 16, 281–298 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Simanshu D. K., Nissley D. V., McCormick F., RAS proteins and their regulators in human disease. Cell 170, 17–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terrell E. M., Morrison D. K., Ras-mediated activation of the Raf family kinases. Cold Spring Harb. Perspect. Med. 9, a033746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez-Vega F., et al., Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 173, 321–337.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caunt C. J., Sale M. J., Smith P. D., Cook S. J., MEK1 and MEK2 inhibitors and cancer therapy: The long and winding road. Nat. Rev. Cancer 15, 577–592 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Canon J., et al., The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Janes M. R., et al., Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Holderfield M., Deuker M. M., McCormick F., McMahon M., Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer 14, 455–467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atkinson V., et al., Dabrafenib plus trametinib is effective in the treatment of BRAF V600-mutated metastatic melanoma patients: Analysis of patients from the dabrafenib plus trametinib named patient program (DESCRIBE II). Melanoma Res. 30, 261–267 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Subbiah V., Baik C., Kirkwood J. M., Clinical development of BRAF plus MEK inhibitor combinations. Trends Cancer 6, 797–810 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Gross A. M., et al., Selumetinib in children with inoperable plexiform neurofibromas. N. Engl. J. Med. 382, 1430–1442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lito P., et al., Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell 25, 697–710 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatzivassiliou G., et al., Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature 501, 232–236 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Heidorn S. J., et al., Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 140, 209–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blasco R. B., et al., c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell 19, 652–663 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karreth F. A., Frese K. K., DeNicola G. M., Baccarini M., Tuveson D. A., C-Raf is required for the initiation of lung cancer by K-Ras(G12D). Cancer Discov. 1, 128–136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohren J. F., et al., Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat. Struct. Mol. Biol. 11, 1192–1197 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Alessi D. R., Cuenda A., Cohen P., Dudley D. T., Saltiel A. R., PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 270, 27489–27494 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Yoshida T., et al., Identification and characterization of a novel chemotype MEK inhibitor able to alter the phosphorylation state of MEK1/2. Oncotarget 3, 1533–1545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishii N., et al., Enhanced inhibition of ERK signaling by a novel allosteric MEK inhibitor, CH5126766, that suppresses feedback reactivation of RAF activity. Cancer Res. 73, 4050–4060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilmartin A. G., et al., GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin. Cancer Res. 17, 989–1000 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Haling J. R., et al., Structure of the BRAF-MEK complex reveals a kinase activity independent role for BRAF in MAPK signaling. Cancer Cell 26, 402–413 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Diedrich B., et al., Discrete cytosolic macromolecular BRAF complexes exhibit distinct activities and composition. EMBO J. 36, 646–663 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park E., et al., Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes. Nature 575, 545–550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liau N. P. D., et al., Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization. Nat. Struct. Mol. Biol. 27, 134–141 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Murai J., et al., Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staker B. L., et al., The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. U.S.A. 99, 15387–15392 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischmann T. O., et al., Crystal structures of MEK1 binary and ternary complexes with nucleotides and inhibitors. Biochemistry 48, 2661–2674 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Iverson C., et al., RDEA119/BAY 869766: A potent, selective, allosteric inhibitor of MEK1/2 for the treatment of cancer. Cancer Res. 69, 6839–6847 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Wallace M. B., et al., Structure-based design and synthesis of pyrrole derivatives as MEK inhibitors. Bioorg. Med. Chem. Lett. 20, 4156–4158 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Heald R. A., et al., Discovery of novel allosteric mitogen-activated protein kinase kinase (MEK) 1,2 inhibitors possessing bidentate Ser212 interactions. J. Med. Chem. 55, 4594–4604 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Sini P., et al., Pharmacological profile of BI 847325, an orally bioavailable, ATP-competitive inhibitor of MEK and Aurora kinases. Mol. Cancer Ther. 15, 2388–2398 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Rice K. D., et al., Novel carboxamide-based allosteric MEK inhibitors: Discovery and optimization efforts toward XL518 (GDC-0973). ACS Med. Chem. Lett. 3, 416–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robarge K. D., et al., Structure based design of novel 6,5 heterobicyclic mitogen-activated protein kinase kinase (MEK) inhibitors leading to the discovery of imidazo[1,5-a] pyrazine G-479. Bioorg. Med. Chem. Lett. 24, 4714–4723 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Khan Z. M., et al., Structural basis for the action of the drug trametinib at KSR-bound MEK. Nature 588, 509–514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brennan D. F., et al., A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature 472, 366–369 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Lavoie H., et al., MEK drives BRAF activation through allosteric control of KSR proteins. Nature 554, 549–553 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Potterton E., Briggs P., Turkenburg M., Dodson E., A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Winter G., xia2: An expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 43, 186–190 (2010). [Google Scholar]
- 41.McCoy A. J., et al., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams P. D., et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Afonine P. V., et al., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huynh H., Soo K. C., Chow P. K., Tran E., Targeted inhibition of the extracellular signal-regulated kinase kinase pathway with AZD6244 (ARRY-142886) in the treatment of hepatocellular carcinoma. Mol. Cancer Ther. 6, 138–146 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Pheneger J., et al., “Characterization of ARRY-438162, a potent MEK inhibitor in combination with methotrexate or ibuprofen in in vivo models of arthritis" Abstract 794, American College of Rheumatology 2006 Annual Scientific Meeting, Washington, DC (2006). https://acr.confex.com/acr/2006/webprogram/Paper5558.html.
- 47.Hoeflich K. P., et al., Intermittent administration of MEK inhibitor GDC-0973 plus PI3K inhibitor GDC-0941 triggers robust apoptosis and tumor growth inhibition. Cancer Res. 72, 210–219 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Cheng Y., Tian H., Current development status of MEK inhibitors. Molecules 22, 1551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barrett S. D., et al., The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg. Med. Chem. Lett. 18, 6501–6504 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Yamaguchi T., Kakefuda R., Tajima N., Sowa Y., Sakai T., Antitumor activities of JTP-74057 (GSK1120212), a novel MEK1/2 inhibitor, on colorectal cancer cell lines in vitro and in vivo. Int. J. Oncol. 39, 23–31 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the BRAF/MEK crystal structures reported here have been deposited in the Protein Data Bank and are available at https://www.rcsb.org/ with the following accession codes: 6V2W (AMPPNP), 7M0T (selumetinib complex), 7M0U (binimetinib complex), 7M0V (cobimetinib complex), 7M0W (pimasertib complex), 7M0X (PD0325901 complex), 7M0Y (trametinib complex), and 7M0Z (CH5126766 complex).