Significance
Prostate cancer is a major cause of cancer-related death in men. Emerging resistance to androgen receptor (AR) signaling inhibition therapy has heightened the need to identify therapeutic vulnerabilities specific to AR-negative prostate cancers. Analysis of CRISPR screen datasets in human prostate cancer cells identified high dependency for the histone demethylase JMJD1C across multiple AR-negative model systems. Integrated analyses revealed that combined AR and JMJD1C inactivation drives up-regulation of the TNFα network which provokes growth suppression. The exquisite sensitivity of AR-negative prostate cancer cells to JMJD1C inhibition may provide a therapeutic option for men with progressive castration-resistant prostate cancer.
Keywords: JMJD1C demethylase, prostate cancer, AR, synthetic lethality
Abstract
Prostate cancer is a leading cause of cancer-related mortality in men. The widespread use of androgen receptor (AR) inhibitors has generated an increased incidence of AR-negative prostate cancer, triggering the need for effective therapies for such patients. Here, analysis of public genome-wide CRISPR screens in human prostate cancer cell lines identified histone demethylase JMJD1C (KDM3C) as an AR-negative context-specific vulnerability. Secondary validation studies in multiple cell lines and organoids, including isogenic models, confirmed that small hairpin RNA (shRNA)–mediated depletion of JMJD1C potently inhibited growth specifically in AR-negative prostate cancer cells. To explore the cooperative interactions of AR and JMJD1C, we performed comparative transcriptomics of 1) isogenic AR-positive versus AR-negative prostate cancer cells, 2) AR-positive versus AR-negative prostate cancer tumors, and 3) isogenic JMJD1C-expressing versus JMJD1C-depleted AR-negative prostate cancer cells. Loss of AR or JMJD1C generates a modest tumor necrosis factor alpha (TNFα) signature, whereas combined loss of AR and JMJD1C strongly up-regulates the TNFα signature in human prostate cancer, suggesting TNFα signaling as a point of convergence for the combined actions of AR and JMJD1C. Correspondingly, AR-negative prostate cancer cells showed exquisite sensitivity to TNFα treatment and, conversely, TNFα pathway inhibition via inhibition of its downstream effector MAP4K4 partially reversed the growth defect of JMJD1C-depleted AR-negative prostate cancer cells. Given the deleterious systemic side effects of TNFα therapy in humans and the viability of JMJD1C-knockout mice, the identification of JMJD1C inhibition as a specific vulnerability in AR-negative prostate cancer may provide an alternative drug target for prostate cancer patients progressing on AR inhibitor therapy.
Prostate cancer is the most common cancer and the second-leading cause of cancer death among men in the United States (1). Androgen deprivation therapy (ADT) has improved the treatment of prostate cancer while also fueling increased incidence of ADT-resistant prostate cancers, a disease state termed castration-resistant prostate cancer (CRPC) (2). ADT resistance mechanisms center on enhanced androgen receptor (AR) signaling via overexpression, genome amplification, and/or mutation of AR. Targeting androgen circuits by AR signaling inhibitors such as enzalutamide and abiraterone is now the standard of care in CRPC and has extended patient survival, although sustained remission is rare (3, 4). Notably, the effective repression of AR signaling by these drugs has been shifting CRPC patients to a more intractable “AR-negative state” (5). This growing population of AR-negative prostate cancer patients with progressive disease (5, 6) has created an urgent need for therapies targeting specific vulnerabilities in AR-negative prostate cancer cells.
Targeting undruggable loss-of-function mutations via synthetic lethal strategies has received increasing attention as a potential strategy to identify context-specific molecular targeted therapies in cancer. A celebrated proof-of-concept example for the targeting of loss-of-function mutations is the clinical effectiveness of poly(ADP)-ribose polymerase inhibitors in BRCA-deficient cancers (7, 8). The advent of genome-wide CRISPR-Cas9 gene-knockout screens has revolutionized the study of genetic interactions in human cells (9, 10), facilitating the potential discovery of additional synthetic lethal pairs and other context-specific vulnerabilities (11).
In this study, CRISPR context dependency analysis (12) in AR-negative prostate cancer cells identified the histone demethylase JMJD1C (KDM3C) as a target for AR-negative prostate cancer. Mechanistic studies identified the repression of tumor necrosis factor alpha (TNFα) signaling as a common point of convergence of AR and JMJD1C in the maintenance of prostate cancer. While TNFα elicits strong cytotoxic and immune modulatory effects on malignant tumors, its use as a biologic therapy in cancer has been limited by deleterious side effects, such as systemic shock and inflammation (13–15). In several prostate cancer models, we show that JMJD1C depletion leads to specific growth suppression of AR-negative cells via activation of the TNFα network, consistent with a synthetic lethal relationship between AR and JMJD1C.
Results
Specific Vulnerabilities Revealed by Analysis of CRISPR Screens.
To identify context-specific candidate targets, we sought to compare CRISPR knockout fitness screens in prostate cancer cell lines with knockouts in other backgrounds. The Cancer Dependency Map (DepMap), the largest public resource of CRISPR knockout screens in cancer cell lines (16), currently contains data from over 700 high-quality screens using the Avana CRISPR library (Materials and Methods); however, only one prostate cancer cell line is present in this dataset. An earlier effort (17) included both LNCaP, an androgen-sensitive prostate cancer cell line with wild-type TP53, and PC3, an androgen-resistant, TP53-negative cell line, among 33 cell lines screened with the earlier-generation GeCKO v2 CRISPR library.
To identify prostate cancer–specific essential genes, we first applied our BAGEL (Bayesian analysis of gene essentiality) pipeline (18, 19) to each screen to evaluate gene essentiality. BAGEL uses gold-standard positive and negative training sets to train a gene essentiality classifier and returns a log Bayes factor (BF) for each gene that, in the context of a typical genome-scale knockout screen in a cell line, represents a blend of statistical confidence and quantitative estimate of knockout fitness.
Direct comparison of CRISPR gene-knockout fitness effects across different experiments can be problematic. Variation in guide RNA (gRNA) sequences, number of reagents per gene, and experimental design can modulate signal and make quantitative comparisons error-prone. To circumvent this issue, we applied a binary classification filter to 719 Avana-screened cell lines in the DepMap database (Materials and Methods). We then calculated the frequency that each gene is identified as essential across these cell lines. Finally, we compared the BAGEL scores of the GeCKO-screened prostate cancer cell lines with the frequency of gene essentiality calculated from the Avana-screened DepMap data (Fig. 1A).
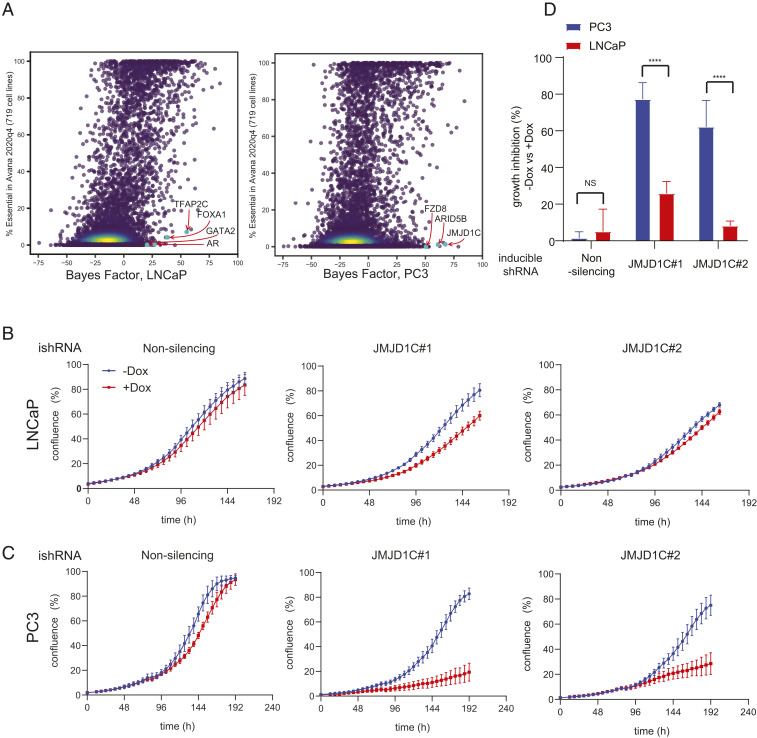
Fig. 1.
CRISPR screening revealed the context-specific dependency in AR/p53–negative PC3 cells. (A) Cell type–specific dependency revealed by BAGEL analysis comparing CRISPR screening data in LNCaP (Left) and PC3 (Right) with 719 DepMap cell lines. (B and C) Growth curves of doxycycline-inducible shRNA expressing LNCaP (B) and PC3 (C), showing specific growth defects in PC3 by JMJD1C knockdown. The mean and SEM are indicated; n = 3 biological replicates. Dox, doxycycline; ishRNA, inducible shRNA. (D) Quantification of the growth assays in B and C. The growth inhibition by doxycycline treatment in each group is shown. The mean and SEM are indicated. NS, not significant. ****P < 0.0001.
To identify context-specific genes, we searched for genes with high BF in prostate cancer cells but low background likelihood of essentiality in Avana screens (Fig. 1A). As expected, androgen-sensitive LNCaP cells showed a high specific vulnerability to knockout of AR as well as the transcription factors involved in hormone signaling (TFAP2C, GATA2) (Fig. 1 A, Left). This cell line also shows dependence on MDM2, MDM4, and PPM1D, which are required to suppress TP53-mediated apoptotic signaling in TP53 wild-type cells (20). By contrast, androgen-insensitive, TP53-mutant PC3 cells show significant differences in their genetic dependencies (Fig. 1 A, Right). Specifically, the Wnt/β-catenin pathway gene FZD8 shows high BFs in PC3 cells but not other cells, consistent with a previous report indicating some metastatic prostate cancers are dependent on FZD8 (21). Interestingly, chromatin remodeling factors ARID5B and JMJD1C, both involved in histone demethylation, are also highly essential in these cells.
Targets shared by both cells may also reflect general characteristics of prostate cancer biology. Pioneer transcription factor FOXA1 and developmental transcription factors HOXB13 and RREB1 are specific to prostate cells relative to cells of other lineages (Dataset S1). Since Wnt-specific vulnerability in p53-null prostate cancer has been elucidated (21), we instead focused on novel chromatin remodeling factors. Among these, JMJD1C possesses putative druggable enzymatic domains (22), and thus we chose JMJD1C for validation using a doxycycline-inducible small hairpin RNA (shRNA) system. The depletion of JMJD1C by two independent hairpins (SI Appendix, Fig. S1 A and B) had marginal effects on the growth of LNCaP cells (Fig. 1 B and D) yet elicited substantial growth suppression in PC3 cells (Fig. 1 C and D).
AR-Negative Prostate Cancer Cells Exhibit Specific Sensitivity to JMJD1C Inhibition.
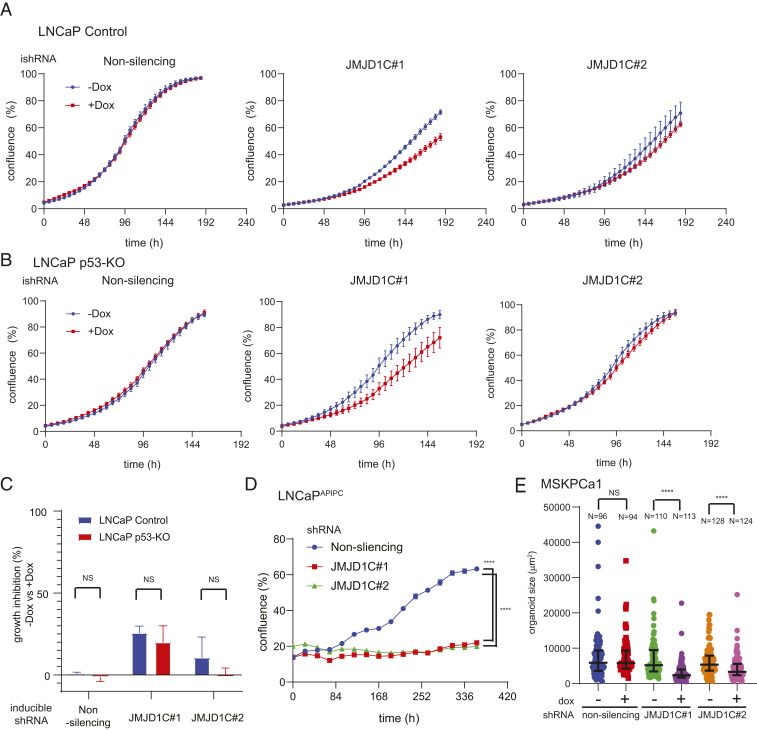
Next, we explored the role of TP53 and AR status in dictating the differential response of LNCaP and PC3 cells to JMJD1C depletion. To assess the relevance of intact p53, we engineered isogenic p53-null LNCaP cells using CRISPR (SI Appendix, Fig. S1C) and demonstrated marginal growth differences upon JMJD1C depletion relative to p53 wild-type LNCaP controls (Fig. 2 A–C). To assess the impact of AR status, we used an inducible AR depletion model of LNCaP (LNCaPAPIPC) described previously (5). As depletion of AR in LNCaPAPIPC is achieved via a doxycycline-inducible shRNA cassette (5), we introduced a doxycycline-inducible shRNA cassette targeting JMJD1C to enable contemporaneous depletion of AR and JMJD1C and afford assessment of the impact of dual depletion versus AR-only depletion (i.e., a nonsilencing control for JMJD1C). In this model, JMJD1C depletion (SI Appendix, Fig. S1D) completely suppressed the growth of AR-depleted LNCaP cells, whereas AR depletion alone (the nonsilencing control) had no effect (Fig. 2D). Similarly, the interaction of AR and JMJD1C was confirmed in a patient-derived prostate cancer organoid model, MSKPCa1, a more clinically relevant model which is intrinsically AR-negative (23) (Fig. 2E and SI Appendix, Fig. S1E). Finally, the AR antagonist enzalutamide sensitized wild-type LNCaP to JMJD1C depletion in both two-dimensional growth and tumor sphere formation assays (SI Appendix, Fig. S2). Together, these results support the view that JMJD1C depletion selectively suppresses the growth of AR-negative prostate cancer cells.
Fig. 2.
AR loss confers selective vulnerability to JMJD1C inhibition. (A and B) Growth curves of doxycycline-inducible shRNA expressing control or p53-KO (knockout) LNCaP. The mean and SEM are indicated; n = 3 biological replicates. (C) Quantification of the growth assays in A and B. The growth inhibition by doxycycline treatment in each group is shown. The mean and SEM are indicated. (D) Growth curves of nonsilencing or JMJD1C shRNA expressing LNCaPAPIPC; n ≥ 3 biological replicates. ****P < 0.0001. (E) MSKPCa1 organoids were cultured in the presence or absence of doxycycline for 24 d. The indicated numbers of organoids from three biological replicates in each group were measured and plotted. Medians and interquartile ranges are indicated. ****P < 0.0001.
AR Loss Confers Selective Vulnerability to the Up-Regulation of TNFα Downstream Genes.
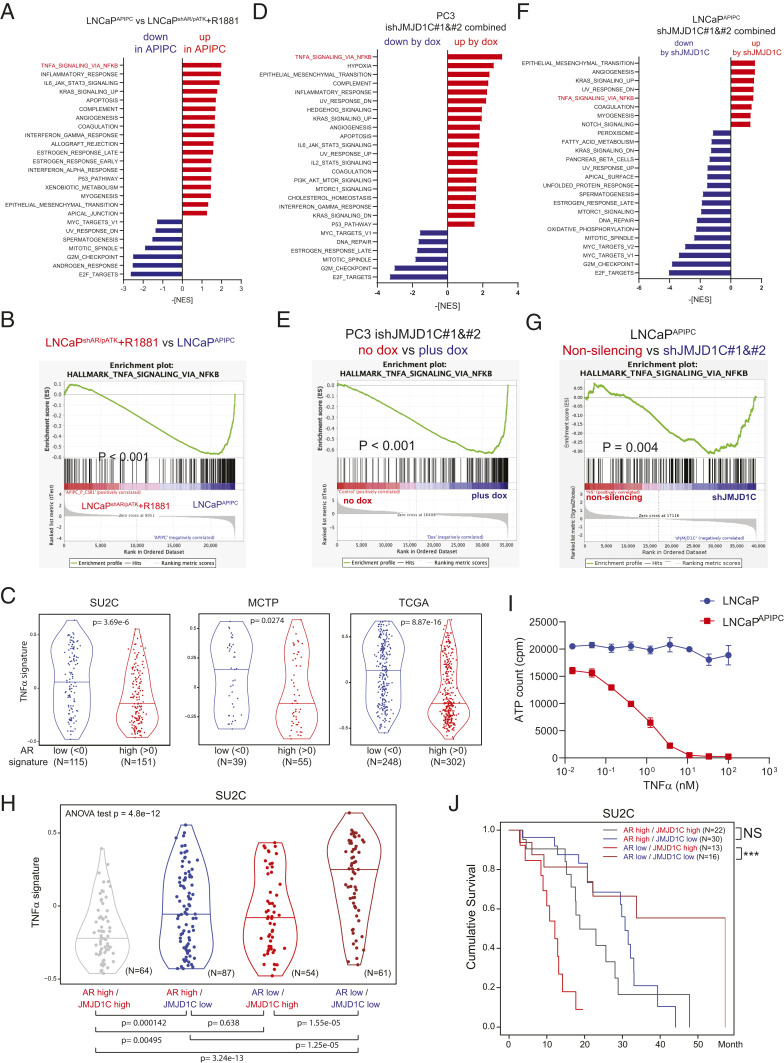
To decipher the mechanisms governing selective growth arrest in JMJD1C-silenced AR-negative prostate cancer cells, we first analyzed the transcriptomic changes brought about by AR loss by comparing AR-negative LNCaP (LNCaPAPIPC) and the parental LNCaP (LNCaPshAR/pATK) treated with an AR agonist (R1881) (5). Gene set enrichment analysis (GSEA) showed that “TNFα signaling via nuclear factor κB (NFκB)” is the most enriched signature up-regulated in AR-negative LNCaP (Fig. 3 A and B). This tendency was confirmed by the observation of clinical datasets, where TNFα signature scores were higher in patients with a low AR signature (Fig. 3C). Next, we conducted whole-transcriptomic analysis to assess the effects of JMJD1C depletion in PC3 and AR-depleted LNCaP cells. GSEA uncovered up-regulation of TNFα signaling via NFκB as one of the most enriched signatures in JMJD1C-depleted PC3 cells (Fig. 3 D and E). Similarly, enrichment of TNFα downstream genes was observed in JMJD1C-depleted AR-negative LNCaP cells (Fig. 3 F and G), but not in the original LNCaP cells (SI Appendix, Fig. S2 A and B). These results indicate that JMJD1C depletion up-regulates TNFα downstream genes in AR-negative prostate cancer cells. To further confirm the cooperative regulation of TNFα circuits by JMJD1C and AR, we analyzed in clinical datasets the JMJD1C signature defined by 117 genes commonly down-regulated by JMJD1C depletion in PC3 and AR-negative LNCaP (SI Appendix, Fig. S3A and Dataset S2), and observed that patients with low AR or JMJD1C signatures showed a modest TNFα signature, whereas patients with a combined low AR and JMJD1C signature showed strong up-regulation of the TNFα signature in three independent cohorts (Fig. 3H and SI Appendix, Fig. S4 B and C).
Fig. 3.
TNFα downstream genes are the point of convergence of JMJD1C and AR. (A) GSEA (hallmark) showing enriched signatures by AR loss comparing LNCaPAPIPC vs. LNCaPshAR/pATK treated with R1881. Signatures with P < 0.05 and false discovery rate (FDR) q value <0.25 are shown. (B) Enrichment plot showing hallmark TNFα signaling via NFκB by AR loss. (C) TNFα signature calculated with GSVA for prostate cancer patients with high (>0) or low (<0) AR signature. (D and F) GSEA (hallmark) showing enriched signatures by JMJD1C knockdown in PC3 (doxycycline treatment 7 d) (D) and in LNCaPAPIPC (14 d after infection) (F). Signatures with P < 0.05 and FDR q value <0.25 are shown. (E and G) Enrichment plots showing hallmark TNFα signaling via NFκB by JMJD1C depletion. (H) TNFα signature calculated with GSVA for prostate cancer patients with high (>0) or low (<0) AR or JMJD1C signature. (I) LNCaP or LNCaPAPIPC cells were treated with the indicated concentrations of TNFα and cultured for 3 d, and live cells were evaluated with CellTiter-Glo. (J) Survival curves of metastatic prostate cancer patients (SU2C) with each signature. P values were calculated with the log-rank test. ***P = 0.000767.
The increased TNFα signature associated with AR loss raised the possibility that prostate cancer cells with AR loss may exhibit increased sensitivity to further up-regulation of the TNFα network. To test this possibility, we treated parental AR-positive LNCaP and AR-negative LNCaP cells with TNFα. We observed dose-dependent killing of AR-negative LNCaP cells with TNFα treatment and no impact on parental AR-positive LNCaP cells (Fig. 3I). Correspondingly, survival analysis for metastatic prostate cancer patients showed that low JMJD1C signature scores correlated with better prognosis only in patients harboring low AR signature scores (Fig. 3J). Together, these findings support the view that activation of the TNFα network may underlie the suppressive growth impact of JMJD1C depletion in AR-negative prostate cancer cells. As such, targeting JMJD1C may confer a selective vulnerability for AR-negative prostate cancers.
Blockade of TNFα Downstream Signaling Attenuates Growth Suppression Induced by JMJD1C Depletion in AR-Negative Prostate Cancer Cells.
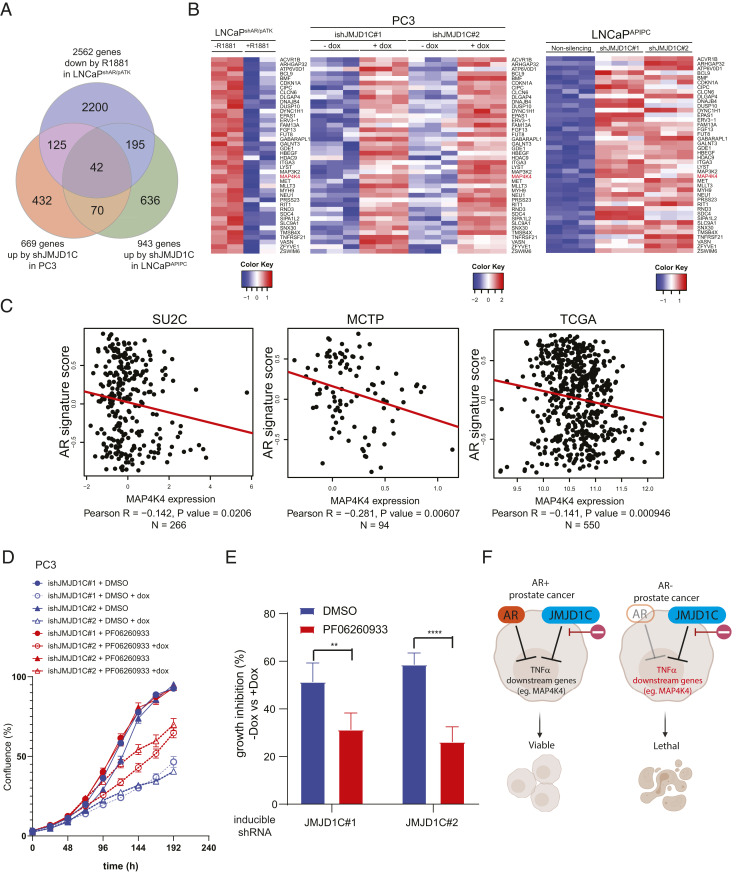
Given that both AR loss and JMJD1C knockdown up-regulate the TNFα network, we hypothesized that genes commonly suppressed by AR and JMJD1C are responsible for the cooperative phenotype. To identify such genes, we intersected genes suppressed by AR agonist treatment (5) and genes up-regulated by JMJD1C depletion in PC3 and AR-negative LNCaP cells (Fig. 4A). Among 42 shared genes, we focused on MAP4K4 as it is a well-established key downstream kinase of TNFα signaling (24, 25) and its up-regulation elicits increased cell death in lung cancer (26) (Fig. 4B). To assess the potential clinical relevance of MAP4K4 in relation to AR status, we interrogated publicly available human prostate cancer datasets, revealing that AR signature correlated negatively with MAP4K4 expression (Fig. 4C). To further test the functional TNFα activation in the AR-negative JMJD1C-depleted context, we assessed the impact of a MAP4K4 selective inhibitor, PF-06260933 (27), revealing partial rescue of the growth suppression induced by JMJD1C depletion (Fig. 4 D and E). Together, these results reinforce the finding that TNFα activation plays a role in the selective sensitivity of AR-negative cells to JMJD1C depletion (Fig. 4F).
Fig. 4.
Blocking TNFα downstream partly reversed the phenotype of JMJD1C depletion. (A) Triangular analysis uncovering common genes suppressed by AR and JMJD1C. (B) Heatmaps of normalized RNA-seq counts showing 42 genes enriched by triangular analysis. (C) Correlation of MAP4K4 messenger RNA expression and AR signature in three prostate cancer patient datasets. (D) Growth curves of doxycycline-inducible shRNA expressing PC3 treated with dimethyl sulfoxide (DMSO) or 1 μM PF-06260933, showing the rescue of the growth defects by JMJD1C knockdown. The mean and SEM are indicated; n = 4 biological replicates. (E) Quantification of the growth assays in D. The growth inhibition by doxycycline treatment in each group is shown. The mean and SEM are indicated. **P = 0.002, ****P < 0.0001. (F) Working model proposing the synthetic lethal relationship between AR and JMJD1C in prostate cancer cells. Created with BioRender.
Discussion
In this study, we identified a JMJD1C dependency driven by AR loss and accompanied by TNFα signaling activation. We showed that JMJD1C depletion specifically suppresses the growth of AR-negative prostate cancer cells using multiple models including isogeneic cell lines and a patient-derived prostate cancer organoid. Whole-transcriptomic analysis revealed that both AR depletion and JMJD1C depletion independently, and in combination, up-regulate the TNFα network. Furthermore, expression signature analysis using prostate cancer patient datasets aligned with the cooperative suppressive function of AR and JMJD1C in TNFα regulation. Isogeneic AR-null models highlighted the specific vulnerability of AR-negative cells to TNFα treatment and, conversely, blocking TNFα downstream signaling with a MAP4K4 inhibitor partially rescued the growth defect of JMJD1C depletion in AR-negative prostate cancer cells. Patients with low AR and JMJD1C signature scores presented the most favorable prognosis. Together, these results show that AR loss confers sensitivity to JMJD1C inhibition, a sensitivity involving TNFα activation.
Our finding that AR loss enhances TNFα signaling is consistent with recent clinical observations where patients refractory to enzalutamide treatment show low AR activity and an activated TNFα signature (28). Furthermore, the ability for JMJD1C to act as a suppressor of TNFα and the NFκB pathways is supported by similar observations in JMJD1C-knockout mice and JMJD1C knockdown in THP-1 cells (29), suggesting that JMJD1C and AR could cooperatively function in the suppression of TNFα downstream genes to prevent overactivation in prostate cancer cells. Cooperation of JMJD1C and AR is further supported by the fact that JMJD1C was originally discovered as a coactivator of AR (30). Partial growth rescue via MAP4K4 blockade in JMJD1C-depleted PC3 cells (Fig. 4E) could be explained by the observation that there are several other TNFα-related genes up-regulated by JMJD1C knockdown, such as CDKN1A, TNFRSF21, a TNF receptor (31), and MAP3K2, which is important in delayed activation of NFκB (32) (Fig. 3B). It is tempting to speculate that targeting MAP4K4 in combination with other targets downstream may fully rescue JMJD1C-dependent growth arrest in AR-negative prostate cancer. Nevertheless, on the basis of alteration of other pathways in JMJD1C-depleted cells (Fig. 3), we cannot exclude the possibility that signaling pathways, beyond the TNFα network, might also contribute to the specific growth defect by JMJD1C blockade in AR-negative prostate cancer cells.
JMJD1C is a member of the KDM3 family of histone demethylases that also includes KDM3A and KDM3B. Previous work has established a role for KDM3A and KDM3B in advanced prostate cancer in the AR-positive state which stands in contradistinction to the specific role of JMJD1C in AR-negative disease. Specifically, KDM3A has been shown to activate AR downstream genes via direct histone modification of AR downstream genes (33), promoting alternative splicing of AR variant 7 (34) and enhancing AR-dependent c-Myc expression (35). KDM3B is required for the growth in AR-expressing androgen-independent prostate cancer cells through regulating the expression of metabolic enzymes such as ARG2 and RDH11 (36). These studies, together with this report, suggest that KDM3 family members play distinctive and critical roles in promoting prostate cancer. Thus, a pan-KDM3 inhibitor may be useful in combating various forms of prostate cancer.
A key finding of this study is the synthetic lethal relationship between AR and JMJD1C. JMJD1C has been identified as a promising target in other cancer types, such as leukemia (37, 38), colorectal cancer (39, 40), and esophageal cancer (41); our findings here expand the potential utility of JMJD1C blockade specifically in AR-negative prostate cancer, which currently has limited treatment options. TNFα treatment has been shown to exhibit efficacy in locally advanced hormone-resistant prostate cancer disease, although its use is limited to local and intratumoral administration due to its significant systemic toxicity (42). Most CRPC cases present with widespread metastatic disease, thus limiting the utility of local TNFα treatment. It is notable that JMJD1C-knockout mice are viable with only male infertility (43), suggesting that JMJD1C blockade may prove to be a safer alternative to TNFα therapy for the treatment of AR-negative metastatic CRPC. In conclusion, the synthetic lethal relationship between AR and JMJD1C may provide a much needed therapeutic option for the growing number of patients suffering from AR-negative prostate cancer.
Materials and Methods
CRISPR Screen Analysis.
Read count data from the Cancer Dependency Map (16) and Aguirre et al. (17) were processed with CRISPRcleanR (44) and BAGEL2 (19) to generate copy-number corrected Bayes factors for each cell line, using the Hart essentials (45, 46) to train the algorithm. For the DepMap data, screens with F1 score <0.8 at BF =3 were discarded as low-quality (n = 719 screens passed the filter). BFs for each gene were then normalized by the number of gRNAs targeting the gene, and genes were classified as essential where normalized BF >3 (corresponding to a posterior probability of gene essentiality >90%), and nonessential otherwise.
Cell Lines and Plasmids.
LNCaP and PC3 cell lines were purchased from the ATCC. Control or p53-knockout LNCaP cells were established using Control CRISPR/Cas9 Plasmid (Santa Cruz Biotechnology; sc-418922) and p53 CRISPR/Cas9 Plasmid (Santa Cruz Biotechnology; sc-416). Briefly, LNCaP cells were transfected with Lipofectamine 3000 (Invitrogen), and green fluorescent protein–positive cells were sorted 48 h after transfection. The bulk population of sorted cells was used for subsequent experiments. LNCaPAPIPC is a kind gift from Peter S. Nelson, Fred Hutchinson Cancer Research Center, Seattle, WA. SMARTvector inducible lentiviral shRNA vectors were purchased from Dharmacon. Target sequences are as follows: nonsilencing, 5′-CAC ACA ACA TGT AAA CCA GGG A-3′; JMJD1C#1, 5′-ACT TCG AAC TGA CAA TGT T-3′; JMJD1C#2, 5′-GGG TCA GTG ATG TAG TTA A-3′. Lentiviruses were packaged in 293T cells using second-generation packaging vectors psPAX2 (Addgene plasmid 12260) and pMD2.G (Addgene plasmid 12259). PC3 and LNCaP cells transduced with lentivirus were selected with 2 μg/mL puromycin.
Tumor Sphere Culture.
LNCaP cells were collected and washed to remove serum, and then suspended in serum-free Dulbecco’s modified Eagle’s medium/F12 supplemented with 20 ng/mL human epidermal growth factor (R&D Systems; 236-EG), 10 ng/mL human basic fibroblast growth factor (Gibco; PHG0261), 2% B27 (Gibco; 17504-044), and 1% N2 supplement (Gibco; 17502-048). The cells were plated in 96-well ultra-low–attachment plates (Corning) at a density of 500 cells per well. Ten days later, solid tumor spheres were counted and quantified using an inverted microscope.
Organoid Culture.
MDKPCa1, a prostate cancer patient–derived organoid, was provided by Yu Chen, Memorial Sloan Kettering Cancer Center, New York, NY, and maintained as described previously (23, 47). Lentiviral transduction was conducted with the modified protocol described previously (48). Briefly, lentivirus vectors were concentrated using a Lenti-X Concentrator (Clontech; PT4421-2) and then the pellet was resuspended in organoid media containing 8 μg/mL polybrene (Millipore; TR1003-G). Organoids were trypsinized, suspended in lentivirus-containing media, and then incubated for 1 h at 37 °C. Organoids were resuspended in Matrigel and plated as described. After transduction (2 to 3 d), the organoids were selected with 1 μg/mL puromycin. Images for organoids were captured using a Leica DFC295 microscope with an HI Plan 20×/0.30 numerical aperture Ph1 objective and LAS v4.13 software. Organoid size was analyzed with ImageJ software.
Western Blot Analyses.
Samples were lysed in 1% LDS buffer (10 mM Tris⋅HCl, pH 8.0, 1% lithium dodecyl sulfate, 10% glycerol) and the protein concentration was quantified with a detergent-compatible protein assay. Lysates (10 to 20 μg per lane) were resolved by sodium dodecyl sulfate–polyacrylamide-gel electrophoresis and transferred onto polyvinylidene fluoride membranes. The antibodies used were rabbit anti-p53 (DO-1 CST18032; Cell Signaling Technology) and mouse anti–β-actin (A2228; Sigma-Aldrich).
RNA Isolation and qRT-PCR.
RNA was isolated by the RNeasy Mini Kit (Qiagen). DNA was removed by treating with DNase from the kit (Qiagen). Complementary DNA was generated using All-in-One RT MasterMix (ABM; G490). qRT-PCR was performed with 1× SYBR Green PCR Master Mix (Thermo Fisher Scientific; 4309155) using primers that had been optimized for their melting curve. Human beta-actin (ACTB) expression was used as an internal control. Primer sets used were as follows. JMJD1C, 5′-AGCAGTATACCAGATGAAGAG-3′ and 5′-TGAATTTTCTGAGTCACTGC-3′; ACTB, 5′-GACGACATGGAGAAAATCTG-3′ and 5′-ATGATCTGGGTCATCTTCTC-3′.
RNA Sequencing and Transcriptomic Analysis.
RNA was isolated by directly adding RLT buffer of the RNeasy Mini Kit (Qiagen). RNA sequencing (RNA-seq) was performed by the Sequencing and ncRNA Program of the Advanced Technology Genomics Core at MD Anderson. Libraries were generated using Illumina’s TruSeq Kit and were sequenced using the Illumina HiSeq 2000 or NextSeq 500 Sequencer. Raw sequencing data (BCL format) were converted to Fastq files using Illumina Casava software (v1.8.2) and aligned to the human reference genome (hg19) using STAR software. The HTSeq-count program was used to generate raw read counts for each gene. The R package DESeq2 was used for data normalization and differential expression analysis. Pathway enrichment analysis was performed using GSEA software based on the normalized counts by DESeq2. The gene expression signature activity scores were calculated in R using the gene set variation analysis (gsva) method of the GSVA v1.24.0 Bioconductor package, taking log2-transformed DESeq2-normalized counts as input (49). The AR signature score, TNFα signature score, and JMJD1C signature score were calculated by using 10 AR activity genes (KLK3, KLK2, TMPRSS2, FKBP5, NKX3-1, PLPP1, PMEPA1, PART1, ALDH1A3, STEAP4), hallmark gene sets (HALLMARK_TNFA_SIGNALING_VIA_NFKB from MSigDB v5.0) (50, 51), and 117 genes commonly down-regulated by JMJD1C depletion in PC3 and LNCaPAPIPC.
Proliferation Assay.
The cell growth of the cell lines was assayed by measuring confluence with IncuCyte (Essen BioScience). Growth curves using IncuCyte were generated by imaging every 6 h with triplicate or quadruplicate replicates. Growth was calculated for each well by subtracting the starting confluence from the final confluence. Growth inhibition was calculated by comparing doxycycline-treated and untreated samples.
TNFα Sensitivity Assay.
Recombinant human TNFα was purchased from Gibco (PHC3015). Cells (1 × 104 per well) were seeded in 96-well plates and treated with TNFα on the following day. After a 3-d incubation, the live cell number was evaluated with the CellTiter-Glo Luminescent Cell Viability Assay (Promega; G7572). Luminescence was counted by CLARIOstar (BMG Labtech).
Clinical Data and Patient Survival Analysis.
RNA-seq gene expression profiles and overall survival data of three clinical prostate cancer datasets were used in the current analysis: the PRAD cohort of The Cancer Genome Atlas (TCGA) (provision 2016-01 from http://gdac.broadinstitute.org/) and MCTP cohort (52) and SU2C cohort (53) from https://www.cbioportal.org/. For TCGA expression analysis, log2-transformed gene-level normalized counts were used. For MCTP and SU2C expression analysis, gene-level z scores were used. AR, JMJD1C, and TNFα signature scores were calculated with gsva as described above. Tumors with negative signature scores (<0) of a pathway were considered as low-activity and those with positive signature scores (>0) were considered as high-activity. Survival analysis with the log-rank test and the generation of Kaplan–Meier curves were done in R.
Statistical Analysis.
GraphPad Prism 8 software was used to conduct statistical analysis of all the data. Two-way ANOVA was used to calculate P values for the comparison of the growth inhibition data obtained by IncuCyte. P values for the organoid size comparison were calculated with the one-way ANOVA Kruskal–Wallis test. P values for comparison of gene signatures in patients were calculated by Student’s t test. The Pearson correlation coefficient was used to study the relationships between variables shown in scatterplots using the cor.test function in R. GSEA was implemented with java web start v2.2.1 from the Broad Institute (50, 54).
Supplementary Material
Acknowledgments
We thank Dr. Yu Chen (Memorial Sloan Kettering Cancer Center, New York, NY) for providing prostate cancer patient–derived organoids. We also thank Dr. Peter S. Nelson (Fred Hutchinson Cancer Research Center, Seattle, WA) for providing the LNCaPAPIPC cell line. We thank Drs. Peter DeWeirdt and John G. Doench (Broad Institute, Cambridge, MA) for visualization tools. We thank the Sequencing and ncRNA Program and the Advanced Technology Genomics Core at MD Anderson (NIH P30 CA16672) and the Texas Medical Center Digestive Disease Center and Core E (NIH P30 DK056338). This work was supported by Prostate Cancer Foundation Challenge Award 17CHAL17 (to R.A.D. and Y.A.W.), NIH 1R01 CA231349-01A1 (to Y.A.W.), and MD Anderson Prostate Cancer Moon Shot (to R.A.D.). T.H. is a Cancer Prevention & Research Institute of Texas Scholar in Cancer Research and is supported by NIH R35 GM130119.
Footnotes
Competing interest statement: T.H. is a consultant for Repare Therapeutics. R.A.D. is a cofounder, advisor, and/or director of Tvardi Therapeutics, Asylia Therapeutics, Stellanova Therapeutics, Nirogy Therapeutics, and Sporos Bioventures. The work of this manuscript was not supported by, nor is related to, these companies.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026324118/-/DCSupplemental.
Data Availability
The RNA-seq data reported in this article have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE166657). All study data are included in the article and/or supporting information. Previously published data were used for this work (5).
References
- 1.Surveillance, Epidemiology, and End Results Program , National Cancer Institute, Cancer statistics factsheets: Common cancer sites. https://seer.cancer.gov/statfacts/html/common.html. Accessed 22 December 2020.
- 2.Attard G., et al., Prostate cancer. Lancet 387, 70–82 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Ryan C. J.et al.; COU-AA-302 Investigators , Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med. 368, 138–148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher H. I.et al.; AFFIRM Investigators , Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 367, 1187–1197 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Bluemn E. G., et al., Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 32, 474–489.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labrecque M. P., et al., Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J. Clin. Invest. 129, 4492–4505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryant H. E., et al., Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Farmer H., et al., Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Shalem O., et al., Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang T., Wei J. J., Sabatini D. M., Lander E. S., Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang A., Garraway L. A., Ashworth A., Weber B., Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discov. 19, 23–38 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Hart T., et al., High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Josephs S. F., et al., Unleashing endogenous TNF-alpha as a cancer immunotherapeutic. J. Transl. Med. 16, 242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts N. J., Zhou S., Diaz L. A. Jr, Holdhoff M., Systemic use of tumor necrosis factor alpha as an anticancer agent. Oncotarget 2, 739–751 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen J., et al., Anti-cancer therapy with TNFα and IFNγ: A comprehensive review. Cell Prolif. 51, e12441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyers R. M., et al., Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguirre A. J., et al., Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov. 6, 914–929 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart T., Moffat J., BAGEL: A computational framework for identifying essential genes from pooled library screens. BMC Bioinformatics 17, 164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim E., Hart T., Improved analysis of CRISPR fitness screens and reduced off-target effects with the BAGEL2 gene essentiality classifier. Genome Med. 13, 2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim E., et al., A network of human functional gene interactions from knockout fitness screens in cancer cells. Life Sci. Alliance 2, e201800278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q., et al., FZD8, a target of p53, promotes bone metastasis in prostate cancer by activating canonical Wnt/β-catenin signaling. Cancer Lett. 402, 166–176 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Xu X., et al., Small molecular modulators of JMJD1C preferentially inhibit growth of leukemia cells. Int. J. Cancer 146, 400–412 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Gao D., et al., Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tesz G. J., et al., Tumor necrosis factor alpha (TNFalpha) stimulates Map4k4 expression through TNFalpha receptor 1 signaling to c-Jun and activating transcription factor 2. J. Biol. Chem. 282, 19302–19312 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Yao Z., et al., A novel human STE20-related protein kinase, HGK, that specifically activates the c-Jun N-terminal kinase signaling pathway. J. Biol. Chem. 274, 2118–2125 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Chen S., et al., SOX2 regulates apoptosis through MAP4K4-survivin signaling pathway in human lung cancer cells. Carcinogenesis 35, 613–623 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Ammirati M., et al., Discovery of an in vivo tool to establish proof-of-concept for MAP4K4-based antidiabetic treatment. ACS Med. Chem. Lett. 6, 1128–1133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alumkal J. J., et al., Transcriptional profiling identifies an androgen receptor activity-low, stemness program associated with enzalutamide resistance. Proc. Natl. Acad. Sci. U.S.A. 117, 12315–12323 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J. Y., et al., Histone Lys demethylase KDM3C demonstrates anti-inflammatory effects by suppressing NF-κB signaling and osteoclastogenesis. FASEB J. 33, 10515–10527 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolf S. S., Patchev V. K., Obendorf M., A novel variant of the putative demethylase gene, s-JMJD1C, is a coactivator of the AR. Arch. Biochem. Biophys. 460, 56–66 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Kasof G. M., et al., Tumor necrosis factor-alpha induces the expression of DR6, a member of the TNF receptor family, through activation of NF-kappaB. Oncogene 20, 7965–7975 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Schmidt C., et al., Mechanisms of proinflammatory cytokine-induced biphasic NF-kappaB activation. Mol. Cell 12, 1287–1300 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Wilson S., Fan L., Sahgal N., Qi J., Filipp F. V., The histone demethylase KDM3A regulates the transcriptional program of the androgen receptor in prostate cancer cells. Oncotarget 8, 30328–30343 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan L., et al., Histone demethylase JMJD1A promotes alternative splicing of AR variant 7 (AR-V7) in prostate cancer cells. Proc. Natl. Acad. Sci. U.S.A. 115, E4584–E4593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan L., et al., Regulation of c-Myc expression by the histone demethylase JMJD1A is essential for prostate cancer cell growth and survival. Oncogene 35, 2441–2452 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saraç H., et al., Systematic characterization of chromatin modifying enzymes identifies KDM3B as a critical regulator in castration resistant prostate cancer. Oncogene 39, 2187–2201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sroczynska P., et al., shRNA screening identifies JMJD1C as being required for leukemia maintenance. Blood 123, 1870–1882 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Izaguirre-Carbonell J., et al., Critical role of Jumonji domain of JMJD1C in MLL-rearranged leukemia. Blood Adv. 3, 1499–1511 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J., et al., KDM3 epigenetically controls tumorigenic potentials of human colorectal cancer stem cells through Wnt/β-catenin signalling. Nat. Commun. 8, 15146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C., et al., Downregulation of histone demethylase JMJD1C inhibits colorectal cancer metastasis through targeting ATF2. Am. J. Cancer Res. 8, 852–865 (2018). [PMC free article] [PubMed] [Google Scholar]
- 41.Cai Y., Fu X., Deng Y., Histone demethylase JMJD1C regulates esophageal cancer proliferation via YAP1 signaling. Am. J. Cancer Res. 7, 115–124 (2017). [PMC free article] [PubMed] [Google Scholar]
- 42.Kramer G., et al., Local intratumoral tumor necrosis factor-alpha and systemic IFN-alpha 2b in patients with locally advanced prostate cancer. J. Interferon Cytokine Res. 21, 475–484 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Kuroki S., et al., JMJD1C, a JmjC domain-containing protein, is required for long-term maintenance of male germ cells in mice. Biol. Reprod. 89, 93 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Iorio F., et al., Unsupervised correction of gene-independent cell responses to CRISPR-Cas9 targeting. BMC Genomics 19, 604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hart T., et al., Evaluation and design of genome-wide CRISPR/SpCas9 knockout screens. G3 (Bethesda) 7, 2719–2727 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hart T., Brown K. R., Sircoulomb F., Rottapel R., Moffat J., Measuring error rates in genomic perturbation screens: Gold standards for human functional genomics. Mol. Syst. Biol. 10, 733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drost J., et al., Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 11, 347–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Lidth de Jeude J. F., Vermeulen J. L., Montenegro-Miranda P. S., Van den Brink G. R., Heijmans J., A protocol for lentiviral transduction and downstream analysis of intestinal organoids. J. Vis. Exp. (98), e52531 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hänzelmann S., Castelo R., Guinney J., GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian A., et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liberzon A., et al., The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grasso C. S., et al., The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abida W., et al., Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 11428–11436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mootha V. K., et al., PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data reported in this article have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE166657). All study data are included in the article and/or supporting information. Previously published data were used for this work (5).