Abstract
We compared the performance of Organon Teknika’s NucliSens and Roche Diagnostic Systems’ Monitor quantitative human immunodeficiency type 1 RNA assays. Both had similar linearity and sensitivity over most of the dynamic range of the assays, although the Monitor assay was superior at the low range of RNA values while the NucliSens assay was more consistent at higher RNA values. NucliSens generally showed less interassay variability.
The advent of techniques that reliably quantify levels of human immunodeficiency virus type 1 (HIV-1) RNA in patient plasma has been pivotal in the development of new insights into in vivo viral dynamics and HIV disease pathogenesis (2). Various commercial RNA quantification techniques differing in sensitivity, dynamic range, and variability have now been introduced into clinical practice (4, 5). In this study we compare performance characteristics of an improved nucleic acid sequence-based analysis (NASBA) assay (NucliSens; Organon Teknika, Durham, N.C.) with the reverse transcription-PCR Amplicor Monitor assay (Roche Diagnostic Systems, Branchburg, N.J.).
(The results of this study were presented in part at the 12th World AIDS Conference, Geneva, held from June 28 to July 3, 1998 [1a].)
Plasma samples were obtained from adult and pediatric patients recruited into clinical trials of antiretroviral therapies. Informed consent was obtained from all patients or their legal guardians prior to enrollment in the studies. Plasma was separated from anticoagulated blood within 6 h of collection in EDTA or acid-citrate-dextrose and stored at −70°C until analyzed. All samples were tested on either the first or second thaw. Spiked plasma sample standards, provided by the Virology Quality Assurance Laboratory (VQAL; Rush-Presbyterian Hospital, Chicago, Ill.; sponsored by the Division of AIDS, NIH), containing a known number of HIV RNA copies (9) were used in direct comparisons of the assays and as external standards in all experiments with patient samples.
Both the Monitor and the NucliSens assays were performed following the manufacturers’ instructions. Because of the nonnormal distribution of RNA concentrations, continuous variables were analyzed following log transformation, usually by nonparametric statistical methods. HIV RNA concentrations that were undetectable or invalid in one or both of the assays were omitted from statistical analysis. All analyses were performed with Statview 4.5 software (Abacus Concepts, Berkeley, Calif.).
Twofold serially diluted plasma samples containing between 250 and 2,000 HIV RNA copies/ml were tested in four or five replicates of both assays. Eight aliquots with nominal RNA concentrations greater than 500 copies/ml gave detectable results with both assays. Of the 10 plasma samples containing fewer than 500 copies/ml, 7 had detectable RNA results with Monitor and 6 had detectable results with NucliSens.
Serial threefold dilutions were made of 4 HIV-1-seropositive patient plasma specimens containing high concentrations of HIV RNA (4.2 × 106 to 6.7 × 106 copies/ml) and tested in both assays. A total of 36 samples were tested. Three samples containing more than 106 HIV-1 RNA copies/ml gave invalid results with Monitor, one sample with a predicted value of 640 copies/ml was undetectable in both assays, and two samples with predicted concentrations of 1,920 and 320 RNA copies/ml were undetectable by NucliSens but gave values of 1,011 and 212 copies/ml with Monitor. The mean overall difference between the results of serially diluted specimens was closer to the true dilution factor and showed less variability when Monitor was used (Table 1). The linearity of Monitor was better than that of NucliSens in the lower half of the dynamic range (<104.4 to 104.75 RNA copies/ml), while the linearities of both assays were comparable in the upper range (Table 1). Log-transformed RNA values from the four dilution series were combined to perform linear regression analysis of the NucliSens- and Monitor-derived results against predicted RNA levels. The curve generated with Monitor provided a very close one-to-one linear fit (y = 1.002x − 0.058, r = 0.997), whereas NucliSens had a linear relationship (y = 0.921x + 0.262, r = 0.964).
TABLE 1.
Comparative linearities of Monitor and NucliSens quantitative HIV-1 RNA assays
| Value typea | Log10 difference (mean ± SD) in HIV-1 RNA copies/ml with each threefold dilutionb
|
|
|---|---|---|
| Monitor | NucliSens | |
| Overall | 0.49 ± 0.19 | 0.45 ± 0.24 |
| High | 0.46 ± 0.14 | 0.44 ± 0.11 |
| Low | 0.51 ± 0.24 | 0.46 ± 0.36 |
The arbitrary cutoff between “high” and “low” putative HIV-1 RNA values was between 26,000 and 54,000 copies/ml in each dilution series.
A true threefold dilution is approximately 0.48 log10.
Spiked plasma samples containing known quantities of HIV-1 RNA (0, 1.5 × 104, 1.5 × 105, 7.5 × 105, or 1.5 × 106 copies/ml), provided by the VQAL, were analyzed in replicate runs with Monitor (88 runs with each standard, with 7.5 × 105 copies/ml as the high standard) and NucliSens (69 runs, with 1.5 × 106 copies/ml as the high standard). Both assays showed a high degree of reproducibility. The interassay variability, given by the standard deviation of the log values of replicate assays, was narrower for NucliSens than for Monitor. The reproducibility of NucliSens improved progressively with higher input copy number, while the interassay variability of Monitor remained constant at around 0.20 log10 from 1.5 × 104 to 7.5 × 105 RNA copies/ml (Table 2). HIV-1 RNA concentrations obtained with Monitor were, on average, 0.27 log10 (close to twofold) greater than those generated by NucliSens P < 0.0001) (Table 2). All samples containing zero copies of HIV RNA were negative, giving a specificity of 100% for both assays.
TABLE 2.
Reproducibility and comparability with VQAL standard
| VQAL standard HIV-1 copies/ml (log10) | Log10 HIV-1 RNA copies/ml (mean ± SD)
|
P | |
|---|---|---|---|
| Monitor (n = 88) | NucliSens (n = 69) | ||
| 15,000 (4.18) | 4.29 ± 0.20 | 4.02 ± 0.18 | <0.0001a |
| 150,000 (5.18) | 5.27 ± 0.18 | 5.00 ± 0.14 | <0.0001a |
| 750,000 (5.88) | 5.88 ± 0.23 | NAc | |
| 1,500,000 (6.18) | NA | 5.96 ± 0.10 | <0.0001b |
Two-tailed P value obtained by unpaired t-test comparison between log10 values obtained by Monitor and NucliSens.
Unpaired t test comparing mean differences between log10 obtained with NucliSens and Monitor and actual input RNA log10 copy number of 6.18 in the case of NucliSens and 5.88 for Monitor.
NA, not applicable.
Quantitative HIV-1 RNA analysis was performed with both the Monitor and the NucliSens assays on plasma samples from 51 HIV-seropositive patients being treated with a variety of antiretroviral drugs or receiving no treatment. Log-transformed HIV-1 RNA values that were above the level of detection limit of both assays (n = 41) were compared before and after adjustment with a regression equation, making use of the nominal log10 RNA concentration in external VQAL standards run concurrently with patient samples (1, 9). The concordance between the assays for RNA detectability was 92%, with three samples which were negative by Monitor measuring 630, 1,100, and 1,200 copies/ml (1,181, 643, and 382 copies/ml, respectively, after adjustment) by NucliSens and one sample which was negative by NucliSens measuring 1,192 copies/ml (384 copies/ml after adjustment) with Monitor. In seven samples where unadjusted HIV RNA concentrates were detectable at less than 1,000 copies/ml with at least one assay, all but one had detectable levels by both methods.
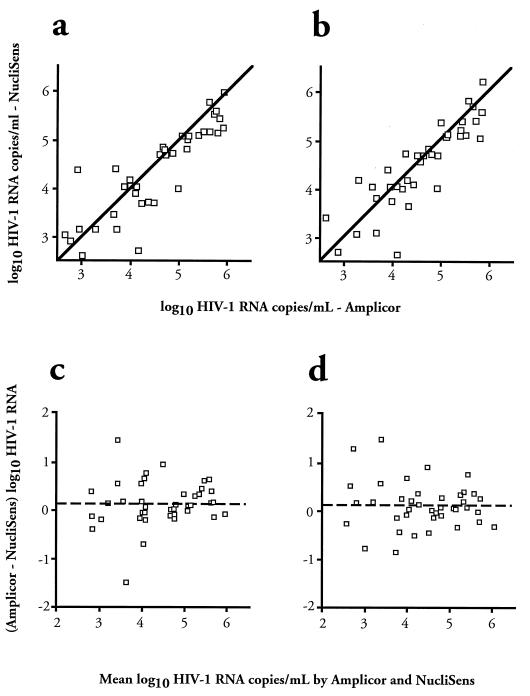
In the 41 samples testing positive by both methods, the median unadjusted HIV RNA value with NucliSens was 2.51 × 104 compared with 3.98 × 104 copies/ml with Monitor (P = 0.016 by Wilcoxon signed rank test). This difference persisted, although to a lesser degree, after adjustment using the results of concurrently run standards (median NucliSens HIV RNA concentration of 3.09 × 104 compared with 3.32 × 104 copies/ml by Monitor; P = 0.081). Simple regression analysis (Fig. 1a and b) showed a close linear relationship between the results obtained with the two assays, with little change following adjustment in the correlation coefficient (r = 0.880 and P < 0.001 after, compared with r = 0.872 and P < 0.001 before adjustment), but much closer approximation to equivalence (y = 0.931x + 0.167, compared with y = 0.815x + 0.685, where y is log NucliSens and x is log Monitor HIV RNA concentration). The disparity between NucliSens and Monitor in these patient samples was greater at lower RNA levels before and after adjustment (Fig. 1c and d).
FIG. 1.
HIV-1 RNA concentrations in patient samples tested by NucliSens plotted against data obtained with Monitor, before (a) and after (b) adjustment by using VQAL standards. The solid line is the line of unity. (c and d) Difference between log10 Monitor and NucliSens data plotted against the mean of the log10 HIV-1 RNA concentrations measured with the two assays, before (c) and after (d) adjustment.
In this study we compared the performance characteristics of NucliSens isothermal HIV-1 RNA amplification with those of Roche Monitor reverse transcription-PCR, which is already in widespread clinical use. The linearities of Monitor and NucliSens assay were comparable from approximately 500 to 1,000,000 RNA copies/ml, although Monitor had superior linearity and sensitivity in the lower range and performed variably at RNA levels greater than 1,000,000 copies/ml. These results resemble those found in comparisons of Monitor with the previous generation NASBA assay, which has a lower detection limit of 1,000 RNA copies/ml (3, 8). Both assays gave correlation coefficients and linear slopes close to unity when compared with the nominal RNA concentrations present in serially diluted samples. Both assays were close to 100% sensitive in detecting HIV-1 RNA in samples containing ≥1,000 copies/ml and had comparable sensitivities between 60 and 70% for RNA concentrations between 250 and 1,000 copies/ml.
Interassay variability assessments made with replicate measurements of standard HIV RNA concentrations were similar for both assays. NucliSens had lower interassay variability, especially at higher RNA concentrations (Table 2). Within both assays, linearity and reproducibility fell away at levels close to the lower detection limit. Direct comparison of the two assays made with seropositive patient samples showed that they correlated closely in their ability to detect viral RNA and in actual quantitative RNA measurement. Following adjustment by regression on the measured concentration of concurrently run VQAL standards, there was some improvement in the agreement between the assays with respect to the absolute HIV-1 RNA concentration, though this was not as marked a correction as has been reported in previous comparisons of Monitor and NASBA (1, 9). On the other hand, following adjustment, the linear regression equation relating log10 NucliSens values to log10 Monitor values much more closely resembled the line of equivalence, with an increase in the slope from 0.82 to 0.93 and a reduction in the intercept from 0.69 to 0.17.
In conclusion, the two assays appear to have similar performance characteristics. The linear dynamic range of NucliSens extends above 106 RNA copies/ml and thus may be more suitable for assessing viral load in infants and children, who usually have higher HIV-1 RNA concentrations than adults (6, 7). In contrast, the linearity, and probably the sensitivity, of the Monitor assay is superior in the lower range.
Acknowledgments
This study was funded in part by NIH contracts AACTG96VD006 and PACTG97PVCL06.
We thank Don Brambilla for helpful discussions.
REFERENCES
- 1.Brambilla D, Leung S, Lew J, Todd J, Herman S, Cronin M, Shapiro D E, Bremer J, Hanson C, Hillyer G V, Sherry G D M, Sperling R S, Coombs R W, Reichelderfer P S. Absolute copy number and relative change in determinations of human immunodeficiency virus type 1 RNA in plasma: effect of an external standard on kit comparisons. J Clin Microbiol. 1998;36:311–314. doi: 10.1128/jcm.36.1.311-314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Dyer J R, Pilcher C D, Shepard R, Schock J, Eron J J, Fiscus S A. Abstr. 42162. 12th World AIDS Conference, Geneva, Switzerland. 1998. [Google Scholar]
- 2.Finzi D, Siliciano R F. Viral dynamics in HIV-1 infection. Cell. 1998;93:665–671. doi: 10.1016/s0092-8674(00)81427-0. [DOI] [PubMed] [Google Scholar]
- 3.Griffith B P, Rigsby M O, Garner R B, Gordon M M, Chacko T M. Comparison of the Amplicor HIV-1 Monitor test and the nucleic acid sequence-based amplification assay for quantitation of human immunodeficiency virus RNA in plasma, serum, and plasma subjected to freeze-thaw cycles. J Clin Microbiol. 1997;35:3288–3291. doi: 10.1128/jcm.35.12.3288-3291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lew J, Reichelderfer P, Fowler M, et al. Determinations of levels of human immunodeficiency virus type 1 RNA in plasma: reassessment of parameters affecting assay outcome. J Clin Microbiol. 1998;36:1471–1479. doi: 10.1128/jcm.36.6.1471-1479.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin H J, Myers L E, Yen-Lieberman B, et al. Multicenter evaluation of methods for the quantitation of plasma HIV-1 RNA. J Infect Dis. 1994;170:553–562. doi: 10.1093/infdis/170.3.553. [DOI] [PubMed] [Google Scholar]
- 6.Mofenson L M, Korelitz J, Meyer W A, et al. The relationship between serum human immunodeficiency virus type 1 (HIV-1) RNA level, CD4 lymphocyte percent, and long-term mortality risk in HIV-1 infected children. J Infect Dis. 1997;175:1029–1038. doi: 10.1086/516441. [DOI] [PubMed] [Google Scholar]
- 7.Shearer W T, Quinn T C, LaRussa P, et al. Viral load and disease progression in infants infected with human immunodeficiency virus type 1. N Engl J Med. 1997;336:1337–1342. doi: 10.1056/NEJM199705083361901. [DOI] [PubMed] [Google Scholar]
- 8.VanDamme A M, Schmit J C, van Dooren S, et al. Quantification of HIV-1 RNA in plasma: comparable results with the NASBA HIV-1 RNA QT and the AMPLICOR HIV Monitor test. J Acquired Immune Defic Syndr. 1996;13:127–139. doi: 10.1097/00042560-199610010-00003. [DOI] [PubMed] [Google Scholar]
- 9.Yen-Lieberman B, Brambilla D, Jackson B, Bremer J, Coombs R, Cronin M, Herman S, Katzenstein D, Leung S, Lin H J, Palumbo P, Rasheed S, Todd J, Vahey M, Reichelderfer P. Evaluation of a quality assurance program for quantitation of human immunodeficiency virus type 1 RNA in plasma by the AIDS Clinical Trials Group virology laboratories. J Clin Microbiol. 1996;34:2695–2701. doi: 10.1128/jcm.34.11.2695-2701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]