Abstract
Randomly amplified polymorphic DNA (RAPD) and pulsed-field gel electrophoresis (PFGE) for the analysis of 13 Moraxella catarrhalis isolates, 11 successive strains isolated from sputa of five children and 2 isolates obtained the same day from twins, were compared. RAPD and PFGE both yielded nine types from the 13 isolates, showing a chronic colonization with one strain in three patients and a successive colonization with different strains in two patients. The promising results obtained with RAPD should be confirmed with a larger number of strains, but RAPD seems as suitable as PFGE for the typing of M. catarrhalis.
Moraxella catarrhalis (Branhamella catarrhalis) is an aerobic gram-negative diplococcus commonly found in the upper respiratory tract (12) which has for a long time been regarded as a nonpathogenic commensal. However, M. catarrhalis is now recognized as a lower-respiratory-tract pathogen (2). The dynamics of M. catarrhalis infection have been studied in patients with chronic obstructive pulmonary disease, using pulsed-field gel electrophoresis (PFGE) to analyze the restriction fragment length polymorphism of chromosomal DNA of clinical isolates (7). To our knowledge, randomly amplified polymorphic DNA (RAPD) analysis, which is less time-consuming, has not been applied to the genotyping of M. catarrhalis. In this study, we compared RAPD analysis with PFGE to analyze the features of M. catarrhalis colonization in five children with chronic obstructive pulmonary disease.
M. catarrhalis isolates were obtained from sputum samples and inoculated onto blood and chocolate agar (bioMérieux, Marcy l’Etoile, France) for 24 to 48 h at 36°C in an atmosphere of 5% CO2 in air. Bacterial species were identified by standard methods (2). Eleven isolates cultured from five children (aged 6 months to 15 years old) and two isolates obtained on the same day from twins (aged 2 years old) were stored at −80°C in glycerol broth until further investigation. The type strain ATCC 25238 was also included. Table 1 lists the patients and bacterial strains studied.
TABLE 1.
Characteristics of the patients and M. catarrhalis strains
| Patient (age) | Strain no. (interval between isolate collection) | RAPD pattern obtained with primer P2 | PFGE pattern obtained with NotI |
|---|---|---|---|
| I (12 yr) | 1 (1 mo) | A | A′ |
| 2 | A | A′ | |
| II (2 yr) | 3 (2 mo) | B | B′ |
| 4 | C | C′ | |
| III (6 mo) | 5 (3 days) | D | D′ |
| 6 | D | D′ | |
| IV (6 mo) | 7 (1 mo) | E | E′ |
| 8 (3.5 mo) | F | F′ | |
| 9 | G | G′ | |
| V (15 yr) | 10 (1.5 mo) | H | H′ |
| 11 | H | H′ | |
| VIa (2 yr) | 12b | J | J′ |
| VIIa (2 yr) | 13b | J | J′ |
Patients VI and VII are twins.
Isolate 12 from patient VI and isolate 13 from patient VII are from specimens obtained on the same day.
RAPD analysis was performed as previously described (8). We used the following six primers (14): 5′-TCACGATGCA-3′ (P1), 5′-GCCCCCAGGGGCACAGT-3′ (P2), 5′-TTATGTAAAACGACGGCCAGT-3′ (P3), 5′-GCAATTAACCCTCACTAAAG-3′ (P4), 5′-GTAATACGACTCACTATAG-3′ (P5), and 5′-GGAAACAGCTATGACCATG-3′ (P6) (Unité de Chimie Organique, Institut Pasteur, Paris, France). The PCR program consisted of 35 cycles (95°C for 1 min, 55°C for 1 min [35°C for primer P1], and 72°C for 1 min for each cycle) in a thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.). At the start of PCR, the first denaturation at 95°C lasted for 3 min and the extension at 72°C lasted for 5 min. The RAPD products were resolved by electrophoresis in a 2% agarose gel and detected by staining with ethidium bromide. Strains were considered different from one another if their patterns differed by one prominent band in three repeated experiments. Small differences in the intensities of major bands or loss of some weak bands was ignored (6). The PFGE procedure was performed as previously described (13). Briefly, overnight cultures were suspended in buffer and mixed with 1.5% low-melting-point agarose to form an agar insert. This plug was first incubated for 24 h at 37°C in a lysis solution containing 0.5% sarcosine and 20 μg of RNase/ml and then in a second solution containing 1% sarcosine and 50 μg of proteinase K/ml for 24 h at 55°C. After digestion with one of five restriction endonucleases (XbaI, NheI, SpeI, SmaI, or NotI) (Gibco-BRL, Gaithersburg, Md.), DNA fragments were separated in a 1% agarose gel by electrophoresis, using the CHEF DR III system (Bio-Rad, Richmond, Calif.) with the switch time ramped from 5 to 20 s over an 18-h period (NotI), at 220 V and at 14°C. The gel was stained with ethidium bromide, and DNA bands were visualized with a UV transilluminator and then photographed. DNA from bacteriophage lambda concatemers was used as a size marker. PFGE patterns were analyzed using the categorization of Tenover et al. (11).
With the primers P1, P4, P5, and P6, patterns with two to five fragments were obtained, establishing only three to six different types from the 13 clinical isolates (data not shown). Moreover, the RAPD procedure with these primers was unsuccessful for some isolates. The primer P3 yielded seven patterns composed of five to eight fragments, of which three permanent bands (690, 490, and 430 bp) were obtained for every isolate, including the type strain ATCC 25238 (data not shown). The primer P2 yielded nine patterns from the 13 clinical isolates. These patterns were composed of 6 to 12 fragments, ranging from 110 to up to 1,114 bp in size (Fig. 1) and correlated perfectly with the nine PFGE types. Several endonucleases (XbaI, NheI, SpeI, SmaI, and NotI) were tested as reported by Kawakami et al. (5) and Klingman et al. (7). XbaI generated patterns composed of more than 25 small fragments (100 kb to less than 25 kb), which were unsuitable for analysis (data not shown). SmaI patterns exhibited five to seven fragments ranging from 50 to 350 kb in size, providing no sufficient differentiation for the isolates of two patients. Moreover, digestion with SmaI was unsuccessful for three other isolates. As expected, the SpeI and NheI patterns were reliable (7). NotI had the most discriminatory power, with patterns of 8 to 10 fragments ranging in size from 50 to 450 kb and yielding nine types from the 13 isolates (Fig. 2). Both the techniques (RAPD analysis with primer P2 and PFGE with NotI) produced the same results, summarized in Table 1. The genotype X of the type strain ATCC 25238 was different from those of clinical isolates. Three different types (A, D, and H) were observed in patients I, III, and V, respectively, each of them retaining the same type in his pair of isolates (1 and 2, 5 and 6, and 10 and 11). Isolates 3 and 4 (from patient II) showed different types, B and C. Isolates 7, 8, and 9 (from patient IV) showed different types, E, F, and G. The isolates 12 and 13 from the twins (patients VI and VII) shared the same type, J.
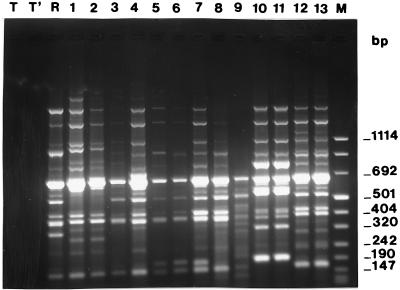
FIG. 1.
RAPD (primer P2) patterns of M. catarrhalis strains. Lanes: T and T′, negative controls; M, size marker; R, strain ATCC 25238; 1 to 13, clinical isolates. Three patterns were obtained from the following pairs of isolates: 1 and 2, 5 and 6, and 10 and 11 (from patients I, III, and V, respectively); two patterns were obtained from isolates 3 and 4 (from patient II); three patterns were obtained from isolates 7, 8, and 9 (from patient IV); and the same pattern was obtained from isolates 12 and 13 (from patients VI and VII [twins]).
FIG. 2.
PFGE (NotI) patterns of M. catarrhalis strains. Lanes: R, strain ATCC 25238; 1 to 13, clinical isolates. Three patterns were obtained from the following pairs of isolates: 1 and 2, 5 and 6, and 10 and 11 (from patients I, III, and V, respectively); two patterns were obtained from isolates 3 and 4 (from patient II); three patterns were obtained from isolates 7, 8, and 9 (from patient IV); and the same pattern was obtained from isolates 12 and 13 (from twins [VI and VII]).
Defining the epidemiology of M. catarrhalis infections has been hampered by the homogeneous appearance of the routine phenotypic markers, which are biochemical patterns and antibiotic susceptibility patterns. Analysis of chromosomal DNA by using restriction endonuclease digestion patterns has been used as a sensitive tool for defining strain relatedness (3, 4, 9, 10). However, its usefulness is limited by interpretation difficulties because the restriction fragment pattern may comprise more than 50 fragments, many of which are poorly resolved. Another typing system using DNA probes has been reported, which showed a great specificity but a limited sensitivity if not combined with restriction enzyme analysis (1). PFGE analysis of restriction fragments allowed the resolution of large DNA fragments and has greatly improved the usefulness of genomic DNA restriction fragment length polymorphism analysis, but the choice of restriction endonucleases is of importance in the case of M. catarrhalis. The enzymes recommended for the genome analysis of low guanine-plus-cytosine contents by PFGE (XbaI, NheI, SpeI, SmaI, and NotI [5]) did not give similar results. XbaI patterns were unsuitable for analysis. SpeI and NheI produced readily comparable banding patterns but required a prolonged electrophoresis period (at least 24 h). SmaI and NotI patterns were obtained with an 18-h electrophoresis, but only NotI had great discriminatory power. PFGE was performed three times for each isolate, and the results were identical each time, indicating the good reproducibility of the technique. Among our 13 clinical isolates, heterogeneity and genetic diversity of DNA patterns were established for the nine different types that could be defined. Each patient harbored a specific strain(s) of M. catarrhalis, but a probable transmission was present between the twins. Intrafamilial spread of the same strain has been reported by Faden et al. (4). We observed a chronic colonization with one strain in three patients and a successive colonization with two or three different strains in two patients. Klingman et al. (7) reported that patients with bronchiectasis were colonized successively by two to four different strains, and the average duration of colonization by the same strain in these patients was 2.3 months. In many circumstances, it may be necessary to use more than one technique to characterize M. catarrhalis strains (9). While PFGE is a time-consuming procedure, the recently introduced RAPD method has the advantages of simplicity and rapidity. The reproducibility of RAPD analysis was acceptable, though variation in the intensities of certain bands was sometimes noted, probably resulting from the DNA preparation (6). We evaluated the effectiveness of the RAPD assay with six different primers to distinguish between M. catarrhalis strains. With the primer P3, the three permanent bands recovered for all the strains (including type strain ATCC 25238) suggest a specific uniform pattern for all M. catarrhalis species. The promising results obtained with the primer P2 should be confirmed on a larger number of strains, but RAPD analysis seems as suitable as PFGE for comparison of M. catarrhalis strains.
Acknowledgments
This work was supported by Délégation à la Recherche Clinique, Assistance Publique-Hôpitaux de Paris (grant 96.003).
REFERENCES
- 1.Beaulieu D, Scriver S, Bergeron M G, Low D E, Parr T R, Jr, Patterson J E, Matlow A, Roy P H. Epidemiological typing of Moraxella catarrhalis by using DNA probes. J Clin Microbiol. 1993;31:736–739. doi: 10.1128/jcm.31.3.736-739.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Catlin B W. Branhamella catarrhalis: an organism gaining respect as a pathogen. Clin Microbiol Rev. 1990;4:293–320. doi: 10.1128/cmr.3.4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickinson D P, Loos B G, Dryja D M, Bernstein J M. Restriction fragment mapping of Branhamella catarrhalis: a new tool for studying the epidemiology of this middle ear pathogen. J Infect Dis. 1988;158:205–208. doi: 10.1093/infdis/158.1.205. [DOI] [PubMed] [Google Scholar]
- 4.Faden H, Harabuchi Y, Hong J J Tonawanda/Williamsville Pediatics. Epidemiology of Moraxella catarrhalis in children during the first 2 years of life: relationship to otitis media. J Infect Dis. 1994;169:1312–1317. doi: 10.1093/infdis/169.6.1312. [DOI] [PubMed] [Google Scholar]
- 5.Kawakami Y, Ueno I, Katsuyama T, Furihata K, Matsumoto H. Restriction fragment length polymorphism (RFLP) of genomic DNA of Moraxella (Branhamella) catarrhalis isolates in a hospital. Microbiol Immunol. 1994;38:891–895. doi: 10.1111/j.1348-0421.1994.tb02142.x. [DOI] [PubMed] [Google Scholar]
- 6.Kersulyte D, Struelens M, Deplano A, Berg D E. Comparison of arbitrarily primed PCR and macrorestriction (pulsed-field gel electrophoresis) typing of Pseudomonas aeruginosa strains from cystic fibrosis patients. J Clin Microbiol. 1995;33:2216–2219. doi: 10.1128/jcm.33.8.2216-2219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klingman K, Pye A, Murphy T F, Hill S L. Dynamics of respiratory tract colonization by Branhamella catarrhalis in bronchiectasis. Am J Respir Crit Care Med. 1995;152:1072–1078. doi: 10.1164/ajrccm.152.3.7663786. [DOI] [PubMed] [Google Scholar]
- 8.Moissenet D, Tabone M-D, Girardet J-P, Leverger G, Garbarg-Chenon A, Vu-Thien H. Nosocomial CDC group IV c-2 bacteremia: epidemiological investigation by randomly amplified polymorphic DNA analysis. J Clin Microbiol. 1996;34:1264–1266. doi: 10.1128/jcm.34.5.1264-1266.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan M G, McKenzie H, Enright M C, Bain M, Emmanuel F X S. Use of molecular methods to characterize Moraxella catarrhalis strains in a suspected outbreak of nosocomial infection. Eur J Clin Microbiol Infect Dis. 1992;11:305–312. doi: 10.1007/BF01962069. [DOI] [PubMed] [Google Scholar]
- 10.Patterson J E, Patterson T F, Farrel P, Hierholzer W J, Jr, Zervos M J. Evaluation of restriction endonuclease analysis as an epidemiologic typing system for Branhamella catarrhalis. J Clin Microbiol. 1989;27:944–946. doi: 10.1128/jcm.27.5.944-946.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenover F C, Arbeit R D, Goering R V, Mickelsen P, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaneechoutte M, Verschraegen G, Claeys G, Weise B, Van den Abeele A M. Respiratory tract carrier rates of Moraxella (Branhamella) catarrhalis in adults and children and interpretation of the isolation of M. catarrhalis from sputum. J Clin Microbiol. 1990;28:2674–2680. doi: 10.1128/jcm.28.12.2674-2680.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vu-Thien H, Moissenet D, Valcin M, Dulot C, Tournier G, Garbarg-Chenon A. Molecular epidemiology of Burkholderia cepacia, Stenotrophomonas maltophilia and Alcaligenes xylosoxidans in a cystic fibrosis center. Eur J Clin Microbiol Infect Dis. 1996;15:876–879. doi: 10.1007/BF01691221. [DOI] [PubMed] [Google Scholar]
- 14.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]