Abstract
A reference library of types of Clostridium difficile has been constructed by PCR ribotyping isolates (n = 2,030) from environmental (n = 89), hospital (n = 1,386), community practitioner (n = 395), veterinary (n = 27), and reference (n = 133) sources. The library consists of 116 distinct types identified on the basis of differences in profiles generated with PCR primers designed to amplify the 16S-23S rRNA gene intergenic spacer region. Isolates from 55% of infections in hospitals in the United Kingdom belonged to one ribotype (type 1), but this type was responsible for only 7.5% of community infections.
Clostridium difficile is the etiologic agent of pseudomembranous colitis (PMC) and is a major cause of nosocomially acquired antibiotic-associated diarrhea (AAD) in the developed world (11). Several typing schemes have been developed to determine the relatedness of strains of C. difficile associated with infection, including serotyping (6, 16), immunoblotting (8), arbitrarily primed PCR (1, 15), pulsed-field gel electrophoresis (PFGE; 5, 9, 15), and PCR ribotyping (4, 7, 13). Collaborative studies have also been undertaken to assess the accordance, relative reliability, and discriminatory power of different schemes (3–5, 9, 10, 12–15). PCR ribotyping has been reported to provide a discriminatory, reproducible, and simple alternative to other typing methods (4). This technique has a number of advantages over other methods; specifically, PCR ribotyping has been shown to be more discriminatory than arbitrarily primed PCR (5) and serotyping (13) and is quicker and simpler than PFGE. PCR ribotyping has one further advantage over PFGE, since some isolates of C. difficile have excessive endogenous nuclease activity that renders them untypeable by PFGE (9, 10, 15). In the present study a library was constructed that comprises 116 distinct types of C. difficile identified on the basis of differences in amplification profiles generated by a modified PCR ribotyping technique (13). It is hoped that the library will facilitate global analysis of the epidemiology and relative virulence of strains of this nosocomial pathogen.
Bacterial isolates and PCR ribotyping.
The Anaerobe Reference Unit of the Public Health Laboratory Service, based at the University Hospital of Wales in Cardiff, has provided a C. difficile typing service to hospitals throughout England and Wales since 1993. A modified PCR ribotyping scheme (13) has been the method of choice for typing isolates from the United Kingdom since 1995.
The identity of isolates of C. difficile submitted for typing was confirmed initially by the assessment of recognized phenotypic criteria (2). Enterotoxin (A) and cytotoxin (B) production were determined by the Tox A TEST immunoassay (TechLab; BioConnections, Leeds, United Kingdom) and Vero cell cytotoxicity (2), respectively.
The 2,030 isolates analyzed in the present study comprised 1,235 isolates from stool samples from hospital patients, 395 isolates from stool samples referred via community practitioners, 150 isolates from the hospital environment, 27 isolates from veterinary sources, 89 isolates from the general environment, 1 isolate from an extraintestinal human site, and 133 reference strains held in the National Collection of Type Cultures, the American Type Culture Collection and the Culture Collection, University of Göteborg, and in the personal collections of C. difficile types held by Delmee and others (6, 16) and other members of the International Study Group on C. difficile (3).
PCR ribotyping was performed in duplicate, with slight modifications to a method described previously (13). Briefly, bacteria were harvested from overnight anaerobic cultures on Fastidious Anaerobe Agar (LabM, Bury, United Kingdom) supplemented with 6% horse blood. Crude template nucleic acid was prepared by resuspension of cells in a 5% (wt/vol) solution of Chelex-100 (Bio-Rad, Hemel Hempstead, United Kingdom) and boiling for 12 min. After the removal of cellular debris by centrifugation (15,000 × g for 10 min), the supernatant (10 μl) was added to a 100-μl PCR mixture containing 50 pmol of each primer 5′-CTGGGGTGAAGTCGTAACAAGG-3′ (positions 1445 to 1466 of the 16S rRNA gene) and 5′-GCGCCCTTTGTAGCTTGACC-3′ (positions 20 to 1 of the 23S rRNA gene), 2 U of Taq polymerase (Pharmacia), and 2.25 mM MgCl2. Reaction mixtures were subjected to 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min. Amplification products were concentrated to a final volume of 25 μl by heating at 75°C for 105 min (13) before electrophoresis (150 mA) in 3% Metaphor agarose (FMC Bioproducts, Rockland, Maine) for 6 h at 8°C. Products were visualized by staining the gel for 20 min in ethidium bromide (0.5 μg ml−1). To enable normalization of all gel patterns, a molecular size standard (100 bp; Advanced Biotechnologies, Epsom, United Kingdom) was run at five-lane intervals.
Library construction.
PCR ribotype profiles were analyzed with GelCompar image analysis software (version 4.0; Applied Maths, Kortrijk, Belgium). The criterion for the proposal of a new library type was the existence of clearly discernible, reproducible (at least six profiles required per type) differences in PCR ribotype pattern from those of all other existing types. The stability, reliability, and homogeneity of the patterns constituting each type have been tested with the cluster correlation algorithm with the unweighted pair group method by using arithmetic averages and fine alignment. The integrity of the library is tested routinely at monthly intervals by blind PCR ribotyping of quality control isolates. PCR ribotype profiles from routine clinical isolates are compared to those profiles which define the library by maximum matching with Pearson correlation.
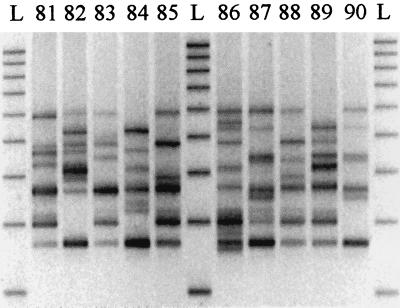
Figure 1 depicts the amplification profile obtained with 10 different PCR ribotypes within the library. At present, of the 2,030 isolates of C. difficile typed by this method, 116 distinct PCR ribotypes have been recognized (Table 1). A representative type strain of each PCR ribotype has been stored on cryobeads (ProLab Diagnostics, Wirral, United Kingdom) and frozen at −80°C.
FIG. 1.
PCR ribotype profiles obtained with strains belonging to PCR ribotypes 81 to 90. Lane L refers to 100-bp ladder (200 bp − 1 kbp).
TABLE 1.
PCR ribotypes of C. difficile depicting type strain, number of isolates obtained to date, and in vitro toxin production
| PCR ribotype | Type strain | Production of toxin A/B | No. of isolates | Commenta |
|---|---|---|---|---|
| 001 | R8366 | +/+ | 846 | Serogroup G |
| 002 | R8375 | +/+ | 60 | Serogroup A2 |
| 003 | R8384 | +/+ | 3 | |
| 004 | R8386 | +/+ | 1 | |
| 005 | R8373 | +/+ | 54 | |
| 006 | R8268 | +/+ | 11 | |
| 007 | R8264 | +/+ | 1 | |
| 008 | R10568 | +/+ | 2 | |
| 009 | R8269 | −/− | 46 | Serogroup I |
| 010 | R8270 | −/− | 110 | Most frequent commu-nity isolate |
| 011 | R7619 | +/+ | 4 | |
| 012 | R6187 | +/+ | 73 | |
| 013 | R5252 | +/+ | 12 | |
| 014 | R11446 | +/+ | 104 | Serogroup H |
| 015 | R6685 | +/+ | 76 | Serogroup G |
| 016 | R10424 | +/+ | 2 | |
| 017 | R7404 | −/+ | 32 | Serogroup F |
| 018 | R6184 | +/+ | 10 | Serogroup A8 |
| 019 | R8637 | +/+ | 1 | |
| 020 | R10079 | +/+ | 84 | Serogroup H |
| 021 | R8763 | +/+ | 5 | Serogroup A1 |
| 022 | R4262 | +/+ | 1 | |
| 023 | R6928 | +/+ | 43 | Weak toxin expression |
| 024 | R6321 | +/+ | 11 | |
| 025 | R7276 | +/+ | 2 | |
| 026 | R10118 | +/+ | 28 | Weak toxin expression |
| 027 | R12087 | +/+ | 1 | CD196 (binary toxin strain) |
| 028 | R9300 | −/− | 1 | |
| 029 | R8438 | +/+ | 3 | |
| 030 | R11004 | −/− | 8 | |
| 031 | R11631 | −/− | 24 | Serogroup K |
| 032 | R6598 | −/− | 2 | |
| 033 | IS58 | −/− | 2 | Serogroup E6 |
| 034 | IS81 | +/+ | 9 | Serogroup A5 |
| 035 | R11812 | −/− | 8 | |
| 036 | CCUG20309 | −/+ | 1 | Strain 8864 |
| 037 | R6641 | +/+ | 1 | |
| 038 | NCTC11206 | −/− | 19 | Serogroup C |
| 039 | R10738 | −/− | 10 | Serogroup A10 |
| 040 | R10917 | −/− | 1 | |
| 041 | R10920 | −/− | 1 | |
| 042 | R11817 | +/+ | 6 | |
| 043 | NCTC11382 | +/+ | 2 | |
| 044 | R10976 | +/+ | 2 | |
| 045 | R10842 | +/+ | 4 | |
| 046 | R10991 | +/+ | 4 | |
| 047 | R10541 | −/+ | 4 | |
| 048 | R10069 | +/+ | 3 | Weak toxin expression |
| 049 | R6320 | +/+ | 8 | |
| 050 | R9414 | +/+ | 9 | |
| 051 | R9549 | −/− | 1 | |
| 052 | R6155 | +/+ | 1 | |
| 053 | IS21 | +/+ | 6 | Serogroup K |
| 054 | IS22 | +/+ | 15 | Serogroup A1 |
| 055 | R11652 | +/+ | 3 | |
| 056 | IS25 | +/+ | 19 | |
| 057 | IS27 | +/+ | 5 | Serogroup K |
| 058 | R10456 | +/+ | 12 | Weak toxin expression |
| 059 | R9304 | +/+ | 1 | |
| 060 | IS40 | −/− | 2 | Serogroup B |
| 061 | R12099 | +/+ | 1 | |
| 062 | R11382 | +/+ | 4 | |
| 063 | IS47 | +/+ | 2 | Serogroup A5 |
| 064 | IS48 | +/+ | 2 | Serogroup A6 |
| 065 | IS49 | −/− | 2 | Serogroup A7 |
| 066 | IS51 | −/− | 4 | Serogroup A9 |
| 067 | IS52 | −/− | 4 | Serogroup A10 |
| 068 | IS56 | −/− | 1 | |
| 069 | IS59 | −/− | 1 | |
| 070 | R9367 | +/+ | 2 | Serogroup K |
| 071 | IS64 | −/− | 1 | Serogroup S1 |
| 072 | R12095 | +/+ | 1 | Serogroup X |
| 074 | IS72 | −/− | 1 | |
| 075 | IS93 | +/+ | 1 | Serogroup A1 |
| 076 | R11548 | +/+ | 4 | Serogroup A8 |
| 077 | R10955 | +/+ | 2 | |
| 078 | R7605 | +/+ | 13 | Weak toxin expression |
| 079 | R7606 | −/− | 1 | |
| 081 | R9764 | +/+ | 9 | |
| 082 | R7638 | −/− | 1 | |
| 083 | R10566 | +/+ | 1 | Serogroup S1 |
| 084 | R8768 | −/− | 6 | |
| 085 | R12098 | −/− | 4 | Serogroup X |
| 086 | R1880 | +/+ | 3 | |
| 087 | R11840 | +/+ | 5 | |
| 088 | R10855 | −/− | 2 | |
| 089 | R8603 | −/− | 1 | |
| 090 | R10737 | +/+ | 1 | |
| 091 | R8643 | −/− | 1 | |
| 092 | R10630 | +/+ | 4 | |
| 093 | R8853 | +/+ | 1 | |
| 094 | R10078 | +/+ | 4 | |
| 095 | R8858 | +/+ | 1 | |
| 096 | R9759 | +/+ | 2 | |
| 097 | R8914 | +/+ | 1 | |
| 098 | R9116 | −/− | 2 | |
| 099 | R7425 | −/− | 1 | |
| 100 | R12104 | −/− | 2 | |
| 101 | R10836 | +/+ | 1 | |
| 104 | R9180 | +/+ | 1 | |
| 106 | R10459 | +/+ | 88 | |
| 107 | R9313 | +/+ | 1 | |
| 110 | R7771 | −/+ | 2 | |
| 111 | R10870 | +/+ | 1 | |
| 112 | R8631 | −/− | 1 | |
| 114 | R11212 | −/− | 1 | |
| 115 | R11244 | +/+ | 6 | Serogroup G |
| 116 | R11347 | +/+ | 1 | |
| 117 | R10071 | +/+ | 1 | |
| 118 | R11394 | +/+ | 1 | |
| 119 | R11805 | −/− | 1 | |
| 120 | R11830 | +/+ | 1 | |
| 121 | R9378 | −/− | 1 | |
| 122 | R9385 | +/+ | 1 | |
| 123 | R11907 | −/− | 1 | |
| 124 | R11919 | −/− | 1 |
Reference where known has been made to Delmee serotype (6).
Strains within the library have also been analyzed by other typing schemes through international collaboration (3). The ribotyping method correlates with other typing schemes and allows subtyping of many of the types produced by other methods (Table 1) (3, 9, 13). In addition, all members of a single type have the same toxin A and toxin B production profiles, a characteristic which is not always exhibited by other typing schemes.
Routine typing of isolates of C. difficile from the United Kingdom.
Of the isolates of C. difficile from patients in hospitals in the United Kingdom (n = 1,235), a single, distinct PCR ribotype (type 1) has been found to be responsible for 55% (n = 682) of all referrals to the Anaerobe Reference Unit. However, it is intriguing that PCR ribotype 1 is detected less frequently (7.5%; n = 30) among isolates referred by community practitioners (n = 395). Type 1, a subtype of serogroup G (3, 13), may be a particularly virulent or transmissible clone of C. difficile or may have been selected by the particular antibiotic regimens used in hospitals in the United Kingdom. Research into the possible clonality of ribotype 1 is currently being undertaken. However, PFGE analysis of PCR ribotype 1 and serogroup G is not a viable option because these isolates produce excessive nuclease activity and are untypeable by this method (9, 10). Isolates which were untypeable by PFGE have also been encountered in nosocomial outbreaks in the United States (15). The 14 isolates described by Samore et al. (15) have been included in the present study and were found to belong to PCR ribotype 1, indicating that this type may also be a potential problem in the United States.
The library contains a number of types that exhibit no toxin A activity but produce active cytotoxicity (Table 1). Some of the strains constituting these types have been isolated from individuals with active AAD or PMC and highlight the limitations of using only toxin A assays for direct detection of C. difficile in stool samples.
The current reference library of fully characterized PCR ribotypes seems ideal for use by groups wishing to compare the performance of other fingerprinting or typing methods and by those studying the various virulence factors attributed to C. difficile. It is hoped that use of the library will facilitate epidemiology and aid virulence studies on this important nosocomial pathogen.
Acknowledgments
We thank the members of the International Study Group on C. difficile for kindly providing strains.
REFERENCES
- 1.Barbut F, Mario N, Frottier J, Petit J C. Use of arbitrary primer PCR for investigating an outbreak of Clostridium difficile-associated diarrhoea in AIDS patients. Eur J Clin Microbiol Infect Dis. 1993;12:724–795. doi: 10.1007/BF02098477. [DOI] [PubMed] [Google Scholar]
- 2.Brazier, J. S. 1993. Role of the laboratory in investigations of Clostridium difficile diarrhoea. Clin. Infect. Dis. 16(Suppl. 4):228–233. [DOI] [PubMed]
- 3.Brazier, J. S., M. E. Mulligan, M. Delmee, S. Tabaqchali, and the International Clostridium difficile Study Group. 1997. Preliminary findings of the International Study Group on Clostridium difficile. Clin. Infect. Dis. 25(Suppl. 2):199–201. [DOI] [PubMed]
- 4.Cartwright C P, Stock F, Beekmann S E, Williams E C, Gill V J. PCR amplification of rRNA intergenic spacer regions as a method for epidemiologic typing of Clostridium difficile. J Clin Microbiol. 1995;33:184–187. doi: 10.1128/jcm.33.1.184-187.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collier M C, Stock F, DeGirolami P C, Samore M H, Cartwright C P. Comparison of PCR-based approaches to molecular epidemiologic analysis of Clostridium difficile. J Clin Microbiol. 1996;34:1153–1157. doi: 10.1128/jcm.34.5.1153-1157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delmee M. Serogrouping of Clostridium difficile strains by slide agglutination. J Clin Microbiol. 1985;21:323–327. doi: 10.1128/jcm.21.3.323-327.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gürtler V. Typing of Clostridium difficile strains by PCR-amplification of variable length 16S-23S rDNA spacer regions. J Gen Microbiol. 1993;139:3089–3097. doi: 10.1099/00221287-139-12-3089. [DOI] [PubMed] [Google Scholar]
- 8.Heard S R, Rasburn B, Matthews R C, Tabaqchali S. Immunoblotting to demonstrate antigenic and immunogenic differences among nine standard strains of Clostridium difficile. J Clin Microbiol. 1986;24:284–287. doi: 10.1128/jcm.24.3.384-387.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyett, A. P., J. S. Brazier, and G. L. O’Neill. 1997. Pulsed-field gel electrophoresis as a method for typing Clostridium difficile in the routine laboratory. Rev. Med. Microbiol. 8(Suppl. 1):S63–S64.
- 10.Kato H, Kato N, Watanabe K, Ueno K, Sakata Y, Fujita K. Relapses or reinfections: analysis of a case of Clostridium difficile-associated colitis by two typing systems. Curr Microbiol. 1996;33:220–223. doi: 10.1007/s002849900103. [DOI] [PubMed] [Google Scholar]
- 11.Lyerly D M, Krivan H C, Williams T D. Clostridium difficile: its disease and toxins. Clin Microbiol Rev. 1988;1:1–18. doi: 10.1128/cmr.1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulligan M E, Peterson L R, Kwok R Y Y, Clabots C R, Gerding D N. Immunoblots and plasmid fingerprints compared with serotyping and polyacrylamide gel electrophoresis for typing Clostridium difficile. J Clin Microbiol. 1988;26:41–46. doi: 10.1128/jcm.26.1.41-46.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Neill G L, Ogunsola F T, Brazier J S, Duerden B I. Modification of a PCR-ribotyping method for application as a routine typing scheme for Clostridium difficile. Anaerobe. 1996;2:205–209. [Google Scholar]
- 14.Rupnik M, Avensani V, Janc M, von Eichel-Streiber C, Delmee M. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J Clin Microbiol. 1998;36:2240–2247. doi: 10.1128/jcm.36.8.2240-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samore M H, Kristjansson M, Venkataraman L, DeGirolami P C, Arbeit R D. Comparison of arbitrarily-primed polymerase chain reaction, restriction enzyme analysis and pulsed-field gel electrophoresis for typing Clostridium difficile. J Microbiol Methods. 1996;25:215–224. [Google Scholar]
- 16.Toma S, Lesiak G, Magus M, Lo H L, Delmee M. Serotyping of Clostridium difficile. J Clin Microbiol. 1988;26:426–428. doi: 10.1128/jcm.26.3.426-428.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]