Abstract
Broad-range PCR has proven to be useful for the detection of bacteria. A set of broad-range PCR primers directed against conserved regions in the 16S rRNA gene was designed to specifically amplify either gram-positive or gram-negative bacteria. The gram type-specific broad-range PCR correctly classified all 62 pathogenic species tested.
Antibiotic treatment of bacterial infections depends on the species of bacteria, with the differentiation between gram positive or gram negative being one of the most important factors. A 100% sensitive and specific method for the identification of the presence of bacteria in usually sterile body fluids would allow for earlier initiation of targeted antibiotic treatment. Recent studies suggest that rapid detection systems can decrease patient mortality rates and costs associated with hospitalization (1). Currently, therapy is usually empiric, involving use of a broad-spectrum antibiotic until culture results are available.
Standard diagnosis of systemic bacterial infection depends on growth in culture, which requires at least 12 to 72 h for detection. The most rapid tests are latex agglutination tests and Gram stains, which are less sensitive than culture and molecular methods (6). Molecular biological methods for detection of nucleic acids have been shown to have greater sensitivity than immunological and staining methods. The use of PCR primers that target DNA regions that are conserved in bacteria for the purposes of DNA sequencing and detection of bacteremia has been described (2). A recent study employed a universal bacterial broad-range PCR in combination with Southern blot hybridization with probes for differentiation of gram-positive and gram-negative species (3). Here, we report a gram type-specific PCR for the differentiation of a wide range of pathogenic gram-positive and gram-negative bacteria. This technique will allow detection and differentiation between gram-positive bacteria and gram-negative bacteria and is more rapid and less time-consuming than Southern blot analysis of the PCR products for gram differentiation.
Pure bacterial isolates were derived from cultures of clinical specimens which were analyzed by conventional procedures at the Institute of Bacteriology, Salzburg, Austria. These specimens included blood, sputum, cerebrospinal fluid, and different kinds of swabs and feces. After a 24-h incubation on solid medium, single colonies were resuspended in 200 μl of phosphate-buffered saline and pelleted by centrifugation. The pellet was then resuspended in 50 μl of a solution containing 20 mM (NH4)2SO4, 75 mM Tris-HCl (pH 9.0), 0.5% Tween 20, 0.5% Triton X-100, and 5 mg of proteinase K/ml and incubated at 60°C for 1 h, followed by an incubation at 95°C for 10 min.
Universal broad-range PCR was carried out by using primers DG74 and 65ab (5′-AACTGGAGGAAGGTGGGGAY-3′) as described previously (3). Position 20 at the 3′-end in the universal primer 65ab is degenerated since there is no single signature which is present in all bacteria. DNA (2 μl) was amplified in a final volume of 30 μl containing 20 mM (NH4)2SO4, 75 mM Tris-HCl (pH 9.0), 0.1% Tween, 2.5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphates, 10 pmol of each primer, and 1 U of Red Hot DNA polymerase (Advanced Biotechnology, Epsom, United Kingdom). To destroy endogenous bacterial DNA present in the reagents, the reaction mixture was exposed to UV light (320 nm) for 5 min in the presence of 38 nM 5-methoxypsoralen before the addition of the DNA (7). The mixture was incubated at 95°C for 5 min, followed by 45 cycles of 95°C for 30 s, 69°C for 30 s, and 72°C for 10 s.
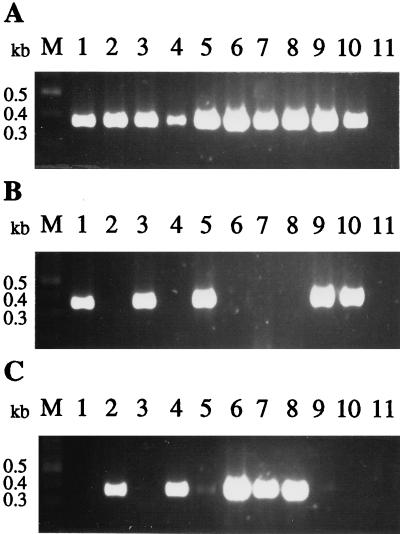
Single oligonucleotide primers were constructed with sequences unique to gram-positive bacteria and gram-negative bacteria, respectively. The gram-positive- and gram-negative-specific primers differ at the 3′-end, where a C residue is present in all gram-positive species analyzed and a G residue is present in all gram-negative species (Fig. 1). For the gram-positive-specific PCR, the primer 143 (5′-GAYGACGTCAARTCMTCATGC-3′) and the universal primer DG74 were used with the same reaction conditions as described for amplification using the universal primers except that the annealing temperature was 65°C and 50 cycles were performed. For the gram-negative-specific PCR, the primer 68d (5′-AYGACGTCAAGTCMTCATGG-3′) and the universal primer DG74 were used under the same reaction conditions as described for amplification using the universal primers except that the concentration of MgSO4 was 1.75 mM and 50 cycles were performed. Amplification with 45 and 50 cycles never gave a PCR product in the negative control reaction. Blood of control patients was spiked with a defined number of CFU of Escherichia coli and Staphylococcus aureus for the determination of the sensitivity of gram type-specific PCRs. For isolation of human genomic DNA from whole blood, erythrocytes were lysed in 150 mM NH4Cl–10 mM NaHCO3–1 mM EDTA. After centrifugation, the DNAs of remaining leukocytes and bacteria were isolated with the DNAzol method according to the manufacturer’s protocol (Molecular Research Center Inc., Cincinnati, Ohio). The sensitivity of the gram-negative-specific PCR was 101 CFU of E. coli per PCR and that of the gram-positive-specific PCR was 103 CFU of S. aureus (data not shown). Processing of blood and the amplification conditions for the gram-positive-specific PCR in low-titer clinical samples might have to be optimized for its application in routine diagnosis. No specific amplification of human or fungal DNA was observed when it was added to the PCR mixtures. The addition of 100 ng of human DNA did not influence PCR performance (data not shown), an important consideration when DNA is isolated directly from clinical samples. With all 116 different bacterial species tested in this study, only one signal was detected either with gram-positive or gram-negative-specific primers, but all samples showed a signal after universal broad-range PCR (Fig. 2, Table 1). Of the 116 clinical isolates, 46 were classified as gram positive by conventional methods and also as gram positive by gram type-specific PCR (Table 1). The balance of the samples were classified as gram negative by routine analysis and also specifically amplified by PCR (Table 1). Figure 2 shows a representative example for the results of the universal and gram type-specific PCRs of a sample of clinical isolates. Faint bands were sometimes seen in gram-negative-specific PCR (Fig. 2C, lanes 5 and 9) with DNA from gram-positive bacteria. If in further studies these bands occur when clinical samples are analyzed it should be sufficient to consider the strongest band.
FIG. 1.
Alignment of gram-positive and gram-negative primer sequences with nucleotide sequences of the 16S rRNA genes of different bacterial species. Residues identical to those of primers are indicated by dashes. Positions which do not match primer sequence are shown in italics. Gram type-specific residues are shown in boldface type. Nucleotide positions corresponding to the E. coli reference sequence are depicted at the top of the figure.
FIG. 2.
Detection of bacterial DNA in clinical isolates by broad-range PCR. (A) Universal PCR with primers DG74 and 65ab. (B) Gram-positive-specific PCR with primers DG74 and 143. (C) Gram-negative-specific PCR with primers DG74 and 68d. PCR products were analyzed by 2% agarose gel electrophoresis and ethidium bromide staining. Lanes M, molecular size markers; lanes 1, Bacillus subtilis; lanes 2, Citrobacter species; lanes 3, Corynebacterium species; lanes 4, Enterobacter cloacae; lanes 5, Enterococcus species; lanes 6: E. coli; lanes 7, K. pneumoniae; lanes 8, Serratia liquefaciens; lanes 9, S. aureus; lanes 10, Streptococcus pneumoniae; lanes 11, negative control.
TABLE 1.
Summary of results of universal PCR, gram-negative-specific PCR, and gram-positive-specific PCR of pathogenic bacteria
| Isolate (no. of different isolates) | Broad-range PCR
|
||
|---|---|---|---|
| Universal | Gram-negative-specific | Gram-positive-specific | |
| Aeromonas caviae (1) | + | + | |
| Aeromonas hydrophila (1) | + | + | |
| Acinetobacter baumannii (2) | + | + | |
| Acinetobacter junii (1) | + | + | |
| Acinetobacter species (3) | + | + | |
| Bacillus cereus (1) | + | + | |
| Bacillus species (1) | + | + | |
| Bacillus subtilis (1) | + | + | |
| Campylobacter jejuni (1) | + | + | |
| Citrobacter freundii (1) | + | + | |
| Citrobacter species (1) | + | + | |
| Clostridium difficile (3) | + | + | |
| Clostridium perfringens (1) | + | + | |
| Clostridium species (1) | + | + | |
| Clostridium subterminale (1) | + | + | |
| Corynebacterium species (2) | + | + | |
| Escherichia coli (6) | + | + | |
| Eikenella corrodens (2) | + | + | |
| Enterobacter aerogenes (3) | + | + | |
| Enterobacter agglomerans (1) | + | + | |
| Enterobacter cloacae (3) | + | + | |
| Enterobacter sakazakii (1) | + | + | |
| Enterobacter species (1) | + | + | |
| Enterococcus species (1) | + | + | |
| Fusobacterium species (1) | + | + | |
| Haemophilus species (2) | + | + | |
| Klebsiella oxytoca (4) | + | + | |
| Klebsiella pneumoniae (3) | + | + | |
| Klebsiella species (3) | + | + | |
| Morganella morganii (2) | + | + | |
| Neisseria sicca (1) | + | + | |
| Neisseria species (1) | + | + | |
| Peptostreptococcus species (2) | + | + | |
| Prevotella melaninogenica (1) | + | + | |
| Prevotella species (1) | + | + | |
| Propionibacterium species (1) | + | + | |
| Proteus mirabilis (1) | + | + | |
| Proteus species (2) | + | + | |
| Pseudomonas aeruginosa (4) | + | + | |
| Pseudomonas species (1) | + | + | |
| Salmonella group C (4) | + | + | |
| Salmonella group D (3) | + | + | |
| Salmonella group E (1) | + | + | |
| Salmonella typhi (2) | + | + | |
| Serratia liquefaciens (1) | + | + | |
| Serratia marcescens (2) | + | + | |
| Staphylococcus aureus (5) | + | + | |
| Staphylococcus epidermidis (4) | + | + | |
| Staphylococcus hominis (1) | + | + | |
| Staphylococcus species (1) | + | + | |
| Stenotrophomonas maltophilia (2) | + | + | |
| Stenotrophomonas species (1) | + | + | |
| Streptococcus agalactiae (1) | + | + | |
| Streptococcus group A (2) | + | + | |
| Streptococcus group B (2) | + | + | |
| Streptococcus group C (1) | + | + | |
| Streptococcus intermedius (2) | + | + | |
| Streptococcus milleri (2) | + | + | |
| Streptococcus mitis (1) | + | + | |
| Streptococcus pneumoniae (4) | + | + | |
| Streptococcus viridans (2) | + | + | |
| Veillonella species (2) | + | + | |
It seems that for the gram-specific broad-range PCR, only one difference in the primer sequence at the last base at the 3′-end is sufficient for gram specificity (Fig. 1). Although the sequence of the target 16S rRNA gene from some of the bacterial species does not match completely the sequence of the gram type-specific primers that we have designed, the specificity of the gram type-specific PCR was not influenced. Surprisingly, three mismatches in the primer sequence to Prevotella melaninogenica do not alter the specificity of the amplification reaction. However, to restore sensitivity, the annealing temperature of the gram-negative-specific PCR for P. melaninogenica had to be lowered by 4°C (data not shown). Therefore, only in the few cases where there is a positive signal in universal PCR but no product with either gram-positive- or gram-negative-specific PCR would a fourth PCR for gram-negative bacteria at 65°C have to be performed.
Many of the bacterial species investigated here, such as Klebsiella pneumoniae and S. aureus, which cause pneumonia and septicemia, respectively, are common pathogens. Other studies have shown that universal broad-range PCR is a rapid diagnostic tool for the detection of bacteria in clinical specimens; however, gram classification was not performed (2, 4, 5, 8). The gram type-specific broad-range PCR could form the basis for the development of a rapid and sensitive procedure for the detection and preliminary classification of bacteria in clinical specimens. This would allow the early choice of specific antibiotic treatment targeted to either gram-positive or gram-negative bacterial species. Conventional microbiological methods would still be required for confirmation of bacterial infection, identification of the species, and determination of the antibiotic susceptibility of any bacteria isolated.
Acknowledgments
These studies were supported by the Children’s Cancer Foundation Salzburg.
We thank Tiina P. Iismaa for critical review of the manuscript and Manfred Müller for the support in this work.
REFERENCES
- 1.Doern G V, Vautour M, Gaudet M, Levy B. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J Clin Microbiol. 1994;32:1757–1762. doi: 10.1128/jcm.32.7.1757-1762.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldenberger D, Künzli A, Vogt P, Zbinden R, Altwegg M. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J Clin Microbiol. 1997;35:2733–2739. doi: 10.1128/jcm.35.11.2733-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kane T D, Alexander J, Johannigman J. The detection of microbial DNA in the blood: a sensitive method for diagnosing bacteremia and/or bacterial translocation in surgical patients. Ann Surg. 1998;227:1–9. doi: 10.1097/00000658-199801000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laforgia N, Coppola B, Carbone R, Grassi A, Mautone A, Iolascon A. Rapid detection of neonatal sepsis using polymerase chain reaction. Acta Paediatr. 1997;86:1097–1099. doi: 10.1111/j.1651-2227.1997.tb14815.x. [DOI] [PubMed] [Google Scholar]
- 6.La Scolea L J, Dryja D. Quantification of bacteria in cerebrospinal fluid and blood of children with meningitis and its diagnostic significance. J Clin Microbiol. 1984;19:187–190. doi: 10.1128/jcm.19.2.187-190.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meier A, Persing D H, Finken M, Böttger E C. Elimination of contaminating DNA within polymerase chain reaction reagents: implications for a general approach to detection of uncultured pathogens. J Clin Microbiol. 1992;31:646–652. doi: 10.1128/jcm.31.3.646-652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilbrin B, van der Heijden I M, Schouls L M, van Emden J D, Hazes J M, Breedveld F C, Tak P P. Detection of bacterial DNA in joint samples from patients with undifferentiated arthritis and reactive arthritis, using polymerase chain reaction with universal 16S ribosomal RNA primers. Arthritis Rheum. 1998;41:535–543. doi: 10.1002/1529-0131(199803)41:3<535::AID-ART20>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]