Abstract
Periodontitis (PD) is characterized by bacterial infection and inflammation of tooth-supporting structures and can lead to tooth loss. PD affects ~47% of the US population over age 30 years and has a heritability of about 50%. Although the host immunoinflammatory response and genetic background play a role, little is known of the underlying genetic factors. We examined natural genetic variation in lipopolysaccharide (LPS)-induced PD across a panel of inbred mouse strains, the hybrid mouse diversity panel (HMDP). We observed a strain-dependent sixfold difference in LPS-induced bone loss across the HMDP with a heritability of 53%. We performed a genomewide association study (GWAS) using FAST-LMM, which corrects for population structure, and identified loci significantly associated with PD. We examined candidate genes at a locus on chromosome 5, which suggested a relationship between LPS-induced bone loss and, together with expression data, identified Cxcl family members as associated with PD. We observed an increase in Cxcl10 protein, as well as immune cells and pro-inflammatory cytokines in C57BL/6J (high bone loss strain) but not in A/J (low bone loss strain) after LPS injections. Genetic deletion of CXCR3 (Cxcl9 and10 receptor) demonstrated a 50% reduction in bone loss and reduced osteoclasts after LPS injections. Furthermore, WT mice treated with AMG-487 (a CXCR3 antagonist) showed a 45% reduction in bone loss and decreased osteoclasts after LPS injections. We conclude that CXCR3 is a strong candidate for modulating the host response in individuals susceptible to PD.
Keywords: GENETIC ANIMAL MODELS, ANIMAL MODELS, DENTAL BIOLOGY, DISEASES AND DISORDERS OF/RELATED TO BONE, THERAPEUTICS, PERIODONTAL DISEASE, PERIODONTITIS
Introduction
Periodontitis (PD) is characterized by a bacterial infection and inflammation that destroys the tissues that surround and support the teeth. If left untreated, PD can result in tooth loss.(1,2) PD affects 47.2% and 70.1% of the population over the ages of 30 and 65 years, respectively.(3) Microorganisms are central to PD pathogenesis, and P. gingivalis (P.g.) is significantly involved in PD infection. Moreover, P.g. is classified as a keystone species in PD disease progression and is consistently found around teeth with PD.(4) In addition to bacteria, environmental and genetic factors contribute to the risk of developing PD. A classic study on tea laborers, with no access to oral hygiene or dental care, highlighted that under similar environmental circumstances, there were wide variations of PD susceptibility, suggesting that PD has a significant genetic component.(5) Moreover, twin studies, after adjustment for environmental and external factors, concluded that approximately 50% of the variance observed in PD is attributed to genetics.(6,7) These studies emphasize that there are inherent host response differences in PD susceptibility and progression.(6,7) When combining host response differences and environmental factors, PD presents as a complex (polygenic) disease.(8,9) Complex trait disorders generally involve many genetic and environmental factors, where each factor can play a small role in trait/disease presentation.(10) Unfortunately, the detailed genetic influences in the pathogenesis of PD are poorly understood.
Genomewide association studies (GWAS) have emerged as a powerful tool to investigate the genetic architecture of complex trait diseases. GWAS allow for the unbiased interrogation of the entire genome in order to identify single-nucleotide polymorphisms (SNPs) associated with disease. To complement human GWAS, animal models can be used and they offer several advantages. Mice specifically, share similar structural, functional, and genetic traits with humans.(11) Moreover, there are powerful molecular and genetic tools, as well as repositories of mouse phenotypic, genotypic, metabolomic, and proteomic databases available to characterize disease pathogenesis.(11–14) Additionally, a major advantage of mouse studies is the ability to dissect disease and signaling pathways through genetic manipulation, transgenic and knockout mice. Several mouse panels, including the hybrid mouse diversity panel (HMDP)(12) and the Collaborative Cross (CC),(15) have been designed to capture the genetic variation present in populations, as well as provide high statistical power and fine mapping of the genome. The HMDP is composed of classic inbred and recombinant inbred mice densely genotyped for SNPs, which provide fine genetic mapping resolution and statistical genotype to phenotype association.(16)
The complexity of PD, the heterogeneous genetic composition of patients, and the difficulty to control environmental parameters pose challenges to clinical genetic studies,(17,18) making animal models an attractive complement to human studies.(19) Indeed, mouse studies on experimental periodontitis induced by Porphyromonas gingivalis (P. gingivalis) colonization reveal a strong genetic component in periodontal disease resistance and susceptibility and demonstrate that genetic determinants affect bacterial colonization, as well as periodontal bone levels.(20,21) These studies provide valuable insight in the heritable aspects of periodontitis as a whole. However, PD is a multifactorial process that involves, among others, bacterial colonization, biofilm organization and establishment, inflammatory host response, periodontal bone loss, and decreased tooth support.(22)
To begin dissecting the genetic influence in these pathogenetic disease processes individually, we explored the heritable nature of periodontal bone loss in response to a controlled inflammatory impact to identify genetic mediators of periodontitis and its potential implications in disease development(23) utilizing a well-characterized animal model that employs localized lipopolysaccharide (LPS) delivery to the periodontal tissues.(24,25)
Materials and Methods
Mice
Seven-week-old male mice of 104 genetically different strains of the HMDP (n ≥ 6/per strain) (Supplemental Table S1) were used according to the guidelines of the Chancellor’s Animal Research Committee of the University of California, Los Angeles, and the Animal Research: Reporting In Vivo Experiments (ARRIVE) protocols for the submission of animal studies were followed.(26) Mice were initially purchased from the Jackson Laboratory (Bar Harbor, ME, USA), bred and housed at UCLA for the duration of the study in a temperature- and light-controlled environment, and fed a standard chow. The number of animals for the GWAS were selected based on a paper that was published by our group.(23) Seven-week-old female B6.129P2-Cxcr3tm1Dgen/J homozygous chemokine receptor Cxcr3 knockout (KO) mice were bred and purchased from the Jackson Laboratory. Mice were maintained and utilized under the same guidelines and environment as described above.
Induction of periodontitis
Inflammatory induced bone loss was performed as previously described.(22) In brief, mice (n ≥ 3/strain) received 2 μL (20 μg) of P. gingivalis-lipopolysaccharide (P.g.-LPS) (ultrapure LPS, signals through TLR4, catalog #tlrl-ppglps, InvivoGen, San Diego, CA, USA) injections in between the first and second maxillary molars on both the right and left sides using a 10-μL Hamilton syringe with a 0.33-gauge needle (Hamilton Company, Reno, NV, USA). Mice received injections twice a week for 6 weeks. Control mice (n ≥ 3/strain) did not receive injections as previously described because there was no statistical difference in bone levels between noninjected and vehicle-injected groups.(22) During the course of injections, mice exhibited no overt clinical signs of soft tissue damage or inflammation. After 6 weeks of injections, mice were euthanized, maxillae were harvested, fixed in 10% buffered formalin for 48 hours, and subsequently stored in 70% EtOH for further analysis. The person performing the injections did not know the content of the syringe or the strain of the mouse.
Micro-computed tomography analysis
Maxillae were scanned using a μ-computed tomography (μ-CT) scanner (SkyScan 1172; SkyScan, Aartelaar, Belgium). Maxillae were scanned at 10-μm voxel size and imaged slices were converted to digital images and communication in medicine (DICOM) format. DICOM files were imported into Dolphin software (Dolphin Imaging, Chatsworth, CA, USA) for linear bone loss measurements. In Dolphin, maxillae were oriented for each molar, first and second, individually. Molars were oriented with the cementoenamel junction (CEJ) perpendicular to the root in the coronal plane. The root was also aligned parallel in the coronal plane. Each molar was oriented in the area corresponding to the middle of the tooth, aligned by the three roots in the axial plane. The distance from the CEJ to the alveolar bone crest (ABC) was recorded for the first molar distal and second molar mesial. Additional measurements, 0.2 mm palatal, were recorded for the first molar distal and second molar mesial. Measurements were recorded for the right and left sides independently and averaged to create a mean value for each mouse. All mice utilized for the duration of this study were scanned, oriented, and analyzed using the same parameters. To quantify the amount of bone loss, the averaged CEJ to ABC distance in the control sites was subtracted from the averaged distances in the LPS-injected sites. The remaining value represented net bone loss.(22)
Factored spectrally transformed-linear mixed modeling
Statistical analysis for the GWAS on LPS-induced periodontal bone loss was performed following previous GWAS studies utilizing the HMDP.(27,28) Genotypes of ~500,000 SNPs were obtained from the mouse diversity array. Only SNPs that presented with a minor allele frequency of >5% and missing genotype frequencies <10% were considered in analysis. The following filtering criteria yielded a final set of ~200,000 SNPs that were considered for analysis. To perform association testing, factored spectrally transformed-linear mixed modeling (FaST-LMM)(29) was performed. FaST-LMM factors in underlying population structure into statistical analysis and has successfully been employed in other GWAS studies utilizing the HMDP.(30–33) FaST-LMM is a linear mixed model method that statistically accounts for population structure in a fast and reproducible manner. To improve power, the kinship matrix was constructed using the SNPs from all the other chromosomes when testing all the SNPs on a specific chromosome. Using these parameters, the SNP gets tested in the regression equation only once. The significance level for the GWAS threshold using the HMDP was determined by the familywise error rate (FWER), which is the probability of detecting one or more false positives across all SNPs/phenotype. These parameters were similar to previous studies utilizing the HMDP.(27,28)
Heritability calculation
Heritability is defined as the fraction of the variance in a trait that is due to genetic factors.(34) To estimate heritability in our GWAS, we utilized the broad sense approach, which, estimates total heritability. To calculate broad sense heritability, an R statistical package was utilized and heritability was estimated based on the reproducibility of trait measurements in different animals of each strain.(35)
Macrophage genomewide expression analysis and correlation to LPS-induced bone loss
A GWAS, evaluating changes in macrophage gene expression in response to E. coli LPS utilizing the HMDP, was utilized to correlate SNPs to our LPS-induced PD model.(36) Ninety-two strains (all males) of the HMDP were obtained from the Jackson Laboratory and housed according to NIH guidelines. Primary macrophages were harvested, divided into two groups (control and LPS-stimulated), and gene expression (RNA) was profiled using Affymetrix HT MG-430A arrays for each group.(36) GWAS association mapping for macrophage gene expression was performed using efficient mixed-model association (EMMA). Macrophage expression data were correlated with the LPS-induced bone loss data using the bicorAndPvalue function from the Weighted Gene Co-expression Network Analysis (WGCNA) R package. Correlations were filtered for p < 10−3.
RNA isolation
Seven-week-old A/J and C57BL/6J mice were injected with one P.g.-LPS injection (2 μL or 20 μg of LPS) in between the first and second and second and third molars. Control mice were not injected. After 4 hours, mice were euthanized as previously described. Immediately after death, under the microscope (Leica Microsystems, Buffalo Grove, IL, USA), mice had approximately a 1.00 mm × 0.50 mm piece of maxillary gingival tissue excised in between the first and second and second and third molars corresponding to the area of LPS injections. Gingival tissues from the right and left sides of two mice were pooled for subsequent RNA isolation. RNA was isolated using a standard TRIzol (Thermo Fisher Scientific, Canoga Park, CA, USA) protocol and RNA quantity and purity was assessed using a NanoDrop 2000 (Thermo Fisher Scientific).
Microarray
RNA samples were prepared for microarray analysis using standard protocols at the UCLA Clinical Genetics Microarray core using the MouseRef-8 v2.0 chip. Gene expression data were analyzed using dChip software (2010.1). Differential gene expression, genes induced by LPS in A/J or C57BL/6J, were filtered using a false discovery rate of 50 and p < 0.05.
Histology
Maxillae were decalcified in 15% ethylenediaminetetraacetic acid (EDTA) for 4 weeks (solution was changed 3×/week). After decalcification, maxillae were paraffin embedded and cut coronally to 5-μm-thick sections using a microtome (McBain Instruments, Chatsworth, CA, USA). Sections were stained with hematoxylin and eosin (H&E) using standard protocols.
To evaluate immune cell populations and cytokines, immunohistochemistry was performed using the following antibodies: anti-NIMP-R14 (neutrophils) (1:250, ab2557, Abcam, Cambridge, UK), anti-CD3 (T cells) (1:100, ab5690, Abcam), and anti-Cxcl10 (15 μg/mL, AF-466-NA, R&D Systems, Minneapolis, MN, USA). After standard deparaffinization protocols, for all antibodies, excluding anti-CD3 and anti-Cxcl10, antigen retrieval was performed using 0.05% trypsin at room temperature for 15 minutes. Primary antibodies were incubated overnight at 4°C in a humidified chamber. Secondary antibodies (1:200 for all primaries) were incubated for 2 hours at room temperature. The immunoreaction was observed using AEC+substrate +chromogen solution (Dako, Carpinteria, CA, USA). For anti-CD3 and anti-Cxcl10, antigen retrieval was performed using 10 mM sodium citrate pH 6.0 overnight at 60°C. Primary and secondary antibodies were incubated as described above. The immunoreaction was observed using DAB peroxidase HRP (Vector Labs, Burlingame, CA, USA). For all stains, slides were digitally imaged using Aperio ImageScope model V11.1.2.752 (Vista, CA, USA). All histological sections used in this study were processed and stained utilizing the same parameters unless otherwise noted.
Cxcr3 knockout
Cxcr3 KO (B6.129P2-Cxcr3tm1Dgen/J homozygous chemokine receptor Cxcr3 knockout) and matched wild-type (WT) mice were randomly divided into Cxcr3 KO control (no LPS), Cxcr3 KO LPS-treated, WT control (no LPS), and WT LPS-treated groups. Mice received 2 LPS injections a week for 6 weeks, total of 12 injections. After LPS treatment, mice were euthanized and maxillae were harvested for further micro-CT and histological analysis. Quantification of linear bone loss was achieved using the same parameters as described above for the analysis of the HMDP.
Histology was performed in the maxilla of Cxcr3 KO and WT mice. Samples were embedded and processed as described above. Tissues were stained for tartrate-resistant acid phosphatase (TRAP, Sigma-Aldrich, St. Louis, MO, USA) to assess osteoclast (OC) counts, anti-Cox-2 (1:250, ab15191, Abcam) to assess general inflammation, and anti-Cxcl10 (15 μg/mL AF-466-NA, R&D Systems). Cells that presented with ≥2 nuclei and were in contact with bone were considered OCs (CITE). Osteoclasts were counted on six tissue sections per mouse and all six slides were averaged to create a total OC value for each mouse (n = 3 mice/group).
CXCR3 antagonist
Seven-week-old male C57BL/6J mice purchased from the Jackson Laboratory were utilized. (±)-AMG-487 (Tocris, R&D Systems), a Cxcr3 antagonist (a small molecular-weight peptide), was utilized to block Cxcr3 in vivo. Mice were divided into three groups: Control (local vehicle injection) + i.p. vehicle injection), LPS + vehicle injection, and LPS + AMG-487. AMG-487 was initially dissolved in a 50% hydroxypropyl-β-cyclodextrin (Sigma-Aldrich) in a sonicating water bath for 2 hours with occasional vortexing. After the AMG-487 powder had completely dissolved, distilled water was added to make a final concentration of 20% hyroxypropyl-β-cyclodextrin solution. Vehicle injections consisted of a 20% hyroxypropyl-β-cyclodextrin solution without AMG-487. At the start of LPS injections, mice received the first injection of AMG-487 at a concentration of 5 mg/g twice a day for the whole duration of the experiment.(37) P.g.-LPS injections were performed as described above. Mice were euthanized after a total of 12 LPS injections (6 weeks). Bone loss measurements, histology, and osteoclast counts (n = 5 slides per mouse, n ≥ 5 mice/group) were performed as described above.
Statistics
All statistical analyses were performed using Prism 5 (GraphPad, La Jolla, CA, USA). For bone loss analysis, measurements were averaged per mouse and subsequently averaged per group (for all experiments, n ≥ 3) to create a mean bone loss value per group (mean ± standard error of the mean). For quantification of TRAP staining, n ≥ 5 slides per mouse were stained and OC numbers were averaged per mouse. Again, each mouse was averaged to create a mean number of OCs per group (mean ± standard error of the mean). Significance levels were evaluated through either a Student’s t test or two-way analysis of variance (ANOVA) followed by a Bonferroni post hoc test with a confidence interval of 95%. Significance levels were as follows: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Results
LPS-induced strain-dependent bone loss across the HMDP
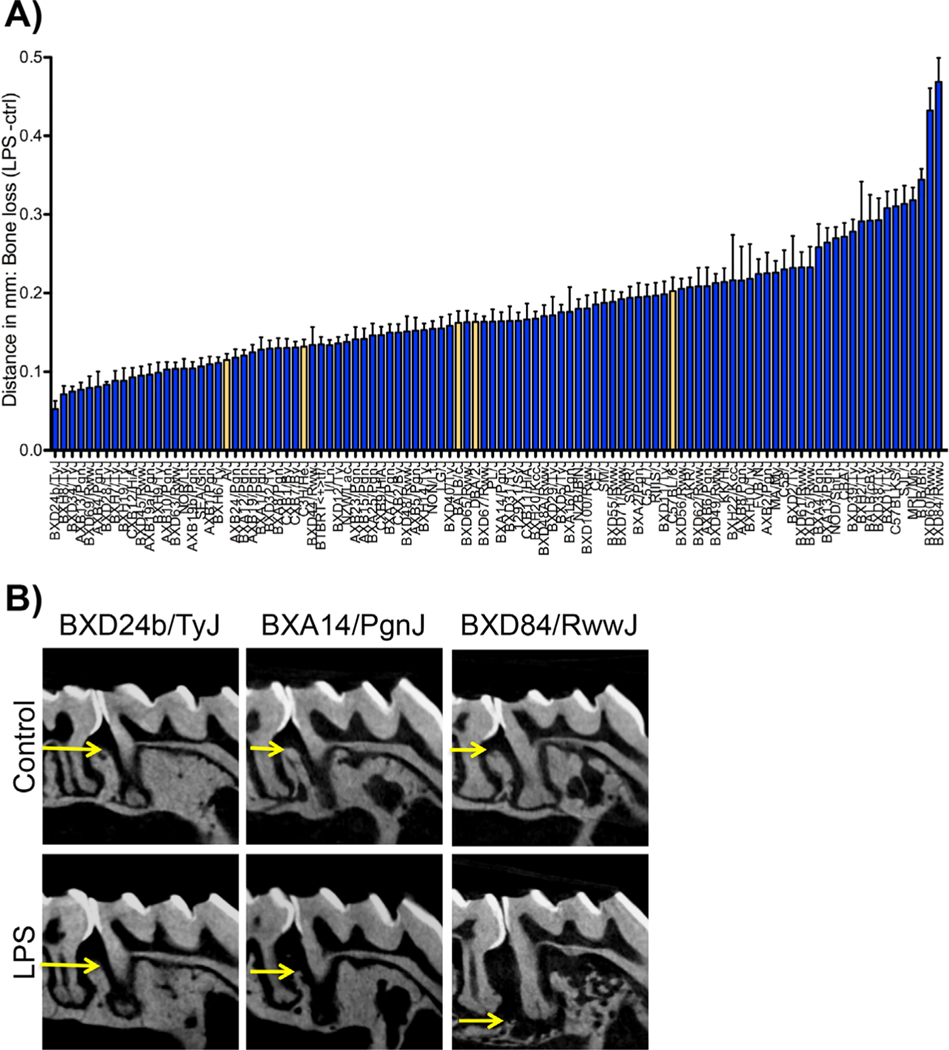
To assess differences in response to P.g. LPS in the HMDP, linear bone loss was quantitated at the injection site (between the first and second molars) after 6 weeks. Bone loss quantitation of 104 strains of the HMDP revealed a strain-dependent bone loss response to P.g. LPS (Fig. 1A). BXD24b/TyJ, a strain derived from a cross between C57BL/6J and DBA/2J, presented with the least amount of bone loss after LPS injections (0.071 ± 0.010). In contrast, BXD84/RwwJ, a strain derived from a cross between DBA/2J and C57BL/6J, presented with the highest amount of bone loss after LPS injections (0.468 ± 0.030) (Fig. 1A, B). Radiographically, representative micro-CT images showed alveolar bone loss in between the first and second molars at the LPS injection site (Fig. 1B).
Fig. 1.
Radiographic evaluation after 6 weeks of P.g.-LPS injections. (A) Graph representing bone loss in mm (LPS-ctrl) in 104 strains of the hybrid mouse diversity panel (HMDP), n ≥ 6 mice/strain. Data are represented as mean ± standard error of the mean (SEM). The yellow bars represent the five parental strains of the HMDP. (B) Representative radiographic images of control and LPS-treated strains of the HMDP. BXD24b/TyJ lost the least amount of bone, whereas BXD84/RwwJ lost the most amount of bone.
Genomewide association of SNPs to LPS-induced bone loss
We calculated the fraction of variance of the PD trait that is due to genetics by comparing the variation within strains (nongenetic) to the total variation across the strains.(35) The broad sense heritability was estimated at about 53%.
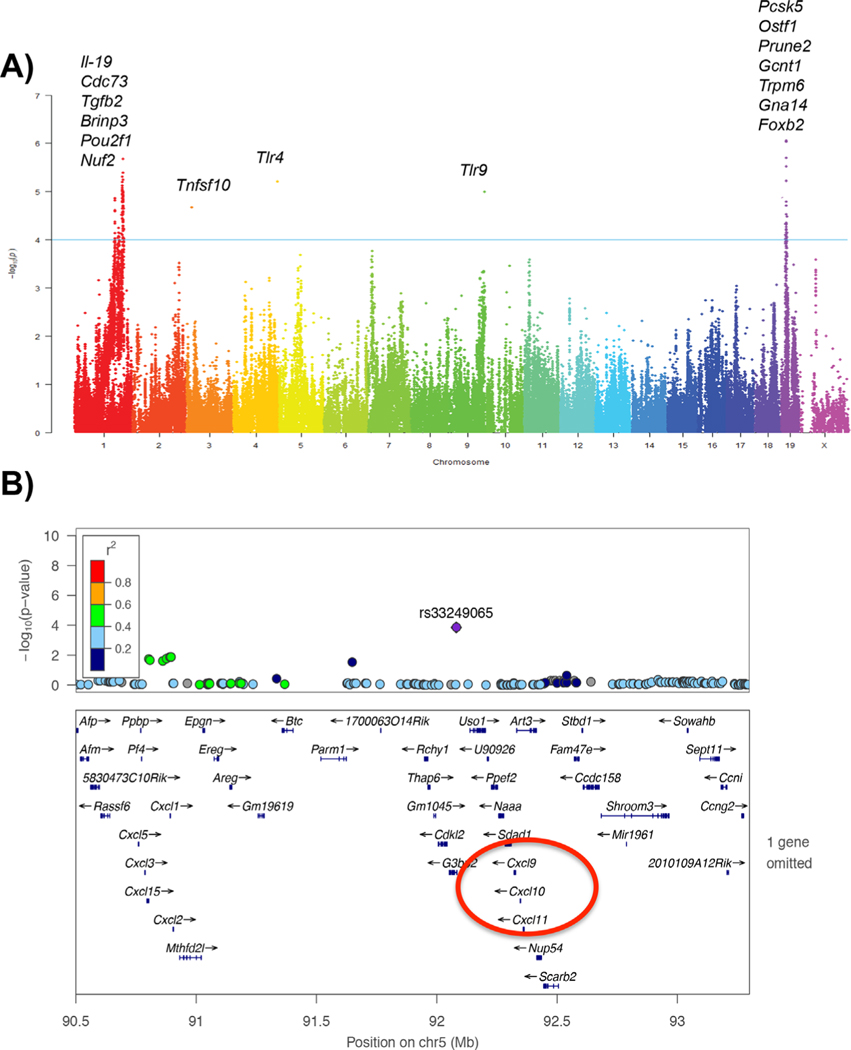
Using association analysis with correction for population structure, we mapped major loci contributing to LPS-induced bone loss in the HMDP. The regions identified as most likely to contain the causal gene(s) were those in strong linkage disequilibrium with the peak SNP (determined by calculated r2 SNP correlations greater than 0.8) and exceeding the FDR cut-off of 5%, corresponding to the association p value of 1.95e-4. A list of all significant associations for LPS-induced bone loss is provided in Supplemental Table S2 and Fig. 2A. In addition, lead SNPs are present in Table 1. A lead SNP was considered the highest peak in a locus. Although many loci fell under statistically significant peaks along these chromosomes, we prioritized rs33249065 located (p value 1-e4) on chromosome 5. Around this SNP, there were several chemokine (C-X-C motif) ligands (CXCL), specifically, Cxcl9 and Cxcl10 (Fig. 2B), as being a suggestive causal gene based on gene and protein expression data described below.
Fig. 2.
Genomewide association for P.g.-LPS-induced bone loss. (A) Manhattan plot for P.g.-LPS-induced bone loss. (B) High-resolution regional plot generated through LocusZoom. Zoom up on Chr 5. The blue horizontal bars denote a gene’s physical location. The linkage disequilibrium (LD) of the highlighted single-nucleotide polymorphism (SNP) at the locus is denoted by the color of the SNP. Highly correlated SNPs would be shown in red (in strong LD with each other), whereas weakly correlated SNPs are shown in navy (correlation represented by r2 color scale, inset).
Table 1.
Lead SNPs Obtained From the GWAS With Annotation of the Nearest Gene
| SNP | Chromosome | BP | p Value | Beta | Gene symbol (nearest gene) |
|---|---|---|---|---|---|
| rs38070275 | 1 | 162956639 | 6.340E-06 | –3.69E-02 | Fmo3 |
| rs31367957 | 1 | 163274880 | 6.340E-06 | −3.69E-02 | Prrx1 |
| rs50462242 | 1 | 164174083 | 9.150E-06 | −3.74E-02 | F5 |
| rs31281184 | 1 | 164353299 | 1.160E-05 | −3.67E-02 | Nme7 |
| rs31392814 | 1 | 165935900 | 1.160E-05 | −3.67E-02 | Pou2f1 |
| rs31791005 | 1 | 164054146 | 1.160E-05 | −3.67E-02 | Sele |
| rs36242840 | 1 | 165186989 | 1.160E-05 | −3.67E-02 | Sft2d2 |
| rs33754148 | 1 | 164933972 | 1.160E-05 | −3.67E-02 | Xcl1 |
| rs31372731 | 1 | 166231877 | 1.940E-05 | −3.81E-02 | Mael |
| rs31399153 | 1 | 165794428 | 2.020E-05 | −3.70E-02 | Cd247 |
| rs32256725 | 1 | 138570700 | 2.450E-05 | −3.22E-02 | Nek7 |
| rs30759208 | 1 | 139058022 | 2.550E-05 | −3.23E-02 | Samsn1 |
| rs33766243 | 1 | 164799184 | 2.760E-05 | −3.54E-02 | Dpt |
| rs31383004 | 1 | 166127042 | 2.760E-05 | −3.54E-02 | Dusp27 |
| rs31406187 | 1 | 165564964 | 2.760E-05 | −3.54E-02 | Sacy |
| rs6245419 | 1 | 163795691 | 3.570E-05 | −3.47E-02 | Kifap3 |
| rs36943249 | 1 | 165394763 | 3.840E-05 | −3.74E-02 | Iqwd1 |
| rs32274763 | 1 | 146689005 | 6.120E-04 | −3.14E-02 | Brinp3 |
| rs27313884 | 2 | 146951244 | 7.960E-04 | 2.96E-02 | Gm114 |
| rs27827801 | 4 | 45257015 | 7.590E-04 | −3.62E-02 | Frmpd1 |
| rs31490744 | 5 | 67462351 | 7.460E-04 | 2.66E-02 | NULL |
| rs31200478 | 7 | 12905344 | 7.500E-04 | 2.85E-02 | Zscan22 |
| rs32401360 | 7 | 12881485 | 7.680E-04 | 2.77E-02 | Zfp128 |
| rs31791117 | 7 | 12926316 | 8.000E-04 | 2.73E-02 | Rps5 |
| rs36682788 | 7 | 16503798 | 8.270E-04 | 2.68E-02 | Grlf1 |
| rs32388382 | 7 | 17010085 | 9.320E-04 | 2.79E-02 | Ppp5c |
| rs33464795 | 9 | 120426474 | 8.040E-04 | 2.81E-02 | Myrip |
| rs26830679 | 11 | 20213844 | 8.810E-04 | −3.12E-02 | Rab1 |
| rs37086609 | 19 | 17748514 | 4.720E-05 | 2.92E-02 | Pcsk5 |
| rs38333681 | 19 | 17335478 | 1.120E-04 | 2.84E-02 | Gcnt1 |
| rs30511916 | 19 | 17024082 | 1.120E-04 | 2.84E-02 | Prune2 |
| rs37426581 | 19 | 18788643 | 3.050E-04 | 2.68E-02 | Trpm6 |
| rs30357164 | 19 | 16508059 | 6.030E-04 | −2.81E-02 | Gna14 |
| rs36505593 | 19 | 18603958 | 8.920E-04 | 2.59E-02 | Ostf1 |
| rs29059597 | 20 | 57836749 | 9.900E-04 | 2.96E-02 | Tnni1 |
SNP = single-nucleotide polymorphism; GWAS = genomewide association study; BP = base pair.
Correlation of genomewide macrophage gene expression to candidate genes in LPS-induced bone loss
It is well documented that macrophages are increased in patients with PD as part of the host immune response to periodontopathogens.(38–41) Therefore, we aimed to correlate our bone loss FaST-LMM association mapping to a previous GWAS utilizing the HMDP assessing macrophage gene expression in response to LPS treatment.(36) Several genes classified as immune response genes, including growth factor receptor bound protein 2-associated protein 3 (Gab3), involved in cytokine signaling pathways and macrophage differentiation, and mitogenactivated protein kinase 7 (Map2k7), which mediates responses to proinflammatory cytokines, were correlated to both macrophage response to LPS and LPS-induced bone loss (Supplemental Table S3). Notably, Cxcl family members (Cxcl15 and Cxcl17) were also correlated (p < 0.05) to both macrophage response to LPS and LPS-induced bone loss (Table 2).
Table 2.
Genes Induced by LPS in Macrophages and That Also Correlated to LPS-Induced Bone Loss
| Probe Set ID | Gene symbol | Bicor | p Value |
|---|---|---|---|
| 1451610_at | Cxcl17 | −0.308918018 | 0.010371218 |
| 1421404_at | Cxcl15 | −0.285037736 | 0.018472638 |
LPS = lipopolysaccharide.
When assessing functional significance of genes correlated to macrophage response to LPS and LPS-induced bone loss through gene ontology (GO), many genes fell under the inflammatory response/cytokine pathway, including Ccr5 and Ccr8 (chemokine receptors), and immune system processes, including Gab3 as previously discussed. The complete list of genes correlated to both macrophage response to LPS and LPS-induced bone loss is in Supplemental Table S3.
Cxcl family members show increased gene expression in a high bone loss strain
To further evaluate differences in mRNA expression levels, in strains with high and low amount of bone loss after LPS injection, we performed microarray analysis utilizing the parental strain with the lowest (A/J) and the highest (C57BL/ 6J) amount of bone loss (Fig. 1A).
Significant differences in mRNA expression were observed between A/J and C57BL/6J 4 hours after LPS treatment (Table 3). Cxcl family members were among the statistically significant differentially expressed genes induced by LPS. The primary genes of interest were genes that were significantly induced in C57BL/6J LPS-treated mice but not induced in A/J LPS-treated mice. For instance, Cxcl9 induction was 38.87-fold difference and Cxcl10 was 19.23-fold difference (Table 3). Both Cxcl chemokines are involved in chemoattraction of immune cells, including monocytes/macrophages, T cells, natural killer cells, and dendritic cells.(42–44) Additionally, the chemokines Ccl4 (5.77-fold difference) and Ccl7 (3.55-fold difference), which are involved in macrophage inflammatory response and monocyte chemoattraction, respectively, were induced in C57BL/6J LPS-treated mice and not in A/J LPS-treated mice, highlighting that several host immune response pathways were significantly induced after LPS treatment in a high bone loss strain. Cxcr3, the receptor for Cxcl9 and Cxcl10, was not significantly induced in C57BL/6J LPS-treated mice and not in A/J LPS-treated mice. The complete list of genes induced by LPS in C57BL/6J but not in A/J can be found in Supplemental Table S4.
Table 3.
Genes Induced by LPS in C57BL/6J Mice But Not in A/J Mice
| Gene symbol | A/J LPS | C57BL/6J LPS | Fold change (FC) | Lower bound of FC | Upper bound of FC | t Statistic | p Value |
|---|---|---|---|---|---|---|---|
| Ccl4 | 180.52 | 1042.14 | 5.77 | 5.06 | 6.71 | 35.385 | 0.0011 |
| Ccl7 | 145.1 | 515.64 | 3.55 | 3.33 | 3.8 | 52.477 | 0.0006 |
| Cxcl10 | 155.66 | 2993.66 | 19.23 | 17.09 | 21.93 | 49.5 | 0.0095 |
| Cxcl9 | 221.42 | 8606.48 | 38.87 | 34.27 | 44.6 | 29.918 | 0.0208 |
LPS = lipopolysaccharide.
Immune and pro-inflammatory markers show increased expression in a high bone loss strain
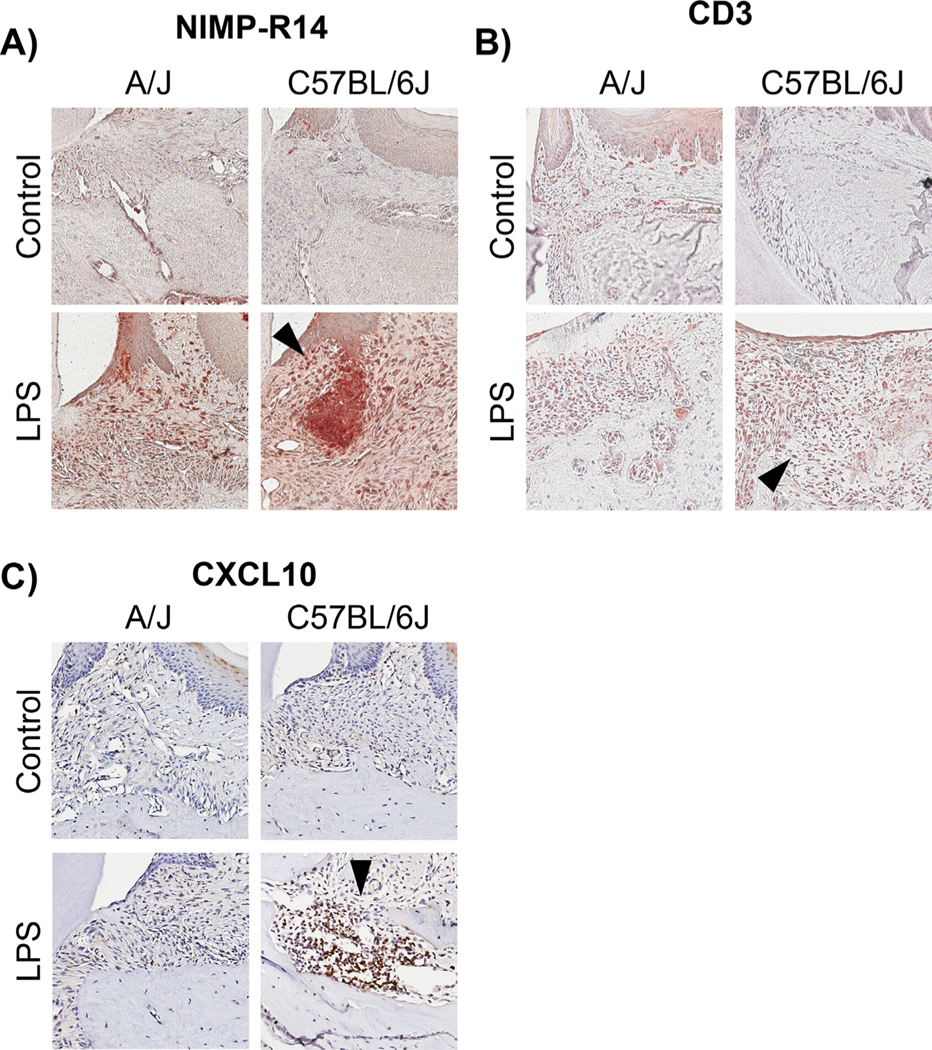
To further characterize differences between A/J, a low bone loss strain, and C57BL/6J, a high bone loss strain, tissue specimens were analyzed for immune and pro-inflammatory cellular markers through immunohistochemistry (IHC) staining. Neutrophil and t cells were assessed in A/J and C57BL/6J mice after LPS treatment because neutrophils and t cells are known to infiltrate into periodontal lesions in response to infection and inflammation.(45,46) When comparing control groups, there was no difference in immunostaining between C57BL/6J control and A/J control groups for both neutrophils and t cells. However, C57BL/6J LPS-treated groups presented with increased expression of neutrophils and t cells (Fig. 3A, B, black arrows) compared with A/J-LPS-treated mice. Furthermore, when staining for CXCL10 protein (chemokine responsible for a wide array of immune response cascades), which was highly associated in our GWAS and upregulated in our gene expression data (microarray), C57BL/6J LPS-treated specimens presented with increased protein expression of CXCL10 (Fig. 3C, black arrows) compared with A/J LPS treated mice. Again, there was no basal difference in CXCL10 protein expression between C57BL/6J control and A/J control groups.
Fig. 3.
Histological assessment of immune cells and cytokine protein expression. (A) Neutrophil immunostaining in A/J control, A/J LPS, C57BL6/J control, and C57BL6/J LPS-treated mice. Note the increased staining in C57BL6/J LPS compared with A/J LPS (black arrow). (B) CD3+ T-cell immunostaining in A/J control, A/J LPS, C57BL6/J control, and C57BL6/J LPS-treated mice. Note the increased staining in C57BL6/J LPS compared with A/J LPS (black arrow). (C) CXCL10 immunostaining in A/J control, A/J LPS, C57BL6/J control, and C57BL6/J LPS-treated mice. Note the increased staining in C57BL6/J LPS compared with A/J LPS (black arrow). All images are at 20×.
To evaluate pro-inflammatory mediators, protein levels of three pro-inflammatory markers, including NF-κB, cyclooxygenase-2 (COX-2), and tumor necrosis factor-alpha (TNF-a), which are known to have increased expression in patients with PD, were assessed.(47–49) C57BL/6J LPS-treated animals showed increased protein expression of all three pro-inflammatory mediators as evident by the brown/red immunoreactivity/ staining (Supplemental Fig. S1A–C, black arrows) compared with A/J LPS-treated mice. For all three pro-inflammatory markers, there was no qualitative difference between C57BL/6J control mice and A/J control mice.
Degradation of the extracellular matrix, caused by the action of matrix metalloproteinase (MMP) enzymes, is a host-mediated response in periodontitis. Therefore, staining for MMP-8 and MMP-13, which are associated with periodontitis in patients,(50,51) was assessed in A/J and C57BL/6J mice. After LPS treatment, C57BL/6J mice presented with increased immunoreactivity and protein expression of both MMP-8 and MMP-13 shown by the brown/red stain (Supplemental Fig. S2A, B, black arrows). When comparing C57BL/6J control groups with A/J control groups, there was no qualitative difference in MMP-8 or MMP-13 protein expression.
Cxcr3 knockout mice present with reduced bone loss after LPS treatment
Based on the GWAS, gene expression, and IHC data, the Cxcl9 and Cxcl10 pathway was further investigated to better understand their involvement in LPS-induced periodontal bone loss. As stated previously, Cxcl9 and Cxcl10 are involved in an array of immune responses, including recruitment of monocytes/macrophages, t cells, natural killer cells, and dendritic cells.(42–44) Furthermore, all three chemokines propagate their responses through the C-X-C motif chemokine receptor 3 (CXCR3). Therefore, to inhibit the function of all three chemokines, we employed a Cxcr3 knockout (KO) mouse and our P.g. LPS-injection model.
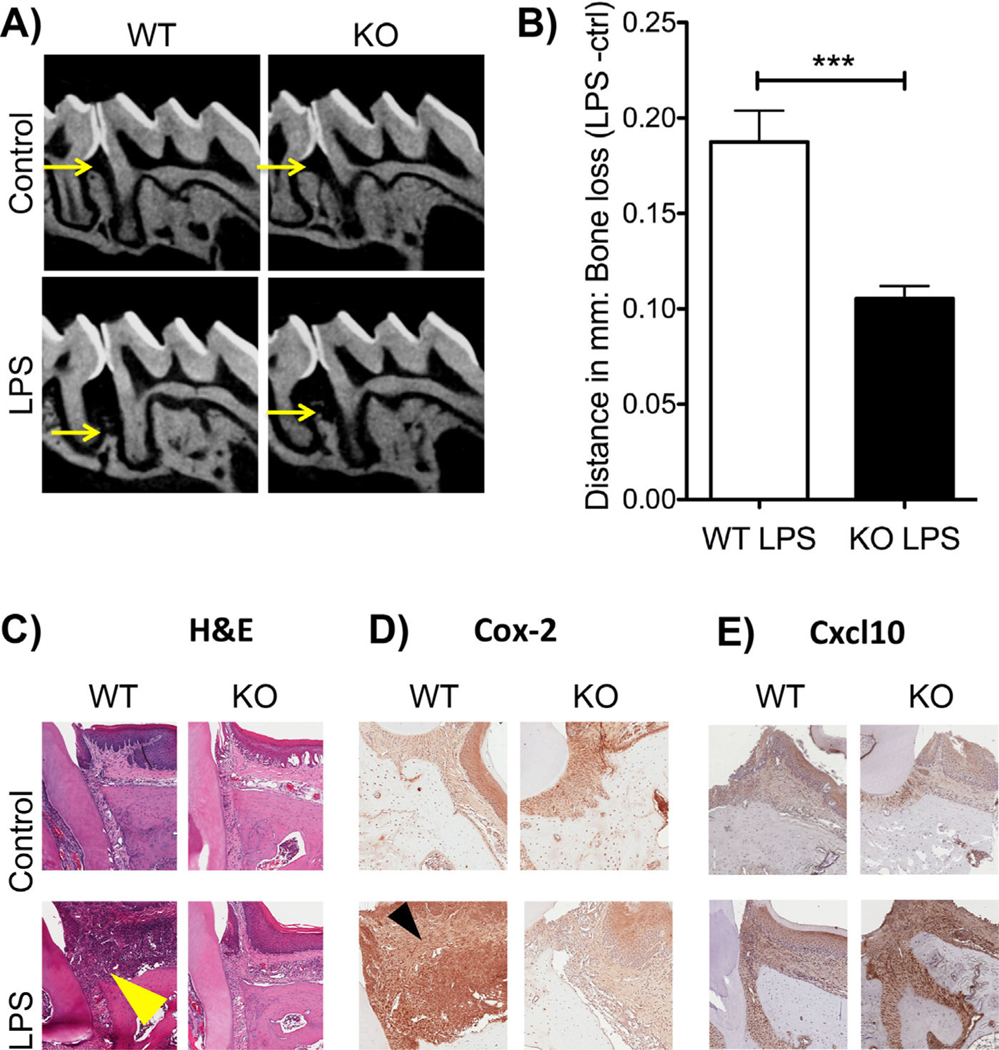
After 12 LPS injections, Cxcr3 KO mice presented with statistically significant less bone loss compared with WT (Fig. 4). Radiographically, WT LPS-treated mice showed a clear reduction in alveolar bone in between the first and second molars compared with Cxcr3 KO mice (Fig. 4A, B).
Fig. 4.
Deletion of Cxcr3 in vivo causes a reduction in bone loss. (A) Representative radiographic images of wild-type (WT) and Cxcr3 knockout (KO) control and LPS-treated mice. Note the increased bone loss in the WT LPS group compared with the KO LPS group. (B) Graph representing the bone loss (ctrl-LPS) of WT and KO mice. Significance was compared using a Student’s t test. n = 3 mice/group, p ≤ 0.05*, p ≤ 0.01**, p ≤ 0.001***. Data represented as mean standard error of the mean (SEM). (C) Hematoxylin and eosin (H&E)-stained tissue sections of WT and KO control and LPS-treated groups. Increased inflammatory infiltrates in the WT LPS group are denoted by the yellow arrow. (D) COX-2 immunostaining in WT and KO control and LPS groups. Increased COX-2 expression (brown stain) is denoted by the black arrow in the WT LPS. (E) Cxcl-10 immunostaining in WT and KO control and LPS groups. Increased Cxcl10 expression in the Cxcxr3 KO LPS-treated mice compared with WT LPS-treated mice (brown stain).
To confirm that the differences observed were in fact due to LPS treatment and not due to inherent bone-quality differences between Cxcr3 KO and WT mice, we assessed initial bone volume/tissue volume (BV/TV) in Cxcr3 KO and WT control animals. For both the maxillae and mesial trabecular bone distal from the growth plate in the femur, there was no statistical difference between BV/TV between Cxcr3 KO and WT mice (Supplemental Fig. S3).
After radiographic assessment of bone loss, Cxcr3 KO and WT mice were further analyzed for histological changes. Through hematoxylin and eosin (H&E) staining, there was an increase in cellular infiltrates observed in the WT LPS-treated group compared with the Cxcr3 KO group (Fig. 4C, yellow arrow). Comparing WT control mice to Cxcr3 KO control mice, there was no difference in cellular infiltrates (purple cells in the epithelial tissue). Further assessment of protein expression of proinflammatory marker COX-2 showed increased staining in WT LPS-treated groups compared with Cxcr3 KO LPS-treated animals. Again, when comparing WT control mice to Cxcr3 KO control mice, there was no overt difference in COX-2 expression. There was increased Cxcl10 expression in the Cxcxr3 KO LPS-treated mice compared with WT mice LPS-treated mice (Fig. 4E).
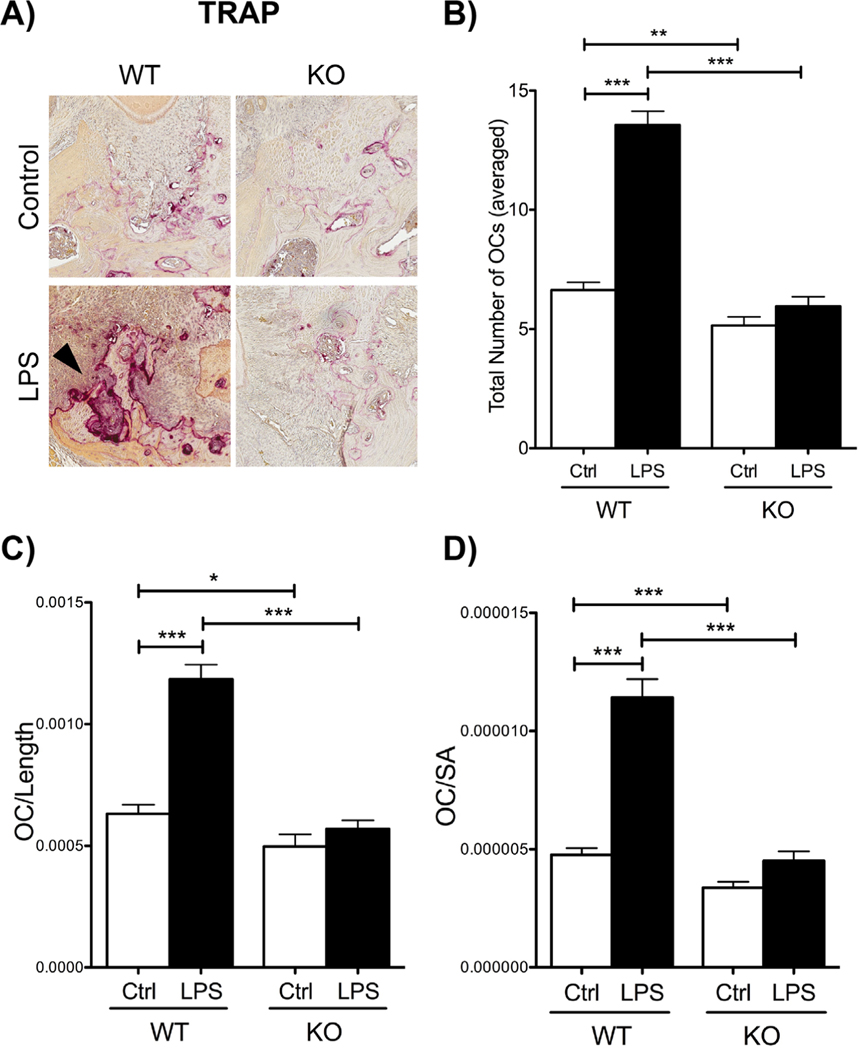
In addition to pro-inflammatory markers, osteoclast numbers were evaluated through TRAP staining after LPS injections between WT and Cxcr3 KO mice (Fig. 5). When comparing WT LPS-treated to Cxcr3 KO LPS-treated, WT mice presented with statistically significantly more TRAP+ cells compared with Cxcr3 KO mice (Fig. 5B). Focusing on control groups, WT control mice presented with significantly more osteoclasts compared with Cxcr3 KO control mice. Furthermore, when normalizing osteoclast numbers to alveolar bone length and surface area considered in analysis, WT LPS-treated mice presented with statistically significantly more osteoclasts per bone length and bone surface area compared with Cxcr3 KO LPS-treated mice (Fig. 5C, D). In addition, when utilizing a ligature-induced periodontitis model, a similar result was obtained (data not shown).
Fig. 5.
Histological assessment of osteoclast numbers in WT and Cxcr3 KO mice. (A) Tartrate-resistant acid phosphatase (TRAP+) staining for osteoclasts. Note the increase in TRAP+ cells in WT LPS-treated mice (black arrow) compared with KO LPS-treated mice. 20 magnification. (B) Graph representing total number of averaged osteoclasts in WT and Cxcr3 KO control and LPS groups. (C) Graph representing osteoclast numbers divided by the length of alveolar bone measured. (D) Graph representing osteoclast numbers divided by the surface area (SA) of the alveolar bone considered in analysis. For all graphs (B–D), significance was compared using a Student’s t test. n = 3 mice/group, p ≤ 0.05*, p ≤ 0.01**, p ≤ 0.001***. Data represented as mean ± standard error of the mean (SEM).
CXCR3 antagonist reduces bone loss in vivo
After LPS injections, Cxcr3 KO mice exhibited a reduction in bone loss and osteoclast numbers compared with WT mice. Therefore, when we chose to investigate if inhibition of Cxcr3 in vivo through a Cxcr3 antagonist would produce similar results, we utilized AMG-487. AMG-487 is a commercially available Cxcr3 antagonist that inhibits CXCR3-cell migration mediated by the chemokines CXCL9 and CXCL10.
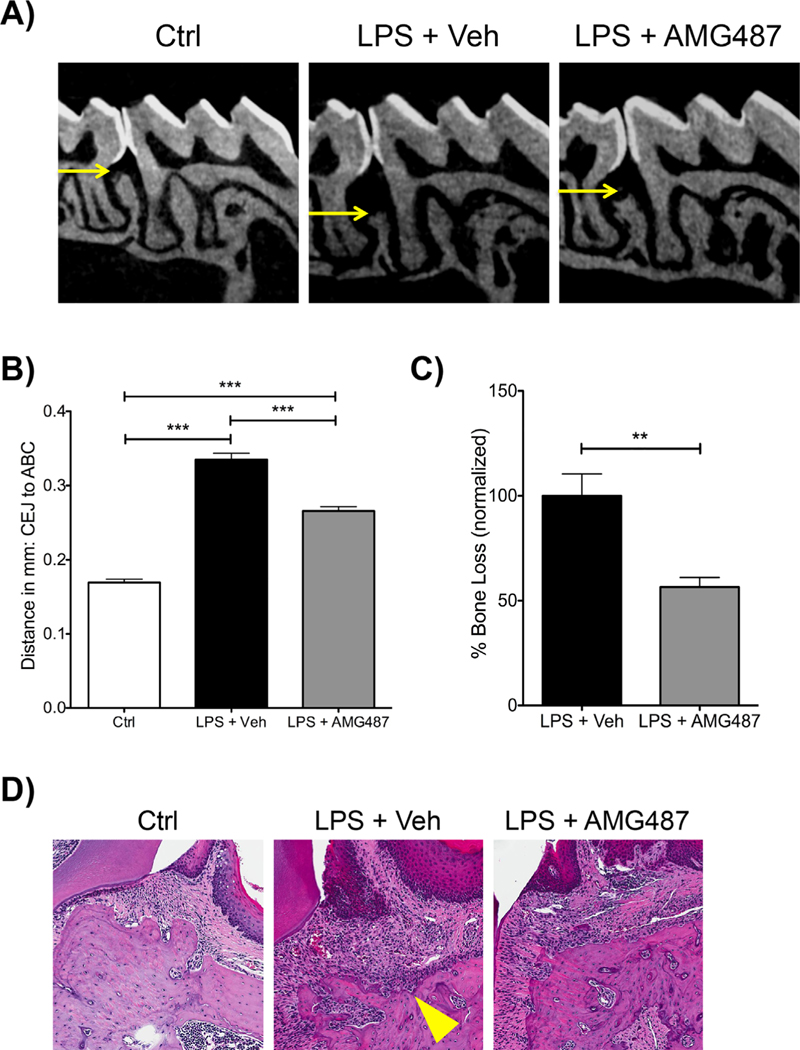
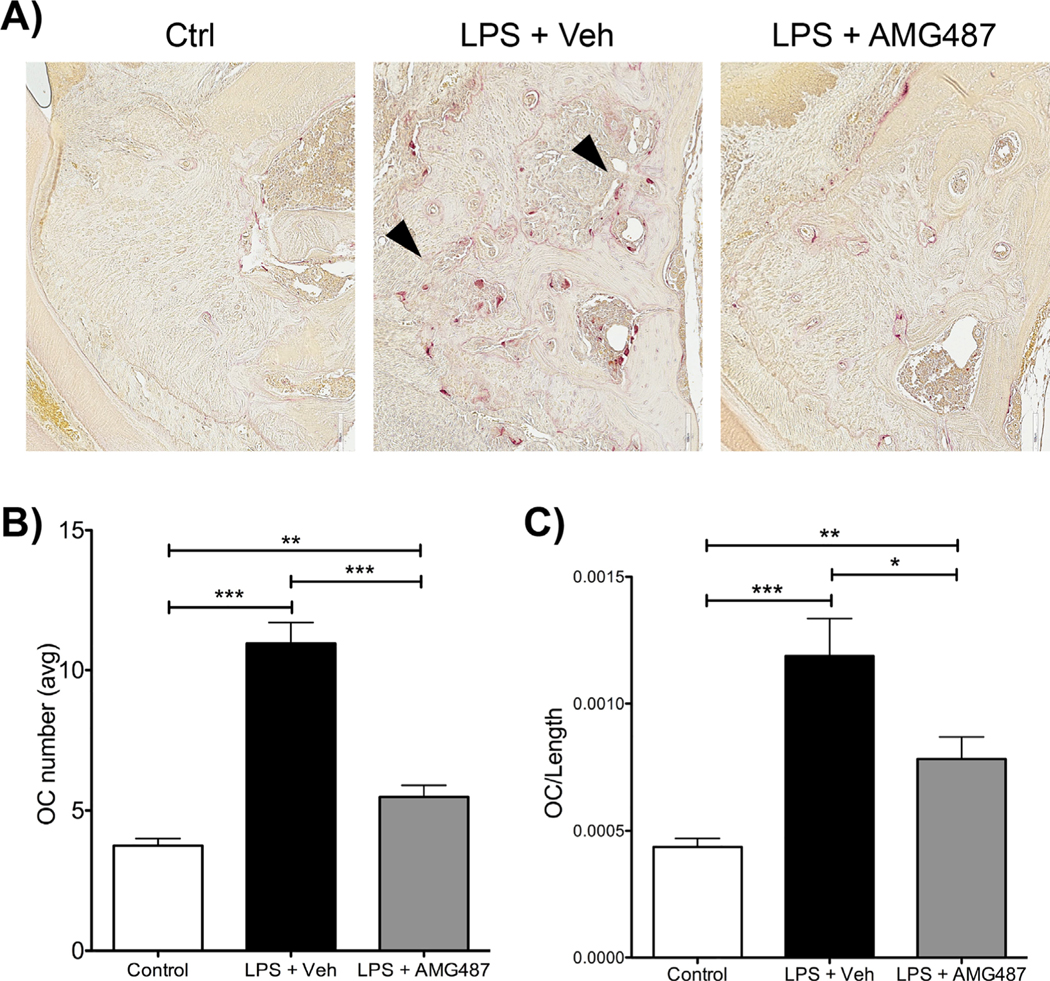
After 12 LPS injections, LPS-injected mice treated with AMG-487 showed a significant reduction in bone loss compared with LPS-treated vehicle-injected mice (Fig. 6A, B). Normalizing bone loss to control, LPS-injected mice treated with AMG-487 showed 45% reduction in bone loss compared with LPS-injected vehicle-treated mice (Fig. 6C). Histologically, after LPS treatment, AMG-487 presented with a qualitative reduction in cellular infiltrates compared with LPS vehicle-treated animals (Fig. 6D). Further assessment of osteoclast numbers showed that after LPS treatment, AMG-487 statistically significantly reduced the total number of TRAP+ cells compared with LPS vehicle-treated mice (Fig. 7A, B). Normalizing osteoclast numbers to bone length showed similar results (Fig. 7C).
Fig. 6.
Systemic delivery of CXCR3 antagonist (AMG-487) reduces bone loss in vivo. (A) Representative radiographic images of control (ctrl), P.g.-LPS + veh injections and P.g.-LPS + AMG-487. Note the reduction in alveolar bone (in between the first and second molars) in the P.g.-LPS + veh group. (B) Graph representing the averaged bone levels in control (Ctrl), P.g.-LPS + veh injections and P.g.-LPS + AMG-487 groups. (C) Graph representing normalized bone loss (control group subtracted) in P.g.-LPS + veh injections and P.g.-LPS + AMG-487 groups. For both graphs (B, C), significance was compared using a Student’s t test. n ≥ 5 mice/group, p ≤ 0.05*, p ≤ 0.01**, p ≤ 0.001***. Data represented as mean ± standard error of the mean (SEM). (D) Hematoxylin and eosin (H&E)-stained slides of control (Ctrl), P.g.-LPS + veh injections and P.g.-LPS + AMG-487 groups. Note the increased cellular infiltrates in the P.g.-LPS + veh injection group (yellow arrow). 20× magnification.
Fig. 7.
Histological assessment of osteoclast numbers after AMG-487 treatment. (A) Tartrate-resistant acid phosphatase (TRAP) staining of control (Ctrl), P.g.-LPS + veh injections and P.g.-LPS + AMG-487 groups. Note the increase in TRAP+ cells in the P.g.-LPS + veh injection group (black arrows). 20 magnification. (B) Graph representing the averaged total number of osteoclasts in control (Ctrl), P.g.-LPS + veh injections, and P.g.-LPS + AMG-487 groups. (C) Graph representing the averaged osteoclast number divided by the alveolar bone length considered in analysis in control (Ctrl), P.g.LPS + veh injections and P.g.-LPS + AMG-487 groups. For both graphs (B, C), significance was compared using a Student’s t test. n ≥ 5 mice/group, p ≤ 0.05*, p ≤ 0.01**, p ≤ 0.001***. Data represented as mean ± standard error of the mean (SEM).
Discussion
PD, as mentioned previously, is a complex disease with genetic and environmental influences, which can be a challenge to dissect in a clinical setting. Through novel resources and technologies, the mouse has become an invaluable tool to interrogate complex trait diseases, including PD.(19,21) Herein, through GWAS, we mapped genetic loci in mice that are associated to PD, and one gene family, including the genes Cxcl9 and Cxcl10, were selected for validation by deleting the Cxcr3 receptor. Furthermore, utilizing Cxcr3 knockout mice and competitive inhibition with a Cxcr3 antagonist, we demonstrated that approximately 50% of the PD phenotype could be rescued in vivo. These findings pave the way for blocking Cxcr3 as a potential therapeutic modality for patients presenting with PD, and the GWAS approach allows for further mechanistic dissection of candidate genes associated to PD.
In an effort to better characterize and understand the genetic underpinning of PD pathogenesis, several groups have utilized a GWAS approach using patient cohorts.(52–55) These studies have highlighted that there is indeed a significant genetic component in PD; however, patient study designs have inherent challenges, including controlling for environmental factors, eg, oral hygiene habits, smoking status, and the presence of other systemic conditions including diabetes and heart disease, which can all have an effect on clinical and research outcomes. Additionally, identifying time of disease onset and standard disease classifications is hard to achieve in patient studies. Even so, patient studies have allowed us to begin to better understand PD pathogenesis. GWAS in humans have begun to lay the foundation for identifying genetic targets associated with disease and have been very effective. In our pool of significantly associated SNPs, several previously identified genes or gene families emerged, which again emphasizes the translational potential of mouse data to human studies, specifically, Toll-like receptor 9 (Tlr9), Toll-like receptor 4 (Tlr4), and several members of the tumor necrosis factor (Tnf) gene family (Tnfsf14 and Tnfsf8),(56–59) which suggests that animal studies can be designed with clinical translation in mind.
Two mouse panels are at the forefront of mouse GWAS studies: the Collaborative Cross and the hybrid mouse diversity panel.(16) Although there are differences in both the CC and the HMDP, both panels perform well for GWAS approaches. Indeed, several groups, including our own, have employed either the CC or the HMDP to further investigate the genetic contribution to PD. Using the CC and an oral infection model of PD, Shusterman and colleagues showed that BALB/cJ mice were highly susceptible and DBA/2J, C57BL/6J, and A/J mice were highly resistant to bacterial-induced PD.(60) Expanding on the CC and their previous study, Shusterman and colleagues observed that the Cxcl4/Cxcl7/Cxcl5 gene cluster was associated with aggressive PD in German and European American populations.(61)
To finely isolate the host response and bypass any genetic influences in host bacterial colonization, we induced PD using P.g. LPS(22) in the HMDP. In our model, control animals were not injected because in our previous study, vehicle injections did not result in measurable bone loss compared with noninjected controls.(22) However, even if that were the case, our data would still be valid because they would reflect heritability to the combined infection/inflammatory/mechanical trauma stimulation.(62) Through our LPS-injection model, we identified several loci with a significance value of 10−5 or higher as associated with our trait. Of all the genes upregulated in our GWAS, we elected to validate the Cxcl family, a suggested gene in the GWAS, because in addition to the significant association in the GWAS, we observed increased gene expression and protein expression (through IHC) in C57BL/6J (high bone loss strain) compared with A/J (low bone loss strain). Interestingly, we were also able to correlate our GWAS findings with a previously performed GWAS assessing macrophage gene expression changes in response to LPS in the HMDP. Through this, we identified several Cxcl family members as associated with both LPS-induced bone loss and macrophage response to LPS, suggesting common pathways involved in the host immunoinflammatory response to bacteria. Most important, we were able to start dissecting the Cxcl pathway to identify a common receptor, CXCR3, and mechanistically interrogate how absence of Cxcr3 affects LPS-induced PD. We believe that the system is overactivated by the increased expression of the ligand and that the systemic administration of the Cxcr3 antagonist competes with the ligand activation of the receptor. For instance, Cxcl9 and Cxcl10 are involved in the development, function, and homeostasis of the immune system. Specifically, Cxcl10 acts as a chemoattractant for monocytes/macrophages, t cells, NK cells, and dendritic cells and promotes T-cell adhesion to endothelial cells.(42,43) The role of Cxrc3 and its ligands, Cxcl9 and Cxcl10, in other systemic diseases in both humans and animals, including diabetes(63–66) and cardiovascular disease,(66–70) have been investigated. For instance, Cxcr3 KO mice showed a delay in diabetes development compared with their WT counterparts. In humans, in a study assessing diabetes and periodontal disease, Cxcr3 gene expression was increased in sites with chronic PD in patients with diabetes and poor glycemic control.(71) Considering both diabetes and PD are characterized by a host immunoinflammatory response, it must be noted that there might be genetic overlap in the susceptibility of these conditions. This opens up an exciting avenue for future research where we might be able to translate clinical diagnostic markers across multiple conditions.
The potential use of peptides to locally modulate host response PD is very exciting, given that current clinical treatment protocols for PD rely primarily on the removal of dental plaque. Although specific bacterial species are known to be highly associated with PD, including P. gingivalis, the oral microbiome is a polymicrobial environment including not only pathogenic bacterial but also healthy microbial species.(72,73) Unfortunately, the current treatment of periodontitis does not take into consideration the host immunoinflammatory response to bacteria. Moreover, the standard treatment and periodontal maintenance protocols undertreats some patients(74–76) and may overtreat others. In the present study, our GWAS approach and candidate gene validation using animal models, here using a Cxcr3 antagonist, allow for clinical translation and targeted treatment options.
In summary, we have identified Cxcl9 and Cxcl10 and their receptor, CXCR3, as associated with PD utilizing a GWAS with the HMDP and a highly reproducible murine model of PD. Furthermore, we have mechanistically interrogated CXCR3’s role in PD through the use of knockout mice, and we have begun to explore possible therapeutic modalities to treat PD by using a Cxcr3 antagonist (AMG-487) in vivo. Our results suggest that modulating the host immune response and specifically monitoring chemokine expression levels could aid in our understanding of PD pathogenesis as well as serve as the foundation for more personalized patient treatment.
Supplementary Material
Acknowledgments
This work was supported by NIH/NIDCR DE023901-01, the UCLA Faculty Development grant, and NIH P01 HL028481 (JL). SH was supported by NIH/NIDCR T90 DE022734-01. We thank the UCLA Tissue Pathology Core Laboratory for the decalcified histological sections.
Authors’ roles: Study design: FQP, ST, PMC, RD, and AJK. Study conduct: FQP and SH. Data collection: FQP, SH, MD, EG, and SL. Data analysis: FQP, SH, EG, SL, and CP. Data interpretation: FQP, ST, and JKL. Drafting manuscript: SH and FQP. Revising manuscript content: SH, MD, EG, SL, FP, ST, PMC, and AJK. Approving final version of the manuscript: SH, EG, MD, RD, SL, CP, ST, PMC, and AJL. SH, CP, and FP take responsibility for the integrity of the data analysis.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Disclosures
All authors state that they have no conflicts of interest.
References
- 1.Albandar JM, Brunelle JA, Kingman A. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988–1994. J Periodontol. 1999;70(1):13–29. [DOI] [PubMed] [Google Scholar]
- 2.Di Benedetto A, Gigante I, Colucci S, Grano M. Periodontal disease: linking the primary inflammation to bone loss. Clin Dev Immunol. 2013;2013:503754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012;91(10):914–20. [DOI] [PubMed] [Google Scholar]
- 4.Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J Dent Res. 2012;91(9):816–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loe H, Anerud A, Boysen H, Morrison E. Natural history of periodontal disease in man. Rapid, moderate and no loss of attachment in SriLankan laborers 14 to 46 years of age. J Clin Periodontol. 1986;13(5):431–45. [DOI] [PubMed] [Google Scholar]
- 6.Michalowicz BS. Genetic and heritable risk factors in periodontal disease. J Periodontol. 1994;65(5 Suppl):479–88. [DOI] [PubMed] [Google Scholar]
- 7.Michalowicz BS, Diehl SR, Gunsolley JC, et al. Evidence of a substantial genetic basis for risk of adult periodontitis. J Periodontol. 2000;71(11):1699–707. [DOI] [PubMed] [Google Scholar]
- 8.Heaton B, Dietrich T. Causal theory and the etiology of periodontal diseases. Periodontol 2000. 2012;58(1):26–36. [DOI] [PubMed] [Google Scholar]
- 9.Loos BG, Papantonopoulos G, Jepsen S, Laine ML. What is the contribution of genetics to periodontal risk? Dent Clin North Am. 2015;59(4):761–80. [DOI] [PubMed] [Google Scholar]
- 10.Marian AJ. Molecular genetic studies of complex phenotypes. Transl Res. 2012;159(2):64–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mouse Genome Sequencing Consortium, Waterston RH, Lindblad-Toh K, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–62. [DOI] [PubMed] [Google Scholar]
- 12.Rau CD, Parks B, Wang Y, et al. High-density genotypes of inbred mouse strains: improved power and precision of association mapping. G3 (Bethesda). 2015;5(10):2021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mouse Genome Informatics [Internet]. Available at: http://www.informatics.jax.org.
- 14.Lusis AJ, Seldin MM, Allayee H, et al. The hybrid mouse diversity panel: a resource for systems genetics analyses of metabolic and cardiovascular traits. J Lipid Res. 2016;57(6):925–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Churchill GA, Airey DC, Allayee H, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36(11):1133–7. [DOI] [PubMed] [Google Scholar]
- 16.Ghazalpour A, Rau CD, Farber CR, et al. Hybrid mouse diversity panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mamm Genome. 2012;23(9–10):680–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinane DF, Hart TC. Genes and gene polymorphisms associated with periodontal disease. Crit Rev Oral Biol Med. 2003;14(6):430–49. [DOI] [PubMed] [Google Scholar]
- 18.Vijayalakshmi R, Geetha A, Ramakrishnan T, Emmadi P. Genetic polymorphisms in periodontal diseases: an overview. Indian J Dent Res. 2010;21(4):568–74. [DOI] [PubMed] [Google Scholar]
- 19.de Vries TJ, Andreotta S, Loos BG, Nicu EA. Genes critical for developing periodontitis: lessons from mouse models. Front Immunol. 2017;8:1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hart GT, Shaffer DJ, Akilesh S, et al. Quantitative gene expression profiling implicates genes for susceptibility and resistance to alveolar bone loss. Infect Immun. 2004;72(8):4471–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker PJ, Dixon M, Roopenian DC. Genetic control of susceptibility to Porphyromonas gingivalis-induced alveolar bone loss in mice. Infect Immun. 2000;68(10):5864–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinrichs JE, Novak MJ. Classification and epidemiology of periodontal diseases. In: Newman MG, Takei HH, Klokkevold PR, Carranza FA, editors. Carranza’s clinical periodontology. 11th ed.St. Louis: Elsevier; 2012. p. 34–64. [Google Scholar]
- 23.Hiyari S, Atti E, Camargo PM, et al. Heritability of periodontal bone loss in mice. J Periodontal Res. 2015;50(6):730–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sartori R, Li F, Kirkwood KL. MAP kinase phosphatase-1 protects against inflammatory bone loss. J Dent Res. 2009;88(12):1125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patil C, Rossa C Jr, Kirkwood KL. Actinobacillus actinomycetemcomitans lipopolysaccharide induces interleukin-6 expression through multiple mitogen-activated protein kinase pathways in periodontal ligament fibroblasts. Oral Microbiol Immunol. 2006;21(6):392–8. [DOI] [PubMed] [Google Scholar]
- 26.Kilkenny C, Browne W, Cuthill IC, et al. Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol. 2010; 160(7):1577–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crow AL, Ohmen J, Wang J, et al. The genetic architecture of hearing impairment in mice: evidence for frequency-specific genetic determinants. G3 (Bethesda). 2015;5(11):2329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salehi P, Myint A, Kim YJ, et al. Genome-wide association analysis identifies Dcc as an essential factor in the innervation of the peripheral vestibular system in inbred mice. J Assoc Res Otolaryngol. 2016;17(5):417–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8(10):833–5. [DOI] [PubMed] [Google Scholar]
- 30.Bennett BJ, Farber CR, Orozco L, et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20(2):281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis RC, van Nas A, Bennett B, et al. Genome-wide association mapping of blood cell traits in mice. Mamm Genome. 2013;24(3–4): 105–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohmen J, Kang EY, Li X, et al. Genome-wide association study for age-related hearing loss (AHL) in the mouse: a meta-analysis. J Assoc Res Otolaryngol. 2014;15(3):335–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park CC, Gale GD, de Jong S, et al. Gene networks associated with conditional fear in mice identified using a systems genetics approach. BMC Systems Biol. 2011;5:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tenesa A, Haley CS. The heritability of human disease: estimation, uses and abuses. Nat Rev Genet. 2013;14(2):139–49. [DOI] [PubMed] [Google Scholar]
- 35.Bennett BJ, Davis RC, Civelek M, et al. Genetic architecture of atherosclerosis in mice: a systems genetics analysis of common inbred strains. PLoS Genet. 2015;11(12):e1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orozco LD, Bennett BJ, Farber CR, et al. Unraveling inflammatory responses using systems genetics and gene-environment interactions in macrophages. Cell. 2012;151(3):658–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walser TC, Rifat S, Ma X, et al. Antagonism of Cxcr3 inhibits lung metastasis in a murine model of metastatic breast cancer. Cancer Res. 2006;66(15):7701–7. [DOI] [PubMed] [Google Scholar]
- 38.Page RC, Kornman KS. The pathogenesis of human periodontitis: an introduction. Periodontol 2000. 1997;14:9–11. [DOI] [PubMed] [Google Scholar]
- 39.Page RC. The role of inflammatory mediators in the pathogenesis of periodontal disease. J Periodont Res. 1991;26(3 Pt 2):230–42. [DOI] [PubMed] [Google Scholar]
- 40.Page RC, Schroeder HE. Spontaneous chronic periodontitis in adult dogs. A clinical and histopathological survey. J Periodontol. 1981;52(2):60–73. [DOI] [PubMed] [Google Scholar]
- 41.Lam RS, O’Brien-Simpson NM, Lenzo JC, et al. Macrophage depletion abates Porphyromonas gingivalis-induced alveolar bone resorption in mice. J Immunol. 2014;193(5):2349–62. [DOI] [PubMed] [Google Scholar]
- 42.Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, Luster AD. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector t cell generation and trafficking. J Immunol. 2002;168(7):3195–204. [DOI] [PubMed] [Google Scholar]
- 43.Angiolillo AL, Sgadari C, Taub DD, et al. Human interferon-inducible protein 10 is a potent inhibitor of angiogenesis in vivo. J Exp Med. 1995;182(1):155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luster AD, Unkeless JC, Ravetch JV. Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature. 1985;315(6021):672–6. [DOI] [PubMed] [Google Scholar]
- 45.Sochalska M, Potempa J. Manipulation of neutrophils by Porphyromonas gingivalis in the development of periodontitis. Front Cell Infect Microbiol. 2017;7:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell L, Millhouse E, Malcolm J, Culshaw S. t cells, teeth and tissue destruction—what do t cells do in periodontal disease? Mol Oral Microbiol. 2016;31(6):445–56. [DOI] [PubMed] [Google Scholar]
- 47.Escalona LA, Mastromatteo-Alberga P, Correnti M. Cytokine and metalloproteinases in gingival fluid from patients with chronic periodontitis. Invest Clin. 2016;57(2):131–42. [PubMed] [Google Scholar]
- 48.Morton RS, Dongari-Bagtzoglou AI. Cyclooxygenase-2 is upregulated in inflamed gingival tissues. J Periodontol. 2001;72(4):461–9. [DOI] [PubMed] [Google Scholar]
- 49.Arabaci T, Cicek Y, Canakci V, et al. Immunohistochemical and stereologic analysis of NF-kappaB activation in chronic periodontitis. Eur J Dent. 2010;4(4):454–61. [PMC free article] [PubMed] [Google Scholar]
- 50.Romanelli R, Mancini S, Laschinger C, Overall CM, Sodek J, McCulloch CA. Activation of neutrophil collagenase in periodontitis. Infect Immun. 1999;67(5):2319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hernandez M, Valenzuela MA, Lopez-Otin C, et al. Matrix metalloproteinase-13 is highly expressed in destructive periodontal disease activity. J Periodontol. 2006;77(11):1863–70. [DOI] [PubMed] [Google Scholar]
- 52.Munz M, Willenborg C, Richter GM, et al. A genome-wide association study identifies nucleotide variants at SIGLEC5 and DEFA1A3 as risk loci for periodontitis. Hum Mol Genet. 2017;26(13):2577–88. [DOI] [PubMed] [Google Scholar]
- 53.Kasbohm E, Holtfreter B, Volker U, et al. Exome variant analysis of chronic periodontitis in 2 large cohort studies. J Dent Res. 2017;96(1):73–80. [DOI] [PubMed] [Google Scholar]
- 54.Sanders AE, Sofer T, Wong Q, et al. Chronic periodontitis genome-wide association study in the Hispanic community health study / study of Latinos. J Dent Res. 2017;96(1):64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Offenbacher S, Divaris K, Barros SP, et al. Genome-wide association study of biologically informed periodontal complex traits offers novel insights into the genetic basis of periodontal disease. Hum Mol Genet. 2016;25(10):2113–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim PD, Xia-Juan X, Crump KE, Abe T, Hajishengallis G, Sahingur SE. Toll-like receptor 9-mediated inflammation triggers alveolar bone loss in experimental murine periodontitis. Infect Immun. 2015; 83(7):2992–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen YC, Liu CM, Jeng JH, Ku CC. Association of pocket epithelial cell proliferation in periodontitis with TLR9 expression and inflammatory response. J Formosan Med Assoc. 2014;113(8):549–56. [DOI] [PubMed] [Google Scholar]
- 58.Chrzeszczyk D, Konopka T, Zietek M. Polymorphisms of toll-like receptor 4 as a risk factor for periodontitis: meta-analysis. Adv Clin Exp Med. 2015;24(6):1059–70. [DOI] [PubMed] [Google Scholar]
- 59.Zhan Y, Zhang R, Lv H, et al. Prioritization of candidate genes for periodontitis using multiple computational tools. J periodontol. 2014;85(8):1059–69. [DOI] [PubMed] [Google Scholar]
- 60.Shusterman A, Durrant C, Mott R, et al. Host susceptibility to periodontitis: mapping murine genomic regions. J Dent Res. 2013; 92(5):438–43. [DOI] [PubMed] [Google Scholar]
- 61.Shusterman A, Munz M, Richter G, et al. The PF4/PPBP/CXCL5 dental gene cluster is associated with periodontitis. J Res. 2017; 96(8):945–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moutsopoulos NM, Konkel JE. Tissue-specific immunity at the oral mucosal barrier. Trends Immunol. 2018;39(4):276–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burke SJ, Karlstad MD, Eder AE, et al. Pancreatic beta-cell production of Cxcr3 ligands precedes diabetes onset. BioFactors. 2016;42(6): 703–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zychowska M, Rojewska E, Pilat D, Mika J. The role of some chemokines from the CXC subfamily in a mouse model of diabetic neuropathy. J Diabetes Res. 2015;2015:750182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu C, Chen X, Shu J, Lee CT. Whole-genome expression analyses of type 2 diabetes in human skin reveal altered immune function and burden of infection. Oncotarget. 2017;8(21):34601–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He J, Lian C, Fang Y, et al. Effect of CXCL10 receptor antagonist on islet cell apoptosis in a type I diabetes rat model. Int J Clin Exp Pathol. 2015;8(11):14542–8. [PMC free article] [PubMed] [Google Scholar]
- 67.Altara R, Mallat Z, Booz GW, Zouein FA. The CXCL10/CXCR3 axis and cardiac inflammation: implications for immunotherapy to treat infectious and non-infectious diseases of the heart. J Immunol Res. 2016;2016:4396368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trott DW, Lesniewski LA, Donato AJ. Selected life-extending interventions reduce arterial CXCL10 and macrophage colony stimulating factor in aged mouse arteries. Cytokine. 2017;96: 102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Corbera-Bellalta M, Planas-Rigol E, Lozano E, et al. Blocking interferon gamma reduces expression of chemokines CXCL9, CXCL10 and CXCL11 and decreases macrophage infiltration in ex vivo cultured arteries from patients with giant cell arteritis. Ann Rheum Dis. 2016;75(6):1177–86. [DOI] [PubMed] [Google Scholar]
- 70.Tavakolian Ferdousie V, Mohammadi M, Hassanshahi G, et al. SerumCXCL10 and CXCL12 chemokine levels are associated with the severity of coronary artery disease and coronary artery occlusion. Int J Cardiol. 2017;233:23–8. [DOI] [PubMed] [Google Scholar]
- 71.Venza I, Visalli M, Cucinotta M, De Grazia G, Teti D, Venza M. Proinflammatory gene expression at chronic periodontitis and periimplantitis sites in patients with or without type 2 diabetes. J Periodontol. 2010;81(1):99–108. [DOI] [PubMed] [Google Scholar]
- 72.Stone VN, Xu P. Targeted antimicrobial therapy in the microbiome era. Mol Oral Microbiol. Epub 2017 Jun 13. DOI: 10.1111/omi.12190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbialcomplexes in subgingival plaque. J Clin Periodontol. 1998;25(2): 134–44. [DOI] [PubMed] [Google Scholar]
- 74.Lindhe J, Nyman S. Long-term maintenance of patients treated for advanced periodontal disease. J Clin Periodontol. 1984;11(8): 504–14. [DOI] [PubMed] [Google Scholar]
- 75.Ramfjord SP, Morrison EC, Burgett FG, et al. Oral hygiene and maintenance of periodontal support. J Periodontol. 1982;53(1): 26–30. [DOI] [PubMed] [Google Scholar]
- 76.McFall WT Jr. Tooth loss in 100 treated patients with periodontal disease. A long-term study. J Periodontol. 1982;53(9):539–49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.