Abstract
Genetic variation accelerates adaptation and resilience and enables the survival of species in their changing environment. Increasing the genetic diversity of crop species is essential to improve their yield and enhance food security. Synthetic directed evolution (SDE) employs localized sequence diversification (LSD) of gene sequence and selection pressure to evolve gene variants with better fitness, improved properties and desired phenotypes. Recently, CRISPR–Cas-dependent and -independent technologies have been applied for LSD to mediate synthetic evolution in diverse species, including plants. SDE holds excellent promise to discover, accelerate and expand the range of traits of the value in crop species. Here, we highlight the efficient SDE approaches for the LSD of plant genes, selection strategies and critical traits for targeted improvement. We discuss the potential of emerging technologies, including CRISPR–Cas base editing, retron editing, EvolvR and prime editing, to establish efficient SDE in plants. Moreover, we cover CRISPR–Cas-independent technologies, including T7 polymerase editor for continuous evolution. We highlight the key challenges and potential solutions of applying SDE technologies to improve the plant traits of the value.
Keywords: synthetic directed evolution (SDE), localized sequence diversification (LSD), CRISPR–Cas systems, retrons, prime editing, trait engineering
Graphical abstract
Genetic variation has shaped life on earth. Human beings have modified our crops over more than the past 10 000 years by selecting desired and improved traits, resulting in crop domestication (1). Modern crops have undergone extensive genetic changes when compared to their wild ancestors. We have also learned that genetic diversity is the basis for crop improvement and food security because it provides a reservoir of largely untapped variation (2). The 20th century witnessed the development of methods to induce genetic variation randomly. During the Green Revolution in the 1960s and 1970s, improved crops generated via random mutagenesis were vital to increasing yield (3).
Later, in the 1980s and 1990s, methods to precisely modify the genome were established by developing site-specific nucleases that facilitate the generation of targeted double-strand breaks (DSBs) in the genome (4). DSBs are then repaired either imprecisely via non-homologous end joining (NHEJ) or precisely by homology-directed repair (HDR) (5). The repair of such induced DSBs can be harnessed for various genetic outcomes in a user-defined manner. Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated nuclease (Cas) systems have been implemented in many species to introduce various changes to the genome, including gene knockouts, gene knock-in or replacement, base editing and epigenome editing (6).
The power of CRISPR–Cas systems has been exploited to engineer traits of interest, including disease resistance, yield improvement and tolerance to abiotic stress (7). The functional analysis of genes and genomes has also fueled identifying targets of interest for crop improvement via CRISPR–Cas systems. However, although to date, CRISPR–Cas systems have largely been implemented to engineer the genome locally (8, 9), induce RNA interference (10, 11) or detect nucleic acids (12, 13), they can also be powerful tools to explore, interrogate and unlock the potential of plant cells.
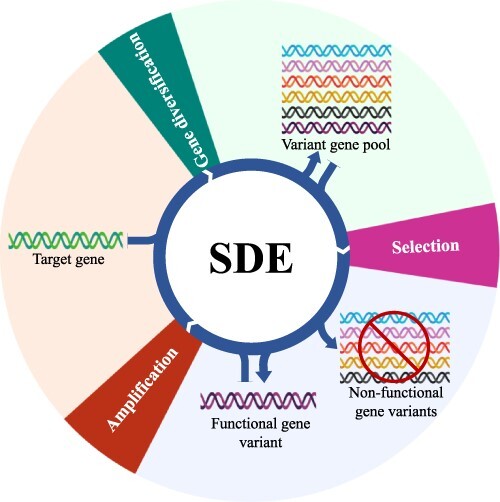
A typical synthetic directed evolution (SDE) cycle involves generating a pool of gene variants, screening while applying specific selective pressure and identifying those gene variants that evoke the desired phenotype; the resulting variants are then amplified and subjected to another cycle of SDE (Figure 1). This process mimics natural selection but in an accelerated form (14). Unlike the rational design of biomolecules, which requires a detailed knowledge of the structure and function of the protein or biomolecule under consideration, in SDE, you usually ‘get what you select for,’ with no prior knowledge required.
Figure 1.

Schematic representation of SDE. Basic SDE involves three key stages: gene diversification, selection and amplification. Initially, the target gene of interest can be employed for localized diversification using several molecular diversification tools. Later, the generated variant gene pool will be subjected to the iterative selection pressure to enrich the variants with superior functional qualities. In the final stage, the functional gene variants can be employed for further continuous gene diversification to amplify the superior functional characteristics.
SDE endows existing biomolecules with new functions, allowing them to accomplish new tasks (15). For example, during the SDE of an enzyme, the enzyme may initially function poorly on a new substrate. However, variants are generated through multiple rounds of sequence diversification and screening until they can use the new substrate. This emerging technology has gained some recognition, as Frances Arnold was awarded the 2018 Nobel Prize in Chemistry for her work on the directed evolution of enzymes (16).
1. Developing technologies for SDE
Because of the ease of selection, the SDE of plant genes has been employed in heterologous hosts, thereby limiting the power of SDE due to the lack of optimal codon usage, proper folding and the complexity of the metabolic environment. Only a few successful cases of SDE have been reported in heterologous hosts (17). There is a pressing need to establish directed evolution in plants in their cellular context to harness the power of this approach. The implementation of a synthetic biology pipeline consisting of the steps (i) design; (ii) build; (iii) test and (iv) learn should allow the successful application of SDE to plant systems (Figure 2).
Figure 2.
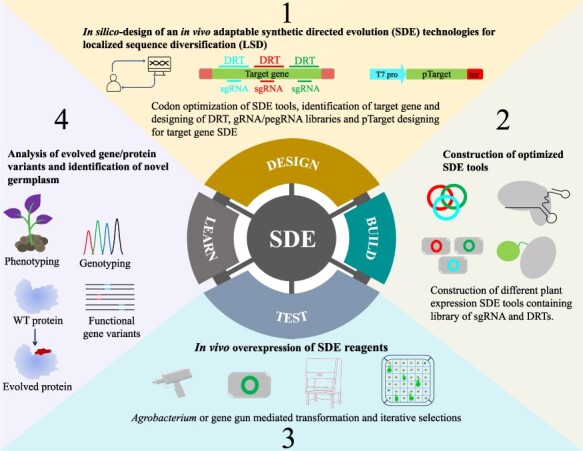
SDE for successful application in plants. The synthetic biology pipeline implementation includes the following steps to allow the successful application of SDE to plant systems. (1) Design, the user has to design the in vivo adaptable SDE tools. This might include the codon optimization of SDE reagents, identification of target gene and designing of DRT, gRNA/pegRNA libraries and pTarget designing for target gene SDE. (2) Build, the optimized SDE tools (CRISPR–Cas dependent and independent) need to be cloned into the efficient plant expression binary vectors and construct the libraries to target the corresponding genes. (3) Test, the Agrobacterium cells harboring the SDE tools will be mobilized into plant tissue via, Agrobacterium-mediated genetic transformation or by gene gun. The selection pressure would be implemented to eliminate the undesired plantlets. (4) Learn, during this stage, the obtained plantlets would be subjected to phenotyping and genotyping. The functional gene variants and evolved proteins will be detected. For continuous gene diversification or to enrich the functional characteristics in the selected plant, the user can proceed for several rounds of design-build-test-learn synthetic evolution cycle.
Developing methods to generate localized sequence diversification (LSD) is the key to SDE applications. SDE has employed many methods geared toward this goal, including mutagenesis via error-prone polymerase chain reaction (PCR), PCR shuffling, recombineering, hypermutator strains or error-prone replication on an orthogonal plasmid (14). CRISPR-based methods harness the power of CRISPR–Cas systems to generate localized sequence variation via NHEJ/HDR (17). Several approaches have been implemented in prokaryotes that support HDR, including multiplexed oligo-nucleotide-mediated genome editing (18). In eukaryotes, base editors fused to the Cas enzyme can provide a means for sequence diversification at targeted break sites (19, 20). Other sequence diversification modalities, including EvolvR, retrons mediated and CRISPR–Cas-independent T7 polymerase-driven continuous editing (TRACE) system, have been harnessed for directed evolution applications in different organisms (21–23).
2. SDE in plants
We adapted CRISPR–Cas systems to establish an in planta SDE platform (24). We selected rice (Oryza sativa) splicing factor 3b (SF3B1) as our initial target for SDE. SF3B1 is a subunit of the SF3B spliceosome complex of the U2 small nuclear ribonucleoprotein (U2SnRNP) complex. The splicing inhibitor drug herboxidiene (GEX1A) and pladienolide B (PB) bind to SF3B1 and block splicing. We reasoned that targeting SF3B1 for SDE might produce variants resistant to GEX1A and PB. We, therefore, designed, constructed and delivered a tiled library of sgRNAs covering the entire SF3B1 coding sequence for targeted mutagenesis. Because SF3B1 is an essential gene, all sgRNAs were designed to introduce DSBs and in-frame mutations may retain normal splicing function, with the hope that one or more gene-editing events would disrupt the binding site of GEX1A or PB. Accordingly, we recovered several mutants, which we termed SGR for SF3B1–GEX1A resistant, that conferred resistance to GEX1A and carried single or multiple amino acid deletions or substitutions.
We then conducted a structural analysis of the binding between GEX1A and the SGR mutants, revealing that these variants indeed cannot bind GEX1A. The plants harboring these SGR SF3B1 mutants were indistinguishable from the wild type, suggesting their spliceosome is fully functional. We also treated wild-type and mutant plants with GEX1A and PB; notably, the SGR mutants were resistant to PB, thus providing compelling evidence that these two drugs share the same target site and that their binding may be similar. Finally, we tested the effects of GEX1A treatment on splicing in the wild-type and the SGR mutants, which revealed that the SGR mutants retain close to full splicing activity; one of the mutants tested, SGR4, appeared completely insensitive to GEX1A (24). This example illustrates the power of SDE to evolve variants that remain functional (in this case: splicing) and also evolve new functions (here, resistance to PB and GEX1A).
Other examples showing the power of directed evolution were recently reported, using a dual base editor to evolve rice acetyl-coenzyme A carboxylase (ACC), whose activity is inhibited by the herbicide haloxyfop, leading to growth retardation. New ACC variants were generated that conferred resistance to haloxyfop by targeting ACC by dual-based editors for continuous C to T and A to G modifications. Indeed, gene-edited seedlings harboring the ACC mutations P1927F and W2125C were resistant to the herbicide, providing another example of the applicability of CRISPR systems to synthetic evolution (19). Recently, Kaung et al. developed enhanced bispyribac sodium herbicide-tolerant rice germplasm containing the P171F variant of acetolactate synthase 1 (ALS1). In their study, the OsALS1 gene was targeted by nCas9 (D10A)-base editors together with the sgRNA library covering the full-length coding region (20).
3. Potential SDE tools for LSD in plants
For SDE in plants, effective technologies and approaches will need to be developed and optimized for LSD, screening, and selection for every target. CRISPR–Cas-based systems can now be used to generate LSD in plants (17). Precisely, a library of single-guide RNAs (sgRNAs) covering a given region of a gene of interest is introduced into Agrobacterium (Agrobacterium tumefaciens) for CRISPR–Cas-mediated gene editing in the target plant tissue (25). As described before, the sequence variation via CRISPR-induced NHEJ is possible when targeting an essential gene by SDE since most mutants will be non-viable (Figure 3A). New CRISPR-based approaches are emerging for LSD in plants. For example, base editing generates edits without DSBs and with the larger editing window size of +50 to −50 relative to PAM sequence (17, 26). Therefore, the complete sequence of a gene may be subjected to base editing by combining a library of sgRNAs with CRISPR-base editors for SDE in plants (Figure 3A).
Figure 3.
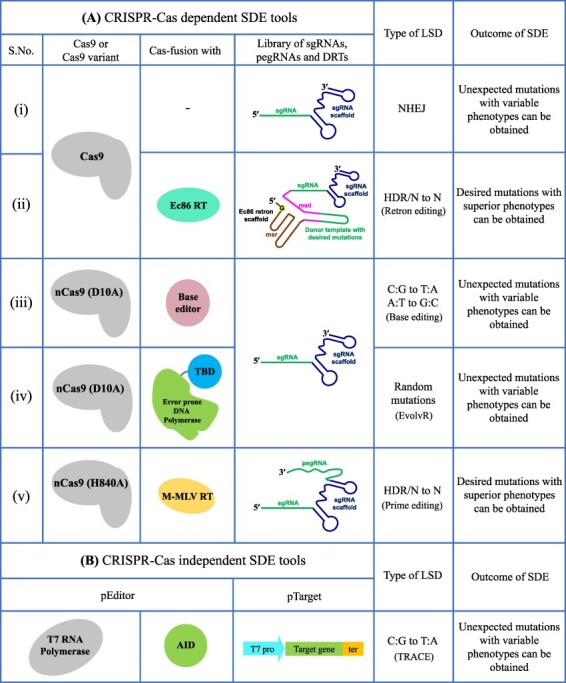
Potential SDE tools for LSD in plants. Target gene LSD can be achieved by CRISPR–Cas-dependent and -independent SDE tools. (A) Several CRISPR–Cas-dependent SDE tools, (i) Cas9 protein together with the library of sgRNAs targeting a complete gene can cause NHEJ-mediated LSD, (ii) Retron editing: Cas9 fusion with Ec86 RT together with the library of retron-DRT-sgRNA targeting the complete gene can produce any N to N conversions or HDR-mediated LSD, (iii) Base editing: nCas9 (D10A) fused with any base editor together with the library of sgRNAs targeting the complete gene will generate C:G to T:A or A:T to G:C conversions near the nick regions, (iv) EvolvR: nCas9 (D10A) fused with error-prone DNA polymerase together with the library of sgRNAs targeting the complete gene can generate random mutations and cause LSD, (v) Prime editing: nCas9 (H840A) and M-MLV RT together with the library of pegRNAs containing the designed mutations can generate any N to N conversions or HDR-mediated LSD in the target gene. (B) CRISPR–Cas-independent TRACE (T7 polymerase-driven continuous editing) can generate sequence diversification in the target gene. The fusion of T7 RNA polymerase and AID base editor (pEditor) will diversify the target gene controlling by T7 promoter (pTarget) via C:G to T:A conversions.
Other possible approaches may include the HDR-based retron technology, which brings the repair templates closer to the DSB site. For example, the E. coli Ec86 retron was coupled with CRISPR and Cas9 to enrich ssDNAs at the targeted locations for DSB repair via HDR in yeast cells (27). Therefore, this method relied on a library of repair templates fused with sgRNA for localized ssDNA production and targeted HDR to generate diversified variants in plants (Figure 3A). Another enzyme-based EvolvR technology uses nCas9 (D10A) with a bacterial error-prone DNA polymerase (PolI3M) and thioredoxin-binding domain together with a target-specific sgRNA to produce continuous LSD in bacteria (23). nCas9 creates a single-stranded nick, from which the nCas9 coupled error-prone DNA polymerase synthesizes new mutant DNA strands. Halperin et al. showed enhanced sequence diversification rates with an editing window of up to +56 to +350 nucleotides from the nick in prokaryotes (23). The design and employing library of sgRNAs targeting complete gene by EvolvR technology can generate various functional gene variants (Figure 3A).
Novel prime editing technology has been successfully used to incorporate genetic information at the target sites into the genome, without DSBs. In prime editing, reverse transcriptase is targeted to nicked DNA, and the prime editing guide RNA (pegRNA) harbors the edited sequence (28). Several groups have reported low editing efficiency when using prime editors in plants (29, 30). We recently applied prime editors for the precise engineering of plant genomes by introducing a single point mutation into the rice ALS gene that had been shown to confer bispyribac resistance in rice (30). We recovered mutations harboring the engineered sequence but with lower efficiency than expected (0.26–2%). We hope that editing efficiency can be raised to over 10% to allow the application of prime editors to SDE. It may be possible to design and construct a pegRNA library to introduce all possible mutations in a specific gene sequence and deliver this library to plant cells; the regeneration of transgenic plants with a simultaneous and constant selective pressure may recover superior variants (Figure 3A). Since the Cas activity is trans, the edited T-DNA-free plants are transgene free. Conventionally, the CRISPR–Cas-edited T-DNA-free plants can be selected by the genetic segregation, back crossing and genotyping methods (3). Therefore, the CRISPR-dependent SDE tools can develop transgene-free plants with the superior phenotypes after the T-DNA segregation step.
CRISPR-independent methods exist: as mentioned before the TRACE, a T7 RNA polymerase fused to activation-induced cytidine deaminase (AID) base editor (pEditor) will bind to the T7 promoter driving the expression of a gene of interest (pTarget) (21). During transcription, the base editor comes close to the DNA intended for editing, leading to base editing over an expanded window of a few hundred base pairs. This system offers a powerful approach for continuous directed evolution (Figure 3B). Unlike CRISPR-dependent tools, the TRACE cannot generate transgene-free plants. In TRACE system, the target gene with in the T-DNA is regulated by T7 promoter in a cis-regulatory manner. So, the T-DNA-free plants are always unedited. However, this technique works best in cell cultures; any testing in whole plants will necessitate engineering the identified edits into the plant genome using one of the CRISPR-based methods described above.
4. Selection strategies
Following sequence diversification, the selection is a crucial stage during which the population of variants is selected to recover variants with strong genotype–phenotype linkage and enable the isolation of evolved variants with the desired function(s). The selection of evolved variants is the key to the successful application of SDE. Although selection may take many forms, the most powerful and easiest form of selection is linked to the survival of the cell or the organism (31). For applications in plants, the directed evolution of herbicide resistance would constitute one of the most straightforward applications of SDE.
Other selection criteria may be applicable, such as changes in color or morphology, yield and resilience to biotic and abiotic factors, but implementing these would be more time-consuming. A number of clones will pass through the selection process, although they do not confer the desired phenotype (false positives), while other clones with the desired genotype nevertheless do not survive the selection stage (false negatives). Since there will be a strong linkage between genotype and phenotype in one population, replication will minimize false positives.
5. Key traits for improvement
SDE can be used to engineer the key traits of value. Engineering herbicide resistance is a clear goal. For example, ‘5-enolpyruvylshikimate-3-phosphate synthase (EPSPS)’ in rice can be targeted to improve the resistance to the herbicide glyphosate. Enhancing crop tolerance against different herbicides is essential to reduce competition with several weed species. Other valuable traits that may be subjected to SDE including an enhanced photosynthesis rate by diversifying ‘ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco)’, abiotic stress resistance by evolving ‘isoprene synthase’ and ‘pyrabactin resistance1/pyr1-like4 (PYL4)’, increased plant protection and productivity by modifying ‘ascorbate peroxidase’, increased number of tillers and yield by evolving ‘carotenoid cleavage dioxygenase 7 (CCD7)’, improved grain yield and immunity against pathogens by diversifying ‘ideal plant architecture 1 (IPA1)’, and for producing more superfoods by raising carotene and anthocyanin contents.
Trait improvement by promoter engineering is one of the important approaches. Promoters are the cis-regulatory noncoding DNA sequences that influence the gene expression levels. Several recent studies demonstrated the role of promoter editing in crop improvement and disease resistance (32, 33). Therefore, the SDE technologies can also be employed for the evolution of a specific promoter by targeting the promoter at multiple locations. The evolved promoters can generate beneficial quantitative variation in the expression of downstream-regulated gene.
6. Conclusion
SDE has revolutionized trait engineering all across a wide community, releasing the laborious and time-consuming fitness burdens from traditional strategies. Whereas limited by the availability of the efficient SDE tools for LSD, the power of trait engineering in plants is still underestimated. We summarized potential SDE tools, selection strategies and essential plant traits that can be improved via SDE in this viewpoint. We envision that SDE will unlock the potential of trait engineering to develop superior crop varieties resilient to climate change with improved yield and quality to support global food security.
Acknowledgments
We would like to thank members of the genome engineering and synthetic biology laboratory at KAUST for critical discussions.
Contributor Information
Gundra Sivakrishna Rao, Laboratory for Genome Engineering and Synthetic Biology, Division of Biological Sciences, 4700 King Abdullah University of Science and Technology (KAUST), Thuwal, 23955-6900, Saudi Arabia.
Wenjun Jiang, Laboratory for Genome Engineering and Synthetic Biology, Division of Biological Sciences, 4700 King Abdullah University of Science and Technology (KAUST), Thuwal, 23955-6900, Saudi Arabia.
Magdy Mahfouz, Laboratory for Genome Engineering and Synthetic Biology, Division of Biological Sciences, 4700 King Abdullah University of Science and Technology (KAUST), Thuwal, 23955-6900, Saudi Arabia.
Funding
KAUST core funding to M.M.
Author contributions
M.M. conceived the idea for the paper and all three authors wrote the manuscript.
Conflict of interest statement.
None declared.
Abbreviations
SDE, synthetic directed evolution; LSD, localized sequence diversification; Pro, promoter; ter, terminator.
References
- 1.Ross-Ibarra J., Morrell P.L. and Gaut B.S. (2007) Plant domestication, a unique opportunity to identify the genetic basis of adaptation. PNAS, 104, 8641–8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doebley J.F., Gaut B.S. and Smith B.D. (2006) The molecular genetics of crop domestication. Cell, 127, 1309–1321. [DOI] [PubMed] [Google Scholar]
- 3.Gao C. (2021) Genome engineering for crop improvement and future agriculture. Cell, 184, 1621–1635. [DOI] [PubMed] [Google Scholar]
- 4.Aouida M., Piatek M.J., Bangarusamy D.K. and Mahfouz M.M. (2014) Activities and specificities of homodimeric TALENs in Saccharomyces cerevisiae. Curr. Genet., 60, 61–74. [DOI] [PubMed] [Google Scholar]
- 5.Knott G.J. and Doudna J.A. (2018) CRISPR-Cas guides the future of genetic engineering. Science, 361, 866–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zafar K., Sedeek K.E.M., Rao G.S., Khan M.Z., Amin I., Kamel R., Mukhtar Z., Zafar M., Mansoor S. and Mahfouz M.M. (2020) Genome editing technologies for rice improvement: progress, prospects, and safety concerns. Front. Genome Ed., 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaidi S.S.-E.-A., Vanderschuren H., Qaim M., Mahfouz M.M., Kohli A., Mansoor S. and Tester M. (2019) New plant breeding technologies for food security. Science, 363, 1390–1391. [DOI] [PubMed] [Google Scholar]
- 8.Ali Z., Shami A., Sedeek K., Kamel R., Alhabsi A., Tehseen M., Hassan N., Butt H., Kababji A., Hamdan S.M.. et al. (2020) Fusion of the Cas9 endonuclease and the VirD2 relaxase facilitates homology-directed repair for precise genome engineering in rice. Commun. Biol., 3, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A.. et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aman R., Mahas A., Butt H., Ali Z., Aljedaani F. and Mahfouz M. (2018) Engineering RNA virus interference via the CRISPR/Cas13 machinery in Arabidopsis. Viruses, 10, 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali Z., Mahas A. and Mahfouz M. (2018) CRISPR/Cas13 as a tool for RNA interference. Trends Plant Sci., 23, 374–378. [DOI] [PubMed] [Google Scholar]
- 12.Aman R., Mahas A. and Mahfouz M. (2020) Nucleic acid detection using CRISPR/Cas biosensing technologies. ACS Synth. Biol., 9, 1226–1233. [DOI] [PubMed] [Google Scholar]
- 13.Ali Z., Aman R., Mahas A., Rao G.S., Tehseen M., Marsic T., Salunke R., Subudhi A.K., Hala S.M. and Hamdan S.M. (2020) iSCAN: an RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res., 288, 198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon A.J., d’Oelsnitz S. and Ellington A.D. (2019) Synthetic evolution. Nat. Biotechnol., 37, 730–743. [DOI] [PubMed] [Google Scholar]
- 15.Arnold F.H. (2018) Directed evolution: bringing new chemistry to life. Angew. Chem.Int. Ed. Engl., 57, 4143–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Service R.F. (2018) Protein evolution earns chemistry Nobel. Science, 362, 142. [DOI] [PubMed] [Google Scholar]
- 17.Butt H., Zaidi S.S., Hassan N. and Mahfouz M. (2020) CRISPR-based directed evolution for crop improvement. Trends Biotechnol., 38, 236–240. [DOI] [PubMed] [Google Scholar]
- 18.Garst A.D., Bassalo M.C., Pines G., Lynch S.A., Halweg-Edwards A.L., Liu R., Liang L., Wang Z., Zeitoun R., Alexander W.G.. et al. (2017) Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol., 35, 48–55. [DOI] [PubMed] [Google Scholar]
- 19.Li C., Zhang R., Meng X., Chen S., Zong Y., Lu C., Qiu J.-L., Chen Y.-H., Li J. and Gao C. (2020) Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol., 38, 875–882. [DOI] [PubMed] [Google Scholar]
- 20.Kuang Y., Li S., Ren B., Yan F., Spetz C., Li X., Zhou X. and Zhou H. (2020) Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms. Mol. Plant, 13, 565–572. [DOI] [PubMed] [Google Scholar]
- 21.Chen H., Liu S., Padula S., Lesman D., Griswold K., Lin A., Zhao T., Marshall J.L. and Chen F. (2020) Efficient, continuous mutagenesis in human cells using a pseudo-random DNA editor. Nat. Biotechnol., 38, 165–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon A.J., Morrow B.R. and Ellington A.D. (2018) Retroelement-based genome editing and evolution. ACS Synth. Biol., 7, 2600–2611. [DOI] [PubMed] [Google Scholar]
- 23.Halperin S.O., Tou C.J., Wong E.B., Modavi C., Schaffer D.V. and Dueber J.E. (2018) CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature, 560, 248–252. [DOI] [PubMed] [Google Scholar]
- 24.Butt H., Eid A., Momin A.A., Bazin J., Crespi M., Arold S.T. and Mahfouz M.M. (2019) CRISPR directed evolution of the spliceosome for resistance to splicing inhibitors. Genome Biol., 20, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gionfriddo M., De Gara L. and Loreto F. (2019) Directed evolution of plant processes: towards a green (r)evolution? Trends Plant Sci., 24, 999–1007. [DOI] [PubMed] [Google Scholar]
- 26.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A. and Liu D.R. (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature, 533, 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharon E., Chen S.A., Khosla N.M., Smith J.D., Pritchard J.K. and Fraser H.B. (2018) Functional genetic variants revealed by massively parallel precise genome editing. Cell, 175, 544–557 e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A.. et al. (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature, 576, 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Li J., Chen J., Yan L. and Xia L. (2020) Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant, 13, 671–674. [DOI] [PubMed] [Google Scholar]
- 30.Butt H., Rao G.S., Sedeek K., Aman R., Kamel R. and Mahfouz M. (2020) Engineering herbicide resistance via prime editing in rice. Plant Biotechnol. J., 18, 2370–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu H., Li C. and Gao C. (2020) Applications of CRISPR-Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol., 21, 661–677. [DOI] [PubMed] [Google Scholar]
- 32.Oliva R., Ji C., Atienza-Grande G., Huguet-Tapia J.C., Perez-Quintero A., Li T., Eom J.-S., Li C., Nguyen H., Liu B.. et al. (2019) Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol., 37, 1344–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodríguez-Leal D., Lemmon Z.H., Man J., Bartlett M.E. and Lippman Z.B. (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell, 171, 470–480.e478. [DOI] [PubMed] [Google Scholar]


