Abstract
Heart malformation is the leading cause of human birth defects, and many of the congenital heart diseases (CHDs) originate from genetic defects that impact cardiac development and maturation. During development, the vertebrate heart undergoes a series of complex morphogenetic processes that increase its ability to pump blood. One of these processes leads to the formation of the finger-like muscular projections called trabeculae. Trabeculae increase cardiac output and permit nutrition and oxygen uptake in the embryonic myocardium prior to coronary vascularization without increasing heart size. Cardiac trabeculation is also crucial for the development of the intraventricular fast conduction system. Alterations in cardiac trabecular development can manifest as a variety of congenital defects such as left ventricular noncompaction. In this review, we discuss the latest advances in understanding the molecular and cellular mechanisms underlying cardiac trabecular development.
Keywords: Cardiac chamber maturation, Cardiac trabeculation, Left ventricle noncompaction
1. Introduction
Heart is the first organ to form and function during embryogenesis. Heart development is a highly orchestrated process involving cardiac specification and differentiation followed by a series of morphogenetic events that ultimately lead to the formation of the multi-chambered functional organ[1–5]. Dysregulation of cardiac development often leads to congenital structural abnormalities that compromise cardiac function and negatively impact life quality of afflicted individuals[6, 7]. Though some congenital heart defects can be fixed with surgical interventions, treatment options for congenital heart diseases in general are very limited[8]. As congenital cardiac defects often arise from genetic defects that affect cardiac development and maturation during embryogenesis, a more complete understanding of the underlying molecular and cellular mechanisms that govern cardiac development will undoubtedly facilitate future development of new therapeutic approaches to repair the damaged hearts[6].
In vertebrate animals, cardiogenesis commences as the cardiac progenitors migrate towards and coalescence at the anterior lateral plate mesoderm to form the bilateral cardiac primordia[2, 3]. Given the spatial arrangement and timing of differentiation, cardiac progenitors are organized into two progenitor cell pools, namely the first heart field (FHF) and the medially positioned second heart field (SHF)[9–12]. Differentiation of cardiac progenitor cells give rise to the formation of diverse cardiac cell types and structures. The first wave of cardiac differentiation occurs as the cells of FHF converge at the ventral midline to form the linear heart tube, additional cardiac cells are then recruited to the heart tube as the late-differentiating SHF extends the linear heart at its arterial and venous poles[13]. Concurrent with addition of the SHF-derived cardiac cells, the linear heart tube jogs leftward and begins cardiac looping. Subsequently, the ventricles undergo a series of morphogenetic processes collectively known as cardiac chamber maturation to enhance the pumping function. As a critical step of chamber maturation process, cardiac trabeculation leads to the formation of the sheet-like muscular structure called cardiac trabeculae that expand into the ventricular lumen[4, 14, 15]. Cardiac trabeculation allows for an increase in cardiac mass and myocardial surface area for blood oxygenation prior to coronary vascularization. Cardiac trabeculation is also required for the establishment of the fast conduction system of the developing ventricle[16, 17]. Failure to form cardiac trabeculae causes embryonic lethality and subtle perturbations of this process could lead to congenital cardiomyopathy[18–20]. In this review, we highlight the latest advances in our understanding of the cellular and molecular mechanism of embryonic chamber maturation.
2. Cellular and molecular basis of cardiac trabeculation
In vertebrate animals, cardiac trabeculation begins right after cardiac looping stage whereby clusters of ventricular cardiomyocytes (CMs) extrude and expand into cardiac jelly to form sheet-like projections. Interestingly, the very first forming cardiac trabecula is often found on the myocardial wall right across from the atrioventricular canal, an area that likely experiences the strongest mechanic forces generated by fluid flow in the heart[21]. After the first trabecular projection forms, the complexity of cardiac trabeculation significantly increases with radially arranged trabecular projections distributing along outer curvature of the ventricle[21]. As such, the ventricular myocardium becomes organized into two visually distinguishable layers - the outer compact layer and the inner trabecular layer. Though the basic morphological changes involved in cardiac trabeculation is clear, it is not until recently that we began to understand the cellular basis of this process. To determine whether the nascent trabeculae are formed as a result of CM delamination, oriented cell division, or a combination of both. Liu et al., performed lineage tracing study and demonstrated that, in the developing zebrafish heart, cardiac trabeculation is primarily driven by CM delamination from the compact myocardium[21]. In contrast, trabecular initiation in mouse hearts appears to involve both oriented cell division and CM delamination, though the relative contribution of these two mechanisms to trabecular formation remains to be further determined[22–24]. At the cellular level, the compact and trabecular CMs are clearly morphologically distinct[21, 25, 26]. The compact CMs exhibit epithelial-like morphology. Consistently, targeted expression of the apical protein Podocalyxin in CMs show a polarized distribution of the GFP tagged Podocalyxin fusion protein on the apical membrane of the compact CMs. EGFP-Podocalyxi, however, is localized all around the cell membrane of the delaminating prospective trabecular CMs, suggesting that cardiac trabeculation likely involves an epithelial-mesenchymal transition[25]. To gain further understanding of the cellular events underlying cardiac trabeculation, Staudt et al., performed combined clonal analysis and live cell imaging of beating hearts. They found that trabeculae are formed by a two-step delamination process whereby the CMs first extended their luminal extensions followed by gradually constricting their apical membrane, thereby moving their cell bodies into the trabecular layer[26]. During this process, the delaminating CMs exhibit morphological changes as well as rearrangements of adhesion junctions, in so doing, these cells remain in tight contact with their neighbors while extruding from the compact layer[27]. Altogether, these studies demonstrated that cardiac trabeculation entails a series of highly coordinated cellular events, including CM apical constriction, depolarization and remodeling of myocardial cell-cell adhesion, that ultimately lead to CM delamination and emergence of trabeculae.
Cardiac trabeculation requires crosstalk at the molecular level between endocardial and myocardial cells. Among the multiple pathways implicated in the control of cardiac trabeculation, Neuregulin/ErbB signaling constitutes one of the most critical nodes for the endocardium-myocardium crosstalk. Neuregulin (Nrg) belongs to the epidermal growth factor (EGF) receptor ligand family[28]. In the developing heart, Nrg is synthesized in the endocardial cells as transmembrane precursor proteins, which are then proteolytically cleaved to release the soluble active form[29]. Binding of Nrg to the receptor tyrosine kinase ErbB4 on the cell surface of the CMs promotes the formation of ErbB4/ErbB2 heterodimeric receptor complex and activates downstream signaling[28]. Nrg/ErbB signaling plays an evolutionary conserved role in cardiac trabeculation. In mouse embryos, knockout of Nrg1, ErbB2 or ErbB4 leads to early embryonic lethality due to defective trabeculation[29–31]. Likewise, loss of Nrg/ErbB2 signaling in zebrafish embryos completely abrogates trabecular formation[21, 32]. By generating chimeric hearts that contained both wildtype and ErbB2 mutant CMs, Liu et al., further demonstrated that ErbB2 functions cell-autonomously in the CMs to promote their delamination and subsequent trabecular emergence[21].
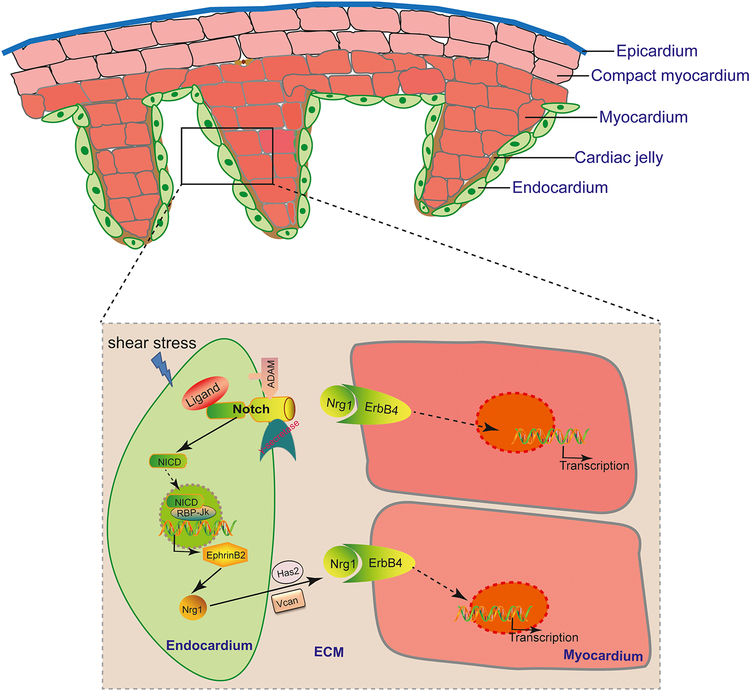
Upstream of Nrg/ErbB signaling, primary cilia-mediated flow sensing is required for trabeculation through activation of Notch signaling in the ventricular endocardium[33]. Notch signaling plays critical roles in diverse developmental and physiological processes. In particular, it has been implicated in regulating many aspects of endothelial biology, such as arteriogenesis and angiogenesis[34]. In the developing heart, the transmembrane Notch ligand and receptor are both expressed in endocardial cells[35, 36]. As Notch ligands and receptors can interact both within the same cell (cis) and across cell boundaries (trans)[37], it remains to be determined whether Notch signaling is activated in cis or trans in the endocardium. Nevertheless, upon ligand binding, Notch is cleaved sequentially by ADAM (a distintegrin and metalloprotease) protease and γ-secretase[38, 39]. This proteolytic cleavage releases the Notch intracellular domain (NICD) from the membrane, allowing its further translocation into the nucleus[40]. In the nucleus, NICD interacts with the transcription factor RBP-Jκ to activate the expression of Notch downstream effectors[40]. Notch signaling is required for cardiac trabeculation in both mouse and zebrafish embryos. Endothelial conditional knockout of Notch receptor Notch1b or RBP-Jκ resulted in impaired murine cardiac trabecular formation[36]. Likewise, loss of Notch signaling in zebrafish embryos led to a complete absence of cardiac trabeculae in the hearts[33]. These studies also indicated that EphrinB2 acts as the critical downstream effector of Notch signaling essential for transcriptional upregulation of Nrg in the endocardium[33, 36] (Fig. 1). Interestingly, the activation of the Notch signaling in the ventricular endocardium coincides with the onset of blood flow. Indeed, blocking cardiac contraction, and hence stopping fluid flow in the heart completely abrogated Notch activation in the ventricular endocardium and subsequent cardiac trabecular formation[33]. In the absence of cardiac contraction, the expression of Notch receptor Notch1b is barely detectable in the ventricular endocardium, suggestion cardiac contraction is required for trabeculation through its role in regulating notch1b transcription. During development, the heart is exposed to many biomechanical forces including those exerted on the wall by blood flow (shear stress), by fluid pressure (cyclic strain), within the wall by cell–cell attachments, and on the wall by extracardiac pressures[20]. Although in vivo and in vitro experiments indicated that the endocardium-specific Notch activation requires functional primary cilia[33], further research is needed to determine whether mechanic forces other than shear stress sensed by primary cilia are involved in regulating cardiac trabeculation. It will also be equally important to unravel the signaling mechanisms through which cells translate biomechanical forces into changes in the signaling events that modulate cardiac wall patterning. Cardiac form and cardiac function are intimately linked, while mechanical forces generated by cardiac contraction are required for cardiac trabeculation and other cardiac developmental processes, cardiac trabeculation has recently been found to serve to enhance contractility for efficient cardiac function and that defects in this process lead to wall-stress induced pathological hypertrophic remodeling, similar to that of adult mammalian hearts subjected to pressure overload[41, 42].
Figure 1. Schematic presentation of molecular crosstalk during cardiac trabeculation.
The ventricular chamber development requires crosstalk between all cardiac tissue layers, namely the epicardium, myocardium, and endocardium, and involves the acellular cardiac jelly. For cardiac trabecular development, primary cilia-mediated flow sensing activates Notch signaling through its role in regulating notch1b transcription. Upon activation, Notch receptor is sequentially processed by ADAM17 and γ-secretase, leading to the release of its intracellular domain (NICD) into the cytoplasm and nucleus. In the nucleus, NICD interacts with transcription factor RBP-Jκ to activate the expression of Notch downstream effector Ephrin B2, which in turn upregulates the expression of the EGF ligand Nrg. Nrg is initially synthesized as transmembrane precursor protein but is subsequently proteolytically cleaved to release the soluble form, which binds to the receptor tyrosine kinase complex ErbB4/ErbB2 expressed on the myocardial cells to activate downstream signaling. Nrg activity and distribution could be modulated by its interaction with ECM components in the cardiac jelly.
Cardiac trabeculation also requires proper establishment of the acellular extracellular matrix, referred to as cardiac jelly, between the myocardial and endocardial layers. Cardiac jelly contains a unique composition of extracellular matrix (ECM) proteins that could impart spatial context for signaling events by influencing the distribution of signaling molecules[43]. For example, binding of Nrg1 immunoglobulin-like domain to ECM components is thought to locally potentiate Nrg1/ErbB signaling activity[44]. Consistent with this idea, mice embryos deficient of cardiac jelly due to loss of hyaluronan synthase-2 (Has2) or versican (Vcan) do not form cardiac trabeculae[45, 46]. Has2 encodes an enzyme required for production of the mucopolysaccharide hyaluronan, and Vcan belongs to aggrecan/versican proteoglycan gene family that encodes large chondroitin sulfate proteoglycan. During cardiac trabeculation, the ECM also appears to be dynamically remodeled by the opposing function of Notch1 and Nrg1 signaling. Notch1 signaling induces ECM degradation, likely creating a local environment permissive for the endocardial cells to form projections through cardiac jelly and establish physical contact with the compact myocardium. In contrast, Nrg1/ErbB2 signaling promotes ECM synthesis necessary for trabecular rearrangement and growth[47]. While proper establishment of cardiac jelly is critical for cardiac trabecular formation, timely degradation of this extracellular structure by secreted matrix metalloproteinases ADAMTS1 (a disintegrin-like and metallopeptidase with thrombospondin type 1 motif, 1) is required for preventing excessive trabeculation[48, 49]. Mice ADAMTS1−/− mutant embryos develop significant increased trabecular muscles. In contrast, aberrant upregulation of ADAMTS1 in the endocardium led to premature breakdown of the cardiac jelly and precocious termination of trabecular development[48].
3. Ventricle chamber growth and differentiation
After the initial trabecular ridges form, trabecular projections increase in length longitudinally and are interconnected radially to form a mesh-like network. In higher vertebrates, trabecular CMs are generally considered to be more differentiated than compact CMs, and the trabecular and compact CMs are marked by the expression of a distinct set of genes[19, 24]. For instance, the trabecular CMs highly express BMP10 and Sphingosine 1-phosphate receptor-1, whereas Tbx20 and Hey2 are enriched in the compact myocardium[24]. In the developing ventricles, some other genes, such as Nppa and Nppb that encode the natriuretic peptides, switches their initial pan ventricular expression to a more trabecular myocardium restricted pattern[50, 51]. The trabecular CMs are also considered to be less proliferative than compact CMs. In support of this idea, clonal analysis demonstrated that the transmural cone-shaped myocyte colonies formed by retrovirus labeled precardiac mesodermal single cell were wider at the epicardial side versus the endocardial side, suggestive of a gradient of transmural CM proliferation[52]. This difference in the proliferative capacity between compact and trabecular myocardium is likely due to the differential expression of p57Kip2 and N-Myc in these two layers. The trabecular myocardium expresses p57Kip2, which encodes a cyclin-dependent kinase inhibitor of the p21 family. Interestingly, this spatial confinement of p57Kip2 expression in the trabecular myocardium is lost in Splotch mutant hearts whereby p57Kip2 expression was significantly derepressed in the compact myocardium. Consequently, the mutants exhibited much reduced myocardium wall thickness and died during mid-gestation[53]. In contrast, the proto-oncogene gene N-Myc was found to be expressed in the compact myocardium. Myocardial ablation of N-Myc significantly reduced cardiac proliferation due to downregulation of N-Myc target genes cyclin D1 and cyclin D2. As a result, the mutant hearts had thin-walled ventricles and displayed precocious myocardial differentiation as evidenced by an upregulation of the adult form of Troponin I, Tnni3[54].
Concomitant with the initiation of trabeculation, cells of the proepicardial organ (PEO) migrate to the post-looped heart to form its outermost layer, the epicardium. In amniotes, the PEO is a transient extracardiac mesothelial cell structure located near the atrioventricular junction of the heart[55–57]. Migration of the PEO cells to form the epicardium requires α4β1 integrin-mediated cell adhesion. Targeted deletion of α4 integrin leads to severely impaired PEO cell migration and a nearly complete absence of the epicardium[58, 59]. While the trabecular formation appears to be minimally affected, the compact myocardium in these mutant hearts is severely reduced in thickness, suggesting that the epicardium is a source of secreted mitogenic factors that regulate compact myocardial growth[58, 59]. One of the well characterized epicardium derived mitogens are fibroblast growth factors (Fgfs). Among the 22 or so Fgf family members, Fgf9 is highly expressed in the epicardium and its expression is under direct control of retinoid acid signaling[60]. Loss of fgf9 function in the mouse embryos leads to decreased myocardial proliferation and reduced compact myocardial wall thickness[60]. Likewise, conditionally knockout Fgf receptors Fgfr1 and Fgfr2 in the myocardium resulted in a similar ventricular wall hypoplasia, further supporting an essential role for epicardium derived FGF signaling in promoting ventricular maturation[60].
In higher vertebrates, the final stage of trabecular development is compaction of the trabeculae towards their base, which transforms the spongy trabecular myocardium into compact and solid muscles[19, 20, 24]. As a result, the base of the trabeculae increases its thickness, and gradually integrates into the myocardial wall proper before it is no longer distinguishable from the original compact myocardium. Failure of trabecular compaction leads to prominent trabeculae, deep inter-trabecular recesses and compromised cardiac function. One of the molecular pathways iteratively utilized to control cardiac development is the Notch signaling pathway. In addition to promoting cardiac trabecular formation, recent study has also implicated Notch signaling in regulating trabecular compaction[61]. During trabecular compaction, the ligand for Notch signaling Jag 1 and Mib1, which encodes the E3 ubiquitin protein ligase required for activation of Jag1 to activate Notch signaling, are both expressed in the myocardium. Histological analysis at E16.5 demonstrated that conditional knockout of Mib1 in the myocardium causes severe hyper-trabeculation and thin compact myocardium. The myocardial wall defects observed in myocardial Mib1 conditional mutants were also evident at the adult stage. Echocardiographic analysis of the adult mutant hearts showed prominent trabeculations with deep intertrabecular recesses[61]. Altogether, these data indicate that inactivation of Notch signaling in the myocardium arrests trabecular compaction that persists all the way into adulthood. The Notch receptor involved in trabecular compaction is likely to be Notch2. In contrast to the restricted activation of Notch1 in the endocardium, Notch2 is primarily activated in the myocardium with dynamic activation pattern. From E9.5 to E10.5, Notch2 is activated throughout the myocardium including both the compact and trabecular layers. At E11.5, however, Notch2 activity becomes confined to the trabecular layer, suggestive a potential role of Notch2 in regulating myocardial wall architecture[62]. Indeed, activating Notch signaling by overexpressing the constitutively active Notch2 in the myocardium resulted noncompaction and hypertrabeculation phenotypes, which is similar to that of inactivation of Notch signaling, suggesting that tightly regulated Notch activity is essential for proper trabecular compaction[62]. Interestingly, trabecular compaction doesn’t occur in lower vertebrates such as the zebrafish and reptiles. To increase the outer myocardial wall thickness, the juvenile zebrafish hearts instead undergo a process called cortical muscle formation, whereby subset of trabecular CMs break through the compact myocardium and subsequently proliferate to form the outermost cortical layer[63].
4. Ventricular conduction system
Cardiac trabeculation is a prerequisite step for the development of the intraventricular conduction system. The formation of the ventricular conduction system (VCS) promotes a transition in the ventricular activation pattern from a primitive base-to-apex pattern to a mature apex-to-base pattern[64]. The electrical signal that triggers cardiac contraction is generated by the specialized pacemaker cells located in the sinoatrial node (SAN). The electrical activity then spread through the atrial myocardium before a temporarily delay in the atrioventricular node (AVN). Subsequently, the VCS receives and then rapidly propagates the impulse to the ventricular apex so that the ventricular myocardium is activated from apex to base. The VCS is capable of conducting the electrical signal more rapidly than the SAN and AVN mainly due to its high levels of Cx40 and Scn5a expression[65]. In higher vertebrates, the VCS is composed of the atrioventricular bundle (AVB), the bundle branches (BBs) and the Purkinje fibers (PFs)[65]. Based on lineage tracing studies performed in different species, all components of the VSC arise from cardiomyogenic progenitors but not neurogenic progenitor cells[16, 17]. Although the exact location of the progenitor cells that give rise to AVB remains to be further determined, the BBs and PFs are formed from subendocardial trabecular CMs of the intraventricular septum and the ventricles, respectively, supporting the “recruitment model” proposed Christoffels and Moorman whereby the conduction cells arise from CM differentiation into conduction phenotype[65]. Clearly, proper development of the VCS is a complex process and requires coordinated growth and differentiation. We direct the interested readers to the reviews by and referenced in Christoffels and Moorman, and Goodyer and Wu[4, 66]. Nevertheless, the induction of conduction phenotype in the subendocardial trabecular cells relies on paracrine signals from the overlying endocardium. Interestingly, the molecular nature of the endocardium derived inductive signals differs between species. In chickens, Endothelin-1 (ET-1) direct underlying trabecular myocytes toward a Purkinje lineage[67], whereas in mammals, ET-1 signaling appears to be dispensable for PFs development, and Nrg1 instead is found to be able to promote Purkinje fiber specification[68, 69]. The effect of Nrg1 signaling on VCS development appears to be mediated by the PEA3 group of ETS family transcription factor Etv1[70]. Etv1 is highly expressed in the His-Purkinje network and its transcript level was significantly increased upon NRG1 treatment. Consistent with its confined cardiac expression in the fast conduction tissues, Etv1 deficiency severely compromised VCS development, causing significant loss of terminal Purkinje cells and thus marked cardiac conduction defects. At the molecular level, Etv1 deficient hearts significantly downregulated the expression of Nkx2–5, Gja5 and Scn5a, which could at least in part contributed to the observed Etv1 mutant VCS development and function defects [70].
5. Left ventricular noncompaction
Left ventricular non-compaction (LVNC) is a rare yet widely recognized cardiomyopathy characterized by prominent trabeculae with deep inter-trabecular recesses and thinning of left ventricular myocardial wall[71, 72]. Due to recent improvement in diagnostic approaches, LVNC has been increasingly diagnosed in adults. The prevalence of this disease is estimated to be around 0.014 to 1.3 percent among patients subjected to echocardiography[73]. While some of the LVNC patients don’t experience any symptoms, others may suffer from life-threatening arrhythmias, thromboembolism, and even cardiac failure[71, 72]. Given the fact that the clinic manifestation of LVNC closely resembles the structural defects associated with abnormal trabecular development, LVNC is generally considered as congenital cardiac disease[74]. Human genetic studies indicate that LVNC has substantial genetic components. Given the prominent role of Mib1 in regulating trabecular compaction in mouse hearts, Luxan et al., sought to determine if MIB1 mutations can be identified in human LVNC patients[61]. By screening a cohort of 100 individuals of southern European ancestry with LVNC, two MIB1 mutations were found to be associated with LVNC. One of the mutations causes an amino acid from valine to phenylalanine at position 943 (V943F), while the other one is a heterozygous mutation that truncates the protein at the MIB1 ankyrin repeats region. Functionally, these two mutations severely comprised the E3 ubiquitin ligase activity of MIB1, thereby strongly dampening Notch activation[61]. LVNC is also associated with mutations in Nkx2.5 and genes involved in nuclear laminar formation, sarcomere formation, calcium handling or mitochondrial function[75]. While increasing numbers of genetic mutations have been identified to be linked to LVNC, we still know little about how these mutations lead to the observed cardiac defects. To unravel the etiology underlying LVNC, several mouse genetic models have been generated. Conditional deletion of Nkx2–5 specifically during the stage of trabecular formation leads to a severe hyper-trabeculation associated with deep inter-trabecular recesses and subendocardial fibrosis[76]. These conditional mutant hearts also exhibited PFs hypoplasia. Furthermore, longitudinal study of cardiac function demonstrates that the adult Nkx2–5 conditional mutant mice exhibit complex ventricular conduction defects, which eventually cause 50% of the mutant mice to heart failure[76], consistent with human studies that LVNC is often associated with arrhythmias.
6. Perspective
Over the last several years, significant progress has been made towards an enhanced understanding of the molecular and cellular mechanisms of cardiac trabeculation, yet several outstanding questions remain to be addressed. One of the interesting questions concerns how primary cilia mediated flow sensing in the endocardial cells leads to an upregulation of Notch receptor Notch1 at the transcriptional level. One possible scenario might involve the activation of mechanosensitive channels, which could in turn increase the transactivation activity of Mef2C through MAPK induced phosphorylation. It also remains unclear how compact CMs are selected to become future trabecular cells. Nrg and its receptors appear to be expressed uniformly in the ventricular endocardium and myocardium, respectively; however, it remains to be determined whether Nrg/ErbB signaling is selectively activated in distinct CMs to initiate cardiac trabeculation. This selective activation of Nrg/ErbB signaling could be imparted by its interaction with extracellular cellular matrix proteins. Alternatively, the spatial regulation of cardiac trabecular initiation can be achieved by the interplay of multiple pathways including the Nrg/ErbB signaling pathway. As aforementioned, cardiac trabeculae serve to increase cardiac mass and have been implicated in VCS formation as cells that give rise to VSC are derived from the trabecular myocardium. Though Nrg1 was found to promote Purkinje fiber specification, it is not clear whether Nrg1 plays an essential role in VSC development. Given the fact that Nrg1 mutants are completely devoid of trabecular formation, a close examination of the role of Nrg1/ErbB2 signaling in VCS development has not been attempted. Trabecular compaction constitutes the final stage of trabecular development. Although growing number of genetic mutations have identified to be associated with clinic LVNC, how these mutations lead to trabecular noncompaction remains to be determined. A detailed study of the cellular basis of trabecular compaction will be required to gain mechanistic insights about how genetic control of trabecular cell behaviors leads to structural remodeling.
Acknowledgements
We would like to thank the members of the Liu and labs for the vivid discussions and valuable input for the review article. We would further like to deeply thank all authors whose work we cited and at the same time apologize to all authors whose papers we could not reference due to space limitations. The Liu lab is supported by NIH/NHLBI R01 grants HL139976 and HL139880, and American Heart Association (AHA) Established Investigator Award 20EIA35320128. This work is also supported by AHA 18TPA34180058, 20EIA35310348 and NIH R35HL155656 to L. Q.
Footnotes
Conflict of interest
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Brade T, Pane LS, Moretti A, Chien KR, Laugwitz KL, Embryonic heart progenitors and cardiogenesis, Cold Spring Harbor perspectives in medicine 3(10) (2013) a013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fishman MC, Chien KR, Fashioning the vertebrate heart: earliest embryonic decisions, Development 124(11) (1997) 2099–117. [DOI] [PubMed] [Google Scholar]
- [3].Harvey RP, Patterning the vertebrate heart, Nature reviews. Genetics 3(7) (2002) 544–56. [DOI] [PubMed] [Google Scholar]
- [4].Moorman AF, Christoffels VM, Cardiac chamber formation: development, genes, and evolution, Physiological reviews 83(4) (2003) 1223–67. [DOI] [PubMed] [Google Scholar]
- [5].Srivastava D, Olson EN, A genetic blueprint for cardiac development, Nature 407(6801) (2000) 221–6. [DOI] [PubMed] [Google Scholar]
- [6].Bruneau BG, The developmental genetics of congenital heart disease, Nature 451(7181) (2008) 943–8. [DOI] [PubMed] [Google Scholar]
- [7].Srivastava D, Building a heart: implications for congenital heart disease, Journal of nuclear cardiology: official publication of the American Society of Nuclear Cardiology 10(1) (2003) 63–70. [DOI] [PubMed] [Google Scholar]
- [8].Holst KA, Said SM, Nelson TJ, Cannon BC, Dearani JA, Current Interventional and Surgical Management of Congenital Heart Disease: Specific Focus on Valvular Disease and Cardiac Arrhythmias, Circulation research 120(6) (2017) 1027–1044. [DOI] [PubMed] [Google Scholar]
- [9].Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S, Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart, Developmental cell 5(6) (2003) 877–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kelly RG, Brown NA, Buckingham ME, The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm, Developmental cell 1(3) (2001) 435–40. [DOI] [PubMed] [Google Scholar]
- [11].Kelly RG, Buckingham ME, Moorman AF, Heart fields and cardiac morphogenesis, Cold Spring Harbor perspectives in medicine 4(10) (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu J, Stainier DY, Zebrafish in the study of early cardiac development, Circulation research 110(6) (2012) 870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Meilhac SM, Buckingham ME, The deployment of cell lineages that form the mammalian heart, Nature reviews. Cardiology 15(11) (2018) 705–724. [DOI] [PubMed] [Google Scholar]
- [14].Sedmera D, Pexieder T, Hu N, Clark EB, Developmental changes in the myocardial architecture of the chick, The Anatomical record 248(3) (1997) 421–32. [DOI] [PubMed] [Google Scholar]
- [15].Sedmera D, Pexieder T, Vuillemin M, Thompson RP, Anderson RH, Developmental patterning of the myocardium, The Anatomical record 258(4) (2000) 319–37. [DOI] [PubMed] [Google Scholar]
- [16].Miquerol L, Beyer S, Kelly RG, Establishment of the mouse ventricular conduction system, Cardiovascular research 91(2) (2011) 232–42. [DOI] [PubMed] [Google Scholar]
- [17].Miquerol L, Moreno-Rascon N, Beyer S, Dupays L, Meilhac SM, Buckingham ME, Franco D, Kelly RG, Biphasic development of the mammalian ventricular conduction system, Circulation research 107(1) (2010) 153–61. [DOI] [PubMed] [Google Scholar]
- [18].Chen H, Shi S, Acosta L, Li W, Lu J, Bao S, Chen Z, Yang Z, Schneider MD, Chien KR, Conway SJ, Yoder MC, Haneline LS, Franco D, Shou W, BMP10 is essential for maintaining cardiac growth during murine cardiogenesis, Development 131(9) (2004) 2219–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang W, Chen H, Qu X, Chang CP, Shou W, Molecular mechanism of ventricular trabeculation/compaction and the pathogenesis of the left ventricular noncompaction cardiomyopathy (LVNC), American journal of medical genetics. Part C, Seminars in medical genetics 163C(3) (2013) 144–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Samsa LA, Yang B, Liu J, Embryonic cardiac chamber maturation: Trabeculation, conduction, and cardiomyocyte proliferation, American journal of medical genetics. Part C, Seminars in medical genetics 163C(3) (2013) 157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu J, Bressan M, Hassel D, Huisken J, Staudt D, Kikuchi K, Poss KD, Mikawa T, Stainier DY, A dual role for ErbB2 signaling in cardiac trabeculation, Development 137(22) (2010) 3867–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Li J, Miao L, Shieh D, Spiotto E, Li J, Zhou B, Paul A, Schwartz RJ, Firulli AB, Singer HA, Huang G, Wu M, Single-Cell Lineage Tracing Reveals that Oriented Cell Division Contributes to Trabecular Morphogenesis and Regional Specification, Cell reports 15(1) (2016) 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Miao L, Li J, Li J, Lu Y, Shieh D, Mazurkiewicz JE, Barroso M, Schwarz JJ, Xin HB, Singer HA, Vincent PA, Zhong W, Radice GL, Wan LQ, Fan ZC, Huang G, Wu M, Cardiomyocyte orientation modulated by the Numb family proteins-N-cadherin axis is essential for ventricular wall morphogenesis, Proceedings of the National Academy of Sciences of the United States of America 116(31) (2019) 15560–15569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wu M, Mechanisms of Trabecular Formation and Specification During Cardiogenesis, Pediatric cardiology 39(6) (2018) 1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jimenez-Amilburu V, Rasouli SJ, Staudt DW, Nakajima H, Chiba A, Mochizuki N, Stainier DYR, In Vivo Visualization of Cardiomyocyte Apicobasal Polarity Reveals Epithelial to Mesenchymal-like Transition during Cardiac Trabeculation, Cell reports 17(10) (2016) 2687–2699. [DOI] [PubMed] [Google Scholar]
- [26].Staudt DW, Liu J, Thorn KS, Stuurman N, Liebling M, Stainier DY, High-resolution imaging of cardiomyocyte behavior reveals two distinct steps in ventricular trabeculation, Development 141(3) (2014) 585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cherian AV, Fukuda R, Augustine SM, Maischein HM, Stainier DY, N-cadherin relocalization during cardiac trabeculation, Proceedings of the National Academy of Sciences of the United States of America 113(27) (2016) 7569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yarden Y, Sliwkowski MX, Untangling the ErbB signalling network, Nature reviews. Molecular cell biology 2(2) (2001) 127–37. [DOI] [PubMed] [Google Scholar]
- [29].Kramer R, Bucay N, Kane DJ, Martin LE, Tarpley JE, Theill LE, Neuregulins with an Ig-like domain are essential for mouse myocardial and neuronal development, Proceedings of the National Academy of Sciences of the United States of America 93(10) (1996) 4833–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lee KF, Simon H, Chen H, Bates B, Hung MC, Hauser C, Requirement for neuregulin receptor erbB2 in neural and cardiac development, Nature 378(6555) (1995) 394–8. [DOI] [PubMed] [Google Scholar]
- [31].Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, Lemke G, Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor, Nature 378(6555) (1995) 390–4. [DOI] [PubMed] [Google Scholar]
- [32].Peshkovsky C, Totong R, Yelon D, Dependence of cardiac trabeculation on neuregulin signaling and blood flow in zebrafish, Developmental dynamics: an official publication of the American Association of Anatomists 240(2) (2011) 446–56. [DOI] [PubMed] [Google Scholar]
- [33].Samsa LA, Givens C, Tzima E, Stainier DY, Qian L, Liu J, Cardiac contraction activates endocardial Notch signaling to modulate chamber maturation in zebrafish, Development 142(23) (2015) 4080–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Alva JA, Iruela-Arispe ML, Notch signaling in vascular morphogenesis, Curr Opin Hematol 11(4) (2004) 278–83. [DOI] [PubMed] [Google Scholar]
- [35].Del Monte G, Grego-Bessa J, Gonzalez-Rajal A, Bolos V, De La Pompa JL, Monitoring Notch1 activity in development: evidence for a feedback regulatory loop, Developmental dynamics: an official publication of the American Association of Anatomists 236(9) (2007) 2594–614. [DOI] [PubMed] [Google Scholar]
- [36].Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama YS, Chen H, Shou W, Ballestar E, Esteller M, Rojas A, Perez-Pomares JM, de la Pompa JL, Notch signaling is essential for ventricular chamber development, Developmental cell 12(3) (2007) 415–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang R, Liu K, Chen L, Aihara K, Neural fate decisions mediated by trans-activation and cis-inhibition in Notch signaling, Bioinformatics 27(22) (2011) 3158–65. [DOI] [PubMed] [Google Scholar]
- [38].Schroeter EH, Kisslinger JA, Kopan R, Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain, Nature 393(6683) (1998) 382–6. [DOI] [PubMed] [Google Scholar]
- [39].Kopan R, Goate A, A common enzyme connects notch signaling and Alzheimer's disease, Genes Dev 14(22) (2000) 2799–806. [DOI] [PubMed] [Google Scholar]
- [40].Artavanis-Tsakonas S, Rand MD, Lake RJ, Notch signaling: cell fate control and signal integration in development, Science 284(5415) (1999) 770–6. [DOI] [PubMed] [Google Scholar]
- [41].Fleming ND, Samsa LA, Hassel D, Qian L, Liu J, Rapamycin attenuates pathological hypertrophy caused by an absence of trabecular formation, Sci Rep 8(1) (2018) 8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ma H, Yu S, Liu X, Zhang Y, Fakadej T, Liu Z, Yin C, Shen W, Locasale JW, Taylor JM, Qian L, Liu J, Lin28a Regulates Pathological Cardiac Hypertrophic Growth Through Pck2-Mediated Enhancement of Anabolic Synthesis, Circulation 139(14) (2019) 1725–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Little CD, Rongish BJ, The extracellular matrix during heart development, Experientia 51(9–10) (1995) 873–82. [DOI] [PubMed] [Google Scholar]
- [44].Li Q, Loeb JA, Neuregulin-heparan-sulfate proteoglycan interactions produce sustained erbB receptor activation required for the induction of acetylcholine receptors in muscle, J Biol Chem 276(41) (2001) 38068–75. [DOI] [PubMed] [Google Scholar]
- [45].Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, Calabro A Jr., Kubalak S, Klewer SE, McDonald JA, Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme, J Clin Invest 106(3) (2000) 349–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mjaatvedt CH, Yamamura H, Capehart AA, Turner D, Markwald RR, The Cspg2 gene, disrupted in the hdf mutant, is required for right cardiac chamber and endocardial cushion formation, Dev Biol 202(1) (1998) 56–66. [DOI] [PubMed] [Google Scholar]
- [47].Del Monte-Nieto G, Ramialison M, Adam AAS, Wu B, Aharonov A, D'Uva G, Bourke LM, Pitulescu ME, Chen H, de la Pompa JL, Shou W, Adams RH, Harten SK, Tzahor E, Zhou B, Harvey RP, Control of cardiac jelly dynamics by NOTCH1 and NRG1 defines the building plan for trabeculation, Nature 557(7705) (2018) 439–445. [DOI] [PubMed] [Google Scholar]
- [48].Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, Shang C, Bayle JH, Shou W, Iruela-Arispe ML, Chang CP, Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis, Developmental cell 14(2) (2008) 298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kuno K, Iizasa H, Ohno S, Matsushima K, The exon/intron organization and chromosomal mapping of the mouse ADAMTS-1 gene encoding an ADAM family protein with TSP motifs, Genomics 46(3) (1997) 466–71. [DOI] [PubMed] [Google Scholar]
- [50].Christoffels VM, Habets PE, Franco D, Campione M, de Jong F, Lamers WH, Bao ZZ, Palmer S, Biben C, Harvey RP, Moorman AF, Chamber formation and morphogenesis in the developing mammalian heart, Dev Biol 223(2) (2000) 266–78. [DOI] [PubMed] [Google Scholar]
- [51].Sergeeva IA, Hooijkaas IB, Van Der Made I, Jong WM, Creemers EE, Christoffels VM, A transgenic mouse model for the simultaneous monitoring of ANF and BNP gene activity during heart development and disease, Cardiovascular research 101(1) (2014) 78–86. [DOI] [PubMed] [Google Scholar]
- [52].Mikawa T, Cohen-Gould L, Fischman DA, Clonal analysis of cardiac morphogenesis in the chicken embryo using a replication-defective retrovirus. III: Polyclonal origin of adjacent ventricular myocytes, Developmental dynamics: an official publication of the American Association of Anatomists 195(2) (1992) 133–41. [DOI] [PubMed] [Google Scholar]
- [53].Kochilas LK, Li J, Jin F, Buck CA, Epstein JA, p57Kip2 expression is enhanced during mid-cardiac murine development and is restricted to trabecular myocardium, Pediatr Res 45(5 Pt 1) (1999) 635–42. [DOI] [PubMed] [Google Scholar]
- [54].Harmelink C, Peng Y, DeBenedittis P, Chen H, Shou W, Jiao K, Myocardial Mycn is essential for mouse ventricular wall morphogenesis, Dev Biol 373(1) (2013) 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Komiyama M, Ito K, Shimada Y, Origin and development of the epicardium in the mouse embryo, Anat Embryol (Berl) 176(2) (1987) 183–9. [DOI] [PubMed] [Google Scholar]
- [56].Viragh S, Challice CE, The origin of the epicardium and the embryonic myocardial circulation in the mouse, The Anatomical record 201(1) (1981) 157–68. [DOI] [PubMed] [Google Scholar]
- [57].Liu J, Stainier DY, Tbx5 and Bmp signaling are essential for proepicardium specification in zebrafish, Circulation research 106(12) (2010) 1818–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sengbusch JK, He W, Pinco KA, Yang JT, Dual functions of [alpha]4[beta]1 integrin in epicardial development: initial migration and long-term attachment, J Cell Biol 157(5) (2002) 873–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yang JT, Rayburn H, Hynes RO, Cell adhesion events mediated by alpha 4 integrins are essential in placental and cardiac development, Development 121(2) (1995) 549–60. [DOI] [PubMed] [Google Scholar]
- [60].Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM, Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo, Developmental cell 8(1) (2005) 85–95. [DOI] [PubMed] [Google Scholar]
- [61].Luxan G, Casanova JC, Martinez-Poveda B, Prados B, D'Amato G, MacGrogan D, Gonzalez-Rajal A, Dobarro D, Torroja C, Martinez F, Izquierdo-Garcia JL, Fernandez-Friera L, Sabater-Molina M, Kong YY, Pizarro G, Ibanez B, Medrano C, Garcia-Pavia P, Gimeno JR, Monserrat L, Jimenez-Borreguero LJ, de la Pompa JL, Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy, Nat Med 19(2) (2013) 193–201. [DOI] [PubMed] [Google Scholar]
- [62].Yang J, Bucker S, Jungblut B, Bottger T, Cinnamon Y, Tchorz J, Muller M, Bettler B, Harvey R, Sun QY, Schneider A, Braun T, Inhibition of Notch2 by Numb/Numblike controls myocardial compaction in the heart, Cardiovascular research 96(2) (2012) 276–85. [DOI] [PubMed] [Google Scholar]
- [63].Gupta V, Poss KD, Clonally dominant cardiomyocytes direct heart morphogenesis, Nature 484(7395) (2012) 479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sedmera D, Reckova M, Bigelow MR, Dealmeida A, Stanley CP, Mikawa T, Gourdie RG, Thompson RP, Developmental transitions in electrical activation patterns in chick embryonic heart, Anat Rec A Discov Mol Cell Evol Biol 280(2) (2004) 1001–9. [DOI] [PubMed] [Google Scholar]
- [65].Christoffels VM, Moorman AF, Development of the cardiac conduction system: why are some regions of the heart more arrhythmogenic than others?, Circ Arrhythm Electrophysiol 2(2) (2009) 195–207. [DOI] [PubMed] [Google Scholar]
- [66].Goodyer WR, Wu SM, Fates Aligned: Origins and Mechanisms of Ventricular Conduction System and Ventricular Wall Development, Pediatric cardiology 39(6) (2018) 1090–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Hall CE, Hurtado R, Hewett KW, Shulimovich M, Poma CP, Reckova M, Justus C, Pennisi DJ, Tobita K, Sedmera D, Gourdie RG, Mikawa T, Hemodynamic-dependent patterning of endothelin converting enzyme 1 expression and differentiation of impulse-conducting Purkinje fibers in the embryonic heart, Development 131(3) (2004) 581–92. [DOI] [PubMed] [Google Scholar]
- [68].Hua LL, Vedantham V, Barnes RM, Hu J, Robinson AS, Bressan M, Srivastava D, Black BL, Specification of the mouse cardiac conduction system in the absence of Endothelin signaling, Dev Biol 393(2) (2014) 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Rentschler S, Zander J, Meyers K, France D, Levine R, Porter G, Rivkees SA, Morley GE, Fishman GI, Neuregulin-1 promotes formation of the murine cardiac conduction system, Proceedings of the National Academy of Sciences of the United States of America 99(16) (2002) 10464–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Shekhar A, Lin X, Liu FY, Zhang J, Mo H, Bastarache L, Denny JC, Cox NJ, Delmar M, Roden DM, Fishman GI, Park DS, Transcription factor ETV1 is essential for rapid conduction in the heart, J Clin Invest 126(12) (2016) 4444–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Jenni R, Rojas J, Oechslin E, Isolated noncompaction of the myocardium, N Engl J Med 340(12) (1999) 966–7. [DOI] [PubMed] [Google Scholar]
- [72].Stollberger C, Finsterer J, Left ventricular hypertrabeculation/noncompaction, J Am Soc Echocardiogr 17(1) (2004) 91–100. [DOI] [PubMed] [Google Scholar]
- [73].Oechslin EN, Attenhofer Jost CH, Rojas JR, Kaufmann PA, Jenni R, Long-term follow-up of 34 adults with isolated left ventricular noncompaction: a distinct cardiomyopathy with poor prognosis, J Am Coll Cardiol 36(2) (2000) 493–500. [DOI] [PubMed] [Google Scholar]
- [74].Angelini A, Melacini P, Barbero F, Thiene G, Evolutionary persistence of spongy myocardium in humans, Circulation 99(18) (1999) 2475. [DOI] [PubMed] [Google Scholar]
- [75].Teekakirikul P, Kelly MA, Rehm HL, Lakdawala NK, Funke BH, Inherited cardiomyopathies: molecular genetics and clinical genetic testing in the postgenomic era, J Mol Diagn 15(2) (2013) 158–70. [DOI] [PubMed] [Google Scholar]
- [76].Choquet C, Nguyen THM, Sicard P, Buttigieg E, Tran TT, Kober F, Varlet I, Sturny R, Costa MW, Harvey RP, Nguyen C, Rihet P, Richard S, Bernard M, Kelly RG, Lalevee N, Miquerol L, Deletion of Nkx2–5 in trabecular myocardium reveals the developmental origins of pathological heterogeneity associated with ventricular non-compaction cardiomyopathy, PLoS Genet 14(7) (2018) e1007502. [DOI] [PMC free article] [PubMed] [Google Scholar]