Abstract
In the past two decades, thousands of non-coding RNAs (ncRNAs) have been discovered, annotated, and characterized in nearly every tissue under both physiological and pathological conditions. Here, we will focus on the role of ncRNAs, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) in ischemic heart disease (IHD), which remains the leading cause of morbidity and mortality in humans—resulting in 8.9 million deaths annually. Cardiomyocyte (CM) proliferation, differentiation, and survival in addition to neovascularization of injured tissues and the prevention of fibrosis are commonly regarded as critically important for the recovery of the heart following myocardial infarction (MI). An abundance of evidence has been accumulated to show ncRNAs participate in cardiac recovery after MI. Because miRNAs are important regulators of cardiac regeneration, the therapeutic potential of at least five of these molecules has been assessed in large animal models of human IHD. In particular, miRNA-based interventions based on miR-132 and miR-92a inhibition in related diseases have displayed favorable outcomes that have provided the impetus for miRNA-based clinical trials for IHD. At the same time, the functional roles of lncRNAs and circRNAs in cardiac regeneration are also being explored. In the present review, we will summarize the latest ncRNA studies aimed at reversing damage to the ischemic heart and discuss the therapeutic potential of targeting miRNAs, lncRNAs, and circRNAs to stimulate cardiac regeneration.
Keywords: non-coding RNAs, noncoding RNAs, miRNAs, long non-coding RNA, circular RNAs, cardiac regeneration, neovascularization, fibrosis, ischemic heart disease
Introduction
Ischemic heart disease (IHD) is the leading cause of death in humans; accounting for 16% of the 55.4 million total deaths worldwide in 2019 [1]. From 2000 to 2019, morbidity caused by IHD rose by more than 2 million, which is the largest increase in morbidity among all causes over the past two decades [1]. Undoubtedly, there is an urgent need to understand the underlying molecular mechanisms of IHD to allow for the development of new therapeutic options for this condition. In particular, the ability to stimulate sufficient cardiomyocyte (CM) regeneration after myocardial infarction (MI) to offset the structural and functional loss of myocardium remains a top priority.
In humans, adult cardiomyocytes largely exit the cell-cycle with a renewal rate of approximately 1% per year at the age of 25 and 0.45% at the age of 75 [2]. Following MI, adult cardiomyocytes fail to efficiently regenerate the ischemic myocardium and the injured tissue becomes fibrotic. The current consensus is that the regeneration of adult mammalian cardiac stem/progenitor cells after MI is negligible and unlikely to contribute to improved recovery of injured tissue. The most prominent CM source for regeneration arises from pre-existing cardiomyocytes and rarely, if at all, from adult cardiac stem/progenitor cells [3–8]. Consequently, low renewal rates and limited CM sources have driven an enormous effort to either expand the potential of endogenous cells to rebuild the injured myocardium or improve CM survival and differentiation from exogenous sources, such as embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), and cardiac fibroblasts (CFs), as a means to enhance cardiac regeneration after MI.
It is also clear that that endothelial cells (ECs) are present in a much higher frequency (>60%) in the non-myocyte population of normal adult mouse hearts when compared to cardiac fibroblasts (<20%) [9]. Stimulating only CM proliferation would likely not lead to effective heart regeneration after MI, as newly-formed CMs will not survive without a parallel increase in ECs to form blood vessels that can supply oxygen and nutrients to nascent tissues [10]. In addition, once an injury occurs, quiescent cardiac fibroblasts begin to activate and deposit excess extracellular matrix (ECM), which results in increased stiffness and reduced compliance of the heart as well as impaired CM function [11, 12]. In short, excessive fibrosis is another independent contributor to the progression of cardiac disease after MI. Together, CM proliferation, differentiation, and survival as well as concomitant neovascularization and prevention of fibrosis are critically important for meaningful recovery of the heart following MI.
With completion of the Human Genome Project (HGP) and the development of next-generation sequencing (NGS) technologies, thousands of ncRNAs have been identified and characterized as microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs). These ncRNAs have been found in nearly every tissue/organ and many of them appear to be important regulators of disease initiation and progression. These molecules have been implicated in a multitude of cellular functions, most notably the regulation of gene expression through transcriptional and post-transcriptional mechanisms. At the same time, ncRNAs have gained attention because of their roles in organ development and disease progression, which underscores their diagnostic and therapeutic potential. For example, miRNA-based drugs are now regarded as one of the most promising means to encourage cardiac regeneration. In addition, the recently discovered lncRNAs and circRNAs are now being scrutinized for their therapeutic and diagnostic potential. In view of that, the present review summarizes recent advancements in cardiac ncRNA research by focusing on the mechanism of action and therapeutic potential of key miRNAs, lncRNAs, and circRNAs for regeneration.
1. Classification and mechanisms of non-coding RNAs
By not strictly following the flow of information outlined in the central dogma of molecular biology, genes encoding ncRNAs produce RNA transcripts that are not translated into proteins. It was estimated that less than 3% of the human genome codes for proteins, compared with more than 80% for ncRNAs, with the remainder comprised of non-transcribed DNA [13–15].
According to the length of the final transcript, ncRNAs were categorized into small non-coding RNAs (<200 nucleotides in length) and long non-coding RNAs (>200 nucleotides in length). Small non-coding RNAs include housekeeping RNAs such as transfer RNAs (tRNAs), some ribosomal RNAs (such as 5S rRNA and 5.8S rRNA), and regulatory RNAs like microRNAs (miRNAs or miRs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and Piwi-interacting RNAs [16].
The best characterized regulatory small non-coding RNAs are miRNAs, which are approximately 22 nucleotides long and highly-conserved ncRNAs. Although produced by cleavage of double-stranded RNA, miRNAs exert their effects as single-stranded RNAs. Typically, miRNAs interact with the 3′ untranslated region (3′ UTR) of target mRNAs to induce their degradation and/or translational repression by incorporating in the RNA-induced silencing complex (RISC) containing Ago and other proteins [17, 18]. In some cases, miRNAs are able to activate translation or regulate transcription [19–21]. Although there are currently about 1917 miRNA hairpin precursors and 2654 mature sequences annotated in the human genome [22], the biological function of many of these ncRNAs remain unknown.
Long non-coding RNAs (lncRNAs) are defined as ncRNAs longer than 200 nts in length without any evidence of coding potential. The majority of lncRNAs are characterized by developmental stage-specific, tissue-specific, and/or cell type-specific distribution when compared to protein-coding genes [23–25]. The known molecular functions of lncRNAs include, but are not limited to, regulation of chromosomal architecture, chromatin organization, transcription, mRNA turnover, translation, and post-translational modification (PTM) (reviewed in [26–30]).
LncRNAs were originally annotated based on their genomic location relative to nearby protein-coding genes. As a result, lncRNAs initially consisted of (i) sense lncRNAs, which are transcribed from the sense strand of a protein-coding gene and overlap with at least one intron and/or exon; (ii) sense intronic lncRNAs, which are located within a single intron of a coding gene; (iii) anti-sense lncRNAs, which are transcribed from the antisense strand of a protein-coding gene and overlap with at least one intron and/or exon; (iv) bidirectional lncRNAs, which are located within 1 kb of the promoter region of a protein-coding gene and transcribed from the antisense strand; (v) enhancer lncRNAs, which are located in the enhancer region of a coding gene and can be transcribed from either the sense or antisense strands; and (vi) intergenic lncRNAs, which are located between two protein-coding genes and far from their promoter regions (> 1kb).
With an increased understanding of lncRNAs, their classification was subsequently based on their mechanism(s) of action. Functionally categorized lncRNAs involved in cardiac regeneration are listed below:
Transcriptional regulators: Transcriptional regulatory lncRNAs combine with RNA-binding proteins (RBPs) and direct the ribonucleoprotein (RNP) complex to a specific genome location to direct downstream gene transcription. An example of a transcriptional regulatory lncRNA is Fendrr, which recruits PRC2 to the promoter regions of Foxf1 and Pitx2 to inhibit their transcription by modulating H3K27 trimethylation and/or H3K4 trimethylation levels [31].
Decoys: Decoy lncRNAs bind and sequester a regulatory protein/RNA to inhibit its function. These regulatory proteins could be transcription factors, chromatin modifiers, other RBPs, or RNA sequences such as miRNAs. Therefore, the miRNA sponge lncRNA is a specific subtype of decoy lncRNA. An example of a decoy lncRNA is the anti-hypertrophic lncRNA Mhrt, which competes with the Brg1 protein for binding to genomic DNA target sites, resulting in suppression of Brg1/BAF complex-mediated chromatin remodeling and hypertrophic-associated gene activation [32].
Enhancers: These lncRNAs are transcribed from the enhancer region of a protein-coding gene and modulate the interaction between the enhancer and promoter regions through chromosomal looping. For example, lncRNA HOTTIP is transcribed from the 5′ end of the HOXA locus with bivalent H3K4me3 and H3K27me3 marks. Through chromosomal looping, the HOTTIP transcript is brought into proximity with target HOXA genes. Meanwhile, HOTTIP recruits WDR5/MLL complexes across these genes, driving H3K4me3 modifications and HOXA gene transcription [33].
Scaffolds: Scaffold lncRNAs combine several proteins simultaneously and serve as an indispensable component to maintain the integrity of the protein complex. For instance, under normal conditions, the lncRNA MALAT1 is associated with phosphorylated serine/arginine (SR) splicing factors in nuclear speckle domains and the nucleoplasm. This ‘scaffold-like’ mechanism enables the normal recruitment of SR splicing factors to pre-mRNAs; thereby, maintaining regular splicing. The absence of MALAT1 greatly increases levels of dephosphorylated SR proteins, resulting in alternative splicing (AS) of the target pre-mRNAs [34].
PTM regulators: These lncRNAs directly or indirectly interact with proteins and regulate modification of target molecules. An example is the dendritic cell (DC)-enriched lncRNA lnc-DC. Lnc-DC is required for DC differentiation and DC-mediated T cell activation by activating the STAT3 transcription factor. Mechanistically, lnc-DC directly binds to STAT3 and maintains its phosphorylation at Tyr705 to prevent dephosphorylation by SHP1 [35].
Other reported mechanisms of action include regulation of chromosomal architecture and interactions, mRNA turnover, and translation, which have been previously reviewed [26–30]. To date, the GENCODE project (version 37) has annotated 17,948 lncRNA genes (~30% of all 60,651 genes) in humans and 13,186 lncRNA genes (~25% of all 53,647 genes) in mice (https://www.gencodegenes.org); however, because of their diverse functions and incomplete annotation, the biological role of most of these lncRNAs is unknown.
CircRNAs are another type of single-strand non-coding RNA, with a unique closed-loop structure. Through a back-splicing mechanism, circRNAs join their 5′ and 3′ end to form a closed circle and their length ranges from a few hundred to thousands of nucleotides (nts) [36]. Although circRNAs are defined as non-coding RNAs, they are primarily derived from protein-coding genes and hundreds of endogenous circRNAs could produce micro-peptides or proteins through an N6-methyladenosine (m6A)-mediated mechanism [37–39]. Moreover, circRNAs have a longer half-life than linear RNAs as a result of exonuclease resistance, and their expression levels are generally comparable to linear RNAs, with several exceptions, such as highly-expressed circRNAs derived from Titin (TTN) and the ryanodine receptor 2 (RYR2) in the human heart [40]. In addition, circRNAs have been reported to display tissue- and cell-specific expression [41, 42] and they primarily function as miRNA sponges, regulators of splicing and transcription, and modifiers of parental gene expression [43].
2. MiRNAs in cardiac regeneration
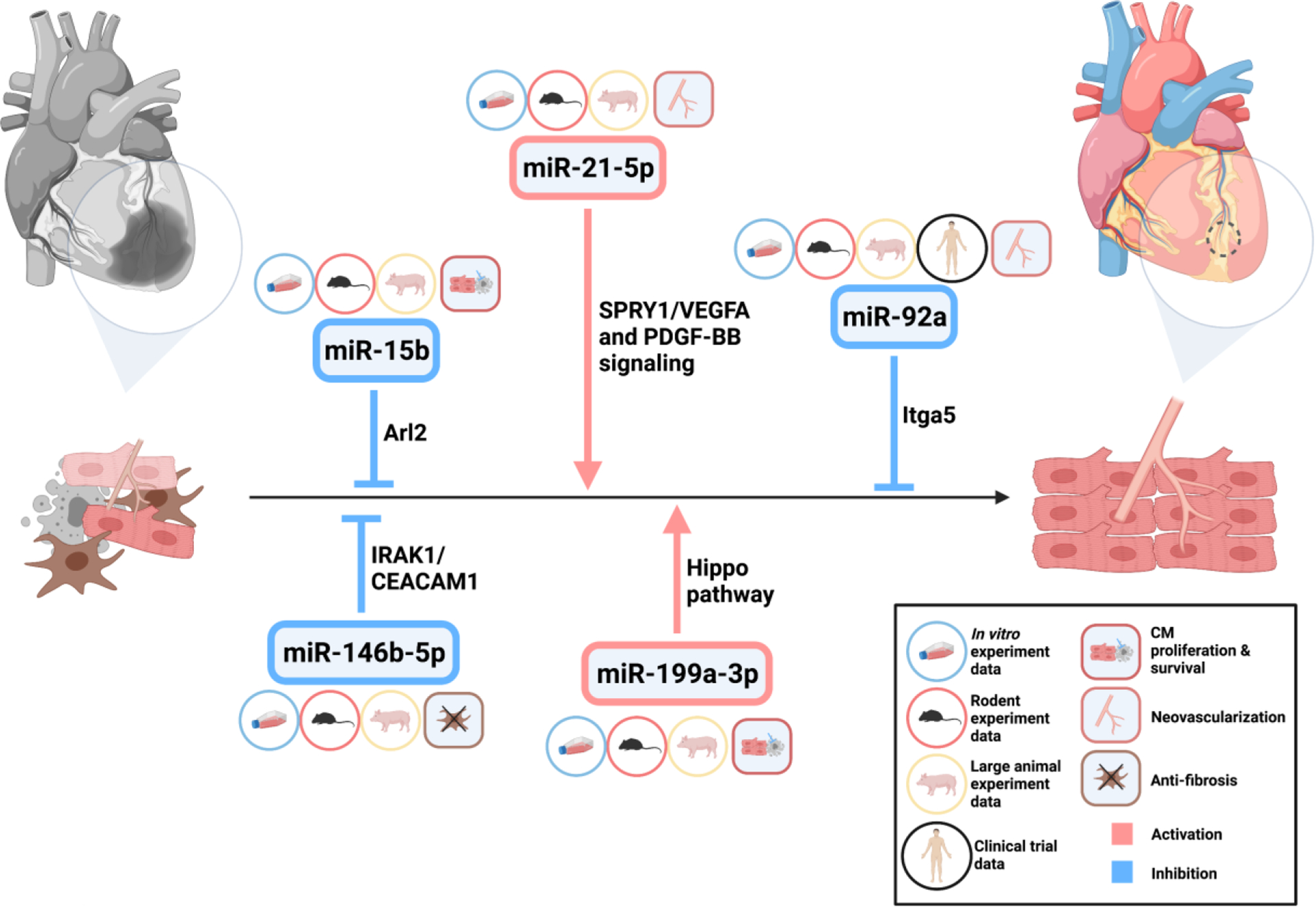
In the past two decades, miRNAs have been shown to play a critical role in multiple processes of cardiac regeneration. Among them, at least five miRNAs including miR-92a, miR-199a, miR-15b, miR-21-5p, and miR-146b-5p have been investigated in a porcine MI model of human IHD. To our knowledge, there are no clinical trials to assess miRNA-based drugs in cardiac regeneration; although, phase 1 clinical trials of miR-132 and miR-92a inhibitors have been completed to test their safety and efficacy in patients with stable heart failure of ischemic origin, and wound healing in healthy volunteers (NCT04045405; NCT03603431) (https://clinicaltrials.gov/ct2/home). These studies displayed favorable clinical data, which will likely encourage clinical trials of miRNA-based medicines for cardiac regeneration. Since the functional roles of miRNAs in CM proliferation, differentiation, and survival as well as in neovascularization and fibrosis after MI have been previously reviewed [10, 44, 45]. To avoid redundancy, this review will focus on the latest advancements of a few miRNAs or miRNA families that have been used in preclinical large-animal experiments and clinical trials.
2.1. MiRNAs in cardiomyocyte proliferation, differentiation, and survival
2.1.1. Pro-proliferative miR-17-92 cluster and paralogs
The miR-17-92 cluster (including miR-17, miR-18a, miR-19a, miR-19b, miR-20 and miR-92a) was initially recognized as a human oncogene (known as OncomiR-1) that was expressed in human B-cell lymphomas [46–48]. This cluster promoted tumor development (along with c-Myc) in a mouse B-cell lymphoma model [47]. MiR-17-92-deficient mice died shortly after birth with lung hypoplasia and a ventricular septal defect [49].
In 2013, our group showed that the miR-17-92 cluster is not only required, but is sufficient, to induce cardiomyocyte proliferation in embryonic, postnatal, and adult mouse hearts [50]. The cardiac-specific overexpression of miR-17-92 in adult cardiomyocytes protects the heart from MI-induced injury and members of the miR-17-92 cluster (miR-19a/19b in particular) are required and sufficient to induce cardiomyocyte proliferation in vitro [50]. Intra-cardiac injection of miR-19a/19b mimics stimulate cardiomyocyte proliferation and cardiac regeneration in response to MI injury by manipulating immune response genes and the anti-proliferative gene Pten [51]. Consistent with these observations, tamoxifen-inducible cardiac-specific Pten deletion significantly promotes CM proliferation and cardiac repair after MI [52]. Consequently, miR-19a/19b appears to be a key mediator of the proliferative function of the miR-17-92 cluster [51].
Another member of this cluster, miR-92a, is a key negative regulator of angiogenesis in ischemic diseases such as MI [53]. Systemic administration of antagomir-92a advanced blood vessel growth and functional recovery of damaged tissue largely by targeting the proangiogenic protein Itga5 in endothelial cells [53]. Furthermore, global knockout of miR-92a significantly improved cardiac function after MI, while cardiac-specific deletion of miR-92a partially rescued cardiac dysfunction caused by MI [54]. Locked nucleic acid-modified antisense miR-92a (LNA-92a) protected cardiomyocytes from hypoxia/reoxygenation–induced cell death in vitro [54]. These data suggest that miR-92a also plays a role in cardiomyocytes during regeneration.
Recent progress has shown that, unlike its function in endothelial cells, miR-92a-3p inhibition ‘derepressed’ different genes in mouse cardiomyocytes, including the metabolism-related genes Abca8b and Cd36, after MI [55]. Abca8b and Cd36 were verified as the direct targets of miR-92a-3p by loss- and gain-of-function studies in CMs in vitro and luciferase reporter assays [55]. Abca8b and CD36-mediated metabolic switching in CMs improves substrate utilization and contributes to cardiac recovery through this pro-survival mechanism after MI [55]. The specific role of miR-92a in neovascularization after MI will be discussed in the neovascularization section (below).
MiR-17-3p has been reported to protect the myocardium against ischemia-reperfusion (IR) injury and is required for exercise-induced cardiac growth in mouse models [56]. AgomiR-mediated miR-17-3p overexpression increased cardiomyocyte proliferation as well as cell size in vitro by targeting the metallopeptidase inhibitor TIMP3 [56]. Another study showed that serum extracellular vesicles (EVs) promoted rat H9C2 cell proliferation with an increased level of miR-17-3p. Inhibition of miR-17-3p also diminished H9C2 proliferation induced by serum EVs [57].
MiR-25 is a member of the miR-17-92 cluster paralog mir-106b-25, which is also implicated in cardiomyocyte proliferation [58]. MiR-25 inhibition in neonatal cardiomyocytes reduced cell proliferation and promoted apoptosis. The effect of miR-25 inhibition on cardiomyocyte survival was significantly attenuated when its predicted target Bim was silenced [58]. Additionally, miR-25 protected cardiomyocytes against oxidative damage by inhibiting the mitochondrial calcium uniporter (MCU) and the mitochondrial apoptosis pathway [59]. Dirkx et al. reported miR-25 silencing by a specific antagomir promoted cardiac remodeling, which was abolished in Hand2 knockout hearts [60]. However, high-throughput functional screening of human miRNAs showed that miR-25 interacted with the 3′ UTR of the SERCA2a mRNA and significantly delayed calcium uptake in cardiomyocytes in vitro. [61]. Consequently, AAV9-mediated miR-25 overexpression caused a reduction of contractile function, while inhibition of miR-25 using an antagomiR significantly improved cardiac contractility in the failing mouse heart [61]. The observed negative effect of miR-25 on myocardial contractility makes its safety and efficacy for cardiac regeneration a question that requires further study. Whether the application of miR-25 favors, or is detrimental to, cardiac regeneration in the long term, needs careful investigation in large animal models.
2.1.2. Pro-proliferative miR-199a and miR-590
Eulalio et al. performed a high-content fluorescence-microscopy-based functional screen of neonatal rat cardiomyocytes for pro-proliferative miRNAs using a whole-genome human miRNA library (875 miRNA mimics) [62]. They used α-actinin, Ki-67, and EdU as proliferative CM markers and 40 miRNAs displayed enhanced EdU incorporation and Ki-67 positivity in both rat and mouse CMs by at least twofold. Among them, two potent RNAs, hsa-miR-199a-3p and has-miR-590-3p, were shown to stimulate substantial cardiac regeneration and almost complete recovery of functional parameters in a mouse MI model [62]. In 2017, the same group proved that a single intracardiac injection of synthetic miRNA-lipid formulation (hsa-miR-199a-3p and hsa-miR-590-3p) was sufficient to improve cardiac regeneration and functional recovery after MI [63].
The most effective miRNAs from the functional screen activated CM proliferation by promoting nuclear translocation of YAP. The Hippo/YAP1 pathway is recognized as an important regulator of cardiac development and regeneration [64–67]. Hippo signal inactivation results in YAP1 activation (i.e., via nuclear translocation) to enhance CM proliferation and cardiac regeneration after injury at both postnatal and adult stages [66, 67]. MiR-199a-3p directly interacts with two targets to activated YAP; the upstream YAP inhibitory kinase TAOK1 and the E3 ubiquitin ligase β-TrCP, which leads to YAP degradation. Alternatively, miR-199a-3p inhibits filamentous actin depolymerization to facilitate YAP nuclear translocation by targeting Cofilin2 [68].
More recently, it was reported that AAV6-mediated delivery of hsa-miR-199a stimulated cardiac regeneration and improved both global and regional myocardial contractility in pig hearts one month after MI [69]. Unfortunately, uncontrolled and long-term activation of this microRNA induced sudden arrhythmic death in most of the treated pigs by week 8. These tachyarrhythmic events were not concurrent with ion channel expression changes in the conduction system, but with infiltration of undifferentiated (i.e., expressing Gata4+, Myog+, and Cald1+) cardiomyocytes in and around the infarct.
Differentiation of these cells may have severely disrupted normal cardiac conduction, resulting in fatal arrhythmias. As an alternative, the unwanted effects of the other strand of miR-199a-5p simultaneously expressed by AAV particles could also be responsible for the adverse outcomes [69, 70]. As a result, it will be worth determining if arrhythmia could be eliminated by more precise administration (e.g., miR-199a-3p alone) or changes in treatment duration and/or dose. In the meantime, these studies show the critical necessity to carefully evaluate the safety of miRNA-based therapeutic interventions in a large animal model prior to initiating clinical trials.
2.1.3. MiR-132 in cardiomyocyte survival
In 2011, Katare et al. reported that secreted miR-132 was the primary mediator of a human saphenous vein-derived pericyte progenitor (SVP)-based cell therapy in the infarcted murine heart [71]. SVP conditioned medium was sufficient to promote endothelial tube formation and inhibit myofibroblast differentiation using the validated miR-132 targets p120RasGAP and MeCP2. MiR-132 inhibition attenuated the beneficial SVP-induced effects on cardiomyocyte contractility, neovascularization, and interstitial fibrosis in infarcted hearts [71].
Hong et al. reported that delivery of exogenous miR-132 protects cardiomyocytes from apoptosis by preventing calcium overload under hypoxic conditions [72]. Ucar et al. also showed that the miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy by directly targeting the anti-hypertrophic and pro-autophagic FoxO3 transcription factor [73]. As a result, preclinical studies of miR-132 inhibition have been performed for the treatment of heart failure in a 56-day post-MI pig heart failure model. Synthetic locked nucleic acid antimiR-132 treatment proved to be safe with favorable dose-dependent pharmacokinetic and pharmacodynamic (PK/PD) relationships, suggesting a translational potential [74].
More recently, a phase 1b randomized, double-blind, placebo-controlled, dose-escalation study has been completed to assess the safety as well as PK/PD parameters of a microRNA-132-3p inhibitor called CDR132L in stable heart failure patients of ischemic origin (Cardior Pharmaceuticals GmbH, NCT04045405) [75]. This study is the first clinical trial of an antisense drug in human heart failure patients. The results indicate that CDR132L was both safe and well-tolerated. The effective dose level of CDR132L started from 1 mg/kg according to a PK/PD dose modeling approach. CDR132L treatment (≥1 mg/kg) provided a median 23.3% reduction of circulating N-terminal pro-brain natriuretic peptide (NT-proBNP) levels, cardiac functional improvements, and fibrosis inhibition [75].
In addition, the long-term therapeutic effects and safety of CDR132L have been estimated in a clinically-relevant pig model of 6-month chronic heart failure following MI [76]. Monthly intravenous (IV) treatments of CDR132L (5 mg/kg) were safe and well-tolerated. Chronic CDR132L treatment reduced circulating NT-proBNP levels, improving cardiac functional recovery and inhibiting adverse remodeling responses [76]. Although the CDR132L study displayed favorable clinical data in chronic heart failure, it should be noted that the role of miR-132 inhibitors in acute cardiac infarction and following heart regeneration is still not clear. Whether miR-132 inhibitors promote cardiac regeneration by enhancing CM survival and/or neovascularization in a large animal, post-MI model warrants further investigation.
2.1.4. MiR-15 family in cardiomyocyte survival
The miR-15 family includes six highly-conserved miRNA members: miR-15a, miR-15b, miR-16-1, miR-16-2, miR-195, and miR-497. The expression levels of this family are reported to be consistently dysregulated in various conditions, including cardiac ischemic injury [77] [78]. For instance, Nishi et al. reported that miR-15b decreased mitochondrial integrity and cellular ATP levels by targeting Arl2 in neonatal rat cardiac myocytes (NRCMs) [79]. In 2011, Hullinger et al. showed that LNA-mediated miR-15b inhibition increased CM resistance to hypoxia-induced cell death in vitro [78]. In animal experiments, systemic delivery of anti-miR-15 significantly reduced infarct size, suppressed adverse remodeling responses, and enhanced cardiac functional recovery in mouse and porcine MI models. The mechanism of action is, at least partly, due to derepression of antiapoptotic genes caused by miR-15 inhibition [78].
Additionally, as components of exosomes, miR-302b-3p and miR-373-3p can contribute to post-MI myocardial repair induced by Cyclin D2 overexpressing human iPSC-derived CMs in a swine MI model by promoting adjacent CM proliferation and survival, which occurs, at least in part, through the Hippo pathway [80]. Whether direct delivery of these two miRNAs in the infarcted region could phenocopy this effect is an interesting question worth pursuing.
2.2. MiRNAs in neovascularization after myocardial infarction
2.2.1. Anti-angiogenic MiR-92a in neovascularization after myocardial infarction
As mentioned earlier, compared to other members of the pro-proliferative miR-17-92 cluster, miR-92a functions predominantly as a negative regulator of angiogenesis [53]. Unlike the pro-angiogenic effects of miR-18a and miR-19a in tumor angiogenesis, miR-92a overexpression in endothelial cells impedes angiogenesis and vessel formation both in vitro and in vivo [48, 53]. Systemic delivery of antagomir-92a advanced blood vessel growth and functional recovery of damaged myocardium by targeting pro-angiogenic protein Itga5 in endothelial cells in mice [53]. In a large-animal model, catheter-based local delivery of miR-92a inhibitor reduced infarct size and improved global and regional myocardial function, which was associated with increased capillary density, decreased inflammation, and cardiac cell death 3 and 7 days after MI when compared with sham-operated control pigs [54]. In accordance with these findings, a long-term large animal study showed that a single intracoronary injection of encapsulated antagomir-92a enhanced vessel growth, induced favorable healing, and prevented adverse remodeling 1 month after MI [81].
In 2018, a synthetic microRNA-92a inhibitor named MRG-110 was screened and developed to promote the growth of new blood vessels and wound healing in acute and chronic excisional wounds using normal pig and db/db mouse models [82]. In each model, the inhibitor MRG-10 significantly induced granulation tissue formation, angiogenesis, and wound healing. Surprisingly, MRG-10 showed comparable, or even more favorable effects, than the positive controls (rhVEGF-165 and rhPDGF-BB) in the chronic excisional wound model. There were no safety concerns noted in either model [82].
At present (2021), a phase 1 clinical trial has been completed to assess the safety, tolerability, pharmacokinetics, and pharmacodynamics of intradermal injection of MRG-110 in acute excisional wound healing (miRagen Therapeutics, Inc, NCT03603431). As expected, the clinical data showed that MRG-110 positively impacted new blood vessel growth and wound healing. MRG-110 was safe and generally well-tolerated when given as three weekly intradermal doses [83]. Further clinical studies are being planned to evaluate the safety and efficacy of this miR-92 inhibitor in the setting of heart failure [83].
In addition, miR-21-5p was reported to participate in exosome-mediated cardiac regeneration after MI by enhancing angiogenesis and CM survival through the PTEN/Akt pathway [84]. Li et al. developed a delivery system in which miR-21-5p was mixed with anti-inflammatory mesoporous silica nanoparticles (MSNs), and then packaged into a hydrogel matrix to allow on-demand miR-21 release upon exposure to an acidic microenvironment. This novel delivery system significantly inhibits the inflammatory response, promotes local neovascularization, and reduces infarct size in a porcine MI model [85].
2.3. MiRNAs in fibrosis after myocardial infarction
2.3.1. MiR-146b-5p in fibrosis after myocardial infarction
In 2013, microRNA-146 was reported to inhibit endothelial activation by suppressing the pro-inflammatory NF-κB pathway and the MAP kinase pathway by targeting endothelial nitric oxide synthase (eNOS) [86]. More recently, Liao et al. reported that expression of miR-146b-5p was significantly upregulated in the plasma of coronary artery chronic obstruction patients as well as in the infarcted mouse myocardium [87]. Elevated miR-146b-5p expression primarily occurs in fibroblasts, endothelial cells, and macrophages under hypoxic stress. Antagomir-mediated inhibition remarkedly suppressed cardiac fibrosis, promoted angiogenesis, increased reparative macrophages, and improved cardiac functional recovery in a mouse MI model by restoring expression of IRAK1 and CEACAM1. Similarly, intramyocardially injection of a miR-146b-5p antagomir significantly improved cardiac fibrosis and ventricular strain in a porcine MI model [87].
Published miRNAs in cardiac regeneration using large-animal MI models are summarized in Figure 1 and Table 1.
Figure 1.
MiRNAs in cardiac regeneration
Table 1.
MicroRNAs in cardiac regeneration in large animal (pig) models
| Name | Target cells | Functional role and mechanism | Intervention approach | Clinical trial | Ref |
|---|---|---|---|---|---|
| miR-15b | CMs | pro-apoptosis; via targeting Arl2 and Bcl2 | LNA antimiR | - | [72] |
| miR-21-5p | M1 macrophages and endothelial cells | anti-inflammation and pro-angiogenesis; via targeting SPRY1/VEGFA and PDGF-BB signaling | agomiR + nanoparticles | - | [79] |
| miR-92a | endothelial cells | anti-angiogenesis; via targeting pro-angiogenic protein Itga5 | antagomir | Yes | [54,75] |
| miR-146b-5p | CFs, endothelial cells and macrophages | pro-fibrotic, anti-angiogenesis and inhibit reparative macrophages; via targeting interleukin 1 receptor associated kinase 1 (IRAK1) and carcinoembryonic antigen related cell adhesion molecule 1 (CEACAM1) | antagomir | - | [81] |
| miR-199a-3p | CMs | pro-proliferative; via targeting Hippo/YAP1 pathway via TAOK1, β-TrCP and Cofilin2 | AAV6 | - | [63] |
3. LncRNAs in cardiac regeneration
Although the role of lncRNAs was less well studied, an increasing number of these molecules have been investigated for their role in cardiac regeneration. Because lncRNAs function very differently, their mechanism of action often remains obscure. Here, we emphasize and summarize the critical lncRNA studies with distinct mechanisms in cardiac regeneration, which will, hopefully, stimulate new lncRNA studies in cardiac regeneration.
3.1. LncRNAs in cardiomyocyte proliferation, differentiation, and survival
3.1.1. Transcriptional regulatory lncRNA CPR in cardiomyocyte proliferation
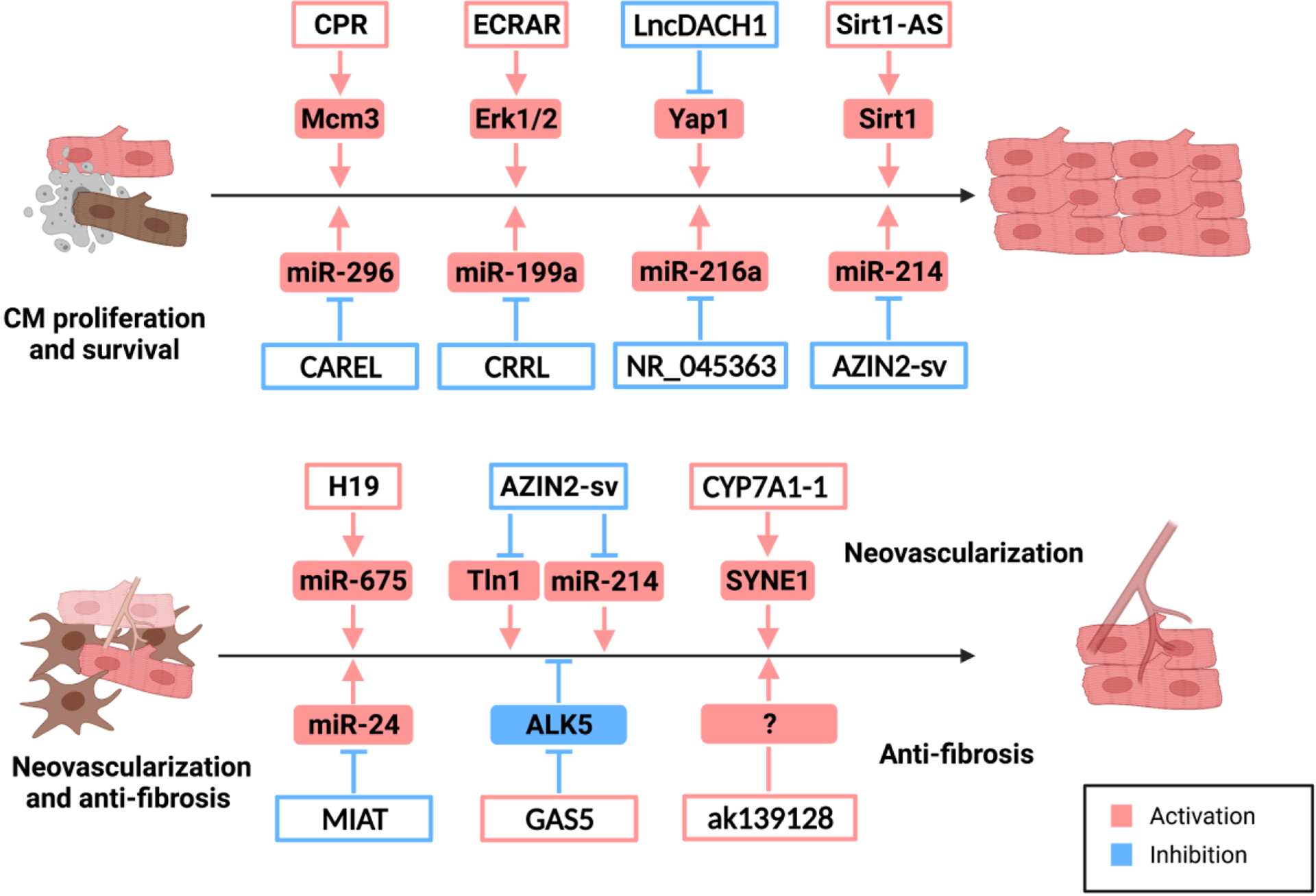
The nuclear-localized lncRNA Cardiomyocyte Proliferation Regulator (CPR) was identified with relative expression analyses of human fetal and adult hearts [88]. CPR has no protein-coding potential, which was demonstrated by bioinformatics analyses and in vitro translation assays. CPR is a cardiac-enriched lncRNA with a higher expression level in the adult heart. In vitro, siRNA-mediated CPR inhibition enhanced CM proliferation in neonatal mouse cardiomyocytes, and its deletion induced CM proliferation in postnatal and adult ventricles of CPR knockout mice. CPR deficiency increased CM proliferation, reduced scar size, and restored myocardial function after MI. In contrast, CM proliferation and cardiac regeneration were suppressed in CPR transgenic mice and AAV9-mediated CPR overexpression hearts subjected to MI [88].
Transcriptome analyses showed that the expression of Mcm3, an important regulatory protein for initiating eukaryotic genome replication, was upregulated several-fold in CPR knockout mouse hearts. Quantitative chromatin immunoprecipitation (CHIP) experiments indicated that lncRNA CPR interacted with DNMT3A and guided the latter to methylate a CpG island in the Mcm3 promoter; resulting in repression of Mcm3 transcription and inhibition of CM proliferation [88]. Additionally, CPR inhibition restricted the guide function of the lncRNA to DNMT3A, derepressed Mcm3 expression, and promoted CM proliferation—providing a promising therapeutic strategy for cardiac regeneration and repair.
3.1.2. Enhancer lncRNA CARMEN in cardiomyocyte differentiation
Ounzain et al. reported that 570 differentially expressed lncRNAs were identified during cardiac differentiation of human cardiac precursor cells (CPCs) using an established in vitro differentiation protocol [89]. One of the 3 most upregulated lncRNAs was a super-enhancer (SE)-associated lncRNA named Cardiac Mesoderm Enhancer-associated Noncoding RNA (CARMEN). ShRNA and antisense GapmeR-mediated CARMEN knockdown negatively impacted cardiac specification and differentiation in mouse and human cardiac precursor cells. Luciferase reporter assays and RNA immunoprecipitation (RIP) experiments suggest that CARMEN functions in cis and interacts with SUZ12 and EZH2 in vitro [89]. Together, the super-enhancer lncRNA CARMEN plays a crucial role in cardiac cell differentiation and homeostasis, while the detailed mechanism and target genes require further elucidation.
3.1.3. MiRNA sponge lncRNA CAREL in cardiomyocyte proliferation
Cai et al. performed a differentially expressed lncRNA screen using postnatal day 1 and day 7 mouse hearts and found that a total of 128 lncRNAs were dysregulated by at least two-fold [90]. Among them, 8 lncRNAs were significantly upregulated more than five-fold. Consequently, a cytoplasm-localized lncRNA called CAREL was further investigated. LncRNA CAREL is a 748-nt transcript with low protein-coding potential. The transcript level of CAREL is increased in mouse hearts from P1 to P10. CAREL-deficiency enhanced cardiomyocyte proliferation and improved heart function after MI in both neonatal and adult mice using intramyocardial administration of an adenoviral shRNA. In contrast, cardiac-specific CAREL overexpression inhibited cardiomyocyte proliferation and neonatal heart regeneration after injury in Myh6-CAREL transgenic mice [90].
Considering the importance of the border zone in endogenous cardiac regeneration after MI, the proliferation assay in this study was performed there instead of the central infarct or in a remote region [91]. Mechanistically, CAREL restored expression of Trp53inp1 and Itm2a as competing endogenous RNAs (ceRNAs) for miR-296-3p. As expected, agomiR-mediated miR-296-3p activation rescued CM proliferation and cardiac function caused by CAREL overexpression after MI in CAREL transgenic mice. Intriguingly, a short, conserved sequence of human CAREL phenocopied the anti-proliferative phenotype in human iPSCs-derived cardiomyocytes, indicating the potential for clinical application following cardiac injury [90].
3.1.4. MiRNA sponge lncRNA CRRL in cardiomyocyte proliferation
In a manner similar to that described for lncRNA CAREL, Chen et al. identified an adult-cardiac upregulated lncRNA NONHSAG007671 (named Cardiomyocyte Regeneration-Related LncRNA or CRRL) through differential expression analyses of human fetal and adult cardiac RNA-sequencing data [92]. The expression of LncRNA CRRL was correlated with the expression of cell cycle-related protein-coding genes. LncRNA CRRL was highly conserved across 5 species, including humans, rats, and mice, with a low protein coding potential. Adenoviral-mediated CRRL knockdown promoted cardiomyocyte proliferation and improved cardiac repair in neonatal and adult rats post-MI by serving as a sponge of miR-199a-3p and derepressing Hopx expression [92].
3.1.5. MiRNA sponge lncRNA NR_045363 in cardiomyocyte proliferation
A highly-conserved cardiomyocyte-enriched lncRNA NR_045363 was found using differential gene expression analysis of human fetal and adult hearts [93]. In vitro, siRNA-mediated NR_045363 knockdown inhibited CM proliferation, while NR_045363 overexpression by an adenovirus enhanced DNA synthesis and cytokinesis in neonatal CMs. Importantly, AAV9-mediated NR_045363 overexpression stimulated CM proliferation and improved cardiac function after MI in neonatal mice. The mechanistic study revealed that NR_045363 functioned as an ceRNA of miR-216a, which restored activity of the JAK2-STAT3 pathway to promote CM proliferation and cardiac regeneration [93]. An independent study showed that siRNA-mediated NR_045363 knockdown induced CM apoptosis by targeting the p53 signaling pathway [94].
3.1.6. MiRNA sponge lncRNA AZIN2-sv in cardiomyocyte proliferation
Sense lncRNA AZIN2-sv is a splice variant of the AZIN2 gene without a detectable protein product. Expression of AZIN2-sv was significantly upregulated in human adult hearts [95]. Loss-and gain-of-function experiments showed that AZIN2-sv negatively regulates endogenous cardiomyocyte proliferation in vitro and in vivo. Mechanistically, through sponging of miR-214, AZIN2-sv restores PTEN activity to inhibit cardiomyocyte proliferation by suppressing the PI3K/Akt pathway [95]. LncRNA AZIN2-sv also functions as a protein modification regulator in neovascularization following MI [96], which is summarized in the neovascularization section below.
3.1.7. Other miRNA sponge lncRNAs in cardiomyocyte proliferation, differentiation, and survival
Other reported miRNA sponge lncRNAs involved in CM proliferation, differentiation, and survival include lnc-NEAT1 (miR-193a, miR-142-3p, and miR-520a), lnc-TUG1 (miR-132-3p), and lnc-SNHG1 (miR-195) [97–101]. Lnc-SNHG1 was also reported to stimulate CM proliferation, enhance angiogenesis, and cardiac recovery after MI by directly binding PTEN and activating the PI3K-Akt pathway [102]. However, how lnc-SNHG1 impacts its protein partner PTEN remains unknown.
3.1.8. LncRNA ECRAR in cardiomyocyte proliferation
A functional lncRNA discovery experiment was performed in human hearts during the fetal-to-adult transition using four RNA-seq datasets with 189 million clear reads [103]. 3,830 novel transcripts were identified as novel lncRNAs with low coding potential. A total of 3,092 lncRNAs were differentially expressed, with 1,343 lncRNAs upregulated and 1,749 lncRNAs downregulated in fetal hearts. Of these, a novel upregulated lncRNA called Endogenous Cardiac Regeneration-Associated Regulator (ECRAR) was significantly induced in the fetal heart by more than 12-fold [103]. Human LncRNA ECRAR is an 851-bp sense lncRNA consisting of four exons and a poly (A) tail, which is located on chromosome 5 overlapping with the protein-coding gene pituitary tumor-transforming 1 (PTTG1). It represents an alternative splice variant of the PTTG1 gene within the retained intron without any open reading frame (ORF), indicating the inability to be translated into protein [103].
In vitro, ECRAR overexpression promotes DNA synthesis, mitosis, and cytokinesis, but does not induce CM hypertrophy in post-natal day 7 rat cardiomyocytes. Adenovirus-based ECRAR overexpression by intramyocardial administration promoted CM proliferation, angiogenesis, and improved cardiac functional recovery in a neonatal rat MI model. In contrast, shRNA-based ECRAR knockdown suppressed post-natal day 1 rat CM proliferation and prevented post-MI cardiac recovery [103]. Mechanistically, ECRAR directly interacted with ERK1/2 and promoted the phosphorylation of the latter, which initiated the transcription of downstream targets of cyclin D1 and cyclin E1, and activated E2F1. The activated E2F1, in turn, enhanced ECRAR transcription. This positive feedback loop propels cell cycle progression and CM proliferation [103].
3.1.9. LncRNA LncDACH1 in cardiomyocyte proliferation
A 2085-nt highly-conserved lncRNA was discovered in the first intron of the DACH1 gene in the mouse and is named lncDACH1 [104]. LncDACH1 is markedly overrepresented during mouse postnatal heart development [105]. Adenovirus-mediated lncDACH1 inhibition activates CM proliferation in P7 and adult mice. In line with this, cardiac-specific lncDACH1 deletion enhances CM proliferation and cardiac recovery after MI surgery in tamoxifen-inducible adult lncDACH1 knockout mice. In contrast, neonatal lncDACH1 transgenic mice show decreased CM proliferation and cardiac recovery after apical resection surgery.
Mechanistically, lncDACH1 inhibits CM proliferation by positively regulating YAP1 phosphorylation and preventing nuclear translocation (i.e., YAP inactivation). This action occurs via lncDACH1-mediated PP1A dephosphorylation, which attenuates the interaction between PP1A and YAP1, and the phosphorylation of the latter [105]. LncDACH1 deficiency-induced CM proliferation was weakened by PP1A siRNA treatment, pharmacological blockers, or the YAP1 inhibitor Verteporfin [105]. Surprisingly, intronic lncRNA LncDACH1 has a minimal influence on DACH1 expression, indicating LncDACH1 functions independent from the host gene. This data also points out a possibly different mechanism of action between intronic lncRNAs and most enhancer lncRNAs, which function in cis.
3.1.10. Messenger RNA stability regulatory lncRNA Sirt1-AS in cardiomyocyte proliferation
It was reported that 30 to 50% of the protein-coding gene loci in the mouse genome show evidence of overlapping antisense transcripts. As natural antisense transcripts (NATs), these lncRNAs may also have functional implications [106, 107]. For example, antisense lncRNA Sirt1-AS has higher expression during embryonic development (E16.5) compared with postnatal days 1 and 28 [108]. LncRNA Sirt1-AS is required and sufficient to promote CM proliferation in vitro as well as in neonatal mouse hearts. In particular, Sirt1-AS overexpression induces CM proliferation and promotes cardiac functional recovery after MI in adult mice. This action occurs through binding the 3′UTR of the Sirt1 mRNA to enhance mRNA stability and abundance [108].
Another highly-conserved antisense lncRNA, uc.40, is reported to be overexpressed in abnormal human embryonic hearts [109]. SiRNA-mediated uc.40 inhibition promotes P19 embryonic cell differentiation to cardiomyocytes by reducing the mRNA level of its sense gene PBX1 [109]. LncRNAs that inhibit or activate cardiomyocyte proliferation are listed in Figure 2.
Figure 2.
LncRNAs in cardiac regeneration
3.2. LncRNAs in neovascularization after myocardial infarction
3.2.1. MiRNA host-gene lncRNA H19 in neovascularization after myocardial infarction
Extracellular vesicles (EVs) are attracting attention due to their apparent contributions to cell therapy-based cardioprotection. LncRNA H19 is reported to be involved in the beneficial effect of Atorvastatin (ATV) in mesenchymal stem cell (MSC)-derived exosomes in an acute myocardial infarction rat model [110, 111]. Huang et al. isolated EVs from control MSCs (MSC-Exo) and ATV-pretreated MSCs (MSCATV-Exo) and delivered them to endothelial cells and CMs in vitro or MI-challenged rat hearts in vivo. Direct control EV delivery had a minimal effect on cardiomyocytes, while MSCATV-Exo enhanced migration, tube formation, and survival of endothelial cells through the H19/miR-675/VEGF/ICAM-1 axis [110, 111].
MiR-675 is a highly conserved miRNA embedded in H19’s first exon and has been implicated in cell differentiation and angiogenesis. The release of miR-675 from H19 was regulated by the stress-response protein HuR [112]. Intriguingly, the exosomes derived from MSCATV-Exo-treated endothelial cells protected cardiomyocytes from hypoxia and serum deprivation (H/SD)-induced apoptosis. As expected, MSCATV-Exo reduced infarct size, induced angiogenesis, and ameliorated cardiac function in a rat MI model. However, endogenous H19/miR-675 may promote CM apoptosis by targeting PA2G4 in Adriamycin-induced dilated cardiomyopathy (DCM) [113]. These contradictory observations of cardiomyocyte survival may be due to context-specific effects and need further investigation. In addition, lncRNA H19 can promote cardiac fibrosis by repressing the DUSP5/ERK1/2 pathway or through sponging of miR-455 [114, 115].
3.2.2. LncRNA AZIN2-sv in neovascularization after myocardial infarction
Besides playing an anti-proliferative role in CMs, AZIN2-sv is a negative regulator of neovascularization in endothelial cells after MI [96]. Adenoviral shRNA-mediated AZIN2-sv knockdown inhibited endothelial apoptosis and promoted vessel formation in vitro. The downregulation of AZIN2-sv induced angiogenesis and improved cardiac functional recovery after MI in vivo [96]. Mechanistically, AZIN2-sv enabled an interaction between Tln1 and PSMC5 to activate PSMC5-mediated, ubiquitination-dependent degradation of Tln1 to inhibit neovascularization. Additionally, the miR-214/PTEN/Akt pathway was also reported to be involved in the regulatory role of AZIN2-sv in neovascularization after MI [96].
3.2.3. Lnc-CYP7A1-1 in neovascularization after myocardial infarction
The delivery of human bone marrow (hBM)-derived MSCs improves cardiac function and suppresses adverse remodeling after MI by increasing angiogenesis and inhibiting fibrosis [116, 117]. The efficacy of transplanted hBM-MSCs diminished with donor age, which impacts the potential treatment of older patients with cardiovascular disease [118–120]. LncRNA lnc-CYP7A1-1 was identified as one of the most significantly induced lncRNAs during differential expression analyses of human bone marrow-MSCs derived from young and old individuals [121]. The application of lnc-CYP7A1-1 to aging hBM-MSCs significantly improved cardiac regeneration and recovery in mouse infarcted hearts with paracrine induction of angiogenesis. The mode of action is possibly mediated by upregulation of the predicted target SYNE1 [121]; however, the actual molecular mechanism needs further investigation.
3.3. LncRNAs in fibrosis after myocardial infarction
3.3.1. MiRNA sponge lncRNA MIAT in fibrosis after myocardial infarction
LncRNA Myocardial Infarction Associated Transcript (MIAT) was discovered in a large-scale case-control association study conferring risk of MI in humans [122]. MIAT transcript levels are induced in a mouse model of MI. Lentiviral shRNA-mediated MIAT knockdown significantly suppresses cardiac fibrosis and improves cardiac function 4 weeks after MI [123]. In vitro, serum or angiotensin II promotes cardiac fibroblast proliferation and collagen accumulation, which is lessened by MIAT knockdown. Although the predicted target miR-24 is upregulated upon MIAT knockdown in vivo and in vitro, whether sponging by miR-24 is the major mechanism of MIAT-mediated cardiac fibrosis needs more careful examination [123].
Another study shows lentiviral shRNA-mediated MIAT knockdown suppresses H9c2 apoptosis under hypoxia/reoxygenation (H/R) conditions. Importantly, MIAT knockdown significantly decreases myocardial infarct size, reduces myocardial apoptosis, and enhances heart function in an ischemia/reperfusion (I/R) mouse model [124]. Cell-specific knockouts may help clarify the main contributor in terms of MIAT-inhibition-mediated cardioprotection in cardiac regeneration and repair after MI. Together, these studies suggest a promising lncRNA-based strategy to combat MI-induced cardiac fibrosis and cardiac dysfunction.
Other previously reported miRNA sponge lncRNAs in fibrosis include lncRNA RNF7 (miR-543) and lncRNA XIST (miR-155-5p) [125, 126]; however, the observations concerning these lncRNAs are mainly based on in vitro studies, which need to be further verified with in vivo experiments. LncRNA GAS5 was reported to inhibit cardiac fibroblast proliferation in atrial fibrillation by downregulating ALK5 (also known as TGF-Beta Receptor Type I) [127]. Hypoxic CM-derived lncRNA ak139128 promoted apoptosis and inhibited proliferation in cardiac fibroblasts through an unknown mechanism [128]. Functional lncRNAs implicated in cardiac regeneration after MI are summarized in Table 2.
Table 2.
LncRNAs in cardiac regeneration*
| Name | Mechanism of action | Functional role | Mechanism | Ref |
|---|---|---|---|---|
| CPR | guide | anti-proliferative | guides DNMT3A to methylate a CpG island in the Mcm3 promoter to inhibit Mcm3 expression | [82] |
| CARMEN | enhancer | promotes cardiac differentiation | exhibits cis-acting function and interacts with SUZ12 and EZH2 | [83] |
| CAREL | miRNA sponge | anti-proliferative | targets miR-296-3p to derepress expression of Trp53inp1 and Itm2a | [84] |
| CRRL | miRNA sponge | anti-proliferative | Sponges miR-199a-3p and restores Hopx expression | [86] |
| NR_045363 | miRNA sponge | anti-proliferative | functions as an ecRNA of miR-216a, which restored activity of the JAK2-STAT3 pathway | [87] |
| AZIN2-SV | miRNA sponge | anti-proliferative | Targets miR-214 to restore PTEN activity | [89,90] |
| protein modification regulator | anti-angiogenesis | Interacts with Tln1 and PSMC5 to activate the degradation of Tln1; miR-214/PTEN/Akt pathway | ||
| ECRAR | Protein modification regulator | pro-proliferative | directly interacts with ERK1/2 and promoted its phosphorylation | [97] |
| LncDACH1 | Protein modification regulator | anti-proliferative | Induces YAP1 phosphorylation and prevents its nuclear translocation via promoting PP1A dephosphorylation | [99] |
| Sirt1-AS | mRNA stability regulator | pro-proliferative | binds the 3’UTR of the Sirt1 mRNA to enhance messenger RNA stability and abundance | [102] |
| H19 | miRNA host-gene | pro-angiogenesis | releases miR-675 | [104,105] |
| Inc-CYP7A1-1 | unknown | pro-angiogenesis | upregulates the predicted target SYNE1 | [115] |
| MIAT | miRNA sponge | pro-fibrotic | sponges miR-24 | [117] |
| GAS5 | unknown | anti-fibrotic | Inhibits ALK5 | [121] |
| IncRNA ak139128 | unknown | anti-fibrotic | - | [122] |
Other reported miRNA sponge lncRNAs involved in CM proliferation, differentiation, and survival include lnc-NEAT1 (miR-193a, miR-142-3p, and miR-520a), lnc-TUG1 (miR-132-3p), and lnc-SNHG1 (miR-195) [91–95]. Lnc-SNHG1 was also reported to stimulate CM proliferation, enhance angiogenesis, and cardiac recovery after MI by directly binding PTEN and activating the PI3K-Akt pathway [96]. However, how lnc-SNHG1 impacts its protein partner PTEN remains unknown.
4. CircRNAs in cardiac regeneration
Like lncRNAs, circRNAs also present a time- and tissue-specific expression pattern compared with protein-coding genes, which may represent potential therapeutic targets. Below, we discuss several representative circRNA studies with distinct mechanisms in cardiac regeneration and other published circRNA studies are summarized at the end.
4.1. Super-enhancer-associated circRNA circNfix in cardiomyocyte proliferation and neovascularization after myocardial infarction
A large-scale circRNA enhancer analysis indicated that compared with typical enhancer–associated circRNAs (TE-circRNAs), super-enhancer-associated circRNAs (SE-circRNAs) showed higher expression, increased tissue-specific distribution, cardiogenesis as well as angiogenesis using an H3K27ac-marked enhancer catalog generated in 23 human tissues [129]. The CM-enriched circRNA circNfix was one of the most differentially expressed SE-circRNAs in neonatal versus adult hearts. Interestingly, the opposite expression trend of circular and linear Nfix suggests an independent biological function of this circRNA [129]. CircNfix expression was significantly increased in hearts and cardiomyocytes during heart development (from E14.5/P0 to p56). Phenotypically, circNfix downregulation promotes CM proliferation in vitro and adult CM proliferation in vivo. AAV9-shRNA mediated circNfix knockdown promotes CM proliferation, angiogenesis, and cardiac functional recovery after MI. In contrast, adenovirus-mediated circNfix overexpression inhibits neonatal cardiac regeneration post-MI [129]. Mechanistically, circNfix strengthens the interaction between Ybx1 with Nedd4l, inducing Ybx1 ubiquitin-dependent degradation, and downregulation of cyclin A2 and B1 expression. As a ceRNA, circNfix could sponge miR-214 to promote Gsk3β expression to repress CM proliferation and angiogenesis [129].
4.2. Mitochondrial fission and apoptosis-related circRNA (MFACR) in cardiomyocyte survival
Mitochondrial fission and apoptosis-related circRNA (MFACR) was identified during circRNA expression analyses in response to anoxia/reoxygenation (A/R) or ischemia/reperfusion (I/R) injury of CMs and mouse hearts, respectively [130]. CircRNA MFACR displays a substantial induction in both conditions as well as resistance to RNase R digestion. Adenoviral siRNA-mediated MFACR knockdown attenuates mitochondrial fission, infarction size, and enhanced functional recovery in a post-MI mouse model. Mechanistically, MFACR sponges miR-652-3p and derepresses its target MTP18, which promotes mitochondrial fission, CM apoptosis, and myocardial dysfunction following MI [130].
4.3. CircFndc3b in neovascularization after myocardial infarction
A highly-conserved 215-nt circRNA circFndc3b was identified using differential expressed circRNA analyses of sham-operated versus MI mouse hearts at post-operative day 3 [131]. Unlike linear Fndc3b mRNA levels, human and mouse CircFndc3b levels are significantly reduced in the left ventricles of ischemic cardiomyopathy patients and post-MI mouse hearts. Intriguingly, downregulation of circFndc3b occurs in endothelial cells and CMs, but not cardiac fibroblasts.
Importantly, circFndc3b overexpression enhances endothelial cell tube formation and suppresses cardiomyocyte apoptosis in vitro. AAV9-mediated CircFndc3b overexpression by intramyocardial injection promotes cardiac functional recovery post-MI in mice, with significant induction of neovascularization and reductions in fibrosis [131]. Mechanistically, circFndc3b does not act as a miRNA sponge, but binds the RBP Fused in Sarcoma (FUS) to activate VEGF signaling. Additional studies are needed to determine how FUS levels are negatively modulated by circFndc3b [131].
Other circRNAs and their targets in CM proliferation, differentiation, and survival after MI include circHIPK3 (miRNA-124-3p; Notch1 and miR-133a) [132, 133], circCDYL (miR-4793-5p) [134], circMGAT1 (miR-34a) [135], circNCX1(miR-133a-3p) [136], circRNA Cdr1as (miR-7a; miR-135a/135b) [137, 138], circRNA Ttc3 (miR-15b) [139], and circAmotl1 (PDK1 and AKT1) [140]. Other circRNAs and their targets functioning in angiogenesis after MI include hsa_circ_0007623 (microRNA-297) [141] and hsa_circ_0003575 (miR-199-3p, miR-9-5p, miR-377-3p and miR-141-3p) [142]. Other reported circRNAs and their targets found in fibrosis after MI include circNFIB (miR-433) [143] and circLAS1L (miR-125b) [144].
5. Conclusions and future perspectives
Numerous miRNAs, lncRNAs, and circRNAs are being investigated with regard to their structure, classification, function, and mechanism of action. While the therapeutic potential of miRNAs is becoming clear, the same potential of lncRNAs and circRNAs for cardiac regeneration in humans remains to be determined.
Because small rodent models are the primary in vivo tools for cardiac regeneration research, the safety and efficacy of novel ncRNA therapeutics based on lncRNAs and circRNAs will require evaluation in large animals prior to the initiation of human clinical trials. At the same time, rodent and cell culture models will be necessary to dissect the diverse biological and molecular mechanisms underlying lncRNA and circRNA function. The combination of genetic manipulation, NGS technologies, identification of ncRNA-interacting partners, and cellular localization will help provide important clues to clarify the functions of specific ncRNAs [30]. In particular, next-generation sequencing-based techniques can be applied to understand the regulatory landscape of these molecules at the whole-genome level. In addition, there are presently many non-coding RNAs with incomplete or nonexistent annotation. Complete annotation is the first step in any non-coding RNA study.
Despite these challenges, further exploration of the role of ncRNAs in cardiac regeneration will result in the near-term development of new therapeutic strategies for human IHD and related diseases.
Acknowledgments
We thank the anonymous reviewer for his/her contribution to this review. We thank members of the Wang laboratory for discussion and support. This work is supported by grants from National Institutes of Health (HL138757, HL125925, and HL093242). YW is supported by an American Heart Association Postdoctoral Fellowship (19POST34370132).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interests
We have no conflicts of interest to declare.
REFERENCES
- 1.Organization, W.H., Global Health Estimates 2020: Deaths by Cause, Age, Sex, by Country and by Region, 2000–2019. 2020.
- 2.Bergmann O, et al. , Evidence for cardiomyocyte renewal in humans. Science, 2009. 324(5923): p. 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Berlo JH and Molkentin JD, An emerging consensus on cardiac regeneration. Nature Medicine, 2014. 20(12): p. 1386–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vagnozzi RJ, et al. , Genetic lineage tracing of Sca-1+ cells reveals endothelial but not myogenic contribution to the murine heart. Circulation, 2018. 138(25): p. 2931–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang J, et al. , Fate mapping of Sca1+ cardiac progenitor cells in the adult mouse heart. Circulation, 2018. 138(25): p. 2967–2969. [DOI] [PubMed] [Google Scholar]
- 6.Neidig LE, et al. , Evidence for minimal cardiogenic potential of stem cell antigen 1–positive cells in the adult mouse heart. Circulation, 2018. 138(25): p. 2960–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, et al. , Cardiac Sca-1+ cells are not intrinsic stem cells for myocardial development, renewal, and repair. Circulation, 2018. 138(25): p. 2919–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soonpaa MH, et al. , Absence of cardiomyocyte differentiation following transplantation of adult cardiac-resident Sca-1+ cells into infarcted mouse hearts. Circulation, 2018. 138(25): p. 2963–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinto AR, et al. , Revisiting cardiac cellular composition. Circulation Research, 2016. 118(3): p. 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters MM, Sampaio-Pinto V, and da Costa Martins PA, Non-coding RNAs in endothelial cell signalling and hypoxia during cardiac regeneration. Biochimica et Biophysica Acta: Molecular Cell Research, 2020. 1867(3): p. 118515. [DOI] [PubMed] [Google Scholar]
- 11.Ottaviano FG and Yee KO, Communication signals between cardiac fibroblasts and cardiac myocytes. Journal of Cardiovascular Pharmacology, 2011. 57(5): p. 513–521. [DOI] [PubMed] [Google Scholar]
- 12.Aghajanian H, et al. , Targeting cardiac fibrosis with engineered T cells. Nature, 2019. 573(7774): p. 430–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.An integrated encyclopedia of DNA elements in the human genome. Nature, 2012. 489(7414): p. 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Djebali S, et al. , Landscape of transcription in human cells. Nature, 2012. 489(7414): p. 101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hangauer MJ, Vaughn IW, and McManus MT, Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet, 2013. 9(6): p. e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romano G, et al. , Small non-coding RNA and cancer. Carcinogenesis, 2017. 38(5): p. 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stefani G and Slack FJ, Small non-coding RNAs in animal development. Nature Reviews: Molecular Cell Biology, 2008. 9(3): p. 219–230. [DOI] [PubMed] [Google Scholar]
- 18.O’Brien J, et al. , Overview of microRNA biogenesis, mechanisms of actions, and circulation. Frontiers in Endocrinology, 2018. 9: p. 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasudevan S and Steitz JA, AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell, 2007. 128(6): p. 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ørom UA, Nielsen FC, and Lund AH, MicroRNA-10a binds the 5′ UTR of ribosomal protein mRNAs and enhances their translation. Molecular Cell, 2008. 30(4): p. 460–471. [DOI] [PubMed] [Google Scholar]
- 21.Bukhari SI, et al. , A specialized mechanism of translation mediated by FXR1a-associated microRNP in cellular quiescence. Molecular Cell, 2016. 61(5): p. 760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozomara A, Birgaoanu M, and Griffiths-Jones S, miRBase: from microRNA sequences to function. Nucleic Acids Research, 2019. 47(D1): p. D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabili MN, et al. , Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & Development, 2011. 25(18): p. 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan L, et al. , Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nature Structural & Molecular Biology, 2013. 20(9): p. 1131. [DOI] [PubMed] [Google Scholar]
- 25.Liu SJ, et al. , Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biology, 2016. 17(1): p. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang KC and Chang HY, Molecular mechanisms of long noncoding RNAs. Molecular Cell, 2011. 43(6): p. 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JT, Epigenetic regulation by long noncoding RNAs. Science, 2012. 338(6113): p. 1435–1439. [DOI] [PubMed] [Google Scholar]
- 28.Devaux Y, et al. , Long noncoding RNAs in cardiac development and ageing. Nature Reviews Cardiology, 2015. 12(7): p. 415. [DOI] [PubMed] [Google Scholar]
- 29.Marchese FP, Raimondi I, and Huarte M, The multidimensional mechanisms of long noncoding RNA function. Genome Biology, 2017. 18(1): p. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao R-W, Wang Y, and Chen L-L, Cellular functions of long noncoding RNAs. Nature Cell Biology, 2019. 21(5): p. 542–551. [DOI] [PubMed] [Google Scholar]
- 31.Grote P, et al. , The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Developmental Cell, 2013. 24(2): p. 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han P, et al. , A long noncoding RNA protects the heart from pathological hypertrophy. Nature, 2014. 514(7520): p. 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang KC, et al. , A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature, 2011. 472(7341): p. 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tripathi V, et al. , The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular Cell, 2010. 39(6): p. 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P, et al. , The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science, 2014. 344(6181): p. 310–313. [DOI] [PubMed] [Google Scholar]
- 36.Guo JU, et al. , Expanded identification and characterization of mammalian circular RNAs. Genome Biology, 2014. 15(7): p. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, et al. , Extensive translation of circular RNAs driven by N 6-methyladenosine. Cell Research, 2017. 27(5): p. 626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pamudurti NR, et al. , Translation of circRNAs. Molecular Cell, 2017. 66(1): p. 9–21. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Legnini I, et al. , Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Molecular Cell, 2017. 66(1): p. 22–37. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan WL, et al. , A landscape of circular RNA expression in the human heart. Cardiovascular Research, 2017. 113(3): p. 298–309. [DOI] [PubMed] [Google Scholar]
- 41.Salzman J, et al. , Cell-type specific features of circular RNA expression. PLoS Genetics, 2013. 9(9): p. e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji P, et al. , Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Reports, 2019. 26(12): p. 3444–3460. e5. [DOI] [PubMed] [Google Scholar]
- 43.Qu S, et al. , Circular RNA: a new star of noncoding RNAs. Cancer Letters, 2015. 365(2): p. 141–148. [DOI] [PubMed] [Google Scholar]
- 44.Dong X, et al. , Non-coding RNAs in cardiomyocyte proliferation and cardiac regeneration: Dissecting their therapeutic values. Journal of Cellular and Molecular Medicine, 2021. 25(5): p. 2315–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boon RA and Dimmeler S, MicroRNAs in myocardial infarction. Nature Reviews Cardiology, 2015. 12(3): p. 135. [DOI] [PubMed] [Google Scholar]
- 46.Ota A, et al. , Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Research, 2004. 64(9): p. 3087–3095. [DOI] [PubMed] [Google Scholar]
- 47.He L, et al. , A microRNA polycistron as a potential human oncogene. Nature, 2005. 435(7043): p. 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dews M, et al. , Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nature Genetics, 2006. 38(9): p. 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ventura A, et al. , Targeted deletion reveals essential and overlapping functions of the miR-17~ 92 family of miRNA clusters. Cell, 2008. 132(5): p. 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J, et al. , mir-17–92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circulation Research, 2013. 112(12): p. 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao F, et al. , Therapeutic role of miR-19a/19b in cardiac regeneration and protection from myocardial infarction. Nature Communications, 2019. 10(1): p. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang T, et al. , Loss of Phosphatase and Tensin Homolog Promotes Cardiomyocyte Proliferation and Cardiac Repair After Myocardial Infarction. Circulation, 2020. 142(22): p. 2196–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonauer A, et al. , MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science, 2009. 324(5935): p. 1710–1713. [DOI] [PubMed] [Google Scholar]
- 54.Hinkel R, et al. , Inhibition of microRNA-92a protects against ischemia/reperfusion injury in a large-animal model. Circulation, 2013. 128(10): p. 1066–1075. [DOI] [PubMed] [Google Scholar]
- 55.Rogg E-M, et al. , Analysis of cell type-specific effects of microRNA-92a provides novel insights into target regulation and mechanism of action. Circulation, 2018. 138(22): p. 2545–2558. [DOI] [PubMed] [Google Scholar]
- 56.Shi J, et al. , miR-17-3p contributes to exercise-induced cardiac growth and protects against myocardial ischemia-reperfusion injury. Theranostics, 2017. 7(3): p. 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Z, et al. , Serum extracellular vesicles promote proliferation of H9C2 cardiomyocytes by increasing miR-17–3p. Biochemical and Biophysical Research Communications, 2018. 499(3): p. 441–446. [DOI] [PubMed] [Google Scholar]
- 58.Qin X, et al. , microRNA-25 promotes cardiomyocytes proliferation and migration via targeting Bim. Journal of Cellular Physiology, 2019. 234(12): p. 22103–22115. [DOI] [PubMed] [Google Scholar]
- 59.Pan L, et al. , MiR-25 protects cardiomyocytes against oxidative damage by targeting the mitochondrial calcium uniporter. International Journal of Molecular Sciences, 2015. 16(3): p. 5420–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dirkx E, et al. , Nfat and miR-25 cooperate to reactivate the transcription factor Hand2 in heart failure. Nature Cell Biology, 2013. 15(11): p. 1282–1293. [DOI] [PubMed] [Google Scholar]
- 61.Wahlquist C, et al. , Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature, 2014. 508(7497): p. 531–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eulalio A, et al. , Functional screening identifies miRNAs inducing cardiac regeneration. Nature, 2012. 492(7429): p. 376–381. [DOI] [PubMed] [Google Scholar]
- 63.Lesizza P, et al. , Single-dose intracardiac injection of pro-regenerative microRNAs improves cardiac function after myocardial infarction. Circulation Research, 2017. 120(8): p. 1298–1304. [DOI] [PubMed] [Google Scholar]
- 64.von Gise A, et al. , YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proceeding of the National Academy of Sciences U S A, 2012. 109(7): p. 2394–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tian Y, et al. , A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Science Translational Medicine, 2015. 7(279): p. 279ra38–279ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heallen T, et al. , Hippo signaling impedes adult heart regeneration. Development, 2013. 140(23): p. 4683–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin Z, et al. , Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circulation Research, 2014. 115(3): p. 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Torrini C, et al. , Common regulatory pathways mediate activity of microRNAs inducing cardiomyocyte proliferation. Cell Reports, 2019. 27(9): p. 2759–2771. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gabisonia K, et al. , MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature, 2019. 569(7756): p. 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lim GB, MicroRNA-directed cardiac repair after myocardial infarction in pigs. Nature Reviews Cardiology, 2019. 16(8): p. 454–455. [DOI] [PubMed] [Google Scholar]
- 71.Katare R, et al. , Transplantation of human pericyte progenitor cells improves the repair of infarcted heart through activation of an angiogenic program involving micro-RNA-132. Circulation Research, 2011. 109(8): p. 894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hong S, et al. , Na+–Ca2+ exchanger targeting miR-132 prevents apoptosis of cardiomyocytes under hypoxic condition by suppressing Ca2+ overload. Biochemical and Biophysical Research Communications, 2015. 460(4): p. 931–937. [DOI] [PubMed] [Google Scholar]
- 73.Ucar A, et al. , The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nature Communications, 2012. 3(1): p. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Foinquinos A, et al. , Preclinical development of a miR-132 inhibitor for heart failure treatment. Nature Communications, 2020. 11(1): p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Täubel J, et al. , Novel antisense therapy targeting microRNA-132 in patients with heart failure: results of a first-in-human Phase 1b randomized, double-blind, placebo-controlled study. European Heart Journal, 2021. 42(2): p. 178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Batkai S, et al. , CDR132L improves systolic and diastolic function in a large animal model of chronic heart failure. European Heart Journal, 2021. 42(2): p. 192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Small EM, Frost RJ, and Olson EN, MicroRNAs add a new dimension to cardiovascular disease. Circulation, 2010. 121(8): p. 1022–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hullinger TG, et al. , Inhibition of miR-15 protects against cardiac ischemic injury. Circulation Research, 2012. 110(1): p. 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nishi H, et al. , MicroRNA-15b modulates cellular ATP levels and degenerates mitochondria via Arl2 in neonatal rat cardiac myocytes. Journal of Biological Chemistry, 2010. 285(7): p. 4920–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao M, et al. , Cyclin D2 Overexpression Enhances the Efficacy of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Myocardial Repair in a Swine Model of Myocardial Infarction. Circulation, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bellera N, et al. , Single intracoronary injection of encapsulated antagomir-92a promotes angiogenesis and prevents adverse infarct remodeling. Journal of the American Heart Association, 2014. 3(5): p. e000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gallant-Behm CL, et al. , A synthetic microRNA-92a inhibitor (MRG-110) accelerates angiogenesis and wound healing in diabetic and nondiabetic wounds. Wound Repair and Regeneration, 2018. 26(4): p. 311–323. [DOI] [PubMed] [Google Scholar]
- 83.Ratti M, et al. , MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as new tools for cancer therapy: first steps from bench to bedside. Targeted Oncology, 2020. 15: p. 261–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qiao L, et al. , microRNA-21–5p dysregulation in exosomes derived from heart failure patients impairs regenerative potential. The Journal of clinical investigation, 2019. 129(6): p. 2237–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li Y, et al. , Injectable hydrogel with MSNs/microRNA-21-5p delivery enables both immunomodification and enhanced angiogenesis for myocardial infarction therapy in pigs. Sci Adv, 2021. 7(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng HS, et al. , MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol Med, 2013. 5(7): p. 1017–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liao Y, et al. , Therapeutic silencing miR-146b-5p improves cardiac remodeling in a porcine model of myocardial infarction by modulating the wound reparative phenotype. Protein Cell, 2021. 12(3): p. 194–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ponnusamy M, et al. , Long noncoding RNA CPR (cardiomyocyte proliferation regulator) regulates cardiomyocyte proliferation and cardiac repair. Circulation, 2019. 139(23): p. 2668–2684. [DOI] [PubMed] [Google Scholar]
- 89.Ounzain S, et al. , CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. Journal of Molecular and Cellular Cardiology, 2015. 89: p. 98–112. [DOI] [PubMed] [Google Scholar]
- 90.Cai B, et al. , The Long Noncoding RNA CAREL Controls Cardiac Regeneration. Journal of the American College of Cardiology, 2018. 72(5): p. 534–550. [DOI] [PubMed] [Google Scholar]
- 91.Ma W, Hua B, and Cai B, Reply: The Effect of Targeting CAREL on Cardiac Regeneration. Journal of the American College of Cardiology, 2018. 72(21): p. 2684–2684. [DOI] [PubMed] [Google Scholar]
- 92.Chen G, et al. , Loss of long non-coding RNA CRRL promotes cardiomyocyte regeneration and improves cardiac repair by functioning as a competing endogenous RNA. Journal of Molecular and Cellular Cardiology, 2018. 122: p. 152–164. [DOI] [PubMed] [Google Scholar]
- 93.Wang J, et al. , A long noncoding RNA NR_045363 controls cardiomyocyte proliferation and cardiac repair. Journal of Molecular and Cellular Cardiology, 2019. 127: p. 105–114. [DOI] [PubMed] [Google Scholar]
- 94.Chen X, et al. , The long noncoding RNA NR_045363 involves cardiomyocyte apoptosis and cardiac repair via p53 signal pathway. Cell Biology International, 2020. 44(9): p. 1957–1965. [DOI] [PubMed] [Google Scholar]
- 95.Li X, et al. , Loss of AZIN2 splice variant facilitates endogenous cardiac regeneration. Cardiovascular Research, 2018. 114(12): p. 1642–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li X, et al. , Inhibition of AZIN2-sv induces neovascularization and improves prognosis after myocardial infarction by blocking ubiquitin-dependent talin1 degradation and activating the Akt pathway. eBioMedicine, 2019. 39: p. 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ren L, et al. , Downregulation of long non-coding RNA nuclear enriched abundant transcript 1 promotes cell proliferation and inhibits cell apoptosis by targeting miR-193a in myocardial ischemia/reperfusion injury. BMC Cardiovascular Disorders, 2019. 19(1): p. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang N, et al. , The long non-coding RNA SNHG1 attenuates cell apoptosis by regulating miR-195 and BCL2-like protein 2 in human cardiomyocytes. Cellular Physiology and Biochemistry, 2018. 50(3): p. 1029–1040. [DOI] [PubMed] [Google Scholar]
- 99.Chen H, Xia W, and Hou M, LncRNA-NEAT1 from the competing endogenous RNA network promotes cardioprotective efficacy of mesenchymal stem cell-derived exosomes induced by macrophage migration inhibitory factor via the miR-142-3p/FOXO1 signaling pathway. Stem Cell Research & Therapy, 2020. 11(1): p. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu HJ, et al. , Long non-coding RNA NEAT1 modulates hypoxia/reoxygenation-induced cardiomyocyte injury via targeting microRNA-520a. Experimental and Therapeutic Medicine, 2019. 18(3): p. 2199–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Su Q, et al. , LncRNA TUG1 mediates ischemic myocardial injury by targeting miR-132-3p/HDAC3 axis. American Journal of Physiology: Heart and Circulatory Physiology, 2020. 318(2): p. H332–H344. [DOI] [PubMed] [Google Scholar]
- 102.Chen Y, et al. , LncRNA SNHG1 Elicits ITS Heart Regeneration Through PTEN/AKT/MYC Positive Feedback Loop. Journal of the American College of Cardiology, 2019. 73(9S1): p. 686–686. [Google Scholar]
- 103.Chen Y, et al. , Long non-coding RNA ECRAR triggers post-natal myocardial regeneration by activating ERK1/2 signaling. Molecular Therapy, 2019. 27(1): p. 29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cai B, et al. , Long Noncoding RNA–DACH1 (Dachshund Homolog 1) Regulates Cardiac Function by Inhibiting SERCA2a (Sarcoplasmic Reticulum Calcium ATPase 2a). Hypertension, 2019. 74(4): p. 833–842. [DOI] [PubMed] [Google Scholar]
- 105.Cai B, et al. , Targeting LncDACH1 promotes cardiac repair and regeneration after myocardium infarction. Cell Death & Differentiation, 2020. 27(7): p. 2158–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Katayama S, et al. , Antisense transcription in the mammalian transcriptome. Science, 2005. 309(5740): p. 1564–1566. [DOI] [PubMed] [Google Scholar]
- 107.Wenric S, et al. , Transcriptome-wide analysis of natural antisense transcripts shows their potential role in breast cancer. Sci Rep, 2017. 7(1): p. 17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li B, et al. , Sirt1 antisense long noncoding RNA promotes cardiomyocyte proliferation by enhancing the stability of Sirt1. Journal of the American Heart Association, 2018. 7(21): p. e009700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li H, et al. , The role of a novel long noncoding RNA TUC40-in cardiomyocyte induction and maturation in P19 cells. The American journal of the medical sciences, 2017. 354(6): p. 608–616. [DOI] [PubMed] [Google Scholar]
- 110.Yang Y-J, et al. , Atorvastatin treatment improves survival and effects of implanted mesenchymal stem cells in post-infarct swine hearts. European Heart Journal, 2008. 29(12): p. 1578–1590. [DOI] [PubMed] [Google Scholar]
- 111.Huang P, et al. , Atorvastatin enhances the therapeutic efficacy of mesenchymal stem cells-derived exosomes in acute myocardial infarction via up-regulating long non-coding RNA H19. Cardiovascular Research, 2020. 116(2): p. 353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Keniry A, et al. , The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nature Cell Biology, 2012. 14(7): p. 659–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Y, et al. , The long non-coding RNA H19 promotes cardiomyocyte apoptosis in dilated cardiomyopathy. Oncotarget, 2017. 8(17): p. 28588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tao H, et al. , Long noncoding RNA H19 controls DUSP5/ERK1/2 axis in cardiac fibroblast proliferation and fibrosis. Cardiovascular Pathology, 2016. 25(5): p. 381–389. [DOI] [PubMed] [Google Scholar]
- 115.Huang Z-W, et al. , Long noncoding RNA H19 acts as a competing endogenous RNA to mediate CTGF expression by sponging miR-455 in cardiac fibrosis. DNA and Cell Biology, 2017. 36(9): p. 759–766. [DOI] [PubMed] [Google Scholar]
- 116.Suzuki G, et al. , Autologous mesenchymal stem cells mobilize cKit+ and CD133+ bone marrow progenitor cells and improve regional function in hibernating myocardium. Circulation Research, 2011. 109(9): p. 1044–1054. [DOI] [PubMed] [Google Scholar]
- 117.Zhao Y, et al. , Mesenchymal stem cell transplantation improves regional cardiac remodeling following ovine infarction. Stem Cells Translational Medicine, 2012. 1(9): p. 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li S-H, et al. , Reconstitution of aged bone marrow with young cells repopulates cardiac-resident bone marrow-derived progenitor cells and prevents cardiac dysfunction after a myocardial infarction. European Heart Journal, 2013. 34(15): p. 1157–1167. [DOI] [PubMed] [Google Scholar]
- 119.Li J, et al. , Young bone marrow Sca-1 cells rejuvenate the aged heart by promoting epithelial-to-mesenchymal transition. Theranostics, 2018. 8(7): p. 1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li J, et al. , Long-term repopulation of aged bone marrow stem cells using young Sca-1 cells promotes aged heart rejuvenation. Aging Cell, 2019. 18(6): p. e13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dong J, et al. , Down-Regulation of Lnc-CYP7A1-1 Rejuvenates Aged Human Mesenchymal Stem Cells to Improve Their Efficacy for Heart Repair Through SYNE1. Frontiers in Cell and Developmental Biology, 2020. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ishii N, et al. , Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. Journal of Human Genetics, 2006. 51(12): p. 1087–1099. [DOI] [PubMed] [Google Scholar]
- 123.Qu X, et al. , MIAT is a pro-fibrotic long non-coding RNA governing cardiac fibrosis in post-infarct myocardium. Scientific Reports, 2017. 7(1): p. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen L, et al. , Targeting MIAT reduces apoptosis of cardiomyocytes after ischemia/reperfusion injury. Bioengineered, 2019. 10(1): p. 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ouyang F, et al. , Long non-coding RNA RNF7 promotes the cardiac fibrosis in rat model via miR-543/THBS1 axis and TGFβ1 activation. Aging (Albany NY), 2020. 12(1): p. 996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang H, et al. , Long non-coding RNA XIST promotes the proliferation of cardiac fibroblasts and the accumulation of extracellular matrix by sponging microRNA-155-5p. Experimental and Therapeutic Medicine, 2021. 21(5): p. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lu J, et al. , Long noncoding RNA GAS5 attenuates cardiac fibroblast proliferation in atrial fibrillation via repressing ALK5. European Reviews for Medical and Pharmacological Sciences, 2019. 23(17): p. 7605–7610. [DOI] [PubMed] [Google Scholar]
- 128.Wang L and Zhang J, Exosomal lncRNA AK139128 derived from hypoxic cardiomyocytes promotes apoptosis and inhibits cell proliferation in cardiac fibroblasts. International Journal of Nanomedicine, 2020. 15: p. 3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang S, et al. , Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation, 2019. 139(25): p. 2857–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang K, et al. , Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death & Differentiation, 2017. 24(6): p. 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Garikipati VNS, et al. , Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nature Communications, 2019. 10(1): p. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bai M, et al. , CircHIPK3 aggravates myocardial ischemia-reperfusion injury by binding to miRNA-124–3p. European Reviews for Medical and Pharmacological Sciences, 2019. 23(22): p. 10107–10114. [DOI] [PubMed] [Google Scholar]
- 133.Si X, et al. , circRNA Hipk3 Induces Cardiac Regeneration after Myocardial Infarction in Mice by Binding to Notch1 and miR-133a. Mol Ther Nucleic Acids, 2020. 21: p. 636–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang M, et al. , Circular RNA (circRNA) CDYL induces myocardial regeneration by ceRNA after myocardial infarction. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research, 2020. 26: p. e923188–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen P, et al. , Circular RNA MGAT1 regulates cell proliferation and apoptosis in hypoxia-induced cardiomyocytes through miR-34a/YAP1 axis. International Journal of Clinical and Experimental Pathology, 2020. 13(10): p. 2474. [PMC free article] [PubMed] [Google Scholar]
- 136.Li M, et al. , A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics, 2018. 8(21): p. 5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Geng H-H, et al. , The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PloS One, 2016. 11(3): p. e0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen C, et al. , The Circular RNA CDR1as Regulates the Proliferation and Apoptosis of Human Cardiomyocytes Through the miR-135a/HMOX1 and miR-135b/HMOX1 Axes. Genetic Testing and Molecular Biomarkers, 2020. 24(9): p. 537–548. [DOI] [PubMed] [Google Scholar]
- 139.Cai L, et al. , Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. Journal of Molecular and Cellular Cardiology, 2019. 130: p. 10–22. [DOI] [PubMed] [Google Scholar]
- 140.Zeng Y, et al. , A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics, 2017. 7(16): p. 3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang Q, et al. , The circular RNA hsa_circ_0007623 acts as a sponge of microRNA-297 and promotes cardiac repair. Biochemical and Biophysical Research Communications, 2020. 523(4): p. 993–1000. [DOI] [PubMed] [Google Scholar]
- 142.Li C-Y, Ma L, and Yu B, Circular RNA hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells proliferation and angiogenesis. Biomedicine & Pharmacotherapy, 2017. 95: p. 1514–1519. [DOI] [PubMed] [Google Scholar]
- 143.Zhu Y, et al. , Upregulation of circular RNA circNFIB attenuates cardiac fibrosis by sponging miR-433. Frontiers in Genetics, 2019. 10: p. 564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sun L.y., et al. , Circ_LAS1L regulates cardiac fibroblast activation, growth, and migration through miR-125b/SFRP5 pathway. Cell Biochemistry and Function, 2020. 38(4): p. 443–450. [DOI] [PubMed] [Google Scholar]


