Abstract
Cancer is driven by both genetic aberrations in the tumor cells and fundamental changes in the tumor microenvironment (TME). These changes offer potential targets for novel therapeutics, yet lack of in vitro 3D models recapitulating this complex microenvironment impedes such progress. Here, we generated several tumor-stroma scaffolds reflecting the dynamic in vivo breast TME, using a high throughput microfluidic system. Alginate (Alg) or alginate-alginate sulfate (Alg/Alg-S) hydrogels were used as ECM-mimics, enabling the encapsulation and culture of tumor cells, fibroblasts and immune cells (macrophages and T cells, of the innate and adaptive immune systems, respectively). Specifically, Alg/Alg-S was shown capable of capturing and presenting growth factors and cytokines with binding affinity that is comparable to heparin. Viability and cytotoxicity were shown to strongly correlate with the dynamics of cellular milieu, as well as hydrogel type. Using on-chip immunofluorescence, production of reactive oxygen species and apoptosis were imaged and quantitatively analyzed. We then show how macrophages in our microfluidic system were shifted from a proinflammatory to an immunosuppressive phenotype when encapsulated in Alg/Alg-S, reflecting in vivo TME dynamics. LC-MS proteomic profiling of tumor cells sorted from the TME scaffolds revealed upregulation of proteins involved in cell-cell interactions and immunomodulation in Alg/Alg-S scaffolds, correlating with in vivo findings and demonstrating the appropriateness of Alg/Alg-S as an ECM biomimetic. Finally, we show the formation of large tumor-derived vesicles, formed exclusively in Alg/Alg-S scaffolds. Altogether, our system offers a robust platform for quantitative description of the breast TME that successfully recapitulates in vivo patterns.
Keywords: biomaterials, tumor microenvironment, breast cancer, microfluidics, 3D disease models, proteomic profiling
Statement of significance
Cancer progression is driven by profound changes in both tumor cells and surrounding stroma. Here, we present a high throughput microfluidic system for the generation and analysis of dynamic tumor-stroma scaffolds, that mimic the complex in vivo TME cell proportions and compositions, constructing robust in vitro models for the study of the TME. Utilizing Alg/Alg-S as a bioinspired ECM, mimicking heparin’s in vivo capabilities of capturing and presenting signaling molecules, we show how Alg/Alg-S induces complex in vivo-like responses in our models. Alg/Alg-S is shown here to promote dynamic protein expression patterns, that can serve as potential therapeutic targets for breast cancer treatment. Formation of large tumor-derived vesicles, observed exclusively in the Alg/Alg-S scaffolds suggests a mechanism for tumor survival.
Graphical Abstract
1. Introduction
Considerable evidence has shown cancer is a systematic disease, driven not only by genetic abnormalities in cancer cells, but by profound changes in the surrounding stroma [1]. Cell-cell and cell-matrix interactions in the tumor microenvironment (TME) are viewed as pivotal for tumor progression [1,2]. The TME in breast cancer is of particular interest, as mechanisms tightly regulating normal functions in the tissue [3,4] are hijacked during cancer progression, resulting in an immunosuppressive and apoptosis-resistant TME [5].
Predominant players in the breast TME include leukocytes, fibroblasts and the extracellular matrix (ECM) [6–8]. Leukocytes account for up to 50% of the tumor mass, primarily macrophages and lymphocytes [6]. Immune cells phenotype in breast cancer were shown to shift with tumor progression, from anti-tumor proinflammatory in early tumor development to immunosuppressive tumor-promoting in progressed tumors [9,10]. Early tumor development is characterized by persistent inflammation, with presence of cytotoxic T cells and M1 macrophages, gradually transitioning into a tumor-tolerant environment, marked by immunosuppressive tumor-associated M2-like macrophages and anergic T cells [9,10]. Fibroblasts, in turn, secrete growth factors and cytokines that orchestrate tumor survival and progression [7]. Finally, the breast cancer extracellular matrix (ECM) was shown to not only provide structural support but to mediate cell-cell and cell-matrix interactions that promote tumor progression [8]. Increased stiffness in breast cancer is associated with ECM remodeling and crosslinking, higher integrin signaling, tumor cell proliferation, reduced responsiveness to chemotherapy and poor prognosis [11].
3D in vitro models offer a platform that can accurately recapitulate the human TME [12], and were shown to result in gene expression patterns that are comparable with in vivo patterns [13,14]. Yet, current 3D culture techniques are of low throughput or offer little control over cell distribution uniformity or spheroid size, restricting the study of the dynamic breast TME [15–19]. Microfluidic 3D systems have emerged in the last decade as valuable tools for the study of the TME. Recent advances in 3D microfluidics enabled the creation of advanced tumor-on-chip [20,21] and organ-on-chip models [22–24], as well as models for migration, extravasation, and immunotherapy evaluation [25–30]. However, these models are not suitable for the high throughput generation of tumor-immune 3D models. Ex vivo microfluidic models of murine and human patients were also reported [31,32]. While these systems offer recapitulation of the in vivo tumor, low reproducibility of spheroid size and cell composition render these models unsuitable for high throughput study.
Here, we present a microfluidic system for high throughput generation, culture, live-imaging and on-chip analysis of human tumor-stroma scaffolds. Using bio-inspired hydrogels, we applied this platform to study the dynamic microenvironment of breast tumor. We generated two human TME scaffold (hTMES) models: (i) Proinflammatory human tumor microenvironment scaffolds (P-hTMES), characteristic of the anti-tumor, proinflammatory breast TME of early tumor development; and (ii) Immunosuppressive human tumor microenvironment scaffolds (I-hTMES), characteristic of the tumor-tolerant breast TME in progressed tumors. The cellular milieu in the models generated in this study follows the tumor and stromal cell proportions and activation states that occur in the in vivo breast TME and include principal cells of the in vivo breast TME, namely macrophages, T cells, fibroblasts and epithelial cells. As ECM, hydrogels made of pristine alginate (Alg) or alginate-alginate sulfate (Alg/Alg-S) were used. Alg has been extensively used for 3D models and biomedical applications [33–37]. Its elasticity is readily adjusted to mimic the in vivo characteristics of the tissue studied, and is biocompatible and immunologically inert [36]. Alg-S is an alginate with the uronic acids sulfated [38], resulting in a heparin/heparan mimic that promotes cell adhesion, tethering and presentation of growth factors and cytokines with equilibrium binding constants that are comparable or higher than those observed with heparin [38], and induce in vivo-like responses [39]. In this study, we demonstrate how the use of Alg/Alg-S hydrogel as an ECM-mimic results in the induction of vivo-like responses.
2. Experimental
2.1. Microfluidic device design and fabrication
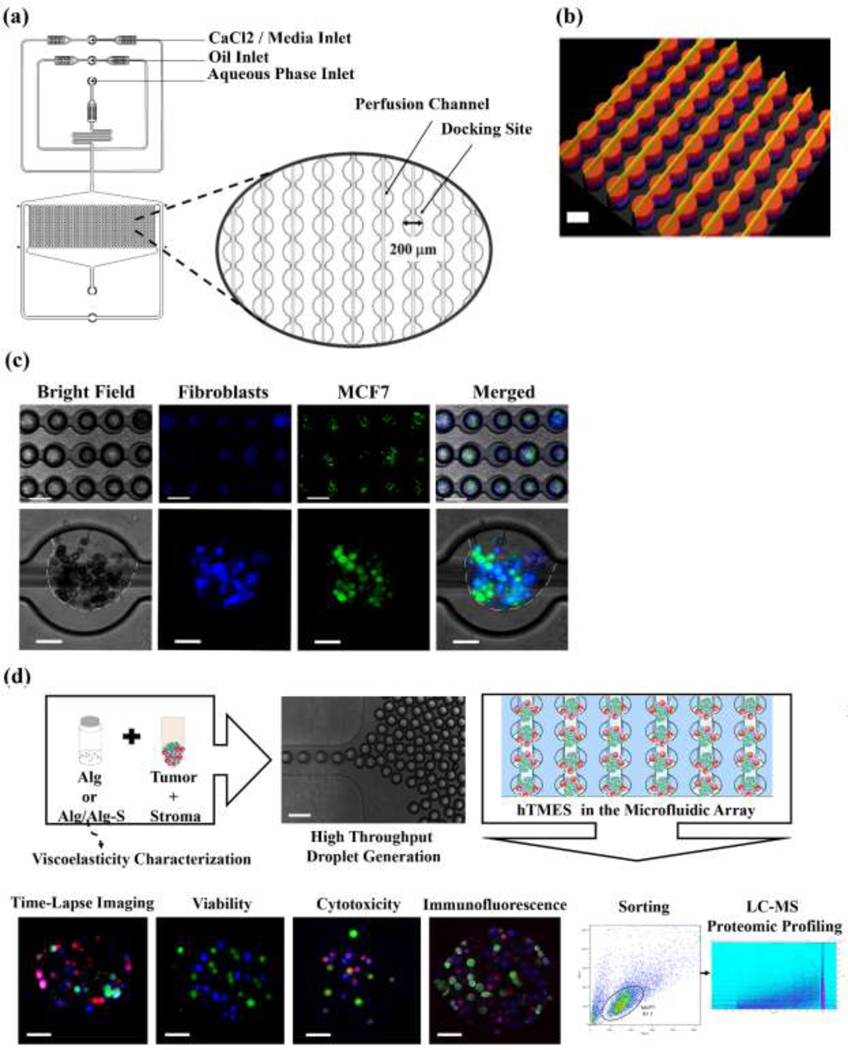
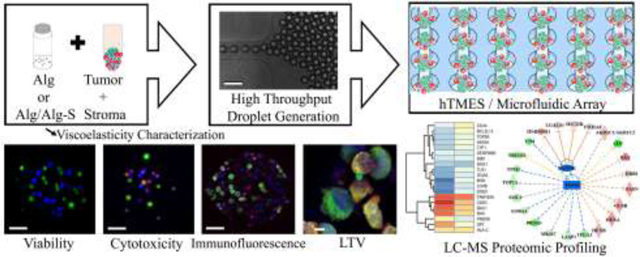
The microfluidic device design is shown in Fig. 1. Fabrication was conducted using standard soft lithography techniques previously described [21,40]. Briefly, a 4”silicon wafer (University wafer, MA) was spin-coated with SU-8 2075 (Microchem, MA) to achieve 120 μm thickness of the array layer, and 40 μm thickness for the upper layer (perfusion channels). Structure integrity and channel dimensions were analyzed using 3D optical profilometer (Bruker, courtesy of Dr. Xiao Wang and Prof. Daniel Irimia, Massachusetts General Hospital, Boston, MA). Droplet generation occurs at a flow-focusing junction between the oil phase and the aqueous phase, with the aqueous includes cells encapsulated in partially crosslinked hydrogel. Generated droplets are trapped in an array composed of 1,000 docking sites, 200 μm in diameter each, arranged in a matrix of 40 rows and 25 columns. An additional inlet is then used for hydrogel crosslinking and media perfusion.
Fig. 1. Microfluidic device design, droplet generation and study work flow.
(a) Schematic representation of the microfluidic design used in this study, with magnification of the array docking sites. (b) Interferometry imaging of the 2-layer fabricated master. (c) Droplet generation, with MCF7 cells labeled with CFSE (green) and CCD1129SK human mammary fibroblasts labeled with CMAC (blue). Upper pannel: 5X magnification, after droplet formation; lower pannel: 20X maginification, after hydrogel crosslinking and oil removal. In the lower panel, the edges of the hydrogel are highlighted in white in the bright field and the merged images. (d) Study workflow. Scaffolds were generated by mixing the cells in a partially cross-linked alginate, and infused into the device for droplet generation and final cross-linking. The scaffolds were then maintained by constant media perfusion, analyzed for viablity, Doxorubicin cytotoxicty, immunofluorescence, and finally collected and sorted for LC-MS proteomic profiling.Scale bar in ((b), and (c),(d) upper panels is 200 μm; scale bar in (c), (d) lower panels is 50 μm.
2.2. Cell culture
MCF7 human breast adenocarcinoma cells, CCD-1129SK human fibroblasts derived from breast tissue removed at bilateral mastectomy, and SC human monocyte/macrophage cells were purchased from American Type Culture Collection (ATCC HTB-22, ATCC CRL-2575, ATCC CRL-9855, respectively, ATCC, USA), and maintained according to manufacturer’s instructions. MCF10a non-tumorigenic human mammary epithelial cell line were generous courtesy of Prof. Vladimir Torchilin of the Department of Pharmaceutical Sciences (Northeastern University, Boston, MA), and maintained according to ATCC instructions. SC were polarized towards an M1 or an M2 macrophage phenotype according to previously published protocols [41,42]. Peripheral blood mononuclear cells (PBMC, Astarte Biologics, WA) were used for T cell collection, expansion and activation, according to a previously published protocol [43]. See Appendix SI, Supplementary Methods for detailed procedure. Activated macrophages and T cells were routinely analyzed for previously established activation markers [41,44,45] by flow cytometry. See Appendix SI, Supplementary Methods for detailed procedure. P-hTMES and I-hTMES were maintained in IMDM media supplemented with 10% FBS, 1% antibiotic-antimycotic, 10 μg x mL−1 human recombinant insulin, 0.05 mM 2-mercaptoethanol, 0.1 mM hypoxanthine (Sigma-Aldrich) and 0.016 mM thymidine, following media optimization experiments (data not shown).
2.3. Rheological characterization
The viscoelastic properties and the elastic modulus of 1% Alg (Pronova LVG up, Dupont, Norway) and Alg/Alg-S, composed of 1% alginate (w/v) containing 10% Alg-S (as percentage of total alginate, w/w, prepared in-house by the group of Prof. Smadar Cohen) were analyzed as previously published [46]. See Appendix SI, Supplementary Methods for a detailed procedure.
2.4. On-chip generation of Alg and Alg/Alg-S TME scaffolds
Alg or Alg/Alg-S hydrogels were prepared by partially cross-linking 4% (w/v) pristine alginate or 4% alginate premixed with 10% alginate sulfate (w/w of total polymer), respectively, with equal volume of 0.8% (w/v) D-Gluconic acid calcium salt (Sigma-Aldrich, MO) by constant stirring at RT for 3 hr. For cell encapsulation and droplet formation, cells and alginate were mixed at equal volumes, resulting in a 1% cell-laden alginate hydrogel, containing 1E7 cells x mL-1. The cells in the partially cross-linked alginate were infused into the device via the aqueous phase inlet at 150 μL x hr-1. Droplets were generated in the flow-focusing junction by flow of oil (mineral oil with 2% Span-80 as surfactant, Sigma-Aldrich) through the continuous phase inlet, at 700 μL x hr-1. After sufficient filling of the array, the infusion of oil and aqueous phase were stopped, and media, supplemented with 150 mM calcium chloride (Sigma-Aldrich) was briefly infused through a third inlet to allow full cross-linking of the hydrogel and removal of oil. For the remaining duration of the experiment, this inlet was used for constant media perfusion of the scaffolds. For the generation of MCF7 scaffolds (MCF7-S), 1E7 cells x mL−1 MCF7 cells were collected and mixed with the hydrogel and scaffolds were generated as previously described. MCF7 and non-immune stroma scaffolds (MCF7-EF-S) were composed of MCF7 cells, MCF10a epithelial cells and CCD1129SK fibroblasts. For the generation of MCF7-EF-S, MCF7 cells, CCD1129SK fibroblasts, and MCF10a epithelial cells were mixed at 3:2 MCF7 to stroma, respectively, with the hydrogel, resulting in 1E7 cells x mL−1 total cell density. On-chip scaffold was proceeded as previously described. For the generation of P-hTMES or I-hTMES, MCF7 cells, MCF10a epithelial cells, CCD-1129SK fibroblasts, SC monocytes following M1 or M2 activation, respectively, and activated or non-activated T cells, respectively, were mixed at 4:4:2 ratio, MCF7 to immune to non-immune stroma, respectively, to reflect the cell ratios in the breast TME reported in vivo [9,47]. The cells were mixed with the hydrogel, resulting in total of 1E7 cells x mL−1, and on-chip scaffold generation was proceeded as previously described. Cells were routinely labeled with live-cell trackers CFSE, CMAC or CMPTX (all purchased from Invitrogen, MA), according to manufacturer’s instructions prior to scaffold generation. All TME scaffold types were maintained with constant media perfusion at 50 μL x hr−1, incubated at 37°C, in a 5% CO2 humidified controlled environment.
2.5. On-chip viability and cytotoxicity assays
Cell viability was monitored following 24 hr and 96 hr culture by perfusion of complete media supplemented with 2 μM ethidium homodimer (EtHD, Invitrogen) as a marker for dead cells. TME scaffolds were imaged by epifluorescence microscopy (Zeiss, Germany). MCF7 cell viability was analyzed in ≥ 50 scaffolds for each TME scaffold type (n=3). Viability was expressed as percentage of live MCF7 cells (out of total MCF7 cells per scaffold) using ImageJ. For analysis of cytotoxicity in 2D culture, MCF7 cells were seeded at 1E5 cells x mL−1 in 96 micro-well plates (Thermo Fisher Scientific, MA) and incubated with complete media supplemented with 0, 5 or 10 μM Doxorubicin (Dox, Abcam, MA) for 24 hr. The cells were then were washed twice and cell viability analyzed by Presto Blue (Invitrogen), according to manufacturer’s instructions using Synergy HT plate reader (BioTek, VT). Viability was calculated for triplets for each Dox concentration (n=3). For analysis of cytotoxicity in MCF7-S, MCF7-EF-S, and I-hTMES, the TME scaffolds were perfused with complete media containing 0, 5 or 10 μM Dox for 24 hr, and then washed by 2 hr perfusion with complete media. Cell viability was monitored before and after Dox treatment, by perfusion of complete media supplemented with 2 μM EtHD, imaged by Zeiss epifluorescence microscopy. MCF7 cell viability was analyzed in ≥ 50 scaffolds for each TME scaffold type and Dox concentration (n=3), expressed as percentage of live cells using ImageJ.
2.6. On-chip Immunofluorescence and Confocal imaging
On-chip immunofluorescence in all experiments except for HLA-DR expression, was preceded by labeling of MCF7 cells with live cell-tracker CMAC prior to encapsulation in the Alg/Alg-S hydrogel. MCF7-S, P-hTMES and I-hTMES scaffolds were generated on-chip as previously described. TME scaffolds were fixed on-chip using fixation buffer (BD Biosciences, CA) perfused at 10 μL x hr−1 for 1 hr at RT, permeabilized by perfusion with 0.1% Triton X-100 (Sigma-Aldrich) in DMEM for 30 min at RT at 10 μL x hr−1, and blocked using 3% BSA in DMEM perfused at 10 μL x hr−1 for 3 hr. Immunolabeling by primary and secondary antibodies was then conducted by sequential perfusion in DMEM at 10 μL x hr−1 for 1 hr at 4°C, followed by overnight incubation at 4°C. Each immunolabeling step was followed by a short wash by pristine DMEM perfusion at10 μL x hr-1. The following antibodies used were purchased from BioLegend: anti-CD3 mAb (mouse anti-human, conjugated with Alexa 594, 1:100); anti-CD86 mAb (mouse anti-human, conjugated with Alexa 488, 1:100); anti-CD163 mAb (rat anti-human, conjugated with Alexa fluor 647, 1:50); and anti-HLA-DR mAb (mouse anti-human, conjugated with Alexa 488, 1:50). The following antibodies used were purchased from Invitrogen: anti-CD86 (rabbit anti-human, 1:100) as primary antibody, followed by labeling with a secondary antibody (goat anti-rabbit, conjugated wit FITC); anti-IL-10 mAb (rat anti-human, conjugated with eFluor 660, 1:50); anti-E-cadherin mAb (rat anti-human, conjugated with eFluor 660, 1:50). Cell nuclei were labeled using Hoechst33342 (1:200, BD Biosciences). For imaging of apoptosis, the TME scaffolds were perfused with Annexin V (conjugated with Alexa 594, Invitrogen) and CellEvent caspase 3/7 green detection reagent (Invitrogen) at 20 μl x mL−1 for 1 hr. Flow rate was then reduced to 5 μL x hr−1 for 2 hr to allow binding. The TME scaffolds were then washed by perfusion with pristine media at 40 μl x mL−1 for 1 hr and then fixed as previously described. Reactive oxygen species (ROS) were labeled by perfusion of the TME scaffolds with complete media supplemented with cell-permeable CellROX green reagent (Invitrogen) at 20 μl x hr−1 for 1 hr. Access reagent was washed by perfusion with pristine media. The TME scaffolds were then fixed as previously described. Imaging was conducted using Zeiss LSM 880 confocal microscopy, using 20X or 40X magnification, 2 μm Z-stacking. ROS production, annexin V binding or protein expression were quantitatively analyzed as fluorescence intensity vs. background using ImageJ.
2.7. On-chip hydrogel dissolution and MCF7 cell sorting
Preceding TME scaffolds generation, MCF7 cells were labeled with live-cell tracker CFSE. On-chip generation of Alg or Alg/Alg-S MCF7-S and P-hTMES was conducted as previously described (n=3). The TME scaffolds were cultured for 24 hr with media perfusion. For cell collection, the TME scaffolds were perfused with complete media containing 4% (w/v) citrate and 5 mM Ethylenediaminetetraacetic acid (EDTA, Sigma-Aldrich) at 5 μl x hr-1. After removal from the device the cells were washed twice with ice-cold PBS. CFSE-labeled MCF7 cells were sorted using Beckman Coulter MoFlo AstriosEQ 4 laser system sorter (The Flow Cytometry Core Facility, Beth Israel Deaconess Medical Center, Boston, MA).
2.8. Preparation of cell lysate, protein digestion and LC-MS/MS analysis
The overall sample processing workflow followed a previously published on-micro SPE tip protocol [48]. For detailed procedure see SI Appendix, Supplementary methods.
2.9. Data and bioinformatics analysis
The Orbitrap Fusion Lumos acquired mass spectrometry data were analyzed with ProteomeDiscoverer (v.2.2, Thermo Fisher Scientific) and searched against a human protein database (UniProt, released March 13, 2018, containing 20,302 reviewed sequences) combined with common contaminant proteins. Orbitrap-detected HCD spectra were searched using the Sequest HT search algorithm with 15 ppm and 0.04 Da mass tolerance for precursor and fragment ions, respectively. The database search was limited to only fully tryptic peptides, with up to two missed cleavage sites per peptide in the search, and the minimum peptide sequence length was set to 5 amino acid residues. Carbamidomethylation of cysteine residues was set as a static modification. N-terminal protein acetylation, methionine oxidation, deamidation on asparagine/glutamine and lysine acetylation were selected as dynamic modifications. For protein identifications, at least one high confidence peptide sequence was required, and a target 1% false discovery rate (FDR) was used for protein identification. Protein quantitation was performed through label-free quantification (LFQ) analysis using ProteomeDiscoverer (v. 2.2, Thermo Fisher Scientific). The FDR ≤ 1% was applied for all peptide-level identifications. Both, unique and razor peptides were used for quantification. Precursor abundance was calculated based on intensity and a normalization based on total peptide amount was used for all samples. Protein ratio calculation was done by summed abundance and a replicate-based resampling was used for LFQ intensity imputation. For differential expression of proteins, the fold-change (FC) was defined as the ratio of replicate averaged normalized LFQ intensities for the test and control case, respectively. Log2-ratios for proteins, were median centered to zero. Positive Log2(FC) values denote highly upregulated proteins in Alg/Alg-S MCF7-S and Alg P-hTMES, in the Alg/Alg-S MCF7-S versus P-hTMES and Alg P-hTMES versus Alg/Alg-S P-hTMES datasets, respectively, and vice versa. The proteomic data set containing Entrez Gene identifiers and fold changes of identified protein was analyzed by the Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, USA). Core analysis with indirect and direct relationships, proteins interactions and pathways, upstream regulatory analysis and functional network identifications were carried out as per manufacturer’s instructions using the human database (Qiagen Inc) [49]. Activation or inhibition of cell functions and upstream regulators were analyzed using IPA, with Z-score ≥ |2| considered statistically significant. The proteomic data set containing UniProtKB accession numbers and fold changes of identified proteins was also analyzed by the StringDB database of known and predicted protein-protein interactions [50]. Core analysis of proteins interactions and pathways (using Gene Ontology biological processes annotation) was conducted, with FDR < 0.05.
2.10. Image acquisition and processing
Imaging was acquired by epifluorescence microscope (Axio Observer.Z1 Microscope, equipped with Hamamatsu digital camera C10600 Orca-R2, Zeiss, Germany) using 5X and 20X objectives, and 2-μm z-stacking. Confocal microscopy (LSM 880 microscope, Zeiss, Germany) was conducted using 20X or 40X magnification, 2-μm z-stacking. Images were processed using Zen (Blue edition, Zeiss, Germany) or Imaris (Bitplane, Switzerland). Quantification of relative fluorescence intensity was conducted using ImageJ.
2.11. Statistical analysis
Viability of MCF7 cells in 2D cell culture was calculated as the mean of mean±S.D. of triplicates for each Dox concentration tested. Viability and cytotoxicity in microfluidic assays were calculated for a minimum of 50 TME scaffolds, n=3. Statistical analysis was conducted using two-way ANOVA, with p-value ≤ 0.05 considered statistically significant. Statistical analysis of relative fluorescence in immunofluorescence assays was conducted by using the Student’s two-tailed T-test (Fig. 7(d), Fig. S6(d) and Fig. S7(b)), or one-way ANOVA (Fig. 6, Fig. 7(a), Fig. S5, and Fig. S6(b)), with p ≤ 0.05 considered statistically significant.
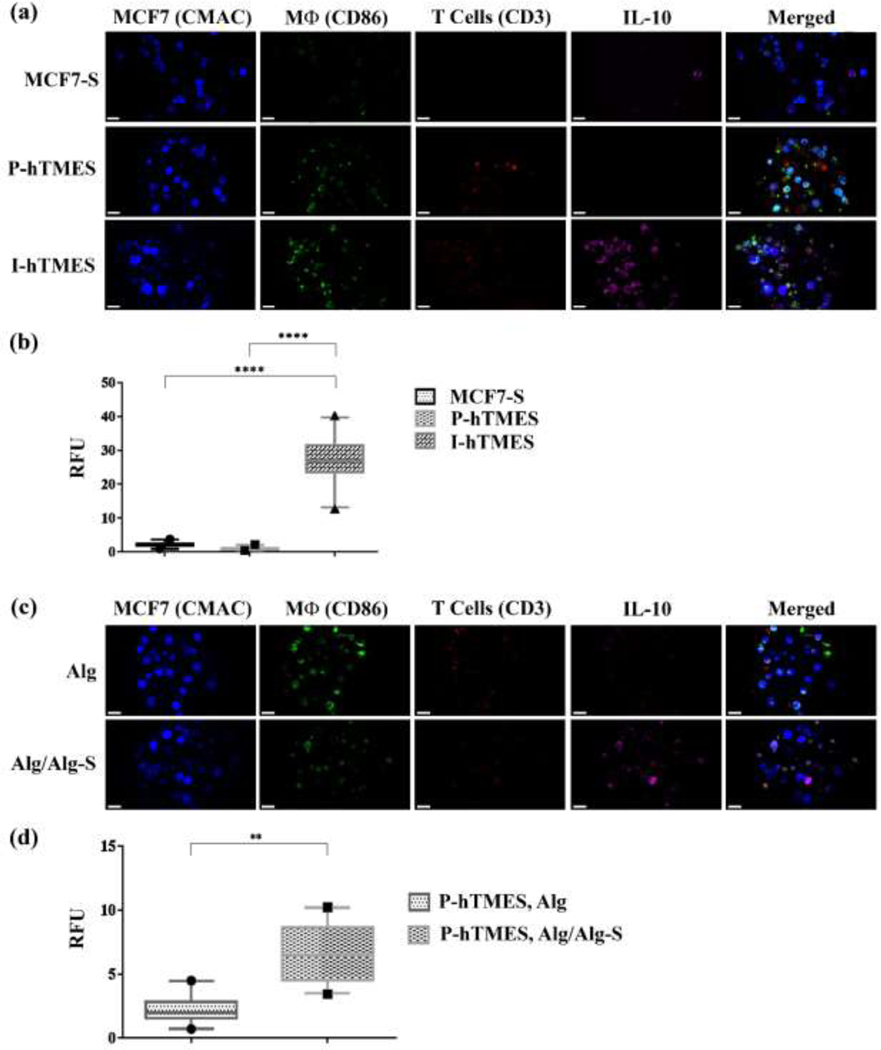
Fig. 7. On-chip analysis of IL-10 expression in MCF7-S, P-hTMES and I-hTMES.
On-chip immunofluorescence for IL-10 expression (magenta); T cells immunolabeled for CD3 (red); macrophages immunolabeled for CD86 (green); and MCF7 labeled with live-cell tracker CMAC (blue). (a) Confocal immunofluorescence imaging of IL-10 expression in MCF7-S, P-hTMES and I-hTMES. (b) Quantitative analysis of IL-10 expression conducted by measuring IL-10 fluorescence intensity vs. background, in n=30 scaffolds for each scaffold type, expressed in relative fluorescence units (RFU). Statistical analysis was conducted for means of fluorescence intensity, using one-way ANOVA, with p ≤ 0.05 considered statistically significant. (c) IL-10 imaging in P-hTMES, encapsulated in Alg (upper panel) or Alg/Alg-S (lower panel), with labeling the same as described in (a). (d) Quantitative analysis of IL-10 production in the P-hTMES following 5 days of incubation was conducted as described in (b), with statistical analysis conducted by using two-tailed Student’s T-test, with p ≤ 0.05 considered statistically significant, in n=20 scaffolds. Box plots are depicted as the 5–95 percentile, where the whiskers show the data range within this percentile, the box denotes the values in the 25th to the 75th percentiles, and the horizontal bar depicting the median. The dots bellow and above the whiskers denote the min and max values measured. Imaging was conducted by Zeiss LSM 880 confocal microscopy, using 20X magnification, 2 μm Z-stacking. Scale bar in (a) and (c) = 20 μm.
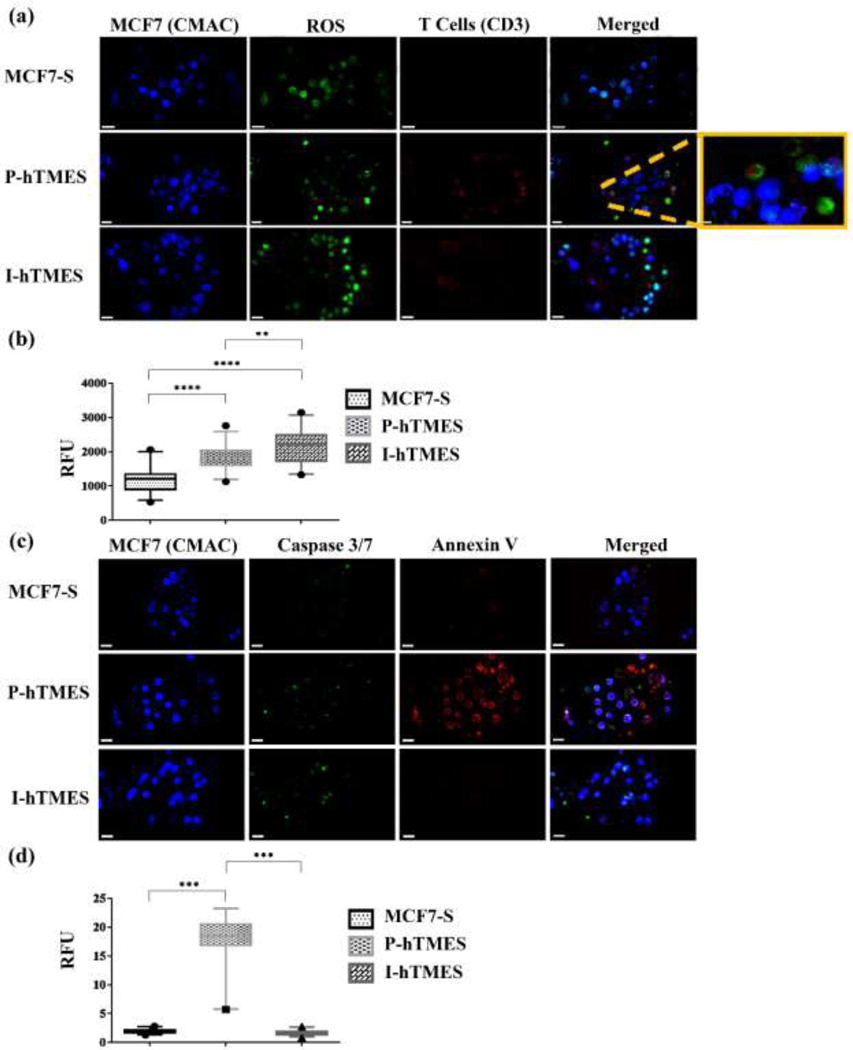
Fig. 6. On-chip analysis of ROS production and apoptosis in MCF7-S, P-hTMES and I-hTMES.
(a) Imaging of ROS productions, with MCF7 cells labeled with live cell-tracker CMAC (blue), T cells immunolabeled for CD3 (red), and ROS detected using a ROS detection reagent (green). (b) ROS production for each scaffold type, quantified by analysis of ROS fluorescence intensity vs. background in n=30 scaffolds for each scaffold type, expressed in relative fluorescence units (RFU). (c) Imaging apoptotic cells, with MCF7 cells labeled with live cell-tracker CMAC (blue); Apoptotic cells labeled by Annexin V (red); and activated caspase 3/7 (green). (d) Apoptosis of MCF7 cells quantitatively analyzed by measuring Annexin V fluorescence intensity vs. background in n=30 scaffolds for each scaffold type, expressed in RFU. Imaging in (a) and (c) was conducted by Zeiss LSM 880 confocal microscopy, using 20X magnification, 2 μm Z-stacking. Scale bar: 20 μm, except for magnified image: 5 μm; Box plots in (b) and (d) are depicted as the 5–95 percentile, where the whiskers show the data range within this percentile, the box denotes the values in the 25th to the 75th percentiles, and the horizontal bar depicting the median. The dots bellow and above the whiskers denote the min and max values measured. Statistical analysis was conducted by using one-way ANOVA for means of fluorescence intensity, with p ≤ 0.05 considered statistically significant.
2. Results
3.1. Microfluidic device design
The microfluidic device design used in this study enabled the on-chip high throughput generation, culture, imaging and analysis of breast tumor - stroma microenvironment in Alg or Alg/Alg-S scaffolds (TME scaffolds) (Fig. 1). The design used in this study was composed of an array layer, featuring a flow-focusing junction for rapid droplet generation and an integrated 1000-docking sites array, each 200 μm in diameter, where the TME scaffolds were trapped and maintained; and an upper layer of 20 μm channels (width), spanning over the array and facilitating efficient perfusion of the TME scaffolds, in addition to perfusion through the array channels (Fig. 1(a)). Successful fabrication was validated by interferometry (Fig. 1(b)). For TME scaffold generation, the cells were mixed with partially cross-linked Alg or Alg/Alg-S and infused into the microfluidic device, through the aqueous phase inlet. Simultaneously, oil flow via the oil inlet, resulted in the rapid generation of water-in-oil droplets that were trapped in the integrated array. The hydrogel-containing droplets were then briefly perfused with complete media supplemented with calcium chloride through a third inlet, for complete cross-linking of the hydrogel and oil removal. During cultivation, the TME scaffolds were maintained by constant media perfusion at 50 μl x hr−1, equivalent to 575 μm x s-1. This flow rate mimics blood flow velocity in solid tumors in vivo, reported to vary from 100 to 800 μm x s−1 [51]. To assess the uniformity of droplets, in terms of droplet size and cell distribution per TME scaffold, imaging and analysis of droplet diameter and cell number per TME scaffold were conducted (Fig. S1 and S2), with average droplet diameter of 144±13 μm (n=100 scaffolds) and 43±15 cells per TME scaffold (100 TME scaffolds, n=2).
3.2. Viscoelasticity characterization of the Alg and Alg/Alg-S hydrogels
Solid tumors were reported to be stiffer compared to their corresponding normal tissue [52]. The Young’s modulus of the normal breast tissue is averaged at 400 Pa, while in the malignant breast tissue it is averaged at 3000 Pa [53]. Increased stiffness is associated with higher integrin signaling, tumor cell proliferation, reduced responsiveness to chemotherapy and poor prognosis [11]. The viscoelastic properties of the two hydrogels, 1% (w/v) Alg, and 1% Alg/Alg-S, containing 10% alginate sulfate as w/w of total alginate polymer, were analyzed before and after crosslinking with 0.2% (w/v) calcium gluconate (SI Appendix, Fig. S3). As expected, the mechanical spectra prior to cross-linking revealed a typical G” > G’ behavior (SI Appendix, Fig. S3(a)), whereas the cross-linked hydrogels exhibited a G’ > G” pattern (SI Appendix, Fig. S3(b)), indicating the formation of a hydrogel. In the analysis of the elastic modulus of the crosslinked Alg and Alg/Alg-S hydrogels, values of 2.9±0.2 kPa and 3.1±0.1 kPa, respectively, were observed (SI Appendix, Table S1), with no significant differences between the samples. These values are in close agreement with the mean elastic modulus values of in vivo breast tumors (~3 kPa) [53]. Thus, the biomaterials chosen for this study were shown to form stable hydrogels and exhibit physical properties that are in line with in vivo properties of the breast cancer solid tumor.
3.3. On-chip generation, viability and cytotoxicity of hTMES models
hTMES models for proinflammatory and immunosuppressive breast TMEs were generated, cultivated and analyzed on-chip, with Alg and Alg/Alg-S hydrogels as biomimetic ECMs (Fig. 2). In the models, cell composition was carefully constructed to mimic the cell types, activation states, and proportions reported in in vivo human breast cancer [9,47]. Two main TME scaffold types were generated using our high throughput microfluidic system: (i) P-hTMES, designed to recapitulate proinflammatory microenvironment of early tumor development, where MCF7 breast adenocarcinoma cells were co-encapsulated with a proinflammatory milieu of M1-activated macrophages and activated T cells, as well as mammary fibroblasts and epithelial cells as the non-immune stroma; and (ii) I-hTMES, designed to mimic the immunosuppressive microenvironment of progressed breast tumor, where MCF7 tumor cells were co-encapsulated with M2-activated macrophages and non-activated T cells, mammary fibroblasts and epithelial cells. Macrophage and T cell differentiation markers were routinely analyzed by flow cytometry using established differentiation markers [44,54,55] (SI Appendix, Fig. S4). The I-hTMES model was based on clinical studies, which have shown higher percentage of M2b and M2c macrophages in advanced breast tumor patients [41,45], compared to both healthy individuals and early-stage patients. Here, M2 differentiation resulted in a mixed M2b/M2c phenotype, characterized by: significantly low CD80 expression compared to M1 macrophages (a general characteristic of M2 polarization, P ≤ 0.0001); high expression of CD86 (compared to non-activated monocytes, P ≤ 0.01, non-significant compared to M1 macrophages), indicating M2b polarization; and high expression of CD163 (compared to non-activated monocytes, P ≤ 0.05), an established M2c marker. Non-immune stromal cells were incorporated into both models, with human mammary fibroblasts, originating from mastectomy samples, and human mammary epithelial cells. MCF7 scaffolds (MCF7-S), composed of MCF7 cells only; and ECF7-EF scaffolds (MCF7-EF-S), where MCF7 cells were co-encapsulated with epithelial cells and fibroblasts only; were generated as controls.
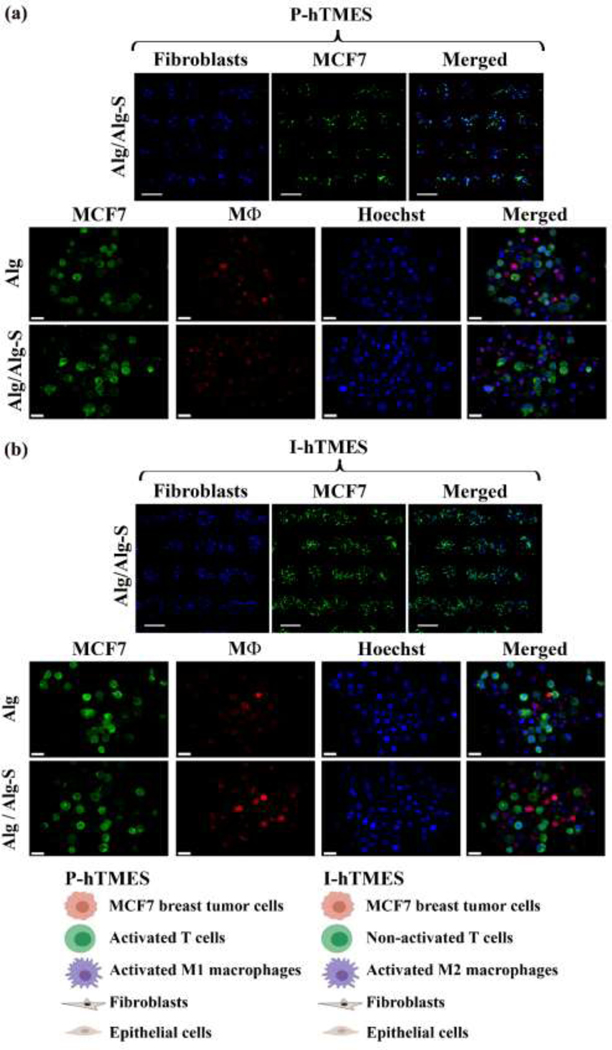
Fig. 2. On-chip generation of Alg and Alg/Alg-S P-hTMES and I-hTMES.
Microscopy images of Alg and Alg/Alg-S (a) P-hTMES and (b) I-hTMES. Imaging shown in upper panels in (a)-(b) was conducted using epifluorescence microscopy (Zeiss Axio Observer.Z1), 5X magnification, using 2 μm Z-stacking, with MCF7 labeled with CFSE (green), and fibroblasts with CMAC (blue), Scale bar: 200 μm; Imaging of scaffolds in two lower panels in (a)-(b) was performed by confocal microscopy (Zeiss LSM 880) with 20X magnification, using 2 μm Z-stacking, with MCF7 labeled with CFSE (green), macrophages with CMPTX (red) and cell nuclei with Hoechst 33342 (blue), Scale bar: 20 μm.
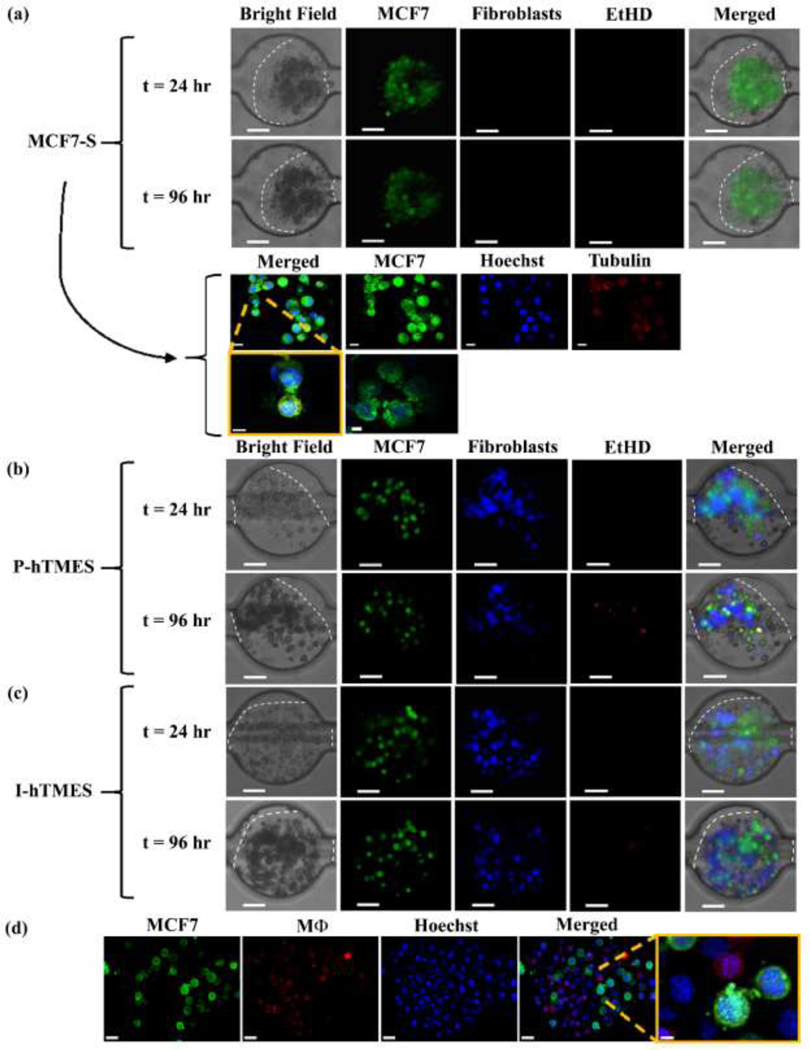
MCF7-S, P-hTMES and I-hTMES were maintained in the microfluidic device with constant media perfusion. During cultivation, some changes in cell morphology across the TME scaffolds were observed over time (described here for day-1 to day-4 culture, Fig. 3 and SI Appendix, Fig. S5). Though no significant changes in total hydrogel area were observed, cells appeared more compact and darker in shade over time, particularly in P-hTMES and I-hTMES, compared to the MCF7-S (Fig. 3). These changes were more pronounced in the Alg/Alg-S hydrogels, as exemplified for I-hTMES in Fig. S5. Examining Alg/Alg-S TME scaffolds, while in the MCF7-S cells appeared to retain their original shape and overall size over time (Fig. 3), and were observed to preserve close cell-cell contacts, the cells in P-hTMES and I-hTMES were smaller, elongated and darker (in bright field) over time. In all TME scaffold types, formation and expansion of cell protrusions, by both tumor cells and stromal cells, was observed (Fig. 3 and SI Appendix, Fig. S5). Cell proliferation was minimal in all TME scaffold types. This is expected due to the nature of the stiff hydrogels used, shown to result in decreased proliferation [56], and mimic low proliferation patterns observed under similar conditions in vivo [57].
Fig. 3. Scaffold morphology in the MCF7-S, P-hTMES and I-hTMES during on-chip culture.
Microscopy imaging of (a) MCF7-S; (b) P-hTMES; and (c, d) I-hTMES in Alg/Alg-S. Imaging conducted using epifluorescence microscopy (Zeiss Axio Observer.Z1) in ((a), two upper panels), (b), and (c), with MCF7 cells labeled with CFSE (green), fibroblasts with CMAC (blue), and dead cells with EtHD (red), using 20X magnification and 2 μm Z-stacking, Scale bar: 50 μm. Confocal imaging (Zeiss LSM 880) in (a), two lower panels, was conducted using 20X magnification, 2 μm Z-stacking, with MCF7 cells labeled with CFSE (green), tubulin labeled with anti-human tubulin mAb (red) and cell nuclei with Hoechst 33342 (blue), scale bar: 10 μm, upper panel, and 5 μm for magnified images (lower panel).Confocal imaging in (d) was conducted using 20X magnification, 2 μm Z-stacking, with MCF7 cells labeled with CFSE (green), macrophages with CMPTX (red), and cell nuclei with Hoechst 33342, Scale bar: 20 um, and 5 μm in the magnified image.
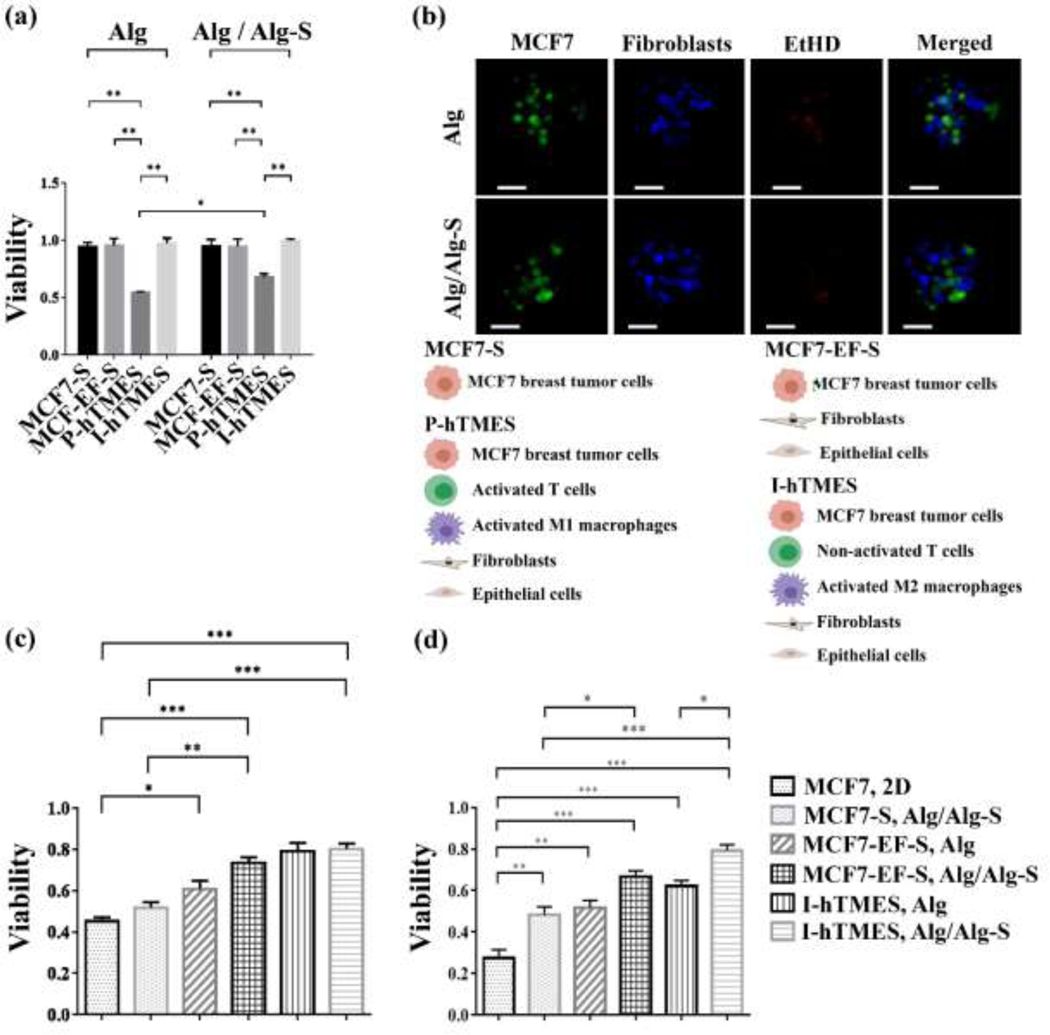
Viability and cytotoxicity in the TME scaffolds were imaged and quantitatively analyzed (Fig. 4). Viability of MCF7 cells in MCF7-S, MCF7-EF-S, P-hTMES and I-hTMES generated in Alg or Alg/Alg-S hydrogels was analyzed. Analysis was conducted for each TME scaffold type in Alg vs. Alg/Alg-S and for different scaffold types in Alg or Alg/Alg-S, hypothesizing that both hydrogel type and cellular milieu affect viability. Following 96 hr culture, viability was significantly lower in Alg P-hTMES compared to all other TME scaffold types (p ≤ 0.01, all, Fig. 4(a)). A similar pattern was observed in Alg/Alg-S P-hTMES, with viability significantly lower (p ≤ 0.01) compared to other TME scaffolds. Of note, a significant difference in viability was observed in Alg versus Alg/Alg-S P-hTMES, with higher viability in Alg/Alg-S scaffolds (p ≤ 0.05, Fig. 4(a-b)). Examining viability over time, viability in both Alg and Alg/Alg-S P-hTMES was observed to drop within the first 48 hr of culture and then remained relatively steady at 96 hr.
Fig. 4. Analysis of on-chip viability and cytotoxicity.
(a) Viability of MCF7 cells in MCF7-S, MCF7-EF-S, P-hTMES and I-hTMES, encapsulated in Alg or Alg/Alg-S following 96 hr incubation with constant media perfusion. (b) Imaging of P-hTMES, in Alg (upper panel) and Alg/Alg-S (lower panel), with MCF7 labeled with CFSE (green), fibroblasts with CMAC (blue), and dead cells with EtHD (red). Imaging was conducted using epifluorescence microscopy, using 20X maginification and 2 μm Z-stacking. Scale bar = 50 μm. (c) MCF7 cells viability following 24 hr perfusion with 5 μM Dox; or (d) 10 μM Dox. MCF7 cell viability is presented as mean viability ± S.D., n=3, calculated as the percentage of live cells in 50 scaffolds for each scaffold type, analyzed using ImageJ. Viability in the 2D culture was analyzed using Presto Blue, mean viability ± S.D., n=3. Statistical analysis in (a) and (c)-(d) was conducted by 2-way ANOVA, and one-way ANOVA, respectively, with p ≤ 0.05 considered statistically significant.
Doxorubicin (Dox) cytotoxicity was analyzed in a 2D monolayer of MCF7 tumor cells and in the TME scaffolds on-chip. Dox is a commonly used chemotherapeutic drug, to which breast cancer cells cultured in 3D culture were previously shown to demonstrate higher resistance compared to 2D monolayer culture [58]. Here, following 5 μM Dox treatment, significant differences in viability between the 2D culture and the MCF7-EF-S and I-hTMES, in both Alg and Alg/Alg-S were observed (p ≤ 0.05 and p ≤ 0.001, respectively, (Fig. 4(c)). Following 10 μM Dox treatment, significant differences in viability between the 2D culture and all TME scaffolds were observed (p ≤ 0.01, Fig. 4(d)). Of note, a significant difference in viability was observed in Alg versus Alg/Alg-S I-hTMES (p ≤ 0.05). Thus, both viability and Dox cytotoxicity were shown to be modulated by both cellular composition (for example viability in P-hTMES versus I-hTMES, or cytotoxicity in MCF7-S vs. MCF7-EF-S and I-hTMES in Alg/Alg-S) and hydrogel type (viability of Alg versus Alg/Alg-S P-hTMES, or cytotoxicity of Alg I-hTMES versus Alg/Alg-S I-hTMES).
3.4. On-chip immunofluorescence in the hTMES models
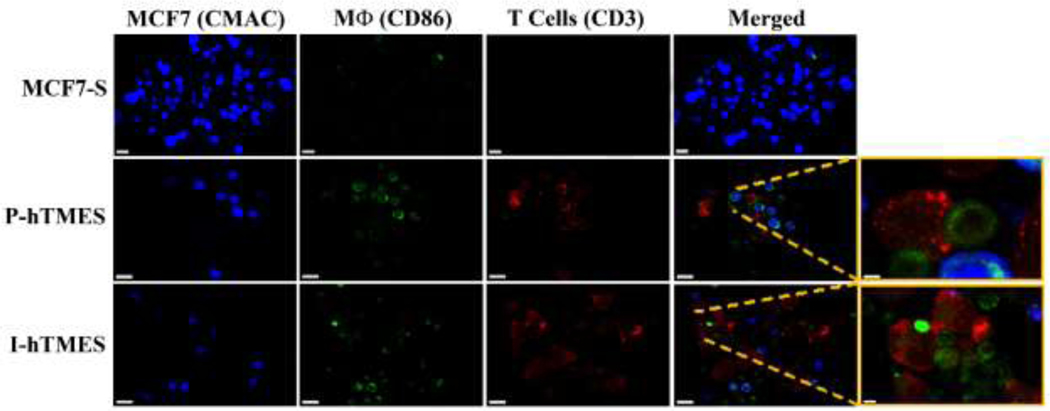
The TME of solid tumors is generally characterized by increased oxidative stress, resistance to apoptosis, and production of immuno-suppressive cytokines such as IL-10. These characteristics were analyzed here by on-chip immunofluorescence and quantitative analysis of the MCF7-S, P-hTMES and I-hTMES, encapsulated in Alg/Alg-S, using confocal microscopy (Figs. 1–7). T cells were immunolabeled for CD3. Differentiated macrophages were immunolabeled for CD86 [41,42,59]. Here, CD86 expression was high and of similar expression levels in both M1 (P-hTMES) and M2 macrophages (I-hTMES) (SI Appendix, Fig. S4), and thus allowed the use of this marker for the detection of differentiated macrophages in both models. Moreover, this facilitated the identification of possible macrophage-T cell interaction sites, as CD86 and CD3 are both localized in the immunological synapse (though not forming direct interactions) [60]. Using confocal microscopy, areas of overlapping CD3 and CD86 labeling were clearly observed in both P-hTMES and I-hTMES (Fig. 5).
Fig.5. On-chip imaging of immune cells in MCF7-S, P-hTMES and I-hTMES.
Imaging was conducted by Zeiss LSM 880 confocal microscopy, using 20X and 40X magnification, 2 μm Z-stacking, with MCF7 cells labeled with live cell-tracker CMAC (blue); T cells immunolabeled for CD3 (red); and Macrophages immunolabeled for CD86 (green). Magnified images on the right show areas of CD3 and CD86 overlap. Scale bar: 20 μm, except for right hand side magnified images: 3 μm for P-hTMES and 5 μm for I-hTMES.
ROS production was imaged and analyzed using a commercially available ROS detection reagent, which is cell-permeable, emitting green fluorescence when oxidized by ROS (Fig. 6(a, b)). ROS was detected in all TME scaffold types, which is in line with previous reports of ROS production in breast tumors [61,62]. In the quantitative analysis of ROS production (by quantification of relative fluorescence), a significant increase in ROS production by P-hTMES and I-hTMES compared to the MCF7-S was observed (p ≤ 0.0001, both, Fig. 6(b)). A significant difference in ROS production was also observed in P-hTMES compared to I-hTMES (p ≤ 0.01), with higher expression in the I-hTMES. These results align with previous works, showing increased oxidative stress in solid tumors [63–65].
Analysis of apoptosis in MCF7-S, P-hTMES and I-hTMES was conducted next, using Annexin V and a detection reagent for caspases 3/7 activation as apoptosis markers (Fig. 6(c, d)). In line with the results obtained in the viability analysis (Fig. 4), apoptotic markers were predominantly observed in the P-hTMES (Fig. 6(c)). In order to quantitatively analyze apoptosis in MCF7 tumor cells, the relative fluorescence intensity of bound annexin V was measured and quantitatively analyzed in the three TME scaffold types (Fig. 6(d)). Significantly higher Annexin V binding was observed in P-hTMES compared to the MCF7-S and I-hTMES (p < 0.001 for both). As caspase 3/7 labeling was inconsistent with annexin V binding, we do not present the analysis of caspase 3/7 activation here. This may be due to non-apoptotic activities previously reported for caspase 3 [66–69], while annexin V remains a reliable marker for early surface change in apoptosis [70].
IL-10 expression was significantly higher in the I-hTMES compared to MCF7-S and P-hTMES, (p ≤ 0.0001 for both, Fig. 7(a, b)). No significant difference in IL-10 expression was observed in MCF7-S versus P-hTMES. IL-10 labeling in I-hTMES was observed to coincide with CD86 labeling, suggesting that IL-10 in the I-hTMES was mainly produced by the M2-activated macrophages. Tumor cells were previously reported to modulate infiltrating macrophages into an M2-like phenotype, with the latter characterized by increased production of immuno-suppressive cytokines such as IL-10 [41,45]. To evaluate re-programming of macrophages into an M2-like phenotype in our system, P-hTMES encapsulated in Alg or Alg/Alg-S were imaged and quantitatively analyzed for IL-10 production following additional five days incubation (Fig. 7(c, d)). IL-10 production was detected in the Alg/Alg-S P-hTMES, and again coincided with CD86 labeling. IL-10 production was significantly higher in Alg/Alg-S P-hTMES compared to Alg P-hTMES (p ≤ 0.01, Fig. 7(d)). Thus, IL-10 production by macrophages in the Alg/Alg-S P-hTMES was induced overtime, suggesting an M1 to M2 phenotype switch in macrophages in Alg/Alg-S scaffolds. As further support for this finding, we analyzed the expression of CD163 and HLA-DR in Alg P-hTMES versus Alg/Alg-S P-hTMES and I-hTMES (Fig. S6). CD163 is a well-established marker for M2 macrophage differentiation [41,44], observed to be highly expressed in our M2-differentiated macrophages (Fig. S4). CD163 levels were significantly higher in the Alg/Alg-S P-hTMES and I-hTMES compared to Alg P-hTMES (p ≤ 0.01 and p ≤ 0.001, respectively, Fig. S6(b)). Attenuation of HLA-DR expression, a validated surface marker for M1 macrophages [41], is a sensitive and reliable marker for immunosuppression [71]. Here, HLA-DR expression was significantly reduced in Alg/Alg-S P-hTMES compared to Alg P-hTMES (p ≤ 0.05, Fig. S6(c, d)). Collectively, these results support an M1 to M2 switch in Alg/Alg-S P-hTMES.
3.5. LC-MS proteomic profiling of MCF7 cells in P-hTMES
Significant differences were observed in the viability of MCF7-S versus P-hTMES, and of Alg versus Alg/Alg-S P-hTMES, indicating both cellular milieu and hydrogel type affect tumor cell survival. Elucidation of differences in protein expression in these models representing early breast TME may shed light on early events promoting tumor progression and potentially serve as valuable tools in future therapeutics. Two sets of comparative LC-MS-based proteomic profiling were conducted in MCF7 cells collected and sorted from on-chip TME scaffolds: (i) Alg/Alg-S MCF7-S versus Alg/Alg-S P-hTMES; and (ii) Alg P-hTMES versus Alg/Alg-S P-hTMES. This way, protein expression patterns that are induced by cellular milieu (set (i)) or hydrogel type (set (ii)) could be differentially detected and analyzed. Bottom-up liquid chromatography-mass spectrometry-based proteomic analysis led to the identification of approx. 2,000 protein groups (at FDR≤1%). Analysis of the datasets was conducted using the Ingenuity Pathway Analysis (IPA) software and the STRING database (STRING-DB).
3.5.1. Proteomic profiling of MCF7 cells in Alg/Alg-S MCF7-S versus P-hTMES
Over 800 proteins were found to be highly differentially expressed in this dataset, with log2 of fold change (log2(FC)) ≥ |1|, calculated as the log2 of the ratio of median protein abundance in MCF7-S to P-hTMES. Based on pathway enrichment analysis by IPA, top canonical pathways detected include the EIF2 signaling pathway, mitochondrial dysfunction and the sirtuin signaling pathway (SI Appendix, Table S2), previously described in human tumors [72–75]. Proteins involved in oxidation, ROS generation and response to oxidative stress differentially expressed in this dataset are summarized in SI Appendix, Table S3. Using IPA diseases and functions analysis, proteins highly associated with apoptosis were differentially expressed in Alg/Alg-S P-hTMES and are summarized in SI Appendix, Table S4.
A relatively large set (> 150) of differentially-expressed proteins involved in modulation of immune response and tumor progression were identified (SI Appendix, Table S5). A few notable proteins involved in immunomodulation, significantly upregulated in Alg/Alg-S P-hTMES include: CD47 (log2(FC)=−1.73, p-value ≤ 0.005), Protein S100-A8 (log2(FC)=−2.22, p-value ≤ 0.1), and NF-kB activating protein (NKAP, log2(FC)=−1.47, p-value ≤ 0.01). In contrast, proteins involved in immuno-recognition and activation, such as HLA-C (Log2(FC)=1.15, p-value ≤ 0.01) were upregulated in MCF7-S. A summary heatmap of several differentially expressed proteins is depicted in SI Appendix, Fig S7(a). Upstream regulators highly upregulated in Alg/Alg-S P-hTMES, as predicted by IPA with z-score ≤ −2, and a p-value << 0.05, were associated with immunosuppression, epithelial to mesenchymal transition (EMT), and tumor progression such as TGF-β1 and Nanog (SI Appendix, Fig. S7(b)). Notable proteins induced by TGF-β1, upregulated in Alg/Alg-S P-hTMES include Integrin alpha-3 (Log2(FC)=−2.02, p-value ≤ 0.001), Vimentin (Log2(FC)=−2.15, p-value ≤ 0.0005) and Proliferation marker protein Ki-67 (Log2(FC)=−0.97, p-value ≤ 0.01). Combined, expression of proteins involved in oxidative stress, immunomodulation, and EMT were upregulated in Alg/Alg-S P-hTMES.
3.5.2. Proteomic profiling of MCF7 cells in Alg versus Alg/Alg-S P-hTMES
Approximately 400 proteins differentially expressed were detected here. While IPA analysis of canonical Pathways enriched revealed similar pathways to the pathways observed previously, pathways involved in cell-cell communication were found to be highly enriched in this dataset (SI Appendix, Table S6). Notably, remodeling of adherens junctions and Clathrin-mediated endocytosis, regulating cell surface signaling were highly enriched. Using STRING-DB hierarchical clustering analysis, clusters of proteins involved in immune response, apoptosis and regulated exocytosis were upregulated in the Alg/Alg-S P-hTMES (SI Appendix, Fig. S7(c)). Upstream regulators predicted by IPA in Alg/Alg-S P-hTMES included proteins previously shown to induce tumor progression, such as C/EBP alpha (encoded by CEBPA) [76] and La-related protein 1 (LARP1), predicted with z-score ≤ −2. IPA diseases and functions and networks analyses detected cell-cell contact, cancer and inflammatory response as the main activated functions in this dataset. Of note, proteins involved in modulation of immune response were differentially expressed in tumor cells collected from Alg/Alg-SP-hTMES compared to Alg P-hTMES. Finally, proteins involved in cell-cell interactions, such as Vinculin, Desmoglein-1 and Desmocollin-1 were all upregulated in Alg/Alg-S P-hTMES (all with log2(FC) ≤ −1, p-value ≤ 0.05).
4. Discussion
In this work, we developed a dynamic, high throughput 3D microfluidic platform for the study of the breast TME, using Alg and Alg/Alg-S hydrogels as ECM biomimetic. The use of Alg/Alg-S and high throughput microfluidics, coupled with the replication of in vivo parameters such as: (i) affinity binding of signaling factors to the Alg/Alg-S that is comparable with in vivo binding to heparin; (ii) hydrogel stiffness that is comparable to the in vivo breast tumor ECM; (iii) perfusion that follows in vivo blood flow velocity in solid tumors; and finally (iv) the co-culture of tumor and stromal cells, in proportions and activation states that replicate in vivo breast TME dynamics; resulted in in vivo-like responses, observed by judging three main categories : (i) Cell morphology and EMT; (ii) Oxidative stress and apoptosis; and (iii) Immunomodulation.
Tumors expressing both epithelial and mesenchymal markers have been reported previously, representing a continuous spectrum of cell states between the two [77–79]. Several observations here indicate EMT phenotypes in Alg/Alg-S P-hTMES and I-hTMES. Morphologically, the epithelial cells in Alg/Alg-S P-hTMES and I-hTMES were observed to adopt an elongated and isolated shape, indicative of mesenchymal transition. Migration was not observed, which may be due to the cells’ inability to breakdown the alginate hydrogel and its density. LC-MS proteomic profiling showed increased expression of proteins involved in remodeling of adherens junctions and Clathrin-mediated endocytosis, which regulate cell surface signaling, in Alg/Alg-S P-hTMES compared to Alg/Alg-S MCF7-S and Alg P-hTMES. Upregulation of proteins involved in remodeling of adherens junctions by LC-MS correlated with the observed generation of cellular protrusions over time in the Alg/Alg-S P-hTMES and I-hTMES (Fig. S5). IPA prediction of TGF-β1 as an upstream regulator (z-score ≤ −2) further indicates EMT patterns. TGF-β1 has been highly correlated with inducing invasion, metastasis and EMT in breast cancer [80,81]. Tumor-associated macrophages isolated from breast cancer tissues were shown to express high levels of CD163, and correlated positively with TGF-β1 expression [82]. Here, high expression of CD163 in Alg/Alg-S P-hTMES and I-hTMES compared to Alg P-hTMES was observed (p ≤ 0.01, Fig. S6), further substantiates a possible EMT phenotype. Induction of EMT was shown to result in the attenuation of surface display of HLA molecules in tumor-associated macrophages in breast carcinomas [83–85]. Similarly, significant downregulation of HLA molecules in Alg/Alg-S P-hTMES compared to Alg P-hTMES was observed by both immunofluorescence and LC-MS proteomic profiling (HLA-DR, P ≤ 0.05, Fig. S6, and HLA-A and HLA-C, p-value ≤ 0.05, SI Appendix, Table S7, respectively). Downregulation of E-cadherin is a well-established EMT marker in breast cancer progression [86]. Significant downregulation in E-cadherin expression was observed in Alg/Alg-S P-hTMES compared to MCF7-S by immunofluorescence (p ≤ 0.05, Fig. S8), and LC-MS proteomic profiling (log2(FC)=0.9, p ≤ 0.05). As EMT is an important mechanism in driving tumor progression, its replication in vitro in our models is evident for their appropriateness to simulate the TME and the study of breast malignancy.
Oxidative stress, mitochondrial dysfunction and apoptosis are well-documented in tumor progression [87–89]. Here, using on-chip immunofluorescence, ROS production was significantly increased in Alg/Alg-S P-hTMES and I-hTMES compared to MCF7-S (p ≤ 0.0001, both). This correlates with mitochondrial dysfunction detected in both LC-MS proteomic profiling datasets and differential expression of proteins involved in oxidation (summarized in SI Appendix, Table S4), ROS production and response to oxidative stress in Alg/Alg-S MCF7-S and P-hTMES (SI Appendix, Table S3). Upregulation of apoptotic proteins by LC-MS proteomic profiling aligns with the higher annexin V binding observed in Alg/Alg-S P-hTMES, indicative of apoptotic membrane flipping (Fig. 6). Comparison of ROS production and apoptosis in P-hTMES and I-hTMES with previous findings in primary tumors reveals similar patterns. Cancer cells are characterized by elevated rates of ROS production, compared to normal cells, due to a loss of redox regulation [90]. In primary breast tumors, high levels of both various antioxidant proteins (such as SOD) and ROS were detected compared to healthy individuals [91,92]. Moreover, increased levels of both were found to be correlated with disease stage [93,94]. Similarly, ROS levels were shown to significantly increase from MCF-S to P-hTMES to I-hTMES. This associates with ROS as a signaling mechanism promoting cell survival in the TME [90], and deserves further study of the I-hTMES. Exposure to high ROS levels has been shown to have contradicting impacts, serving as both a signaling mechanism inducing metabolic changes that induce cells survival, but can also result in cell arrest and apoptosis [95]. In primary breast adenocarcinomas, expression of both anti-apoptotic proteins such as Bcl-2, as well as pro-apoptotic proteins such as Bax were detected [89]. Similarly, the induced expression of proapoptotic proteins such as Bcl2-L-13 in Alg/Alg-S P-hTMES by proteomic analysis (log2(FC)=−1.29, p-value ≤ 0.01) and higher apoptosis observed by immunofluorescence (Fig. 6(c), (d)) is indicative of analogous mechanisms. Immunofluorescence for apoptotic markers such as membrane flipping, detected by annexin V binding, and caspase 3/7 activation yielded contradicting results. While annexin V binding proved consistent with viability analysis, analysis of caspase 3 activation did not align with annexin V binding. This may be attributed to previously reported non-apoptotic activities for activated caspase 3, such as tumor proliferation and monocyte differentiation [66,68,69,96]. Indeed, further study into the potential role of caspase 3 activation may be of value in understanding of its role in tumor progression.
Significant evidence for immunomodulation in our models was gathered. First, the upregulation in IL-10 expression in Alg/Alg-S P-hTMES correlates with previous reports on IL-10 expression in vivo. In primary breast tumors, high expression of IL-10 was significantly higher compared to normal breast tissues [97], and high IL-10 expression was associated with higher tumor grade [98,99]. Tumor cells were previously shown to induce transition from M1 to M2 phenotype in infiltrating macrophages, characterized by increase in the expression of immunosuppressive cytokines, such as IL-10, supporting tumor progression and immuno-evasion [41,99,100] A similar trend was observed here, with significant increase in IL-10 expression in Alg/Alg-S P-hTMES (Fig. 7), further supported by significant increase in CD163 expression and downregulation of HLA-DR (SI Appendix, Fig. S6), both previously associated with immunosuppression and tumor progression [101]. Upregulation of proteins involved in Immunomodulation was also detected by LC-MS proteomic profiling, summarized in SI Appendix, Table S5. One notable example is NF-kB activating protein (NKAP), recently reported to promote tumor growth and induce immunosuppression in gliomas [102]. Specifically, NKAP was reported to alter the polarization and infiltration of tumor associated macrophages. Here, though changes in cell mobility were not observed, an M1 to M2 polarization was observed by the aforementioned changes in IL-10, CD163, and HLA-DR expression (Fig. 7 and S6, respectively). Combined, these observations may indicate a similar role for NKAP in promoting tumor progression. Two particular examples indicate the role of Alg/Alg-S in promoting cell-cell interactions in the models presented here: (i) upregulation of monocarboxylate transporter 4 (SLC16A, or MCT4) in Alg/Alg-S P-hTMES versus both Alg/Alg-S MCF7-S and Alg P-hTMES (log2(FC)=−2.18 and −1.1, respectively, p ≤ 0.05, both). MCT4 is a lactate–proton symporter which supports glycolytic tissues by pumping out lactate and H+ [103]. In turn, the acidification of the extracellular space induces M2 polarization of macrophages [104]. It was found to be highly expressed in breast tumor cells compared to normal mammary cells, both in vitro and in vivo, induced particularly in areas under oxidative stress. Similarly, its high expression in the Alg/Alg-S P-hTMES can indicate a similar mechanism in inducing M2 polarization; (ii) Upregulation of both HLA-A and HLA-C in Alg P-hTMES versus Alg/Alg-S P-hTMES (log2(FC)=1.07 and 0.9, respectively, p ≤ 0.05, both), indicating immuno-evasion mechanisms activated in the Alg/Alg-S scaffolds, similar to in vivo tumor progression. Collectively, our results suggest that signaling in the TME Alg/Alg-S scaffolds is dynamic, adaptive and mimics previously reported in vivo responses in breast cancer. The observations presented here cannot be explained by differences in hydrogel stiffness (non-significant statistically). It is more likely that the activity of alginate-sulfate in capturing and binding signaling molecules makes the matrix more active towards cells. As supporting evidence, formation of large tumor-derived vesicles (LTV, 2.3±0.3 μm in mean diameter (n=10)), previously reported to inhibit immune response [105] and promote tumor progression and metastasis [105–107], was observed in Alg/Alg-S scaffolds (Fig. S9). This, coupled with the upregulated expression of proteins involved in secretion and regulated exocytosis in the Alg/AlgS P-hTMES by proteomic profiling (SI Appendix, Table S7) suggest a possible mechanism for tumor survival.
System limitations include: (i) slight deviation from Gaussian distribution in cell number per scaffold (Fig. S2). Variability is expected due to flow patterns developing in the geometries employed here. However, the system presented here still offers a rapid and efficient way for high throughput generation of droplets of high uniformity; (ii) exclusion of endothelial cells and adipocytes, shown to play important roles in the breast TME [2,108,109]. However, incorporating stromal cells in in vivo proportions and activation states, application of constant perfusion, and controlling hydrogel stiffness, allowed us to present a meaningful model of the breast TME; (iii) the microfluidic design presented here enables a single experimental condition per device, however this can be easily solved by alternative designs, as demonstrated in previous studies [110,111]. Existing models [29,31,32,112] offer limited control over size or cell composition of the cell constructs, or use ex vivo tissues that offer limited viability. Here, we were able to generate complex and dynamic in vitro models of the breast TME, using high throughput microfluidics, that offer control over scaffold size, composition and stiffness. Our results confirmed increased expression of proteins involved in tumor progression and immune-modulation that were previously reported in vivo, demonstrating Alg/Alg-S as a powerful ECM-mimic.
DATA AVAILABILITY
All the data supporting the findings of this study are available within the article and its supplementary information files and from the corresponding author upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository[113] with the dataset identifier PXD021509.
Supplementary Material
Acknowledgments
The authors are grateful for the support provided by the National Institutes of Health grants under awards numbers 1R33CA223908-01 and 1R01GM127714-01A1 (awarded to T.K.), R01GM120272, R01CA218500, and R35GM136421 (awarded to A.R.I.), and by the National Science Foundation under Grant No. G00005964 (awarded to T. K.). The authors would like to thank Northeastern University for providing seed funding support through the Tier 1 program (A.R.I. and T.K.). I.B.F gratefully acknowledges the Negev fellowship from Kreitman School. This work was done in partial fulfillment of the requirements for a Ph.D. degree (I.B.F) at the Avram and Stella Goldstein-Goren Department of Biotechnology Engineering, Ben-Gurion University of the Negev, Israel. Prof. Cohen holds the Claire and Harold Oshry Professor Chair in Biotechnology. We thank the Institute for Chemical Imaging of Living Systems at Northeastern University and the confocal imaging core facility at Beth Israel Deaconess medical center for consultation and imaging support.
Footnotes
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Korkaya H, Liu S, Wicha MS, Breast cancer stem cells, cytokine networks, and the tumor microenvironment, J Clin Invest. 121 (2011) 3804–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mao Y, Keller ET, Garfield DH, Shen K, Wang J, Stromal cells in tumor microenvironment and breast cancer, Cancer Metastasis Rev. 32 (2013) 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Li M, Liu X, Robinson G, Bar-Peled U, Wagner K, Young WS, Hennighausen L, Furth PA, Mammary-derived signals activate programmed cell death during the first stage of mammary gland involution, Proc Natl Acad Sci USA. 94 (1997) 3425–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Brady NJ, Chuntova P, Schwertfeger KL, Macrophages: Regulators of the Inflammatory Microenvironment during Mammary Gland Development and Breast Cancer, Mediators Inflammation. 2016 (2016) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Law AMK, Lim E, Ormandy CJ, Gallego-Ortega D, The innate and adaptive infiltrating immune systems as targets for breast cancer immunotherapy, Endocr Relat Cancer. 24 (2017) R123–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sica A, Allavena P, Mantovani A, Cancer related inflammation: The macrophage connection, Cancer Lett. 267 (2008) 204–215. [DOI] [PubMed] [Google Scholar]
- [7].Shiga K, Hara M, Nagasaki T, Sato T, Takahashi H, Takeyama H, Cancer-Associated Fibroblasts: Their Characteristics and Their Roles in Tumor Growth, Cancers. 7 (2015) 2443–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kaushik Shelly, Pickup Michael W. and Weaver Valerie M., From Transformation to Metastasis: Deconstructing the Extracellular Matrix in Breast Cancer, Cancer Metastasis Rev. 35 (2016). 655–667- [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].DeNardo DG, Coussens LM, Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression, Breast Cancer Res. 9 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Whiteside TL, The tumor microenvironment and its role in promoting tumor growth, Oncogene. 27 (2008) 5904–5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pickup MW, Mouw JK, Weaver VM, The extracellular matrix modulates the hallmarks of cancer, EMBO reports. 15 (2014) 1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Costa EC, Moreira AF, de Melo-Diogo D, Gaspar VM, Carvalho MP, Correia IJ, 3D tumor spheroids: an overview on the tools and techniques used for their analysis, Biotechnol Adv. 34 (2016) 1427–1441. [DOI] [PubMed] [Google Scholar]
- [13].Ghosh S, Spagnoli GC, Martin I, Ploegert S, Demougin P, Heberer M, Reschner A, Three-dimensional culture of melanoma cells profoundly affects gene expression profile: A high density oligonucleotide array study, J Cell Physiol. 204 (2005) 522–531. [DOI] [PubMed] [Google Scholar]
- [14].Luca AC, Mersch S, Deenen R, Schmidt S, Messner I, Schäfer K, Baldus SE, Huckenbeck W, Piekorz RP, Knoefel WT, Krieg A, Stoecklein NH, Impact of the 3D microenvironment on phenotype, gene expression, and EGFR inhibition of colorectal cancer cell lines, PLOS ONE. 8 (2013) e59689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Premaratne ID, Toyoda Y, Celie K, Brown KA, Spector JA, Tissue Engineering Models for the Study of Breast Neoplastic Disease and the Tumor Microenvironment, Tissue Eng. Part B Rev. (2020) 423–442. [DOI] [PubMed] [Google Scholar]
- [16].Liu C, Lewin Mejia D, Chiang B, Luker KE, Luker GD, Hybrid collagen alginate hydrogelas a platform for 3D tumor spheroid invasion, Acta Biomater. 75 (2018) 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Phan-Lai V, Florczyk SJ, Kievit FM, Wang K, Gad E, Disis ML, Zhang M, Three-Dimensional Scaffolds to Evaluate Tumor Associated Fibroblast-Mediated Suppression of Breast Tumor Specific T Cells, Biomacromolecules (Online). 14 (2013) 1330–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Toyoda Y, Celie K, Xu JT, Buro JS, Jin J, Lin AJ, Brown KA, Spector JA, A 3-Dimensional Biomimetic Platform to Interrogate the Safety of Autologous Fat Transfer in the Setting of Breast Cancer, Ann Plast Surg. 80 (2018) S223–S228. [DOI] [PubMed] [Google Scholar]
- [19].Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, Liu IH, Chiou S, Salahudeen AA, Smith AR, Deutsch BC, Liao L, Zemek AJ, Zhao F, Karlsson K, Schultz LM, Metzner TJ, Nadauld LD, Tseng Y, Alkhairy S, Oh C, Keskula P, Mendoza-Villanueva D, De La Vega, Francisco M., Kunz PL, Liao JC, Leppert JT, Sunwoo JB, Sabatti C, Boehm JS, Hahn WC, Zheng GXY, Davis MM, Kuo CJ, Organoid Modeling of the Tumor Immune Microenvironment, Cell. 175 (2018) 1972–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sontheimer-Phelps A, Hassell BA, Ingber DE, Modelling cancer in microfluidic human organs-on-chips, Nat Rev Cancer. 19 (2019) 65–81. [DOI] [PubMed] [Google Scholar]
- [21].Sabhachandani P, Sarkar S, Mckenney S, Ravi D, Evens AM, Konry T, Microfluidic assembly of hydrogel-based immunogenic tumor spheroids for evaluation of anticancer therapies and biomarker release, J Control Release. 295 (2019) 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yoon No D, Lee K, Lee J, Lee S, 3D liver models on a microplatform: well-defined culture, engineering of liver tissue and liver-on-a-chip, Lab Chip. 15 (2015) 3822–3837. [DOI] [PubMed] [Google Scholar]
- [23].Shrestha J, Razavi Bazaz S, Aboulkheyr Es H, Yaghobian Azari D, Thierry B, Ebrahimi Warkiani M, Ghadiri M, Lung-on-a-chip: the future of respiratory disease models and pharmacological studies, Crit Rev Biotechnol. 40 (2020) 213–230. [DOI] [PubMed] [Google Scholar]
- [24].Purwada A, Jaiswal MK, Ahn H, Nojima T, Kitamura D, Gaharwar AK, Cerchietti L, Singh A, Ex vivo engineered immune organoids for controlled germinal center reactions, Biomaterials. 63 (2015) 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pavesi A, Tan AT, Koh S, Chia A, Colombo M, Antonecchia E, Miccolis C, Ceccarello E, Adriani G, Raimondi MT, Kamm RD, Bertoletti A, A 3D microfluidic model for preclinical evaluation of TCR-engineered T cells against solid tumors, JCI insight. 2 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lee SWL, Adriani G, Ceccarello E, Pavesi A, Tan AT, Bertoletti A, Kamm RD, Wong SC, Characterizing the Role of Monocytes in T Cell Cancer Immunotherapy Using a 3D Microfluidic Model, Front Immunol. 9 (2018) 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Park D, Son K, Hwang Y, Ko J, Lee Y, Doh J, Jeon NL, High-Throughput Microfluidic 3D Cytotoxicity Assay for Cancer Immunotherapy (CACI-IMPACT Platform), Front Immunol. 10 (2019) 1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ayuso JM, Truttschel R, Gong MM, Humayun M, Virumbrales-Munoz M, Vitek R, Felder M, Gillies SD, Sondel P, Wisinski KB, Patankar M, Beebe DJ, Skala MC, Evaluating natural killer cell cytotoxicity against solid tumors using a microfluidic model, Oncoimmunology. 8 (2019) 1553477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Parlato S, De Ninno A, Molfetta R, Toschi E, Salerno D, Mencattini A, Romagnoli G, Fragale A, Roccazzello L, Buoncervello M, Canini I, Bentivegna E, Falchi M, Bertani FR, Gerardino A, Martinelli E, Natale C, Paolini R, Businaro L, Gabriele L, 3D Microfluidic model for evaluating immunotherapy efficacy by tracking dendritic cell behaviour toward tumor cells, Sci. Rep 7 (2017) 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Boussommier-Calleja A, Atiyas Y, Haase K, Headley M, Lewis C, Kamm RD, The effects of monocytes on tumor cell extravasation in a 3D vascularized microfluidic model, Biomaterials. 198 (2019) 180–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Aref AR, Campisi M, Ivanova E, Portell A, Larios D, Piel BP, Mathur N, Zhou C, Coakley RV, Bartels A, Bowden M, Herbert Z, Hill S, Gilhooley S, Carter J, Cañadas I, Thai TC, Kitajima S, Chiono V, Paweletz CP, 3D microfluidic ex vivo culture of organotypic tumor spheroids to model immune checkpoint blockade, Lab Chip. 18 (2018) 3129–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, Bowden M, Deng J, Liu H, Miao D, He MX, Walker W, Zhang G, Tian T, Cheng C, Wei Z, Palakurthi S, Bittinger M, Vitzthum H, Kim JW, Merlino A, Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids, Cancer Discov. 8 (2018) 196–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Glicklis R, Shapiro L, Agbaria R, Merchuk Jose C., Cohen S, Hepatocyte behavior within three-dimensional porous alginate scaffolds, Biotechnol. Bioeng. 67 (2000) 344–353. [DOI] [PubMed] [Google Scholar]
- [34].Shapiro L, Cohen S, Novel alginate sponges for cell culture and transplantation, Biomaterials. 18 (1997) 583–590. [DOI] [PubMed] [Google Scholar]
- [35].Leor J, Aboulafia-Etzion S, Dar A, Shapiro L, Barbash I, Battler A, Granot Y, Cohen S, Bioengineered cardiac grafts: A new approach to repair the infarcted myocardium?, Circulation. 102 (2000) III56–61. [DOI] [PubMed] [Google Scholar]
- [36].Lee KY, Mooney DJ, Alginate: properties and biomedical applications, Prog Polym Sci. 37 (2012) 106–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pawar SN, Edgar KJ, Alginate derivatization: A review of chemistry, properties and applications, Biomaterials. 33 (2012) 3279–3305. [DOI] [PubMed] [Google Scholar]
- [38].Freeman I, Kedem A, Cohen S, The effect of sulfation of alginate hydrogels on the specific binding and controlled release of heparin-binding proteins, Biomaterials. 29 (2008) 3260–3268. [DOI] [PubMed] [Google Scholar]
- [39].Felder S, Masasa H, Orenbuch A, Levaot N, Shachar Goldenberg M, Cohen S, Reconstruction of the ovary microenvironment utilizing macroporous scaffold with affinity-bound growth factors, Biomaterials. 205 (2019) 11–22. [DOI] [PubMed] [Google Scholar]
- [40].Sabhachandani P, Motwani V, Cohen N, Sarkar S, Torchilin V, Konry T, Generation and functional assessment of 3D multicellular spheroids in droplet based microfluidics platform, Lab Chip. 16 (2016) 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sousa S, Brion R, Lintunen M, Kronqvist P, Sandholm J, Mönkkönen J, Kellokumpu-Lehtinen P, Lauttia S, Tynninen O, Joensuu H, Heymann D, Määttä JA, Human breast cancer cells educate macrophages toward the M2 activation status, Breast Cancer Res. 17 (2015) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Genin M, Clement F, Fattaccioli A, Raes M, Michiels C, M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide, BMC Cancer. 15 (2015) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Trickett A, Kwan YL, T cell stimulation and expansion using anti-CD3/CD28 beads, J Immunol Methods. 275 (2003) 251–255. [DOI] [PubMed] [Google Scholar]
- [44].Wang L, Zhang S, Wu H, Rong X, Guo J, M2b macrophage polarization and its roles in diseases, J Leukoc Biol. 106 (2019) 345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hung C, Chen F, Lin Y, Tsai M, Wang S, Chen Y, Chen Y, Hou M, Altered monocyte differentiation and macrophage polarization patterns in patients with breast cancer, BMC Cancer. 18 (2018) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bondalapati S, Ruvinov E, Kryukov O, Cohen S, Brik A, Rapid End-Group Modification of Polysaccharides for Biomaterial Applications in Regenerative Medicine, Macromol Rapid Commun. 35 (2014) 1754–1762. [DOI] [PubMed] [Google Scholar]
- [47].Girdhari R, Weimin L, 3D scaffolds in breast cancer research, Biomaterials. 81 (2016) 135–156. [DOI] [PubMed] [Google Scholar]
- [48].Li S, Plouffe BD, Belov AM, Ray S, Wang X, Murthy SK, Karger BL, Ivanov AR, An Integrated Platform for Isolation, Processing, and Mass Spectrometry-based Proteomic Profiling of Rare Cells in Whole Blood, Mol Cell Proteom. 14 (2015) 1672–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Krämer A, Green J, Pollard J, Tugendreich S, Causal analysis approaches in Ingenuity Pathway Analysis, Bioinformatics. 30 (2014) 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV, STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets, Nucleic Acids Res. 47 (2019) D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yuan F, Dellian M, Fukumura D, Leunig M, Berk DA, Torchilin VP, Jain RK, Vascular permeability in a human tumor xenograft: molecular size dependence and cutoff size, Cancer Res. 55 (1995) 3752–3756. [PubMed] [Google Scholar]
- [52].Acerbi I, Cassereau L, Dean I, Shi Q, Au A, Park C, Chen YY, Liphardt J, Hwang ES, Weaver VM, Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration, Integr Biol. 7 (2015) 1120–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lopez JI, Kang I, You W, McDonald DM, Weaver VM, In situ force mapping of mammary gland transformation, Integr Biol. 3 (2011) 910–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Raggi F, Pelassa S, Pierobon D, Penco F, Gattorno M, Novelli F, Eva A, Varesio L, Giovarelli M, Bosco MC, Regulation of human macrophage M1–M2 polarization balance by hypoxia and the triggering receptor expressed on myeloid cells-1, Front Immunol. 8 (2017) 1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Spiller KL, Anfang RR, Spiller KJ, Ng J, Nakazawa KR, Daulton JW, Vunjak-Novakovic G, The role of macrophage phenotype in vascularization of tissue engineering scaffolds, Biomaterials. 35 (2014) 4477–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Banerjee A, Arha M, Choudhary S, Ashton RS, Bhatia SR, Schaffer DV, Kane RS, The influence of hydrogel modulus on the proliferation and differentiation of encapsulated neural stem cells, Biomaterials. 30 (2009) 4695–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kalli M, Stylianopoulos T, Defining the role of solid stress and matrix stiffness in cancer cell proliferation and metastasis, Front oncol. 8 (2018) 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lovitt CJ, Shelper TB, Avery VM, Evaluation of chemotherapeutics in a three-dimensional breast cancer model, J. Cancer Res. Clin. Oncol. 141 (2015) 951–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Na Y, Yoon Y, Son D, Seok S, Cyclooxygenase-2 inhibition blocks M2 macrophage differentiation and suppresses metastasis in murine breast cancer model, PLOS ONE. 8 (2013) e63451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Dustin ML, The immunological synapse, Cancer immunology research. 2 (2014) 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Okoh VO, Garba NA, Penney RB, Das J, Deoraj A, Singh KP, Sarkar S, Felty Q, Yoo C,Jackson RM, Roy D, Redox signalling to nuclear regulatory proteins by reactive oxygen species contributes to oestrogen-induced growth of breast cancer cells, Br J Cancer. 112 (2015) 1687–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Johar Ramesh, Sharma Rohit, Kaur Amanpreet, Mukherjee Tapan K., Role of Reactive Oxygen Species in Estrogen Dependant Breast Cancer Complication, Anticancer Agents Med Chem. 16 (2016) 190–199. [DOI] [PubMed] [Google Scholar]
- [63].Moloney JN, Cotter TG, ROS signalling in the biology of cancer, Semin. Cell Dev. Biol. 80 (2018) 50–64. [DOI] [PubMed] [Google Scholar]
- [64].Vander Heiden MG, Cantley LC, Thompson CB, Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation, Science. 324 (2009) 1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhang Y, Choksi S, Chen K, Pobezinskaya Y, Linnoila I, Liu Z, ROS play a critical role inthe differentiation of alternatively activated macrophages and the occurrence of tumor-associated macrophages, Cell research. 23 (2013) 898–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Mukai M, Kusama T, Hamanaka Y, Koga T, Endo H, Tatsuta M, Inoue M, Cross talk between apoptosis and invasion signaling in cancer cells through caspase-3 activation, Cancer Res. 65 (2005) 9121–9125. [DOI] [PubMed] [Google Scholar]
- [67].Liu Y, Sun B, Zhao X, Gu Q, Liu Z, Dong X, Che N, Mo J, Basal caspase-3 activity promotes migration, invasion, and vasculogenic mimicry formation of melanoma cells, Melanoma Res. 23 (2013) 243–253. [DOI] [PubMed] [Google Scholar]
- [68].Heshiki W, Tomihara K, Yamazaki M, Arai N, Nakamori K, Noguchi M, Constitutive activation of caspase-3 in non-apoptotic oral squamous cell carcinoma cells, J Cancer Sci Ther. 7 (2015) 75–80. [Google Scholar]
- [69].Donato AL, Huang Q, Liu X, Li F, Zimmerman MA, Li C, Caspase 3 promotes surviving melanoma tumor cell growth after cytotoxic therapy, J Invest Dermatol. 134 (2014) 1686–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Sankari SL, Masthan K, Babu NA, Bhattacharjee T, Elumalai M, Apoptosis in cancer-an update, Asian Pac J Cancer Prev. 13 (2012) 4873–4878. [DOI] [PubMed] [Google Scholar]
- [71].Ma XC, Hattori T, Kushima R, Terata N, Kodama M, Expression of HLA-class II antigen in gastric carcinomas: its relationship to histopathological grade, lymphocyte infiltration and five-year survival rate, Acta Oncol. 33 (1994) 187. [DOI] [PubMed] [Google Scholar]
- [72].Chen EI, Mitochondrial dysfunction and cancer metastasis, J Bioenerg Biomembr. 44 (2012) 619–622. [DOI] [PubMed] [Google Scholar]
- [73].Costa-Machado LF, Fernandez-Marcos PJ, The sirtuin family in cancer, Cell Cycle. 18 (2019) 2164–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Elangovan S, Ramachandran S, Venkatesan N, Ananth S, Gnana-Prakasam JP, Martin PM, Browning DD, Schoenlein PV, Prasad PD, Ganapathy V, Thangaraju M, SIRT1 is essential for oncogenic signaling by estrogen/estrogen receptor alpha in breast cancer, Cancer Res. 71 (2011) 6654–6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Rouschop KM, Dubois LJ, Keulers TG, van den Beucken T, Lambin P, Bussink J, van der Kogel, Albert J., Koritzinsky M, Wouters BG, PERK/eIF2a signaling protects therapy resistant hypoxic cells through induction of glutathione synthesis and protection against ROS, Proc Natl Acad Sci USA. 110 (2013) 4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Konopka B, Szafron LM, Kwiatkowska E, Podgorska A, Zolocinska A, Pienkowska-Grela B, Dansonka-Mieszkowska A, Balcerak A, Lukasik M, Stachurska A, Timorek A, Spiewankiewicz B, El-Bahrawy M, Kupryjanczyk J, The significance of c.690G>T polymorphism (rs34529039) and expression of the CEBPA gene in ovarian cancer outcome, Oncotarget. 7 (2016) 67412–67424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Bae YK, Choi JE, Kang SH, Lee SJ, Epithelial-Mesenchymal Transition Phenotype Is Associated with Clinicopathological Factors That Indicate Aggressive Biological Behavior and Poor Clinical Outcomes in Invasive Breast Cancer, J Breast Cancer. 18 (2015) 256–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Li W, Kang Y, Probing the fifty shades of EMT in metastasis, Trends Cancer. 2 (2016) 65–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Yamashita N, Tokunaga E, Iimori M, Inoue Y, Tanaka K, Kitao H, Saeki H, Oki E,Maehara Y, Epithelial paradox: clinical significance of coexpression of E-cadherin and vimentin with regard to invasion and metastasis of breast cancer, Clin Breast Cancer. 18 (2018) e1003–e1009. [DOI] [PubMed] [Google Scholar]
- [80].Pang M, Georgoudaki A, Lambut L, Johansson J, Tabor V, Hagikura K, Jin Y, Jansson M, Alexander J, Nelson CM, TGF-β1-induced EMT promotes targeted migration of breast cancer cells through the lymphatic system by the activation of CCR7/CCL21-mediated chemotaxis, Oncogene. 35 (2016) 748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Fuxe Jonas and Mikael CI Karlsson, TGF-β-induced epithelial-mesenchymal transition: a link between cancer and inflammation, Semin Cancer Biol. 22 (2012) 455–461-461. Academic Press. [DOI] [PubMed] [Google Scholar]
- [82].Shabo I, Stål O, Olsson H, Doré S, Svanvik J, Breast cancer expression of CD163, a macrophage scavenger receptor, is related to early distant recurrence and reduced patient survival, Int J Cancer. 123 (2008) 780–786. [DOI] [PubMed] [Google Scholar]
- [83].Blum JS, Wearsch PA, Cresswell P, Pathways of antigen processing, Annu Rev Immunol.31 (2013) 443–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Dongre A, Weinberg RA, New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer, Nat Rev Mol Cell Biol. 20 (2019) 69–84. [DOI] [PubMed] [Google Scholar]
- [85].Helm O, Held-Feindt J, Grage-Griebenow E, Reiling N, Ungefroren H, Vogel I, Krüger U, Becker T, Ebsen M, Röcken C, Kabelitz D, Schäfer H, Sebens S, Tumor-associated macrophages exhibit pro- and anti-inflammatory properties by which they impact on pancreatic tumorigenesis, Int J Cancer. 135 (2014) 843–861. [DOI] [PubMed] [Google Scholar]
- [86].Sarrió D, Rodriguez-Pinilla S, Hardisson D, Cano A, Moreno-Bueno G, Palacios J, Epithelial-Mesenchymal Transition in Breast Cancer Relates to the Basal-like Phenotype, Cancer Res. 68 (2008) 989. [DOI] [PubMed] [Google Scholar]
- [87].Księżakowska-Łakoma K, Żyła M, Wilczyński JR, Mitochondrial dysfunction in cancer, Prz Menopauzalny. 13 (2014) 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Pagano G, Aiello Talamanca A, Castello G, Cordero MD, d’Ischia M, Gadaleta MN, Pallardó FV, Petrović S, Tiano L, Zatterale A, Oxidative stress and mitochondrial dysfunction across broad-ranging pathologies: toward mitochondria-targeted clinical strategies, Oxid Med Cell Longev. 2014. (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zapata JM, Krajewska M, Krajewski S, Huang R, Takayama S, Wang H, Adamson E, Reed JC, Expression of multiple apoptosis-regulatory genes in human breast cancer cell lines and primary tumors, Breast Cancer Res Treat. 47 (1998) 129–140. [DOI] [PubMed] [Google Scholar]
- [90].Glasauer A, Chandel NS, Targeting antioxidants for cancer therapy, Biochem Pharmacol. 92 (2014) 90–101. [DOI] [PubMed] [Google Scholar]
- [91].Bogdanović V, Turšijan S, Đorđević M, Nikolić A, Mrđanović J, Jakimov D, Trajković V, Bogdanović G, Ivković-Kapicl T, Dugandžija T, Activity of lactate dehydrogenase and superoxide dismutase in the circulation of patients with breast carcinoma, Arch Oncol. 16 (2008) 39–41. [Google Scholar]
- [92].Portakal O, zkaya, Erden nal M, Bozan B, Ko an M, Sayek I, Coenzyme Q10 concentrations and antioxidant status in tissues of breast cancer patients, Clin Biochem. 33 (2000) 279–284. [DOI] [PubMed] [Google Scholar]
- [93].Khanzode SS, Muddeshwar M, Khanzode SD, Dakhale GN, Antioxidant enzymes and lipidp eroxidation in different stages of breast cancer, Free Radic Res. 38 (2004) 81–85. [DOI] [PubMed] [Google Scholar]
- [94].Pande D, Negi R, Karki K, Khanna S, Khanna RS, Khanna HD, Oxidative damage markers as possible discriminatory biomarkers in breast carcinoma, Transl Res. 160 (2012) 411–418. [DOI] [PubMed] [Google Scholar]
- [95].Nicco C, Laurent A, Chereau C, Weill B, Batteux F, Differential modulation of normal and tumor cell proliferation by reactive oxygen species, Biomed Pharmacother. 59 (2005) 169–174. [DOI] [PubMed] [Google Scholar]
- [96].Sordet O, Rébé C, Plenchette S, Zermati Y, Hermine O, Vainchenker W, Garrido C, Solary E, Dubrez-Daloz L, Specific involvement of caspases in the differentiation of monocytes into macrophages, Blood. 100 (2002) 4446–4453. [DOI] [PubMed] [Google Scholar]
- [97].Ohara M, Yamaguchi Y, Matsuura K, Murakami S, Arihiro K, Okada M, Possible involvement of regulatory T cells in tumor onset and progression in primary breast cancer, Cancer Immunol Immunother. 58 (2009) 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Llanes-Fernández L, Álvarez-Goyanes RI, Arango-Prado MdC, Alcocer-González JM, Mojarrieta JC, Pérez XE, López MO, Odio SF, Camacho-Rodríguez R, Guerra-Yi ME, Madrid-Marina V, Tamez-Guerra R, Rodríguez-Padilla C, Relationship between IL-10 and tumor markers in breast cancer patients, Breast J. 15 (2006) 482. [DOI] [PubMed] [Google Scholar]
- [99].Liu C, Liu J, Wang J, Liu Y, Zhang F, Lin W, Gao A, Sun M, Wang Y, Sun Y, B7-H3 expression in ductal and lobular breast cancer and its association with IL-10, Mol Med Rep. 7 (2013) 134–138. [DOI] [PubMed] [Google Scholar]
- [100].Cardillo MR, Ippoliti F, IL-6, IL-10 and HSP-90 expression in tissue microarrays from human prostate cancer assessed by computer-assisted image analysis, Anticancer Res. 26 (2006) 3409–3416. [PubMed] [Google Scholar]
- [101].Dunne MR, Phelan JJ, Michielsen AJ, Maguire AA, Dunne C, Martin P, Noonan S, Tosetto M, Geraghty R, Fennelly D, Sheahan K, Ryan EJ, O’Sullivan J, Characterising the prognostic potential of HLA-DR during colorectal cancer development, Cancer Immunol Immunother. 69 (2020) 1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Gu G, Gao T, Zhang L, Chen X, Pang Q, Wang Y, Wang D, Li J, Liu Q, NKAP alters tumor immune microenvironment and promotes glioma growth via Notch1 signaling, J Exp Clin Cancer Res. 38 (2019) 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Choi S, Kim M, Yoon Y, Koh D, Kim M, Cho S, Kim K, Hur M, Hypoxia-induced RelA/p65 derepresses SLC16A3 (MCT4) by downregulating ZBTB7A, Biochim Biophys Acta Gene Regul Mech. 1862 (2019) 771–785. [DOI] [PubMed] [Google Scholar]
- [104].Pearce EJ, Everts B, Dendritic cell metabolism, Nat Rev Immunol. 15 (2015) 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Szajnik M, Czystowska M, Szczepanski MJ, Mandapathil M, Whiteside TL, Tumor-Derived Microvesicles Induce, Expand and Up-Regulate Biological Activities of Human Regulatory T Cells (Treg), PLOS ONE. 5 (2010) e11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J, Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells, Nat Cell Biol. 10 (2008) 619–624. [DOI] [PubMed] [Google Scholar]
- [107].Di Vizio D, Kim J, Hager MH, Morello M, Yang W, Lafargue CJ, True LD, Rubin MA, Adam RM, Beroukhim R, Demichelis F, Freeman MR, Oncosome Formation in Prostate Cancer: Association with a Region of Frequent Chromosomal Deletion in Metastatic Disease, Cancer Res. 69 (2009) 5601–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Ono M, Molecular links between tumor angiogenesis and inflammation: inflammatory stimuli of macrophages and cancer cells as targets for therapeutic strategy, Cancer Sci. 99 (2008) 1501–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Lanigan F, O’Connor D, Martin F, Gallagher WM, Molecular links between mammary gland development and breast cancer, Cell Mol Life Sci. 64 (2007) 3159–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kang W, Sarkar S, Lin ZS, McKenney S, Konry T, Ultrafast parallelized microfluidic platform for antimicrobial susceptibility testing of Gram positive and negative bacteria, Anal Chem. 91 (2019) 6242–6249. [DOI] [PubMed] [Google Scholar]
- [111].Berger Fridman I, Ugolini GS, Vandelinder V, Cohen S, Konry T, High throughput microfluidic system with multiple oxygen levels for the study of hypoxia in tumor spheroids, Biofabrication. 13 (2021) [DOI] [PubMed] [Google Scholar]
- [112].Zervantonakis IK, Hughes-Alford S, Charest JL, Condeelis JS, Gertler FB, Kamm RD, Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function, Proc Natl Acad Sci USA. 109 (2012) 13515–13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu Q, Wang R, Hermjakob H, 2016 update of the PRIDE database and its related tools, Nucleic Acids Res. 44 (2016) D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data supporting the findings of this study are available within the article and its supplementary information files and from the corresponding author upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository[113] with the dataset identifier PXD021509.