Abstract
Background:
Advances in imaging technology have the potential to transform the early diagnosis and treatment of hepatocellular carcinoma (HCC) through quantitative image analysis. Computational ‘radiomic’ techniques extract biomarker information from images which can be used to improve diagnosis and predict tumor biology.
Aims:
We performed a systematic review of literature on radiomic features in HCC diagnosis and prognosis, with a focus on reporting metrics and methodologic standardization.
Methods:
A systematic review was performed of all full-text articles published from inception through December 1, 2019. Standardized data extraction and quality assessment metrics were applied to all studies.
Results:
A total of 54 unique studies were included for analysis. Radiomic features demonstrated good discriminatory performance to differentiate HCC from other solid lesions (c-statistics 0.66–0.95), predict microvascular invasion (c-statistic 0.76–0.92), predict early recurrence after hepatectomy (c-statistics 0.71–0.86), and predict prognosis after locoregional or systemic therapies (c-statistics 0.74–0.81). Common stratifying features for diagnostic and prognostic radiomic tools included analyses of imaging skewness, analysis of the peritumoral region, and feature extraction from the arterial imaging phase. The overall quality of the included studies was low, with common deficiencies in both internal and external validation, standardized imaging segmentation, and lack of comparison to a gold standard.
Conclusions:
Quantitative image analysis demonstrates promise as a non-invasive biomarker to improve HCC diagnosis and management. However, standardization of protocols and outcome measurement, sharing of algorithms and analytic methods, and external validation are necessary prior to widespread application of radiomics to HCC diagnosis and prognosis in clinical practice.
Keywords: HCC, radiogenomics, MRI, early detection, prognosis, biomarker
Graphical Abstract
Introduction:
Hepatocellular carcinoma (HCC) is the fourth most common cause of cancer-associated death worldwide and the fastest-growing cause of cancer death in the United States.1,2 The rising mortality associated with HCC is driven in part by limitations in the screening and early detection of HCC. Most HCC is diagnosed at an advanced stage when curative treatment options are limited.3 There are few available diagnostic and risk stratification tools to prioritize at-risk populations for surveillance and early detection of HCC. The Liver Imaging Reporting and Data System (LI-RADS or LR) criteria was established to provide standardized criteria for the radiographic diagnosis of HCC.4 However, validation of these criteria remains limited and there are two categories of indeterminate nodules (i.e. LR3 and LR4) for which there is uncertainty regarding diagnostic approach.5 Biopsy for the diagnosis of HCC is not currently routinely recommended by guidelines due to the risk of tumor seeding, bleeding and sampling error.6 Similarly, the only validated noninvasive prognostic markers for HCC are tumor staging and alpha-fetoprotein levels, both of which have significant limitations in approximating tumor biology.7,8 This is particularly important in light of recent data showing variation in tumor growth patterns, with one-fourth of HCC having rapid growth patterns and over one-third having indolent growth.9,10 There remains an unmet need for non-invasive biomarkers to aid in the early detection of HCC and prediction of tumor behavior. ‘Radiomics,’ a term that describes the ‘omics’ approach for analysis of imaging data, has emerged as a novel tool for of the diagnosis and prognosis of HCC.11 Radiomics leverages advanced computing tools to extract deeper and more granular data from imaging.12 Quantitative image features predictive of tumor behavior, treatment response, and overall outcomes have been identified in other malignancies including breast, pancreatic, and lung cancer.13–16
Quantitative Imaging
Advanced image analysis is frequently divided into two categories: semantic and quantitative. The term ‘semantic’ refers to radiologist-derived image features such as the presence of internal arteries, hypodense halos, and tumor-liver difference.17,18 The clinical utility of semantic imaging features has been limited by labor-intensive extraction process and concerns about suboptimal inter- and intra-observer reliability (k-0.50–0.70).19 ‘Quantitative’ imaging features, also known as agnostic features, by comparison, are computer-derived mathematically extracted quantitative image characteristics of the tissue of interest.20 These are extracted by analytic software and can be categorized into morphologic (shape) and statistical (first-order, second-order, and higher-order) features based on complexity.20–23 Varying analytic approaches have been used for radiomics studies including traditional regression analysis or machine learning approaches to measure the association between voxel data and a clinical outcome of interest.
Radiomic analysis involves five primary steps, as outlined in Figure 1: image acquisition, tumor segmentation, feature extraction, feature selection, and model creation.24 Image acquisition refers to the process of collecting and reconstructing imaging studies in a manner that minimizes variations in extracted numerical data.25 Tumor segmentation refers to the selection of regions of interest (ROI) around tumoral tissue. This can be performed either manually or with the assistance of semi-automatic contour selection tools.26 Tumor segmentation is a major source of inter-reader variability and can introduce biases in quantification.27 Feature extraction refers to the application of specialized software to derive quantitative descriptions of the voxel patterns in each image, generating thousands of individual variables. The distribution of the voxel intensity values is considered first-order features and the spatial relationship of the voxels, also known as texture analysis, is considered second-order features.28 Feature Selection is the process of using supervised or unsupervised statistical analysis to identify the variables most predictive of the desired outcome measure. Because of the large number of variables, radiomic studies must also perform considerable dimensionality reduction to reduce the risk of overfitting.29 Model Creation: refers to the creation of a nomogram or multivariable model using the most successful radiomic variables. Generally, the best-performing models incorporate a combination of established clinical and pathological biomarkers with radiomic data. In this systematic review, we aimed to evaluate the role of radiomic tools for diagnosis and prognosis of HCC.
Figure 1:
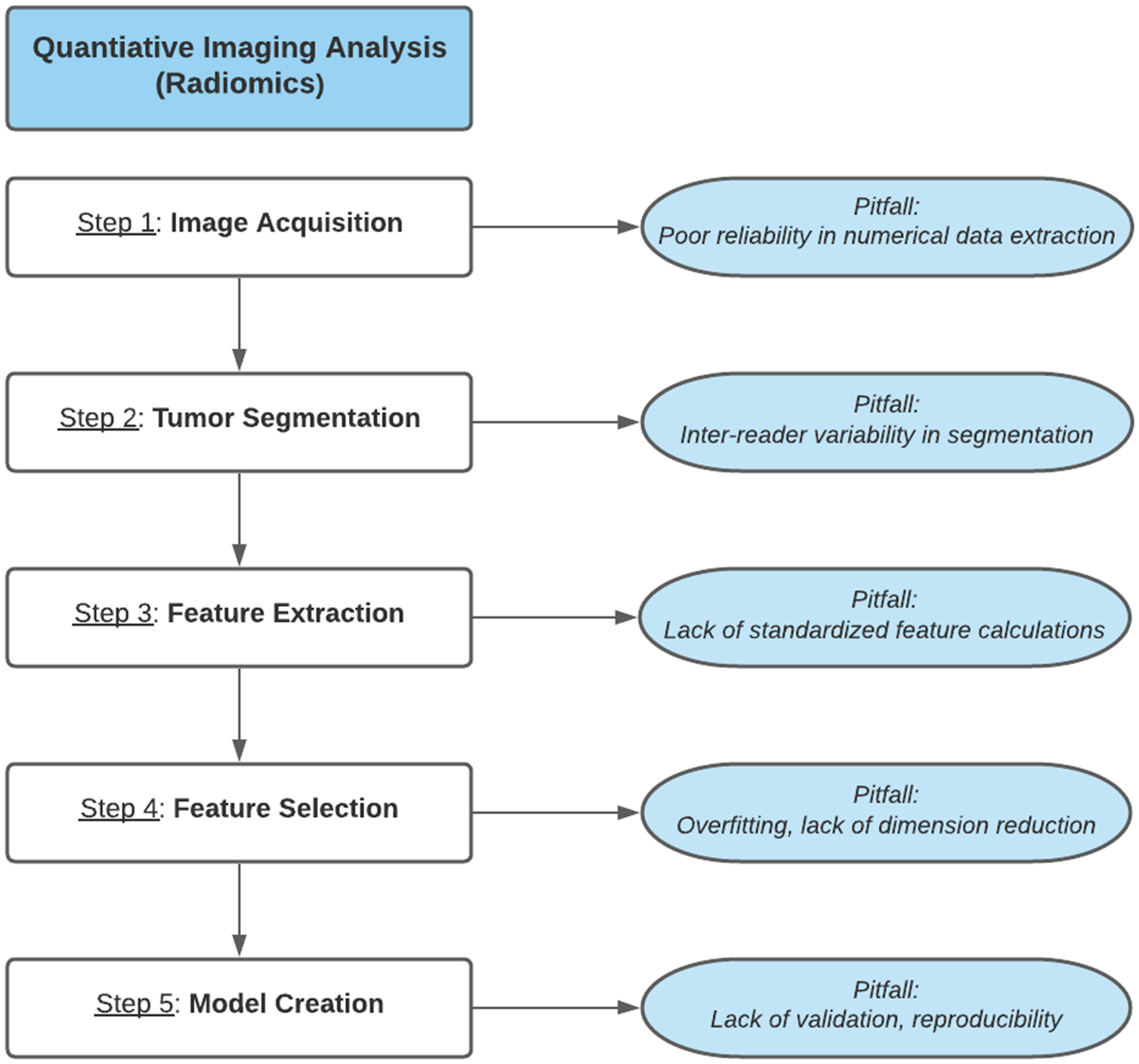
Radiomics analysis workflow with common pitfalls
Methods:
With the assistance of a trained librarian (WT), we performed a systematic review of the literature, concordant with the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines.30 The literature review captured studies from inception to December 1, 2019 in PubMed Legacy, Embase.com, Scopus.com, Web of Science Core Collection (SCI-EXPANDED, SSCI, A&HCI, CPCI-S, CPCI-SSH, BKCI-S, BKCI-SSH, ESCI, CCR-EXPANDED), Cochrane CENTRAL via Wiley, and clinicaltrials.gov. The full search strategy including MESH headings is listed in Supplementary Methods. References of selected articles were also reviewed to identify additional articles. Duplicates were eliminated automatically. Inclusion criteria included those that met the following criteria:
Evaluated HCC using an MRI or CT-based radiomics approach AND
Provided information relating to diagnosis (detection, characterization) or
Provided information relating to prognosis (microvascular invasion, response to therapy, survival, recurrence rate)
Exclusion criteria included the following: (1) studies not available in English with translation, (2) non-peer reviewed articles (3) ultrasound based studies (4) studies without an outcome measure (5) studies exclusively focused on semantic imaging features. This search identified a total of 754 unique records, 116 articles were assessed for eligibility of which 54 met inclusion criteria. Figure 2 describes our selection process and reasons for study exclusion.
Figure 2:
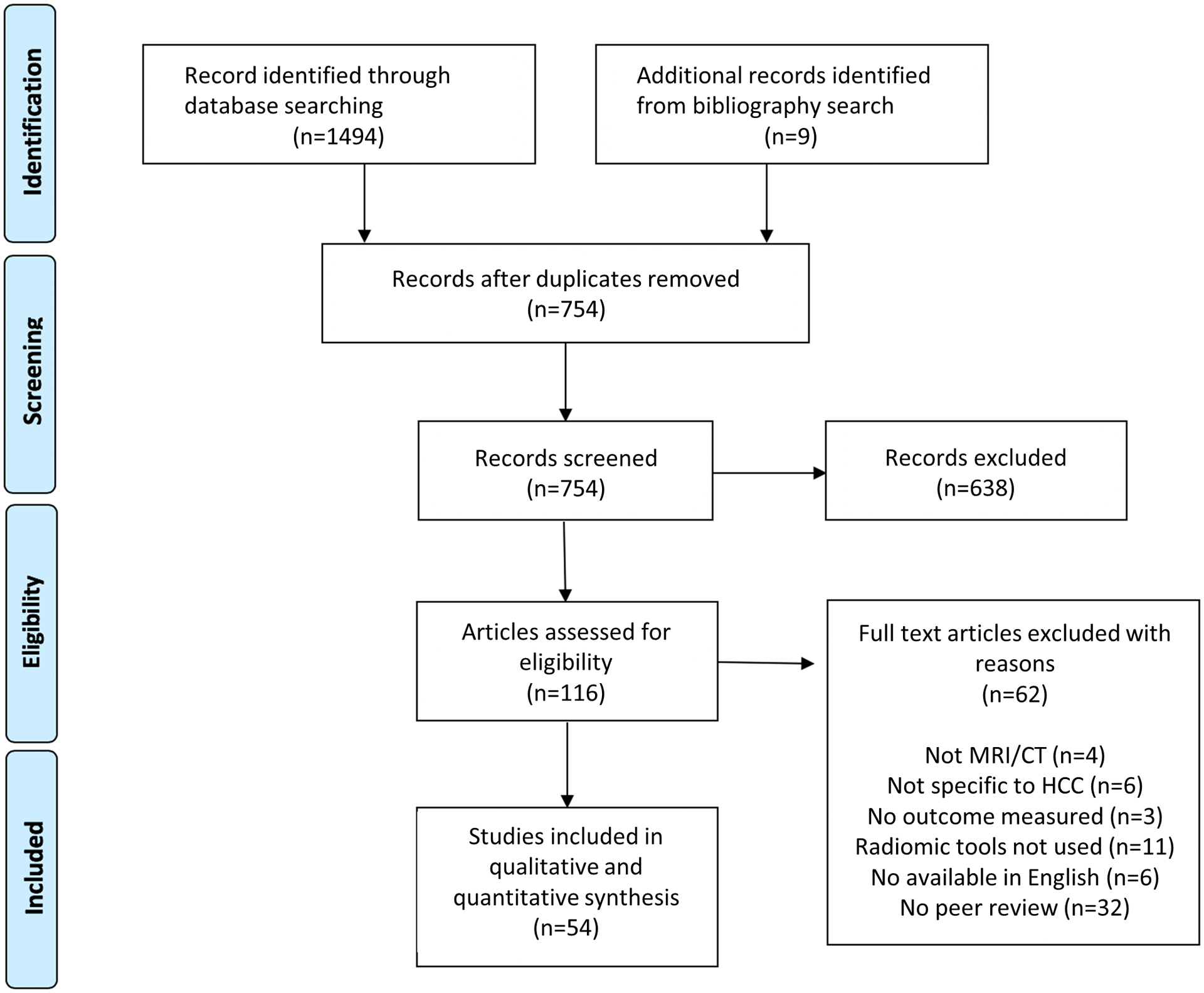
Literature search algorithm for generation of MRI and CT-based radiomic studies
Two authors (E.H.T., B.M.) independently reviewed all papers for eligibility. Studies were categorized into five groups: diagnosis, prognosis, microvascular invasion (MVI), pathologic correlates, and treatment response. Data extraction using standardized forms was then performed by three authors (E.H.T., E.C., B.M.) independently and discrepancies were resolved by consensus. Data were intended for meta-analysis; however, due to large differences in techniques/methodologies between the studies this was not possible.
Quality Assessment:
Two reviewers (E.H.T. and B.M.) applied the radiomics quality score (RQS) to assess radiomic studies on the basis of 16 components (score range 0–36). Each reviewer individually scored studies and discrepant results were adjudicated by consensus. The score is based on clinical utility, feature reduction, image protocol quality, multivariable analysis, gold standard comparison, cut-off analysis, discrimination statistics, multiple segmentations, biological correlates, calibration statistics, validation, prospective study, multiple timepoints, phantom studies, open science data, and cost-effectiveness analysis.31
Results
HCC Diagnosis
Of the 54 studies met eligibility criteria, 9 evaluated aspects of HCC diagnosis (Table 1). Four studies primarily focused on distinguishing hepatic hemangiomas from HCC.32–35 Mokrane et al evaluated 178 patients with indeterminate nodules and sought to categorize the nodules as high- or low-risk for HCC. They demonstrated an AUC of 0.74 in a training cohort and 0.66 in a validation cohort using CT scans.36 Dankerl et al demonstrated that radiomic tools could outperform radiologists at predicting lesion histology (benign vs malignant) with an accuracy of 75.1% compared with a range of 52–74% for radiologists depending on level of experience.37 Stocker et al also compared radiomic features against human radiologists, and demonstrated that a combination of 13 arterial phase features outperformed radiologists at distinguishing HCC from non-malignant tumors.38 The majority of diagnostic studies have relied on textural features alone, however, even simple textural features have not been directly comparable between protocols. Asayama et al demonstrated that the noncancerous hepatic parenchyma of livers with HCC exhibits a consistent pattern of high kurtosis and low skewness on MRI, compared to patients without HCC, indicating that patients with HCC have features in the background parenchyma that can predict risk of HCC.39 Rosenkrantz et al found that high MRI skewness (a measure of lateral histogram distortion) in an indeterminate liver lesion was associated with the progression of indeterminate lesions to malignant HCC.40
Table 1:
Studies evaluating radiomic tools for early diagnosis in hepatocellular carcinoma.
| Author | CT/MRI | N (Train / Valid) | Extraction Tool | Specific Outcome Measured | Statistical Result | Clinical Model | RQS |
|---|---|---|---|---|---|---|---|
| Dankerl 2013 | CT | 372 | CADx | Differentiation of benign vs. malignant lesion (nodule vs. HCC) | AUC 0.75 for textural features AUC 0.91 for texture + semantic |
No | 5 |
| Song 2019 | CT | 84 | Omni-Kinetic | Differentiation of benign vs. malignant lesion (HCC vs. HH vs. FNH vs. HA) | AUC 0.927 for textural features | No | 9 |
| Stocker 2018 | MRI | 108 | Matlab | Differentiation of benign vs. malignant lesion | AUC 0.92 arterial phase | No | 7 |
| Li 2017 | MRI | T: 112 V:50 |
Internal | Differentiation of HH from HCC | AUC 0.73 for GLCM Energy-mean | No | 10 |
| Oyama 2019 | MRI | T: 50,50 V: 50 |
Matlab | Differentiation of HH from HCC | AUC 0.95 textural features | No | 9 |
| Wu 2019 | MRI | 369 | Internal | Differentiation of HH from HCC | AUC 0.89 textural features | No | 8 |
| Mokrane 2019 | CT | T: 142 V: 36 |
Internal | Categorize indeterminate nodule as high-risk or low-risk for HCC | AUC 0.74 for training cohort AUC 0.66 for validation cohort |
No | 10 |
| Asayama 2016 | MRI | 84 | Internal | Comparison of individual textural features of non-cancerous parenchyma between those with and without HCC | p = 0.0006 for kurtosis p = 0.0152 for skewness |
No | 6 |
| Rosenkrantz 2015 | MRI | 20 | Internal | Progression of hypovascular nodule to likely HCC on subsequent MRI | AUC 0.68 for skewness | No | 7 |
CT: computed tomography; MRI: magnetic resonance imaging; AUC: area under the curve; HCC: hepatocellular carcinoma; HH: hepatic hemangioma; FNH: focal nodular hyperplasia; HA: hepatic adenoma
Prediction of Prognosis
Seventeen studies evaluated HCC prognosis following hepatectomy (Table 2). These were primarily performed with CT imaging; five studies involved MRI. Eight studies evaluated prediction of early recurrence,41–48 and eight evaluated overall survival and recurrence-free survival,49–56 and one evaluated post hepatectomy acute liver failure.57 Radiomic models predicted early recurrence with AUCs that varied between 0.71 and 0.86. When only second-order textural features were included, skewness was the most commonly identified feature predictive of outcomes. Oh et al reported that skewness predicted overall survival with a HR of 10.96 (95% CI: 3.21–37.46), compared with microvascular invasion with a HR of 2.12 (95% CI: 1.06–4.25).49 Defour et al performed multivariable analysis of textural features in the portal-venous phase and found skewness to be associated with overall survival with a HR of 438.7 (95% CI: 2.44–78,968.25).52 The majority of studies used higher-order radiomic features. Kim et al evaluated 168 patients using a 3-dimensional technique which extracted 3,903 radiomic features per patient and found that high-order feature analysis performed similarly to a combined clinical model (age, hepatitis C, alcohol use, cirrhosis, tumor capsule, and microvascular invasion) in predicting early recurrence.45 The authors also demonstrated that the inclusion of 3mm of peritumoral tissue improved risk prediction over segmenting the tumor alone.45 Nine studies compared their radiomic tools against clinical models or created a combined model using both radiomic and clinical features. In all cases, the combined model was equal or superior to the clinical model alone. The characteristics of these studies are described in Supplementary Table 1. In one of the largest studies, Zhou et al compared a clinical model (based on serum alpha fetoprotein, vascular invasion, and non-smooth tumor margin) against a combined model for prediction of early recurrence (ER) in 214 patients and patients with HCC had differential background liver texture. The addition of a 21-feature radiomics signature improved the clinical model AUC from 0.781 to 0.836 when clinical features were used in combination with radiomic data.43
Table 2:
Studies evaluating radiomic tools for prediction of microvascular invasion in hepatocellular carcinoma.
| Author | CT /MRI |
N (Train/Valid) | Extraction Tool | Segment Tool | Specific Outcome Measured | Statistical Result | Clinical Model | RQS |
|---|---|---|---|---|---|---|---|---|
| Bakr 2017 | CT | 28 | Internal | Manual ROI | Prediction of microvascular invasion | AUC 0.76 Texture analysis of MVI | Semantic Model | 6 |
| Ma 2019 | CT | T: 110 V: 47 |
Matlab | Manual ROI | Prediction of microvascular invasion (compares portal venous phase vs. arterial phase) | AUC 0.793 Portal Venous Phase for MVI | Clinical Model | 10 |
| Zheng 2017 | CT | 120 | Matlab | Semi-Automatic ROI | Prediction of microvascular invasion (compares tumors < 5cm vs. > 5cm) | AUC 0.80 for single feature (angle co-occurrence matrix) if < 5cm AUC 0.75 for single feature (local binary pattern) if > 5cm |
Clinical Model | 6 |
| Xu 2019 | CT | T: 350 V: 145 |
Python | Semi-Automatic VOI | Prediction of microvascular invasion (combined clinical + agnostic + radiomic model) | AUC 0.909 training/validation AUC 0.889 test |
Clinical Model | 11 |
| Feng 2019 | MRI | T: 110 V: 50 |
Internal | Manual VOI | Prediction of microvascular invasion using both intra-tumoral and peritumoral regions | AUC 0.850 training AUC 0.833 validation |
No | 12 |
| Zhang 2019 | MRI | T: 194 V: 73 |
Matlab | Manual ROI | Prediction of microvascular invasion (radiomic score compared against nomogram) | AUC 0.784 training for rad signature AUC 0.820 validation for rad signature |
Clinical Model | 12 |
| Zhu 2019 | MRI | 142 | Omni-Kinetics | Manual ROI | Prediction of microvascular invasion (arterial phase vs. portal venous phase) | AUC 0.765 training for arterial AUC 0.773 validation for arterial |
Clinical Model | 11 |
CT: computed tomography; MRI: magnetic resonance imaging; AUC: area under the curve; ROI: region of interest
Prediction of Microvascular Invasion
Microvascular invasion (MVI) is among the strongest predictors of outcomes following liver transplantation or hepatectomy for HCC.58,59. Seven studies evaluated radiomics as a tool for prediction of MVI on explant following hepatectomy.60–66 (Table 3) These studies reported AUCs ranging from 0.76 to 0.91. Six of the studies evaluated their result against a clinical model, and in all cases the combined model performed comparably or better than the clinical model. Xu et al, in the largest study to date, evaluated CT scans from 495 patients and found that a combination of clinical, radiologic, and radiomic features predicted histologic MVI with an AUC of 0.909 in the training/validation and 0.889 in an separate test set.63 Clinical features included aspartate aminotransferase (AST) and alpha fetoprotein (AFP), while radiologist-derived features included non-smooth tumor margin, extrahepatic growth, ill-defined pseudo-capsule, and peritumoral arterial enhancement, as well as the presence of a previously published radio-genomic venous invasion signature. The authors also compared the use of the 3-dimensional VOI of the tumor only against a volume which extends 5mm in every direction from the tumor. Although MVI occurs primarily at the periphery of tumors, the inclusion of peritumoral tissue in the VOI did not improve on the prediction of MVI.63 To create a simple decision tool, Zhang et al published a nomogram for the prediction of MVI which includes a radiomic score and alpha fetoprotein, tumor type, peritumoral enhancement, arterial rim and internal arteries.65 This nomogram outperformed a clinical and radiologic model with an AUC of 0.858 vs. 0.729. In the limited studies examining MRI radiomic tools, the arterial phase of the image predicted MVI more effectively than venous phase.63
Table 3:
Studies evaluating radiomic tools for prognosis in hepatocellular carcinoma.
| Author | CT/MRI | N (Train/Valid) | Extraction Tool | Segment Tool | Specific Outcome Measured | Statistical Result | Clinical Model | RQS |
|---|---|---|---|---|---|---|---|---|
| Akai 2018 | CT | 127 | TexRAD | Manual ROI | Model categorizes as high risk or low risk for OS and DFS | P < 0.0001 for OS from Kaplan Meier LR | No | 10 |
| Chen 2017 | CT | 61 | Matlab | Manual ROI | Prediction of OS and RFS with individual features | P = 0.001 for OS from Kaplan Meier LR | No | 9 |
| Defour 2018 | CT | 47 | TexRAD | Manual ROI | Prediction of OS and RFS with individual textural features | P = 0.0084 of kurtosis in MV of OS | No | 6 |
| Kiryu 2017 | CT | 122 | TexRAD | Manual ROI | Prediction of OS and RFS with individual textural features | P < 0.001 of entropy in Kaplan-Meier LR of OS | No | 7 |
| Peng 2018 | CT | T: 113 V: 64 |
IBEX | Semi-automatic ROI | Radiomic score used to categorize as high risk or low risk for OS and DFS | P < 0.0001 of model in Kaplan-Meier LR of OS | Clinical Model | 13 |
| Guo 2019 | CT | T: 93 V: 40 |
Python | Semi-automatic VOI | Radiomic model as a predictor of RFS | 0.743 Training for RFS 0.705 Validation for RFS |
Clinical Model | 10 |
| Zheng 2019 | CT | T: 212 V: 107 |
Matlab | Manual ROI | Radiomic score and radiomic-score based nomograms used to predict OS | 0.714 Training for OS 0.71 Validation for OS |
Clinical Model | 12 |
| Cai 2019 | CT | T: 80 V: 32 |
Internal | Semi-automatic VOI | Radiomic score used to predict post-hepatectomy acute liver failure | 0.822 training for post-hepatectomy acute liver failure 0.762 validation for post-hepatectomy acute liver failure |
Clinical Model | 10 |
| Oh 2019 | CT | 81 | TexRAD | Manual ROI | Prediction of DFS with individual textural features | P < 0.001 for skewness (SSF2.0) in MV of DFS | No | 9 |
| Ning 2019 | CT | T: 225 V: 100 |
Matlab | Semi-automatic VOI | Prediction of early recurrence after hepatectomy | 0.817 Training for ER 0.719 Validation for ER |
Clinical Model | 9 |
| Shan 2019 | CT | T: 109 V: 47 |
Internal | Manual ROI | Prediction of early recurrence after hepatectomy (models compare peritumoral and tumoral features against tumor enhancement) | 0.80 Training for ER 0.79 Validation for ER |
No | 11 |
| Zhou 2017 | CT | 214 | Matlab | Manual ROI | Prediction of early recurrence after hepatectomy (summary model used) |
0.836 for ER | Clinical Model | 11 |
| Hui 2018 | MRI | 50 | Matlab | Manual ROI | Prediction of early recurrence after hepatectomy (individual radiomic features only) | 0.82 for S(0,3) SumofSqs for ER 0.84 for S(4,0) SumVarnc |
No | 10 |
| Kim 2019 | MRI | T: 129 V: 39 |
Python | Semi-automatic VOI | Prediction of early recurrence after hepatectomy (peritumoral model) | 0.716 for clinical + radiomic model in predicting ER | No | 9 |
| Zhang 2019 | MRI | 100 | Internal | Semi-automatic VOI | Prediction of early recurrence after hepatectomy (individual radiomic features only, <3cm vs. > 3cm) | 0.867 skewness + entropy | No | 10 |
| Zhang 2019 | MRI | T: 108 V: 47 |
Internal | Semi-automatic VOI | Prediction of early recurrence after hepatectomy | 0.757 Training for ER 0.728 Validation for ER |
Clinical Model | 12 |
| Ahn 2019 | MRI | 179 | Internal | Manual ROI | Prediction of early recurrence after hepatectomy (combines agnostic and radiomic) | 0.83 for radiomic + agnostic features for ER | No | 6 |
CT: computed tomography; MRI: magnetic resonance imaging; AUC: area under the curve; ROI: region of interest; OS: overall survival; LR: log rank; DFS: disease free survival; MV: multivariate; ER: early recurrence
Prediction of Pathologic and Molecular Correlates
Ten studies used radiomic features to visually identify the pathologic and genetic correlates of HCC. These include p53 mutation status, Ki-67, and CD8+ T-cell invasion. (Supplementary Table 2).67–76 In a landmark study by Kuo et al, the authors demonstrated an association between radiomic textural features and a doxorubicin drug response gene signature previously shown to be predictive of tumor stage.69 Chen et al demonstrated among 207 patients that radiomic features including the peritumoral region were associated with a validated ‘immunoscore.’ This score characterizes the tumor infiltrating lymphocyte population, and theoretically reflects the immune phenotype of the tumor microenvironment.67
Treatment Response
An additional 11 studies evaluated treatment response, primarily following local-regional therapy (LRT) (Supplementary Table 3).77–87 These studies had the most variability in quality, with a median RQS of 7. Most studies were focused on single textural features and just two studies involved clinical models for comparison. Kim et al demonstrated in 88 patients that a combination of clinical (Child-Pugh score, serum alpha fetoprotein, and tumor size) and radiomic features (surface area-to-volume ration, kurtosis, median, size zone variability) can predict post-TACE overall survival with a HR of 19.88 and 95% CI of 6.37–62.02.79 These findings were also seen in other studies, in which radiomic features extracted from pre-treatment imaging (CT or MRI) for prediction of treatment response after TACE were compared to post-treatment response evaluation.80,87 Mule et al found post-Sorafenib overall survival correlated significantly with individual textural features.83
Assessment of Methodology
Radiomic methods varied significantly between studies. Among quantitative imaging studies, no two groups used the same extraction tool or segmentation process. A majority of studies used proprietary investigator-developed tools which are not publicly available. Study outcomes and reporting methods were heterogeneous. More recently, groups have begun to transition to using software packages, such as Matlab, for data extraction. There was wide variation in the number of features extracted, ranging from 5 to 3,903. Recent studies have also begun to transition from 2 dimensional to 3 dimensional ‘volume of interest’ models as large-scale data analysis becomes streamlined. The use of manual and semi-automatic ROI selection tools also varied significantly between studies, and inter-rater reliability of ROI selection was rarely performed. Indistinct nodules represent a challenge because minor changes in ROI selection can substantially influence the radiomic signature generated. A minority of studies performed internal validation experiments against a portion of their data set, but there were no examples of external validation using imaging derived from outside institutions.
Radiomics Quality Scores
The range of radiomics quality scores reflect the large degree of heterogeneity which currently exists within the field. The median RQS was 9 and the range was 5–13 out of a possible 36 points. The most notable limitations were in studies of cost effectiveness analysis, phantom use, open publication of methods, and prospective study protocol. Quality adherence was highest for feature reduction and discrimination statistics. Notably, the quality has improved over time and studies performed in 2019 consistently scored higher than prior years, primarily through the incorporation of validation cohorts, although most were internal and not external validation with some continued risk of overestimation of model performance.
Discussion
Quantitative image analysis has the potential to transform the early detection and management of HCC. Because high-resolution cross-sectional imaging is already widely available, radiomics has the ability to improve HCC management more rapidly than novel molecular biomarkers. We found radiomic tools to date have been studied primarily for their ability to predict overall survival and early recurrence following hepatectomy and have demonstrated good predictive accuracy, with AUCs exceeding 0.80; however, many of these models have not been tested in validation cohorts including none being externally validated. Fewer studies evaluated response to non-surgical treatments or association with molecular biomarkers, although the ones to date have also demonstrated promising accuracy. As methodology has improved, studies have progressed from simple textural features to thousands of three-dimensional higher-order variables. Studies to date have been limited by small, single-center studies with heterogeneous methods and lack of validation cohorts.
The largest gaps in the use of radiomic technology are in early detection and diagnosis. Only 9 of 54 radiomic studies focused on aspects of HCC diagnosis. Those studies were of variable quality and performed simple radiologic tasks such as distinguishing hepatic hemangiomas from HCC. The next frontier for HCC radiomics will be to assist radiologists with liver nodule risk stratification. This may initially involve the automation of LI-RADS classification, a task which is relatively simple but burdensome for abdominal radiologists. Subsequent tools might also assist in the differentiation of LR3 and LR4 lesions into malignant and benign categories, reducing the number of follow-up imaging studies required to diagnose true HCC from indeterminate nodules. This is particularly important in light of evolving data quantifying potential physical harms related to false-positive and indeterminate surveillance tests.88,89 In addition, further studies of post-treatment survival or recurrence will be needed in response to the increasingly wide array of HCC treatments available. Replication of existing radiographic diagnostic and treatment criteria (e.g. LIRADs and modified Response Evaluation Criteria in Solid Tumors [mRECIST]) using radiomics, may be iteratively followed by eventual replacement of these criteria with more sophisticated and accurate radiomic based models. Ultimately, radiomic models may also assist in guiding the selection of appropriate systemic or local-regional treatments based on an individual’s radiologic, clinical, and genomic profiles. Alternatively, radiomic features could inform an overall treatment strategy for a patient with HCC, rather than treatment of an individual tumor, such as the decision of whether to pursue liver transplantation, or systemic versus locoregional therapy. Evaluation of treatment response and analysis of longitudinal imaging to evaluate how changes in imaging over time may predict future clinical events are relatively unexplored areas that could benefit from more objective analyses. The addition of novel molecular tracers and hepatocyte-specific contrast agents may offer a promising synergistic strategy, improving the capacity of radiomic tools to identify HCC at an early stage.
There are several requisite steps before radiomics can be considered ready for use in clinical applications. Automation of the manual segmentation and extraction process will be essential prior to a transition into real world use. Tools capable of providing consistent and accurate ROI selections are needed to reduce inter-reader variability in tumor segmentation. This would also streamline the currently labor-intensive workflow and allow radiomic models to provide an automatic readout that augments radiologist expertise without increasing time spent. Automated segmentation would also address the challenges in patients with multiple tumors with varying features and underlying tumor biology. Complex models capable of automatically segmenting the entirety of the patient’s imaging, such as convolutional neural networks, would be necessary to providing a holistic radiomic analysis. A second critical step will be the development of consensus around feature extraction methods. Currently the field of radiomics is limited by the fact that no two studies can be directly compared against one another. Proprietary feature extraction tools result in thousands of quantitative variables that have no meaning outside of the context of a single research study. This reduces the ability to perform external validation and prevents the development of cumulative knowledge around specific radiomic feature types. A rigorous approach to standardization, methods-sharing, and increased transparency will be critical to the expansion of radiomics beyond single-institution proof-of-concept studies. To create large-scale training datasets in HCC would require the creation of a centralized image biorepository of HCC scans across many institutions. The NCI’s National Biomedical Image Archive (NBIA) program provides a national image database which seeks to accelerate quantitative imaging resources and has been used to generate open-source datasets in lung, breast, and head and neck cancers.90 Data sharing in HCC radiomics would enable cross-center validation of models and longitudinal adjustment with follow-up data available over time. Automated deidentification of imaging data would be necessary for compliance with patient privacy regulations (e.g. the Health Insurance Portability and Accountability Act), and several existent software packages exist that can reliably deidentify images prior to sharing. External validation is the most critical first step towards realizing the potential of radiomics in the management of HCC, and should be included if feasible in all published radiomic models.
Although early results are encouraging, the limitations of radiomic studies in the current era are substantial. Standardizing analytical methods and image acquisition techniques will be critical to reproducibility across institutions. The Quantitative Imaging Network (QIN) and Radiologic Society of North America are developing consensus protocols and digital phantoms that can help bring radiomics into the realm of clinical utility.91 Test-retest studies of stable phantom objects within a given scanner have estimated reproducibility in only approximately 30% of MRI features, while multi-scanner phantom studies have shown feature reproducibility ranging from 15–85%.92,93 MRI, in particular, is subject to fundamental intensity inhomogeneity across static fields, as well as large amounts of motion artifact, noise, and machine-to-machine variation in acquisition parameters.94 As a result, voxel intensity is often not directly comparable between MRI images and the reproducibility of feature extraction has thus far been poor.95 Quantitative texture analysis is sensitive to scanner variability, and minor changes between institutions could create major distortions in model output. Many of the studies in this review are from Asian cohorts, which have a higher frequency of non-cirrhotic HCC. Derived textural features may differ between Asian and Western cohorts, due to differences in underlying disease etiology and fibrosis burden. Finally, the extraction of high-dimensional data from a small sample results in a high risk of overfitting during model creation and high false-positive rate.96 It is notable that only 2 of 32 models reporting ROC curves in our study had an AUC below 0.70, suggesting possible bias in reporting and over-fitting of data. The reduction of radiomic features to a smaller set of consistently evaluated variables would improve reliability across studies. Although high-throughput imaging data has great promise, the field of radiomics has not yet conclusively demonstrated the capacity to accurately reflect tissue biology. To reach clinical relevance, radiomics will need to develop rigorous cross-center standardization protocols and evidence of a reproducible, generalizable outcome across multiple contexts.97,98 Larger cohorts are needed to improve model performance by reducing overfitting while retaining dimensionality of the models.
Conclusions:
Quantitative image analysis has the potential to transform the early detection and management of HCC. There is a critical need for non-invasive techniques to assist in both diagnostic and prognostic decision-making. Early work in radiomics has demonstrated substantial promise, particularly in the prediction of microvascular invasion and post-hepatectomy outcomes. There are, however, fundamental issues which prevent the clinical application of this technology. Unrecognized errors can introduce bias and unrecognized variability in quantitative analysis. Increased standardization, external validation of models, and rigorously designed prospective studies will be essential to the growth and maturation of radiomics in HCC.
Supplementary Material
Supplementary Table 1: Studies comparing radiomic and clinical models.
Supplementary Table 2: Studies evaluating radiomic tools for prediction of pathologic features
Supplementary Table 3: Studies evaluating radiomic tools for prediction of post-treatment response
Grants and Financial Support:
E.H.T. and J.L. were supported in part by a University of Michigan Training in Gastrointestinal Epidemiology T32 grant (NIDDK T32DK062708). G.L.S, A.S.L. and N.D.P. were supported in part by U01CA230669 from the National Cancer Institute. A.G.S. and N.D.P. were supported in support in part by U01CA230694 from the National Cancer Institute.
Financial disclosures:
Emily Harding-Theobald, Jeremy Louissaint, Edward Cuaresma, Bharat Maraj, Mishal Mendiratta-Lala, Whitney Townsend: No financial disclosures
Amit Singal: Consultant: Wako Diagnostics, Glycotest, Exact Sciences, Roche, GRAIL, Genentech, Bayer, Eisai, Exelixis, AstraZeneca, BMS, and TARGET Pharmasolutions.
Grace Su: U.S. Patent 9,036,883 issued May 19, 2015 “System and Methods for Detecting Liver Disease”. Equity interest in Prenovo and Applied Morphomics
Anna Lok: Advisory board: Epigenomics
Neehar Parikh: Consultant: Bristol-Myers Squibb, Exelixis, Freenome, Eli Lilly; Advisory Board: Eisai, Bayer, Exelixis, Wako Diagnostics, Genentech; Research Grants: Genentech, Bayer, Target Pharmasolutions, Exact Sciences, Glycotest
Abbreviations:
- HCC
hepatocellular carcinoma
- MRI
magnetic resonance imaging
- CT
computed tomography
- RQS
radiomic quality score
- ROI
region of interest
- AFP
alpha-feto protein
- HH
Hepatic Hemangioma
- FNH
Focal Nodular Hyperplasia
- HA
Hepatic Adenoma
- AUC
Area Under Curve
- N
Number of Patients
- VOI
Volume Of Interest
- T
Training Set
- V
Validation Set
References:
- 1.Kim HS, El-Serag HB. The Epidemiology of Hepatocellular Carcinoma in the USA. Curr Gastroenterol Rep. 2019;21(4):17. [DOI] [PubMed] [Google Scholar]
- 2.Tapper EB, Parikh ND. Mortality due to cirrhosis and liver cancer in the United States, 1999–2016: observational study. BMJ. 2018;362:k2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsumoto MM, Mouli S, Saxena P, et al. Comparing Real World, Personalized, Multidisciplinary Tumor Board Recommendations with BCLC Algorithm: 321-Patient Analysis. Cardiovasc Intervent Radiol. 2021. [DOI] [PubMed] [Google Scholar]
- 4.Tang A, Singal AG, Mitchell DG, et al. Introduction to the Liver Imaging Reporting and Data System for Hepatocellular Carcinoma. Clin Gastroenterol Hepatol. 2019;17(7):1228–1238. [DOI] [PubMed] [Google Scholar]
- 5.van der Pol CB, Lim CS, Sirlin CB, et al. Accuracy of the Liver Imaging Reporting and Data System in Computed Tomography and Magnetic Resonance Image Analysis of Hepatocellular Carcinoma or Overall Malignancy-A Systematic Review. Gastroenterology. 2019;156(4):976–986. [DOI] [PubMed] [Google Scholar]
- 6.Marrero JA, Kulik LM, Sirlin CB, et al. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018;68(2):723–750. [DOI] [PubMed] [Google Scholar]
- 7.Parikh ND, Scaglione S, Li Y, et al. A comparison of staging Systems for Hepatocellular Carcinoma in a multicenter US cohort. Clinical gastroenterology and hepatology: the official clinical practice journal of the American Gastroenterological Association. 2018;16(5):781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parikh ND, Mehta AS, Singal AG, Block T, Marrero JA, Lok AS. Biomarkers for the early detection of hepatocellular carcinoma. Cancer Epidemiology and Prevention Biomarkers. 2020;29(12):2495–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rich NE, John BV, Parikh ND, et al. Hepatocellular carcinoma demonstrates heterogeneous growth patterns in a multicenter cohort of patients with cirrhosis. Hepatology. 2020;72(5):1654–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nathani P, Gopal P, Rich N, et al. Hepatocellular carcinoma tumour volume doubling time: a systematic review and meta-analysis. Gut. 2021;70(2):401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambin P, Leijenaar RT, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nature reviews Clinical oncology. 2017;14(12):749–762. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, Wang S, Dong D, et al. The Applications of Radiomics in Precision Diagnosis and Treatment of Oncology: Opportunities and Challenges. Theranostics. 2019;9(5):1303–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Zhu Y, Burnside ES, et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 gene assays. Radiology. 2016;281(2):382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Zhu Y, Burnside ES, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ breast cancer. 2016;2(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toyama Y, Hotta M, Motoi F, Takanami K, Minamimoto R, Takase K. Prognostic value of FDG-PET radiomics with machine learning in pancreatic cancer. Scientific reports. 2020;10(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Liu Z, He L, et al. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non—small cell lung cancer. Radiology. 2016;281(3):947–957. [DOI] [PubMed] [Google Scholar]
- 17.Paul R, Schabath M, Balagurunathan Y, et al. Explaining Deep Features Using Radiologist-Defined Semantic Features and Traditional Quantitative Features. Tomography. 2019;5(1):192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho ES, Choi JY. MRI features of hepatocellular carcinoma related to biologic behavior. Korean J Radiol. 2015;16(3):449–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fowler KJ, Tang A, Santillan C, et al. Interreader Reliability of LI-RADS Version 2014 Algorithm and Imaging Features for Diagnosis of Hepatocellular Carcinoma: A Large International Multireader Study. Radiology. 2018;286(1):173–185. [DOI] [PubMed] [Google Scholar]
- 20.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizzo S, Botta F, Raimondi S, et al. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larkin TJ, Canuto HC, Kettunen MI, et al. Analysis of image heterogeneity using 2D Minkowski functionals detects tumor responses to treatment. Magn Reson Med. 2014;71(1):402–410. [DOI] [PubMed] [Google Scholar]
- 23.Yang F, Ford JC, Dogan N, et al. Magnetic resonance imaging (MRI)-based radiomics for prostate cancer radiotherapy. Translational andrology and urology. 2018;7(3):445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki K Overview of deep learning in medical imaging. Radiol Phys Technol. 2017;10(3):257–273. [DOI] [PubMed] [Google Scholar]
- 25.Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol. 2016;61(13):R150–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braman NM, Etesami M, Prasanna P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017;19(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rios Velazquez E, Aerts HJ, Gu Y, et al. A semiautomatic CT-based ensemble segmentation of lung tumors: comparison with oncologists’ delineations and with the surgical specimen. Radiother Oncol. 2012;105(2):167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Just N Improving tumour heterogeneity MRI assessment with histograms. British journal of cancer. 2014;111(12):2205–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke R, Ressom HW, Wang A, et al. The properties of high-dimensional data spaces: implications for exploring gene and protein expression data. Nat Rev Cancer. 2008;8(1):37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. Annals of internal medicine. 2009;151(4):W65–94. [DOI] [PubMed] [Google Scholar]
- 31.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. [DOI] [PubMed] [Google Scholar]
- 32.Li Z, Mao Y, Huang W, et al. Texture-based classification of different single liver lesion based on SPAIR T2W MRI images. BMC Med Imaging. 2017;17(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oyama A, Hiraoka Y, Obayashi I, et al. Hepatic tumor classification using texture and topology analysis of non-contrast-enhanced three-dimensional T1-weighted MR images with a radiomics approach. Sci Rep. 2019;9(1):8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Liu A, Cui J, Chen A, Song Q, Xie L. Radiomics-based classification of hepatocellular carcinoma and hepatic haemangioma on precontrast magnetic resonance images. BMC Med Imaging. 2019;19(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song S, Li Z, Niu L, et al. Hypervascular hepatic focal lesions on dynamic contrast-enhanced CT: preliminary data from arterial phase scans texture analysis for classification. Clin Radiol. 2019;74(8):653 e611–653 e618. [DOI] [PubMed] [Google Scholar]
- 36.Mokrane FZ, Lu L, Vavasseur A, et al. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol. 2020;30(1):558–570. [DOI] [PubMed] [Google Scholar]
- 37.Dankerl P, Cavallaro A, Tsymbal A, et al. A retrieval-based computer-aided diagnosis system for the characterization of liver lesions in CT scans. Acad Radiol. 2013;20(12):1526–1534. [DOI] [PubMed] [Google Scholar]
- 38.Stocker D, Marquez HP, Wagner MW, et al. MRI texture analysis for differentiation of malignant and benign hepatocellular tumors in the non-cirrhotic liver. Heliyon. 2018;4(11):e00987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asayama Y, Nishie A, Ishigami K, et al. Heterogeneity of non-cancerous liver parenchyma on gadoxetic acid-enhanced MRI: an imaging biomarker for hepatocellular carcinoma development in chronic liver disease. Clin Radiol. 2016;71(5):432–437. [DOI] [PubMed] [Google Scholar]
- 40.Rosenkrantz AB, Pinnamaneni N, Kierans AS, Ream JM. Hypovascular hepatic nodules at gadoxetic acid-enhanced MRI: whole-lesion hepatobiliary phase histogram metrics for prediction of progression to arterial-enhancing hepatocellular carcinoma. Abdom Radiol (NY). 2016;41(1):63–70. [DOI] [PubMed] [Google Scholar]
- 41.Ning P, Gao F, Hai J, et al. Application of CT radiomics in prediction of early recurrence in hepatocellular carcinoma. Abdom Radiol (NY). 2020;45(1):64–72. [DOI] [PubMed] [Google Scholar]
- 42.Shan QY, Hu HT, Feng ST, et al. CT-based peritumoral radiomics signatures to predict early recurrence in hepatocellular carcinoma after curative tumor resection or ablation. Cancer Imaging. 2019;19(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, He L, Huang Y, et al. CT-based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdom Radiol (NY). 2017;42(6):1695–1704. [DOI] [PubMed] [Google Scholar]
- 44.Hui TCH, Chuah TK, Low HM, Tan CH. Predicting early recurrence of hepatocellular carcinoma with texture analysis of preoperative MRI: a radiomics study. Clin Radiol. 2018;73(12):1056 e1011–1056 e1016. [DOI] [PubMed] [Google Scholar]
- 45.Kim S, Shin J, Kim DY, Choi GH, Kim MJ, Choi JY. Radiomics on Gadoxetic Acid-Enhanced Magnetic Resonance Imaging for Prediction of Postoperative Early and Late Recurrence of Single Hepatocellular Carcinoma. Clin Cancer Res. 2019;25(13):3847–3855. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Liu X, Zhang H, et al. Texture Analysis Based on Preoperative Magnetic Resonance Imaging (MRI) and Conventional MRI Features for Predicting the Early Recurrence of Single Hepatocellular Carcinoma after Hepatectomy. Acad Radiol. 2019;26(9):1164–1173. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z, Jiang H, Chen J, et al. Hepatocellular carcinoma: radiomics nomogram on gadoxetic acid-enhanced MR imaging for early postoperative recurrence prediction. Cancer Imaging. 2019;19(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahn SJ, Kim JH, Park SJ, Kim ST, Han JK. Hepatocellular carcinoma: preoperative gadoxetic acid-enhanced MR imaging can predict early recurrence after curative resection using image features and texture analysis. Abdom Radiol (NY). 2019;44(2):539–548. [DOI] [PubMed] [Google Scholar]
- 49.Oh J, Lee JM, Park J, et al. Hepatocellular Carcinoma: Texture Analysis of Preoperative Computed Tomography Images Can Provide Markers of Tumor Grade and Disease-Free Survival. Korean J Radiol. 2019;20(4):569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akai H, Yasaka K, Kunimatsu A, et al. Predicting prognosis of resected hepatocellular carcinoma by radiomics analysis with random survival forest. Diagn Interv Imaging. 2018;99(10):643–651. [DOI] [PubMed] [Google Scholar]
- 51.Chen S, Zhu Y, Liu Z, Liang C. Texture analysis of baseline multiphasic hepatic computed tomography images for the prognosis of single hepatocellular carcinoma after hepatectomy: A retrospective pilot study. Eur J Radiol. 2017;90:198–204. [DOI] [PubMed] [Google Scholar]
- 52.Brenet Defour L, Mule S, Tenenhaus A, et al. Hepatocellular carcinoma: CT texture analysis as a predictor of survival after surgical resection. Eur Radiol. 2019;29(3):1231–1239. [DOI] [PubMed] [Google Scholar]
- 53.Kiryu S, Akai H, Nojima M, et al. Impact of hepatocellular carcinoma heterogeneity on computed tomography as a prognostic indicator. Sci Rep. 2017;7(1):12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng J, Qi X, Zhang Q, et al. A radiomics nomogram for preoperatively predicting prognosis of patients in hepatocellular carcinoma. Translational Cancer Research. 2018;7(4):936–946. [Google Scholar]
- 55.Guo D, Gu D, Wang H, et al. Radiomics analysis enables recurrence prediction for hepatocellular carcinoma after liver transplantation. Eur J Radiol. 2019;117:33–40. [DOI] [PubMed] [Google Scholar]
- 56.Zheng BH, Liu LZ, Zhang ZZ, et al. Radiomics score: a potential prognostic imaging feature for postoperative survival of solitary HCC patients. BMC Cancer. 2018;18(1):1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cai W, He B, Hu M, et al. A radiomics-based nomogram for the preoperative prediction of posthepatectomy liver failure in patients with hepatocellular carcinoma. Surg Oncol. 2019;28:78–85. [DOI] [PubMed] [Google Scholar]
- 58.Mazzaferro V, Llovet JM, Miceli R, et al. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol. 2009;10(1):35–43. [DOI] [PubMed] [Google Scholar]
- 59.Lim KC, Chow PK, Allen JC, et al. Microvascular invasion is a better predictor of tumor recurrence and overall survival following surgical resection for hepatocellular carcinoma compared to the Milan criteria. Ann Surg. 2011;254(1):108–113. [DOI] [PubMed] [Google Scholar]
- 60.Bakr S, Echegaray S, Shah R, et al. Noninvasive radiomics signature based on quantitative analysis of computed tomography images as a surrogate for microvascular invasion in hepatocellular carcinoma: a pilot study. J Med Imaging (Bellingham). 2017;4(4):041303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma X, Wei J, Gu D, et al. Preoperative radiomics nomogram for microvascular invasion prediction in hepatocellular carcinoma using contrast-enhanced CT. Eur Radiol. 2019;29(7):3595–3605. [DOI] [PubMed] [Google Scholar]
- 62.Zheng J, Chakraborty J, Chapman WC, et al. Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma Using Quantitative Image Analysis. J Am Coll Surg. 2017;225(6):778–788 e771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu X, Zhang HL, Liu QP, et al. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol. 2019;70(6):1133–1144. [DOI] [PubMed] [Google Scholar]
- 64.Feng ST, Jia Y, Liao B, et al. Preoperative prediction of microvascular invasion in hepatocellular cancer: a radiomics model using Gd-EOB-DTPA-enhanced MRI. Eur Radiol. 2019;29(9):4648–4659. [DOI] [PubMed] [Google Scholar]
- 65.Zhang R, Xu L, Wen X, et al. A nomogram based on bi-regional radiomics features from multimodal magnetic resonance imaging for preoperative prediction of microvascular invasion in hepatocellular carcinoma. Quant Imaging Med Surg. 2019;9(9):1503–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu YJ, Feng B, Wang S, et al. Model-based three-dimensional texture analysis of contrast-enhanced magnetic resonance imaging as a potential tool for preoperative prediction of microvascular invasion in hepatocellular carcinoma. Oncol Lett. 2019;18(1):720–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen S, Feng S, Wei J, et al. Pretreatment prediction of immunoscore in hepatocellular cancer: a radiomics-based clinical model based on Gd-EOB-DTPA-enhanced MRI imaging. Eur Radiol. 2019;29(8):4177–4187. [DOI] [PubMed] [Google Scholar]
- 68.Gao F, Yan B, Chen J, Wu M, Shi D. Pathological grading of Hepatocellular Carcinomas in MRI using a LASSO algorithm. Paper presented at: Journal of Physics: Conference Series2018. [Google Scholar]
- 69.Kuo MD, Gollub J, Sirlin CB, Ooi C, Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Interv Radiol. 2007;18(7):821–831. [DOI] [PubMed] [Google Scholar]
- 70.Li Y, Yan C, Weng S, et al. Texture analysis of multi-phase MRI images to detect expression of Ki67 in hepatocellular carcinoma. Clin Radiol. 2019;74(10):813 e819–813 e827. [DOI] [PubMed] [Google Scholar]
- 71.Liao H, Zhang Z, Chen J, et al. Preoperative Radiomic Approach to Evaluate Tumor-Infiltrating CD8(+) T Cells in Hepatocellular Carcinoma Patients Using Contrast-Enhanced Computed Tomography. Ann Surg Oncol. 2019;26(13):4537–4547. [DOI] [PubMed] [Google Scholar]
- 72.Wang HQ, Yang C, Zeng MS, et al. Magnetic resonance texture analysis for the identification of cytokeratin 19-positive hepatocellular carcinoma. Eur J Radiol. 2019;117:164–170. [DOI] [PubMed] [Google Scholar]
- 73.Wu M, Tan H, Gao F, et al. Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur Radiol. 2019;29(6):2802–2811. [DOI] [PubMed] [Google Scholar]
- 74.Wu H, Chen X, Chen J, et al. Correlations between P53 Mutation Status and Texture Features of CT Images for Hepatocellular Carcinoma. Methods Inf Med. 2019;58(1):42–49. [DOI] [PubMed] [Google Scholar]
- 75.Xia W, Chen Y, Zhang R, et al. Radiogenomics of hepatocellular carcinoma: multiregion analysis-based identification of prognostic imaging biomarkers by integrating gene data-a preliminary study. Phys Med Biol. 2018;63(3):035044. [DOI] [PubMed] [Google Scholar]
- 76.Zhou W, Zhang L, Wang K, et al. Malignancy characterization of hepatocellular carcinomas based on texture analysis of contrast-enhanced MR images. J Magn Reson Imaging. 2017;45(5):1476–1484. [DOI] [PubMed] [Google Scholar]
- 77.Cozzi L, Dinapoli N, Fogliata A, et al. Radiomics based analysis to predict local control and survival in hepatocellular carcinoma patients treated with volumetric modulated arc therapy. BMC Cancer. 2017;17(1):829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fu S, Chen S, Liang C, et al. Texture analysis of intermediate-advanced hepatocellular carcinoma: prognosis and patients’ selection of transcatheter arterial chemoembolization and sorafenib. Oncotarget. 2017;8(23):37855–37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim J, Choi SJ, Lee SH, Lee HY, Park H. Predicting Survival Using Pretreatment CT for Patients With Hepatocellular Carcinoma Treated With Transarterial Chemoembolization: Comparison of Models Using Radiomics. AJR Am J Roentgenol. 2018;211(5):1026–1034. [DOI] [PubMed] [Google Scholar]
- 80.Kloth C, Thaiss WM, Kargel R, et al. Evaluation of Texture Analysis Parameter for Response Prediction in Patients with Hepatocellular Carcinoma Undergoing Drug-eluting Bead Transarterial Chemoembolization (DEB-TACE) Using Biphasic Contrast-enhanced CT Image Data: Correlation with Liver Perfusion CT. Acad Radiol. 2017;24(11):1352–1363. [DOI] [PubMed] [Google Scholar]
- 81.Lee SH, Hayano K, Sahani DV, Zhu AX, Yoshida H. Kinetic textural biomarker for predicting survival of patients with advanced hepatocellular carcinoma after antiangiogenic therapy by use of baseline first-pass perfusion ct. Paper presented at: International MICCAI Workshop on Computational and Clinical Challenges in Abdominal Imaging2014. [Google Scholar]
- 82.Li M, Fu S, Zhu Y, et al. Computed tomography texture analysis to facilitate therapeutic decision making in hepatocellular carcinoma. Oncotarget. 2016;7(11):13248–13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mule S, Thiefin G, Costentin C, et al. Advanced Hepatocellular Carcinoma: Pretreatment Contrast-enhanced CT Texture Parameters as Predictive Biomarkers of Survival in Patients Treated with Sorafenib. Radiology. 2018;288(2):445–455. [DOI] [PubMed] [Google Scholar]
- 84.Park HJ, Kim JH, Choi SY, et al. Prediction of Therapeutic Response of Hepatocellular Carcinoma to Transcatheter Arterial Chemoembolization Based on Pretherapeutic Dynamic CT and Textural Findings. AJR Am J Roentgenol. 2017;209(4):W211–W220. [DOI] [PubMed] [Google Scholar]
- 85.Reiner CS, Gordic S, Puippe G, et al. Histogram Analysis of CT Perfusion of Hepatocellular Carcinoma for Predicting Response to Transarterial Radioembolization: Value of Tumor Heterogeneity Assessment. Cardiovasc Intervent Radiol. 2016;39(3):400–408. [DOI] [PubMed] [Google Scholar]
- 86.Yuan C, Wang Z, Gu D, et al. Prediction early recurrence of hepatocellular carcinoma eligible for curative ablation using a Radiomics nomogram. Cancer Imaging. 2019;19(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu JY, Zhang HP, Tang ZY, et al. Value of texture analysis based on enhanced MRI for predicting an early therapeutic response to transcatheter arterial chemoembolisation combined with high-intensity focused ultrasound treatment in hepatocellular carcinoma. Clin Radiol. 2018;73(8):758 e759–758 e718. [DOI] [PubMed] [Google Scholar]
- 88.Atiq O, Tiro J, Yopp AC, et al. An assessment of benefits and harms of hepatocellular carcinoma surveillance in patients with cirrhosis. Hepatology. 2017;65(4):1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Konerman MA, Verma A, Zhao B, Singal AG, Lok AS, Parikh ND. Frequency and outcomes of abnormal imaging in patients with cirrhosis enrolled in a hepatocellular carcinoma surveillance program. Liver Transplantation. 2019;25(3):369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. Journal of digital imaging. 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zwanenburg A, Abdalah M, Ashrafinia S, et al. Results from the image biomarker standardisation initiative. Radiotherapy and Oncology. 2018. [Google Scholar]
- 92.Kickingereder P, Neuberger U, Bonekamp D, et al. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro-oncology. 2018;20(6):848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, et al. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology. 2018;288(2):407–415. [DOI] [PubMed] [Google Scholar]
- 94.Vovk U, Pernus F, Likar B. A review of methods for correction of intensity inhomogeneity in MRI. IEEE transactions on medical imaging. 2007;26(3):405–421. [DOI] [PubMed] [Google Scholar]
- 95.Schwier M, van Griethuysen J, Vangel MG, et al. Repeatability of multiparametric prostate MRI radiomics features. Scientific reports. 2019;9(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lewis S, Hectors S, Taouli B. Radiomics of hepatocellular carcinoma. Abdominal Radiology. 2020:1–13. [DOI] [PubMed] [Google Scholar]
- 97.Wakabayashi T, Ouhmich F, Gonzalez-Cabrera C, et al. Radiomics in hepatocellular carcinoma: a quantitative review. Hepatology international. 2019;13(5):546–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Santos JMMM, Oliveira BC, Araujo-Filho JdAB, et al. State-of-the-art in radiomics of hepatocellular carcinoma: a review of basic principles, applications, and limitations. Abdominal Radiology. 2020;45(2):342–353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Studies comparing radiomic and clinical models.
Supplementary Table 2: Studies evaluating radiomic tools for prediction of pathologic features
Supplementary Table 3: Studies evaluating radiomic tools for prediction of post-treatment response


