Abstract
Metaproteomics is becoming widely used in microbiome research for gaining insights into the functional state of the microbial community. Current metaproteomics studies are generally based on high-throughput tandem mass spectrometry (MS/MS) coupled with liquid chromatography. In this paper, we proposed a deep-learning-based algorithm, named DeepFilter, for improving peptide identifications from a collection of tandem mass spectra. The key advantage of the DeepFilter is that it does not need ad hoc training or fine-tuning as in existing filtering tools. DeepFilter is freely available under the GNU GPL license at https://github.com/Biocomputing-Research-Group/DeepFilter.
Significance:
The identification of peptides and proteins from MS data involves the computational procedure of searching MS/MS spectra against a predefined protein sequence database and assigning top-scored peptides to spectra. Existing computational tools are still far from being able to extract all the information out of MS/MS data sets acquired from metaproteome samples. Systematical experiment results demonstrate that the DeepFilter identified up to 12% and 9% more peptide-spectrum-matches and proteins, respectively, compared with existing filtering algorithms, including Percolator, Q-ranker, PeptideProphet, and iProphet, on marine and soil microbial metaproteome samples with false discovery rate at 1%. The taxonomic analysis shows that DeepFilter found up to 7%, 10%, and 14% more species from marine, soil, and human gut samples compared with existing filtering algorithms. Therefore, DeepFilter was believed to generalize properly to new, previously unseen peptide-spectrum-matches and can be readily applied in peptide identification from metaproteomics data.
Keywords: peptide identification, deep learning, tandem mass spectrometry, CNN
Graphical Abstract

1. Introduction
Metaproteomics focuses on the entire protein complement recovered directly from complex microbial communities, like aqueous ecosystems, terrestrial systems, and eukaryotic host microbiomes [1, 2, 3, 4]. Understanding the functionality of microbial communities is essential. For example, the gut microbiome was known to play a crucial role in health by benefiting the immune system and helping control digestion [5, 6, 7]. The microbial activities can be inferred from the total proteins of its constituent microorganisms. Mass spectrometry (MS)-based metaproteomics has emerged as a discovery method for analyzing the proteome of a microbial community in a high-throughput fashion. In shotgun MS-based metaproteomics, proteins are digested into peptides using high-performance liquid chromatography (HPLC), then ionized, isolated, fragmented, and detected in the mass analyzer as they elute from the HPLC. The central component of computational metaproteomics data analysis is database searching. This is where measured tandem mass spectra (MS/MS), of unknown microbial peptides, are compared with theoretical tandem mass spectra predicted from a database of proteins encoded in metagenomes. Peptide-spectrum match (PSM) scores are calculated by the comparisons between each MS/MS and in-silico digested peptides from the protein database. The peptide in the top-scoring PSM is used as a candidate for the query MS/MS. The candidate PSMs are filtered with a score threshold to generate a set of confident PSMs at a designed false discovery rate (FDR).
In the database matching procedure, it is crucial to choose an appropriate PSM scoring function which plays two important roles. First, the scoring function is used to rank candidate peptides for a single spectrum, producing a top-scoring PSM for each spectrum. Second, the scoring function is used to rank the PSMs from different spectra. The second ranking task is intrinsically more complicated than the first ranking task due to the variance of spectra. A perfect scoring function for ranking top-scoring PSM per spectrum may not be a perfect scoring function for ranking PSMs from different spectra because PSM scoring may not be well-calibrated from one spectrum to the other. To solve this issue, a variety of approaches have been developed to learn PSM scoring functions for the second ranking task after the initial PSM scoring. These approaches can be categorized into two types. The first type is based on statistical modeling [8]. For instance, PeptideProphet [9] determines the confidence of identified PSMs by a probability-based model using Bayes’ Law. Other statistical modeling methods, such as linear discriminant analysis [10] and Bayes classifier [11] were also used as discriminant functions. Ivanov et al. [12] designed a multiple-parameter scoring scheme to find PSM outliers to estimate the distributions of PSMs with information from the experimental spectra. iProphet [13] implemented five models based on the number of sibling searches, replicate spectra, sibling experiments, sibling ions, and sibling modifications to filter PSMs. The second type of PSM filtering algorithms discriminate true PSMs from false ones based on machine learning [14, 15, 16, 17, 18, 19, 20], Percolator [16], Q-ranker [19], and CRanker [20] belong to the second type. They train Supporting Vector Machines to classify PSMs. Other machine learning models, such as decision tree [14], random forest [15], Bayesian network models [17], and logistic regression [18] were also applied to re-rank or re-score PSMs with different strategies in constructing training data sets and feature extraction.
Although the above-mentioned methods improved the number of identified PSMs for single organism proteome, there are still more than 50% of spectra without correctly assigned peptides in MS-based metaproteomics [21, 22, 23]. One reason for that is the large metaproteomic protein databases, which may contain millions of predicted proteins spanning thousands of organisms in complex communities [24, 25]. And the scores of random matches generally follow a probabilistic distribution with a small tail towards high scores. A spectrum should have a high score for a correctly matched peptide. As a result, when the databases of candidate peptides increase in size, the probability of an incorrect random match that scores higher than the correct match increases as well. Therefore, a more sensitive ranking strategy is needed for ranking PSMs from different spectra with the properties of spectra and peptide sequences being taken into account. Another drawback of the existing PSM filtering algorithms is that they often do not generalize well across different metaproteome samples and experimental conditions, such as different instrument platforms, etc. For solving this issue, ad hoc training is required when the samples and experimental conditions change. Therefore, it will be hard to justify the confidence of results when training data is from the one needs to be inferred.
In this study, we propose a deep learning model, called DeepFilter, to re-rank PSM candidates after the database search for shotgun metaproteomics. DeepFilter has two key contributions. First, it can learn the mapping patterns between spectra and peptide sequences and combine them with the features known to relate to the PSM score distribution. These automatically extracted features enabled DeepFilter to produce substantially higher numbers of PSM, peptide, and protein identifications in complex metaproteomics data sets than the existing algorithms benchmarked here. Second, DeepFilter eliminates the ad hoc training and can be applied to analyze different metaproteome samples without fine-tuning and still obtain substantial improvements over existing tools. The rest of the paper is organized as follows. In section 2, we elaborate on the architecture of DeepFilter and the whole workflow, including training data set construction, spectrum peak charge detection, and feature extraction. In section 3, systematic experiments on five real-world metaproteomes and single organism proteome were used to demonstrate that our method outperformed the other state-of-the-art approaches. In section 4, we visualized the learned features in DeepFilter by using class activation mappings. In section 5, we concluded that DeepFilter not only achieves higher identification performance but also can be generalized to different metaproteomic studies.
2. Materials and Methods
The workflow of building DeepFilter is shown in Fig. 1, which includes five steps: training data set construction (Part A), charge detection for experimental spectra (Part B), theoretical isotopic envelope generation (Part C), 11 features extraction (Part D), and feature/spectrum encoding (Part E). In the following sections, We will explain the details of each component. DeepFilter is freely available under the GNU GPL license at https://github.com/Biocomputing-Research-Group/DeepFilter, where step-by-step installation and usage were provided. In short, DeepFilter needs the mass spectra data in ms2 format and the database searching results by Comet in pin format, and generates re-ranked PSMs in a tab-separated values file. Note that DeepFilter needs 20 GB GPU memory for training.
Figure 1.
The workflow of building DeepFilter
2.1. Training data construction
There are ten data sets used in our experiments. The summary of spectrum numbers is in Tab. 1. Marine 1, 2, and 3 are metaproteomes of marine microbial communities [26]. Soil 1, 2, and 3 are metaproteomes of soil microbial communities [27]. P1 and P2 are metaproteomoes of mock community [28]. HG is the metaproteome of human hut microbial community [29]. Marine data sets and one of soil data sets were used to construct our training data set, and others are for benchmarking.
Table 1.
The total numbers of MS/MS of nine metaproteome data sets.
| Marine 1 |
Marine 2 |
Marine 3 |
Soil 1 | Soil 2 | Soil 3 | P1 | P2 | HG | |
|---|---|---|---|---|---|---|---|---|---|
| # of spectra | 138,682 | 143,344 | 127,075 | 391,249 | 489,785 | 409,202 | 356,160 | 351,658 | 668,162 |
P1 and P2 are two mock metaproteome samples
HG is the human gut metaproteome sample.
Since the metaproteome data sets do not have ground-truth PSMs, we used existing algorithms to generate a set of PSMs as positive data points. We used Comet [30] to collect a set of top-scoring PSM candidates for each spectrum, and re-scored these PSMs by different filters, including Percolator [16], Q-ranker [19], PeptideProphet [13], and iProphet [13]. We chose the one that identified the highest number of identified PSMs with FDR controlled by the target-decoy search [31]. In our experiments, Percolator performed best and was chosen for generating positive PSMs. We generated a training data set for each Marine data set. Take Marine 1 for example. After the Comet search and the Percolator filtering, top-5 scoring PSM candidates for each spectrum were collected. The PSM candidates with posterior error probabilities (calculated by Percolator) larger than 0.93 were removed. A total of 243,928 PSM candidates were left. For the top-ranked PSM candidates, if they were target PSMs from the matched protein database, we labeled them as positive PSMs. For the rest PSMs, including all decoy PSMs from the decoy protein database and non-top-ranked target PSMs, we labeled them as negative PSMs. The number of positive and negative PSMs for three training data sets are shown in the Tab. 2.
Table 2.
The numbers of postive and negative PSMs in training data sets.
| Marine 1 | Marine 2 | Marine 3 | Soil 1 | |
|---|---|---|---|---|
| # of positive PSMs | 79,057 | 77,916 | 79,496 | 252,829 |
| # of negative PSMs | 160,562 | 153,692 | 132,051 | 1,702,530 |
2.2. Charge detection for experimental mass spectra
To provide more information to the spectrum encoder, each experimental spectrum was deconvoluted with charge states assigned to each fragment peak. Not all the MS data comes with charge information. Here, we developed a charge detection method based on Patterson routine algorithm [32]. We assigned charges up to +3 and, for the fragment ions with charges more than +3, we put them into one group without further categorization. The equation of charge detection is shown in Equ. 1, where ΔM represents the mathematical inverse of charge state which is being evaluated, Mi represents the m/z of nearby fragment peaks, and f(Mi) is the intensity of corresponding peak. The details of determining the charge state for each fragment peak is described in Algorithm 1.
| (1) |
| Algorithm 1: Charge detection | |
|---|---|
|
|
2.3. Isotope envelope generation for peptide sequences
In addition to using the most abundant theoretical peaks of peptide sequences as in Comet and other database searching tools, we also generated the theoretical spectrum with isotope envelopes for each fragment ions. Here, we modified our open-source tool, Sipros [33], to obtain the isotope envelopes for the peptide sequences in the training PSMs. For each ion, we sorted the isotopes in descending order based on their abundances and keep the isotopes until the cumulative isotopic abundance no less than 98%. We then clustered fragment ions into six groups by considering 3 charge states, i.e., +1, +2, and +3, and two ion types, i.e., b-ion and y-ion.
2.4. Input representations of PSMs and engineered features
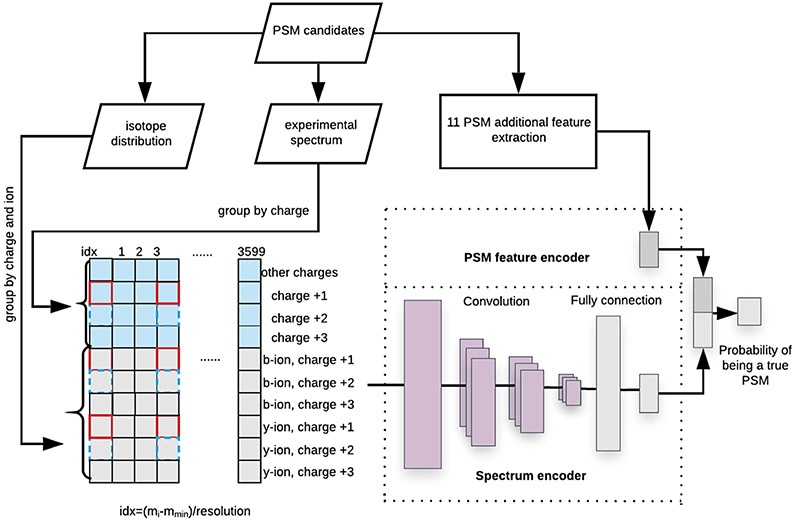
Each input PSM was converted into a matrix, where the peaks in the experimental spectrum and theoretical spectrum were discretized based on their m/z values, and grouped based on their charge states and ion types. This PSM matrix was fed into a spectrum encoder based on the CNN (Convolutional Neural Network) model, which uses convolution kernels to construct a shared weight architecture. Inspired by the existing filtering algorithms [16, 19, 9], we extracted 11 features for each input PSM and encoded them by a PSM feature encoder based on a fully connected layer. The details of these two input representations are described as follows.
Spectrum representation.
Our spectrum representation is a matrix constructed by peaks. The column index indicates the m/z value, and the row index indicates the ion types and the charge states. An example is shown in Fig. 2. We used 0.5 Da as a resolution parameter and considered the m/z values ranged from 100 Da to 1900 Da. We then constructed an 10×3600 matrix, where the first 4 rows are for the fragment ions in charge +1, +2, +3, and above in the experimental spectrum, and the rest 6 rows are for the predicted b-ions and y-ions in charge +1, +2, +3 from the peptide sequence, respectively. The column index was calculated as index =(mi-mmin)/resolution, where mi is the m/z value of i th peak, mmin is the minimum m/z value considered, which is 100 Da. If the m/z value of a peak is 421 Da, then its intensity value is filled in the cell in 642nd column. The intensities in the experimental spectrum were used to fill the first four rows, and the abundances of theoretical spectrum were used to fill the rest six rows. An L2 normalization was applied to the input matrix before the CNN model calculation.
Figure 2.
The architecture of DeepFilter model.
PSM feature representation.
In addition to the features from CNN model, DeepFilter also used another 11 features extracted from the initial PSM score, the observed spectrum, and the peptide sequence for each input PSM. These features are shown in Tab. 3.
Table 3.
11 PSM features used in DeepFilter
| 1 | Xcorr | Cross correlation between theoretical and observed spectra |
| 2 | ΔCn | Fractional difference between current and second best XCorr |
| 3 | Fractional difference between current and the fifth best XCorr | |
| 4 | Mass | The observed mass [M + H]+ |
| 5 | ΔM | The difference in calculated and observed mass |
| 6 | abs(ΔM) | The absolute value of the difference in calculated and observed mass |
| 7 | pepLen | The length of the matched peptide, in residues |
| 8 | enzInt | Number of missed internal enzymatic (tryptic) sites |
| 9-11 |
charge 1-3 |
Three Boolean features indicating the charge state |
2.5. DeepFilter model architecture
The architecture of our DeepFilter model is in Fig. 2. It has two encoders, i.e., spectrum encoder and PSM feature encoder. The representations from the spectrum encoder and PSM feature encoder were concatenated together into a 1024-dimension vector and fed into a fully connected layer with the softmax activation function. The output is the probability from 0 to 1 to indicate how likely a PSM candidate is a true match. We used a modified cross-entropy loss multiplied by the weights that indicate the probability of PSM being a true match. The detail of each encoder and loss function are described as follows.
Spectrum encoder.
The spectrum encoder consists of four dilated convolutional layers and two fully connected layers. To grab features between experimental and theoretical representations within the same charge state, we set the dilation rate to be 3 as highlighted in the red boxes of the kernels of CNN in Fig. 2. For the four dilated convolutional layers, we used 16 kernels for each layer, and in each convolutional layer, we used different kernel sizes, which are (3,7), (2,5), (2,6), (2,6), respectively. We used max-pooling with (1,2) kernel size after each convolution operation. To speed up the calculation and avoid the over-fitting issue, we applied batch-normalization for each convolutional layer and added a dropout layer after the last convolutional layer, with the dropout rate being 0.5. In the first fully connected layer, the input dimension is 3504, and the hidden units are 1024. For the second fully connected layer, the input dimension is 1024, and the dimension of output vector is 512, which is activated by ReLU function, and used as the representation of spectrum for the next PSM classification task.
PSM feature encoder.
The 11 PSM features were given to the PSM feature encoder made of a single fully connected layer. The input dimension for this layer is 11 with ReLU as the activation function. And the output is a 512-dimension vector. This vector was used as the representation of 11 PSM features.
Loss function.
The scoring model is a binary classifier. We applied a modified cross-entropy loss function as in Equ. 2 by incorporating the posterior error probability ( pep ) calculated by Percolator. pi is the predicted probability that i th PSM is a correct match, and pepi is the posterior error probability. This modified loss function achieved a better number of identified PSMs than the classical cross-entropy loss did (data is not shown here).
| (2) |
Training DeepFilter.
Here, we would like to emphasize some important techniques for training DeepFilter. DeepFilter was implemented using PyTorch version 1.4.0 and trained in a workstation with 8 GeForce RTX 2080 Ti GPUs. We randomly split the data sets into training and validation data sets with a ratio of 9 to 1. For the training data sets, we set the mini-batch size to 256. We applied backward propagation to get the gradient in each mini-batch and save the model as a checkpoint when the performance improved based on the accuracy calculated based on the validation data sets. We set the epochs as 150 to ensure the convergence and performed Adam optimizer, whose learning rate and weight decay were set to 1e-4.
3. Experiments and Results
3.1. Experimental design
We evaluated the performance of DeepFilter using three metaproteome data sets from soil communities [27], three metaproteome data sets from marine communities [26], and one E. coli proteome data set. The summary of these data sets is in Tab. 1. These (meta)proteomes were all measured using the Multidimensional Protein Identification Technology (MudPIT) approach [34] on an LTQ Orbitrap Elite mass spectrometer (Thermo Scientific). Their matched metagenomes were used to construct a soil protein database with 3.4 million target proteins and a marine protein database with 392,000 target proteins. The mock community protein database contains 123,100 target proteins, and the human gut protein database has more than 4.9 million target proteins. The MS data and protein databases for marine and soil metaproteome are available from the ProteomeXchange Consortium via the PRIDE repository with the data set identifier of PXD007587. Details on these benchmarking data sets are described in our previous study [35]. The MS data and protein databases for human gut and mock community are provided through the PRIDE repository PXD006118 [28] and PXD013386 [29], respectively.
DeepFilter was compared with four state-of-the-art filtering algorithms, including Percolator [16], Q-ranker [19], PeptideProphet [8], and iProphet [13]. We did not compare with other filtering algorithms because they are either not available or outperformed by the tools mentioned above for metaproteome analysis. Percolator, Q-ranker, and PeptideProphet used the PSMs scored by Comet. iProphet used the PSMs scored by PeptideProphet. Because iProphet used the features at peptide and protein levels, which may cause the machine learning to share information among PSMs for discrimination and destroy the independence among the PSMs [36], so we employed the iProphet without the features at the peptide and protein levels, including the number of sibling ions (NSI), the number of sibling modifications (NSM), and the number of sibling peptides (NSP). We also used iProphet by enabling above features and the results are in Supplementary Table 2 in the supplementary document. This version of iProphet achieved comparable results to DeepFilter at the PSM level but gave less protein identifications.
Benchmarking datasets were searched using Comet 2018.01 rev. 2. The database searching results were filtered by Percolator version 3.03.0, Q-ranker from Crux toolkit version 3.2, PeptideProphet, and iProphet from TPP v5.2.0 with default configuration settings, respectively. The following parameters were used: precursor mass tolerance set to 0.09 Da, fragment mass tolerance set to 0.01 Da, peptide mass range set from 700 Da to 7000 Da, Trypsin/P used for enzyme, and the allowed number of missing cleavages set to three. The protein FASTA files were from the PRIDE repository, where the studies provided both the mass spectrum data and the protein databases. The PSM filtering was executed on a desktop computer with one 2.3 GHz Intel(R) Xeon(R) Gold 5118 CPU and 32 GB memory.
The performance metrics include the number of identified target PSMs, peptides, and proteins with FDR controlled at different levels (where FDR was estimated by the target-decoy strategy) [37]. For each observed spectrum, only the top-scoring PSM was used for estimating FDR. The score threshold was adjusted to reach a user-defined FDR. The FDR is calculated as
| (3) |
For peptide and protein level FDRs, we adjust the PSM level score threshold to control FDRs. For example, suppose the protein FDR is higher than the user-defined value. In that case, we will use a high score threshold to remove more decoy PSMs, which will reduce the more decoy proteins than target proteins, thus lower the protein FDR. We applied the same FDR control strategy for all the tested tools.
3.2. Performance comparison of DeepFilter on marine microbial complex
A rotation training and testing were applied here. More specifically, each marine data set was used to create a training data set for DeepFilter, and the remaining marine data sets were used to test the performance between DeepFilter and the other five existing filtering tools. Details on the execution of these algorithms are described in the supplementary document. Although these MS data sets were all from marine microbial communities, the metaprotome samples were extracted at the different times and dates, and the protein/peptide compositions were different among these marine data sets. Fig. 3 shows the compositions of peptides in marine data sets reported by Comet and the ones used in training DeepFilter. The three marine data sets share some peptides but only a small portion, which means that a significant amount of spectra in the test data sets were not seen in the training data sets. This experiment shows how well DeepFilter can be generalized to unseen data but similar to the ones used in the training.
Figure 3.
The composition of peptide for PSMs in marine data sets
(a) The composition of peptide for PSMs in marine data sets
(b) The composition of peptide for positive PSMs in marine data sets
The filtering results are shown in Tab. 4, where the bold entry and underline entry represents the best and second best results, respectively. Tab. 4 demonstrates that DeepFilter achieves the highest identifications of PSMs, peptides, and proteins. The improvements of DeepFilter over the second best were 11.8% more PSMs, 10.3% more peptides, and 9.9% more proteins at 1% FDR on average. We also found that the DeepFilter model using Marine 3 as the training data set obtained a slightly better improvement compared to the ones using Marine 1 and Marine 2, which may cause by the larger number of positive PSMs in Marine 3, as observed in Fig. 3(b). Our DeepFilter model trained on Soil 1 also obtained more identifications compared to baseline methods, although the improvement is not as significant as the DeepFilter models trained on the marine datasets. This may be caused by the difference between training and test data distributions. To show the discrepancy between Soil and Marine metaproteome samples, we did a taxonomic analysis by searching filtered proteins with FDR controlled at 1% against the NCBI database. We used Protein-Protein BLAST version 2.11.0+ with default parameters except only keeping one query result with the best E-value. The overlap of taxonomic compositions is shown in Fig. 4. The percentage of the shared identified species within marine and soil samples are 70% and 60%, separately on average. In contrast, only 5% species are shared between soil and marine samples. Even with a low number of shared species, the DeepFilter model trained on Soil 1 still obtained more identifications compared to baseline methods, which shows how well DeepFilter can be generalized to unseen data. For marine samples, the overlap of identified PSMs, peptides, and proteins by the best DeepFilter model, Comet, and the second-best baseline are shown in Supplementary Figure 4. Comet, DeepFilter, and other baseline methods share a significant portion of identifications. On average, 5353 PSMs, 3181 peptides, and 1035 proteins are identified only by DeepFilter, whereas 2974 PSMs, 1602 peptides, 173 proteins identified only by the second-best tool.
Table 4.
Identification performance using marine metaproteomes at FDR 1%
| Baseline | DeepFilter | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| C | P | Q | PP | I | M1 | M2 | M3 | S1 | |
| # PSM identification at PSM FDR 1% | |||||||||
| Marine 1 | 34425 | 37951 | 36472 | 33061 | 33358 | - | 41423 | 43597 | 41170 |
| Marine 2 | 31822 | 34741 | 33899 | 30670 | 30846 | 38927 | - | 39421 | 37165 |
| Marine 3 | 38490 | 41714 | 40832 | 37072 | 37304 | 44664 | 44273 | - | 44073 |
| # Peptide identification at Peptide FDR 1% | |||||||||
| Marine 1 | 21334 | 23597 | 23007 | 20961 | 20961 | - | 25012 | 26790 | 25387 |
| Marine 2 | 22004 | 24150 | 23589 | 21597 | 21696 | 26582 | - | 26816 | 25767 |
| Marine 3 | 25085 | 27522 | 26674 | 24653 | 24661 | 29300 | 29007 | - | 29127 |
| # Protein identification at Protein FDR 1% | |||||||||
| Marine 1 | 6676 | 7312 | 7221 | 6458 | 6458 | - | 7687 | 7956 | 7781 |
| Marine 2 | 7033 | 7715 | 7617 | 7039 | 5375 | 8313 | - | 8740 | 8109 |
| Marine 3 | 7457 | 8209 | 8151 | 7354 | 7433 | 8851 | 8367 | - | 8690 |
Baseline searching algorithms & filters: C, Comet only; P, Comet & Percolator; Q, Comet & Q-ranker; PP, Comet & PeptideProphet; I, Comet,PeptideProphet & iProphet
DeepFilter models trained by M1 (Marine 1), M2 (Marine 2), M3 (Marine 3) and S1 (Soil 1)
The best entry was in bold and the next best from baseline methods was underlined.
Figure 4.
Phylogenetic tree of the species only found by DeepFilter from the marine metaproteome samples.
(a) The taxonomic composition overlap within marine metaproteome data sets
(b) The taxonomic composition overlap within soil metaproteome data sets
(c) The taxonomic composition overlap between marine and soil metaproteome data sets
3.3. Performance comparison of DeepFilter on soil microbial complex
To further investigate the generalization ability of DeepFilter, we conducted the performance comparison of DeepFilter trained by marine data sets and one soil data set then tests on the rest marine and soil metaproteome data sets. Given the different microbe compositions between marine and soil microbial communities, the numbers of common peptides/proteins will be even less than the ones we have in Fig. 3. DeepFilter was trained on one marine data set and was applied to soil data sets without ad hoc training or fine-tuning. If DeepFilter can still outperform other filtering tools, this means DeepFilter can be well generalized to unseen MS data even without ad hoc training or fine-tuning.
The results are shown in Tab. 5, where bold and underlined entries represent the best and the second best results. Among the six algorithms tested, DeepFilter always generated the highest number of identified PSMs, peptides, and proteins at 1% FDR. The improvements of DeepFilter over the second best were 6.5% more PSMs, 6.3% more peptides, and 6.9% more proteins at 1% FDR, by using marine metaproteome data sets for training. Therefore, DeepFilter was believed to have well modeled the matching between experimental spectra and peptide sequences. We also did an experiment by using soil 1 as training data set and found that DeepFilter improved the PSM/peptide/protein identifications, which gave us up to 8.2% more PSMs, 9.1% more peptides, and 8.8% proteins.
Table 5.
Identification performance using three soil metaproteomes at FDR 1%
| Baseline | DeepFilter | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| C | P | Q | PP | I | M1 | M2 | M3 | S1 | |
| # PSM identification at PSM FDR 1% | |||||||||
| Soil 1 | 79505 | 88037 | 86433 | 73821 | 75360 | 92221 | 91745 | 94011 | - |
| Soil 2 | 75693 | 84623 | 82773 | 71281 | 73331 | 89465 | 88093 | 88372 | 91384 |
| Soil 3 | 72454 | 81331 | 79211 | 68067 | 70121 | 86809 | 87017 | 87233 | 88015 |
| # Peptide identification at Peptide FDR 1% | |||||||||
| Soil 1 | 26068 | 29304 | 29163 | 25288 | 25403 | 30111 | 30006 | 30923 | - |
| Soil 2 | 23500 | 26989 | 26116 | 23478 | 22775 | 28968 | 27883 | 28923 | 29338 |
| Soil 3 | 20423 | 23275 | 23673 | 19863 | 19922 | 25006 | 25018 | 25116 | 25392 |
| # Protein identification at Protein FDR 1% | |||||||||
| Soil 1 | 6938 | 7756 | 7684 | 6821 | 6819 | 8069 | 8011 | 8184 | - |
| Soil 2 | 6913 | 7519 | 7498 | 6848 | 6879 | 8041 | 7727 | 8031 | 8169 |
| Soil 3 | 5644 | 6183 | 6462 | 5473 | 5577 | 6976 | 6980 | 6998 | 7029 |
Baseline searching algorithms & filters: C, Comet only; P, Comet & Percolator; Q, Comet & Q-ranker; PP, Comet & PeptideProphet; I, Comet,PeptideProphet & iProphet
DeepFilter models trained by M1 (Marine 1), M2 (Marine 2), M3 (Marine 3) and S1 (Soil 1)
The best entry was in bold and the next best from baseline methods was underlined.
For soil samples, the overlap of identified PSMs, peptides, and proteins by the best DeepFilter, Comet, and the second-best baseline are shown in Supplementary Figure 5. Similar to the marine data sets, Comet, DeepFilter, and other baseline methods share a significant portion of identifications. There were more identification results only reported by DeepFilter compared to other methods. On average, 920 proteins are identified only by DeepFilter at FDR 1%, whereas 234 proteins are identified only by the second-best tool.
3.4. Performance comparison of DeepFilter on human gut microbial complex
To investigate if DeepFilter performs well in the metaproteome with a large database, we tested DeepFilter on the human gut microbial complex with a protein database consisting of 5 million target proteins [29]. First, we used a small portion of mass spectra from the human gut metaproteome to test which DeepFilter model, trained by different data sets, performed best. We selected the one trained by Soil 1 data set, which achieved slightly better performance than other data sets. The identification results are shown in Tab. 6. Although DeepFilter did not achieve the same level improvements for human gut samples as in marine and soil samples, it still identified 6.2% more PSMs, 6.7% more peptides, and 4% more proteins compared to the best baseline method.
Table 6.
Performance comparison on human gut metaproteome at FDR 1%
| PSM | Peptide | Protein | |
|---|---|---|---|
| Comet | 231919 | 160472 | 35085 |
| Percolator | 249371 | 171183 | 36183 |
| Q-ranker | 239467 | 168731 | 35707 |
| PeptideProphet | 211706 | 148840 | 33566 |
| iProphet | 211706 | 148840 | 33566 |
| DeepFilter | 264875 | 182698 | 37644 |
The best entry was in bold and the next best from baseline methods was underlined.
For the human gut sample, the overlap of identified PSMs, peptides, and proteins by the best DeepFilter, Comet, and the second-best baseline are shown in Supplementary Figure 6. Similar to the marine and soil data sets, Comet, DeepFilter, and other baseline methods share a significant portion of identifications. 23620 PSMs, 15976 peptides, and 2914 proteins were identified by only DeepFilter at FDR 1%, whereas 4889 PSMs, 2845 peptides, and 480 proteins were identified only by the second-best tool.
3.5. Performance comparison of DeepFilter on mock community
We also tested the DeepFilter model using a mock community data set to see if DeepFilter is effective for a microbial complex with only a few species (30 species). We chose two of the “P” type communities from [28]. The “P” means the data sets have the same protein contents. Here, we labeled them as P1 and P2. The identification results for these two data sets are shown in Tab. 7. Note that the results by the DeepFilter model were trained on another mock community data set, P3, from [28], and this DeepFilter model was slightly better than the models trained on marine and soil samples (Data not shown). DeepFilter identified 6.3% more PSMs, 6.7% more peptides, and 7.7% more proteins on average at FDR 1% than the second-best baseline method. Therefore, DeepFilter can also perform well in the metaproteome samples with a few species. For mock community samples, the overlap of identification results for PSM/peptide/protein levels by the best DeepFilter, Comet, and the second-best post-processing tools are shown in Supplementary Figure 7. Up to 8311 PSMs, 3176 peptides, and 742 proteins were identified only by DeepFilter at FDR 1%, whereas up to 1254 PSMs, 363 peptides, and 51 proteins were identified only by the second-best tool.
Table 7.
Identification performance using mock community metaproteome at FDR 1%
| Baseline | DeepFilter | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| C | P | Q | PP | I | |||||
| # PSM identification at PSM FDR 1% | |||||||||
| P1 | 95098 | 101563 | 98461 | 93669 | 93669 | 107970 | |||
| P2 | 103405 | 111018 | 103798 | 102639 | 102639 | 118029 | |||
| # Peptide identification at Peptide FDR 1% | |||||||||
| P1 | 26773 | 28706 | 27334 | 25642 | 25642 | 30587 | |||
| P2 | 39424 | 42042 | 41203 | 33820 | 33820 | 44901 | |||
| # Protein identification at Protein FDR 1% | |||||||||
| P1 | 7157 | 7670 | 7603 | 6884 | 6884 | 8316 | |||
| P2 | 9417 | 10134 | 9897 | 9557 | 9557 | 10838 | |||
Baseline searching algorithms & filters: C, Comet only; P, Comet & Percolator; Q, Comet & Q-ranker; PP, Comet & PeptideProphet; I, Comet,PeptideProphet & iProphet
The best entry was in bold and the next best from baseline methods was underlined.
3.6. Computation time
Tab. 8 presents the computation time when applying the DeepFilter and other filtering algorithms on different data sets. DeepFilter was running on a workstation with 8 GeForce RTX 2080 Ti GPUs, each with 12 GB memory. Baseline algorithms were executed on a desktop computer with one 2.3 GHz Intel Xeon Gold 5118 CPU and 32 GB memory. DeepFilter can finish the filtering in around 10 minutes with GPU acceleration.
Table 8.
Computation time for nine metaproteomes (precise to second)
| Marine 1 |
Marine 2 |
Marine 3 |
Soil 1 |
Soil 2 |
Soil 3 |
HG | P1 | P2 | |
|---|---|---|---|---|---|---|---|---|---|
| DeepFilter | 204 | 218 | 210 | 578 | 476 | 432 | 1,547 | 231 | 256 |
| Percolator | 126 | 134 | 127 | 359 | 277 | 266 | 902 | 145 | 157 |
| Q-ranker | 235 | 251 | 242 | 671 | 550 | 528 | 1,639 | 247 | 260 |
| PeptideProphet | 67 | 68 | 182 | 159 | 142 | 136 | 473 | 96 | 108 |
| iProphet | 297 | 314 | 300 | 835 | 691 | 654 | 2,217 | 429 | 319 |
P1 and P2 are two mock metaproteome samples
HG is the human gut metaproteome sample.
4. Discussion
4.1. Analysis of the taxonomy information from protein identification results
To show the impact of DeepFilter on the taxonomy analysis, we used the protein-protein blast to search the protein identification results from different data sets against the NCBI database. The summary of the numbers of species identified is shown in Tab. 9. For the marine metaproteome data sets, up to 7% more species were found by DeepFilter compared to the second-best. DeepFilter found 10.21% more species for the soil metaproteome data sets. For the mock communities, all 30 species were found by the tested methods. For the human gut metaproteome, DeepFilter can discover more than 14% species than the second-best one.
Table 9.
The number of species searched using protein identification results at FDR 1%.
| Comet | Percolator | Q-ranker | PeptideProphet | iProphet | DeepFilter | |
|---|---|---|---|---|---|---|
| Marine 1 | 709 | 765 | 756 | 705 | 706 | 805 |
| Marine 2 | 723 | 804 | 711 | 739 | 738 | 860 |
| Marine 3 | 643 | 671 | 650 | 637 | 637 | 643 |
| Soil 1 | 777 | 864 | 819 | 785 | 778 | 934 |
| Soil 2 | 632 | 705 | 645 | 606 | 623 | 777 |
| Soil 3 | 343 | 377 | 374 | 329 | 330 | 395 |
| P1 | 30 | 30 | 30 | 30 | 30 | 30 |
| P2 | 30 | 30 | 30 | 30 | 30 | 30 |
| Human gut | 2012 | 2148 | 2071 | 1987 | 1990 | 2454 |
The species found only by DeepFilter for marine, soil, and human gut metaproteome samples have their lineage shown in Fig. 4.1-4.1. These phylogenetic trees include 104 taxa for marine samples, 160 taxa for soil samples, and 306 taxa for the human gut samples. The taxa, which have the greatest number of identified proteins for marine, soil, human gut, and mock communities, are shown in Fig. 4.1-4.1. From these figures, we found that DeepFilter identified the largest or comparable numbers of proteins for each species.
4.2. Analysis of the significance of the spectrum encoder
To evaluate the spectrum encoder’s significance, we compared the performance of DeepFilter with the spectrum encoder disabled. The models trained on Marine 2 were used here. Fig. 11 shows the improvements of DeepFilter over the second-best one from baseline methods at PSM FDR 1%. We can see that without the spectrum encoder, the PSM identifications of DeepFilter were slightly better than the existing tools but dropped significantly compared to the one with the spectrum encoder. The improvement decline was not even among different data sets. By looking at the number of unique peptides that are not shared by multiple protein sequences, fewer unique peptides give fewer protein identifications, which is the main reason for the uneven improvement decline.
Figure 11.
Performance comparison of DeepFilter with/without the spectrum encoder.
(a) The numbers of identified proteins at 1% FDR in the three out of five most abundant species for P1.
(b) The numbers of identified proteins at 1% FDR in the three out of five most abundant species for P2.
4.3. Analysis of the features learned in DeepFilter
To mine the patterns and visualize the features learned by our DeepFilter, we adopted a class activation mapping (CAM) generation technique [38] to interpret the learning decision of DeepFilter. In the image analysis, CAM is used to show the input image regions that contribute to prediction process. In our experiments, we applied CAM in the spectrum representation to visualize the patterns that help predict correct PSMs.
Fig. 12 presents the CAMs for a target and a decoy PSM, respectively. The color mapping in these figures shows the weight from zero to one, as the legend indicates. The background color represents the learning weights from CNN for different regions. The red color regions make the PSMs more likely to be true (positive) PSMs if there are non-zero input values. In contrast, the blue color regions make the PSMs more likely to be false (negative) PSMs. The white points represent the peaks from the experimental and theoretical mass spectra. The CAMs have ten rows, each of which represents the peaks grouped by different charge states and different ion types as used in Section 2.4. The first four rows are for experimental spectra, and the rest six rows are for theoretical spectra. The actual spectra are shown in Supplementary Figure 9. In Fig. 12, we can see that the isotopic envelopes of fragment ions from experimental spectra are mostly inside the red regions. Given that the isotopic envelopes indicate high-quality peaks, if the theoretical spectra also contain these ions, DeepFilter will tend to label the input PSM as a positive PSM; otherwise, DeepFilter will tend to label it as a negative PSM. For example, in the CAM of a target PSM in Fig. 12, there are several isotopic envelope mappings covered by the red region, which means there is a strong connection between the experimental spectrum and the theoretical spectrum. However, in the CAM of a decoy PSM in Fig. 12, the red region covers the parts where no isotopic envelope mappings exist. The above analysis showed that our DeepFilter models lean towards a true PSM if the matching fragment ions are of high intensity and have a detectable isotope pattern.
Figure 12.
Class activation mappings of one target and one decoy PSMs.
We examined the PSMs reported only by the DeepFilter in the hope of finding any pattern of these PSMs and why the DeepFilter performed better than the baseline methods. It turns out there are no obvious characteristics of these PSMs. We also visualized the score distribution of PSMs from the DeepFilter and the Percolator (Supplementary Figure 8), from which we can found that both distributions of the PSMs from the Percolator and the DeepFilter are mixture distributions. For the PSMs reported only by the DeepFilter at 1% FDR, a large number of them have scores over 0.5 (Supplementary Figure 8(c)).
4.4. Analysis of the identifications by DeepFilter in terms of false-discovery rate
In this study, we used the decoy-database approach to assess the confidence of identifications. Although the decoy-database approach is currently the gold standard in shotgun proteomics experiments, what might appear to be a good result could be, in fact, the product of overfitting [39]. Here, we used a modified decoy method, termed a semi-labeled decoy approach [39], to estimate if DeepFilter generated confident results. The semi-labeled decoy approach relies on labeled decoys and unlabeled decoys, where the latter serve as an internal error reference that helps to statistically deal with overfitting. In this experiment, we generated two types of decoys, i.e., PR and MR sequences, for each target sequence. A PR peptide is generated by first swapping the two outermost amino acids, then treating pairs of the remaining amino acids as units and reversing their order. An MR peptide is generated by first swapping the two outermost amino acids, then dividing the remaining portion in half and reversing each of the halves separately. We randomly chose one of the decoys as labeled decoys. Tab. 4 shows the DeepFilter results on the Marine 2 data set. The results from the DeepFilter trained on Marine 3 appear to be consistent between the two types of decoys. For statistically estimating the overfitting issue, the number of unlabeled decoys identified follows the binomial distribution under the hypothesis that the results are not overfitted. Thus, the overfitting p -value can be approximated by , where X is a random variable indicating the number of identified unlabeled decoys, Bin is the binomial distribution function, s is the number of identified unlabeled decoys by DeepFilter, n is the total number of identifications, p is the expected fraction of unlabeled decoys (i.e., given FDR). By re-analyzing Tab. 10, we believe that the results from the DeepFilter can be taken with confidence (P≫0.05) without overfitting issue.
Table 10.
The identified PSMs/peptides using semi-labeled decoys by the DeepFilter on marine 2 data set.
| Labeled Decoys | Unlabeled Decoys | Total Target | Total | |
|---|---|---|---|---|
| PSM | 207 | 192 | 39,421 | 39,820 |
| Peptide | 137 | 13,3 | 26,816 | 27,086 |
5. Conclusion
In this study, a CNN-based deep learning model, called DeepFilter, was designed to filter PSM candidates after database searching. It can automatically learn the features from experimental spectra and peptide sequences and combine with other engineered features to predict if PSMs are correct matches or not. Unlike the existing filtering tools, we did not apply a semi-supervised fashion or fine-tune the filter using a subset of the working data. Instead, we trained DeepFilter on a separate data set and tested its performance on other metaproteome data and single organism proteome data. The experimental results demonstrate that DeepFilter achieved the highest or comparable numbers of identified PSMs, peptides, and proteins. Therefore, DeepFilter was believed to generalize properly to new, previously unseen PSMs. In the future, we will further improve DeepFilter by training it on a composite data set with mass spectra from various microbes, such as those in the human intestines. We will also investigate its performance on other microbial communities that are available in Proteomics Identifications database [40].
Supplementary Material
Figure 5.

Phylogenetic tree of the species only found by DeepFilter from the soil metaproteome samples.
Figure 6.

Phylogenetic tree of the species only found by DeepFilter from the human gut metaproteome sample.
Figure 7.

The numbers of identified proteins at 1% FDR in the three out of five most abundant species for marine communities.
Figure 8.
The numbers of identified proteins at 1% FDR in the three out of five most abundant species for soil communities.
(a) The numbers of identified proteins at 1% FDR in the three out of five most abundant species for Marine 1.
(b) The numbers of identified proteins at 1% FDR in the three out of five most abundant species for Marine 2.
(c) The numbers of identified proteins at 1% FDR in the three out of five most abundant species for Marine 3.
Figure 9.
The numbers of identified proteins at 1% FDR in the three out of five most abundant species for human gut community.
(a) The numbers of identified proteins at 1% FDR in the three out of five most abundant species for Soil 1.
(b) The numbers of identified proteins at 1% FDR in the three out of five most abundant species for Soil 2.
(c) The numbers of identified proteins at 1% FDR in the three out of five most abundant species for Soil 3.
Figure 10.
The numbers of identified proteins at 1% FDR in the three out of five most abundant species for mock communities.
Highlights.
First use of deep-learning models to create a fast algorithm to identify peptides using MS-based metaproteomics data.
Proposed models outperformed current algorithms.
The proposed models trained by marine metaproteome samples can be well generalized to soil metaproteome samples.
Ad hoc training or fine-tuning is not needed in the proposed models.
Acknowledgments
Research reported in this publication was supported by the National Library of Medicine of the National Institutes of Health under award number R15LM013460.
Footnotes
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability Statement
The raw MS data and protein databases are available from the PRIDE repository with the following data set identifier PXD007587, PXD006118, and PXD013386. The datasets generated during the current study are available in the GitHub repository at https://github.com/Biocomputing-Research-Group/DeepFilter.
References
- [1].Zwittink RD, van Zoeren-Grobben D, Martin R, van Lingen RA, Jebbink LJG, Boeren S, Renes IB, van Elburg RM, Belzer C, Knol J, Metaproteomics reveals functional differences in intestinal microbiota development of preterm infants, Molecular & Cellular Proteomics 16 (9) (2017) 1610–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Timmins-Schiffman E, May DH, Mikan M, Riffle M, Frazar C, Harvey H, Noble WS, Nunn BL, Critical decisions in metaproteomics: achieving high confidence protein annotations in a sea of unknowns, The ISME journal 11 (2) (2017) 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Liu D, Keiblinger KM, Schindlbacher A, Wegner U, Sun H, Fuchs S, Lassek C, Riedel K, Zechmeister-Boltenstern S, Microbial functionality as affected by experimental warming of a temperate mountain forest soil—a metaproteomics survey, Applied soil ecology 117 (2017) 196–202. [Google Scholar]
- [4].Penzlin A, Lindner MS, Doellinger J, Dabrowski PW, Nitsche A, Renard BY, Pipasic: similarity and expression correction for strain-level identification and quantification in metaproteomics, Bioinformatics 30 (12) (2014) i149–i156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Alcock J, Maley CC, Aktipis CA, Is eating behavior manipulated by the gastrointestinal microbiota? evolutionary pressures and potential mechanisms, Bioessays 36 (10) (2014) 940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Holmes E, Li JV, Marchesi JR, Nicholson JK, Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk, Cell metabolism 16 (5) (2012) 559–564. [DOI] [PubMed] [Google Scholar]
- [7].Zhang X, Chen W, Ning Z, Mayne J, Mack D, Stintzi A, Tian R, Figeys D, Deep metaproteomics approach for the study of human microbiomes, Analytical Chemistry 89 (17) (2017) 9407–9415. [DOI] [PubMed] [Google Scholar]
- [8].Nesvizhskii AI, Keller A, Kolker E, Aebersold R, A statistical model for identifying proteins by tandem mass spectrometry, Analytical chemistry 75 (17) (2003) 4646–4658. [DOI] [PubMed] [Google Scholar]
- [9].Keller A, Nesvizhskii AI, Kolker E, Aebersold R, Empirical statistical model to estimate the accuracy of peptide identifications made by ms/ms and database search, Analytical chemistry 74 (20) (2002) 5383–5392. [DOI] [PubMed] [Google Scholar]
- [10].Ding Y, Choi H, Nesvizhskii AI, Adaptive discriminant function analysis and reranking of ms/ms database search results for improved peptide identification in shotgun proteomics, Journal of proteome research 7 (11) (2008) 4878–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Choi H, Nesvizhskii AI, Semisupervised model-based validation of peptide identifications in mass spectrometry-based proteomics, Journal of proteome research 7 (01) (2008) 254–265. [DOI] [PubMed] [Google Scholar]
- [12].Ivanov MV, Levitsky LI, Lobas AA, Panic T, Laskay UA, Mitulovic G, Schmid R, Pridatchenko ML, Tsybin YO, Gorshkov MV, Empirical multidimensional space for scoring peptide spectrum matches in shotgun proteomics, Journal of proteome research 13 (4) (2014) 1911–1920. [DOI] [PubMed] [Google Scholar]
- [13].Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, Mendoza L, Moritz RL, Aebersold R, Nesvizhskii AI, iprophet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates, Molecular & cellular proteomics 10 (12) (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Elias JE, Gibbons FD, King OD, Roth FP, Gygi SP, Intensity-based protein identification by machine learning from a library of tandem mass spectra, Nature biotechnology 22 (2) (2004) 214–219. [DOI] [PubMed] [Google Scholar]
- [15].Ulintz PJ, Zhu J, Qin ZS, Andrews PC, Improved classification of mass spectrometry database search results using newer machine learning approaches, Molecular & Cellular Proteomics 5 (3) (2006) 497–509. [DOI] [PubMed] [Google Scholar]
- [16].Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ, Semi-supervised learning for peptide identification from shotgun proteomics datasets, Nature methods 4 (11) (2007)923–925. [DOI] [PubMed] [Google Scholar]
- [17].Klammer AA, Reynolds SM, Bilmes JA, MacCoss MJ, Noble WS, Modeling peptide fragmentation with dynamic bayesian networks for peptide identification, Bioinformatics 24 (13) (2008) i348–i356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gonnelli G, Stock M, Verwaeren J, Maddelein D, De Baets B, Martens L, Degroeve S, A decoy-free approach to the identification of peptides, Journal of proteome research 14 (4) (2015) 1792–1798. [DOI] [PubMed] [Google Scholar]
- [19].Spivak M, Weston J, Bottou L, Kall L, Noble WS, Improvements to the percolator algorithm for peptide identification from shotgun proteomics data sets, Journal of proteome research 8 (7) (2009) 3737–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liang X, Xia Z, Jian L, Niu X, Link A, An adaptive classification model for peptide identification, BMC genomics 16 (S11) (2015) S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Muth T, Benndorf D, Reichl E, Rapp E, Martens L, Searching for a needle in a stack of needles: challenges in metaproteomics data analysis, Molecular BioSystems 9 (4) (2013) 578–585. [DOI] [PubMed] [Google Scholar]
- [22].Heyer R, Schallert K, Zoun R, Becher B, Saake G, Benndorf D, Challenges and perspectives of metaproteomic data analysis, Journal of biotechnology 261 (2017) 24–36. [DOI] [PubMed] [Google Scholar]
- [23].Yao Q, Li Z, Song Y, Wright SJ, Guo X, Tringe SG, Tfaily MM, Paša-Tolic′ L, Hazen TC, Turner BL, et al. , Community proteogenomics reveals the systemic impact of phosphorus availability on microbial functions in tropical soil, Nature ecology & evolution 2 (3) (2018) 499–509. [DOI] [PubMed] [Google Scholar]
- [24].Ahn T-H, Chai J, Pan C, Sigma: strain-level inference of genomes from metagenomic analysis for biosurveillance, Bioinformatics 31 (2) (2015) 170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Haider B, Ahn T-H, Bushnell B, Chai J, Copeland A, Pan C, Omega: an overlap-graph de novo assembler for metagenomics, Bioinformatics 30 (19) (2014) 2717–2722. [DOI] [PubMed] [Google Scholar]
- [26].Bryson S, Li Z, Pett-Ridge J, Hettich RL, Mayali X, Pan C, Mueller RS, Proteomic stable isotope probing reveals taxonomically distinct patterns in amino acid assimilation by coastal marine bacterioplankton, Msystems 1 (2) (2016) e00027–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Butterfield CN, Li Z, Andeer PF, Spaulding S, Thomas BC, Singh A, Hettich RL, Suttle KB, Probst AJ, Tringe SG, et al. , Proteogenomic analyses indicate bacterial methylotrophy and archaeal heterotrophy are prevalent below the grass root zone, PeerJ 4 (2016) e2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kleiner M, Thorson E, Sharp CE, Dong X, Liu D, Li C, Strous M, Assessing species biomass contributions in microbial communities via metaproteomics, Nature communications 8 (1) (2017) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Long S, Yang Y, Shen C, Wang Y, Deng A, Qin Q, Qiao L, Metaproteomics characterizes human gut microbiome function in colorectal cancer, NPJ biofilms and microbiomes 6 (1) (2020) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Eng JK, Jahan TA, Hoopmann MR, Comet: an open-source ms/ms sequence database search tool, Proteomics 13 (1) (2013) 22–24. [DOI] [PubMed] [Google Scholar]
- [31].Elias JE, Gygi SP, Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry, Nature methods 4 (3) (2007) 207–214. [DOI] [PubMed] [Google Scholar]
- [32].Senko MW, Beu SC, McLafferty FW, Automated assignment of charge states from resolved isotopic peaks for multiply charged ions, Journal of the American Society for Mass Spectrometry 6 (1) (1995) 52–56. [DOI] [PubMed] [Google Scholar]
- [33].Hyatt D, Pan C, Exhaustive database searching for amino acid mutations in proteomes, Bioinformatics 28 (14) (2012) 1895–1901. [DOI] [PubMed] [Google Scholar]
- [34].Washburn MP, Wolters D, Yates JR, Large-scale analysis of the yeast proteome by multidimensional protein identification technology, Nature biotechnology 19 (3) (2001) 242–247. [DOI] [PubMed] [Google Scholar]
- [35].Guo X, Li Z, Yao Q, Mueller RS, Eng JK, Tabb DL, Hervey WJ IV, Pan C, Sipros ensemble improves database searching and filtering for complex metaproteomics, Bioinformatics 34 (5) (2018) 795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Granholm V, Noble WS, Kall L, On using samples of known protein content to assess the statistical calibration of scores assigned to peptide-spectrum matches in shotgun proteomics, Journal of proteome research 10 (5) (2011) 2671–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jeong K, Kim S, Bandeira N, False discovery rates in spectral identification, BMC bioinformatics 13 (S16) (2012) S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A, Learning deep features for discriminative localization, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 2921–2929. [Google Scholar]
- [39].Barboza R, Cociorva D, Xu T, Barbosa VC, Perales J, Valente RH, França FM, Yates JR, Carvalho PC, Can the false-discovery rate be misleading?, Proteomics 11 (20) (2011) 4105–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. , The pride database and related tools and resources in 2019: improving support for quantification data, Nucleic acids research 47 (D1) (2019) D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw MS data and protein databases are available from the PRIDE repository with the following data set identifier PXD007587, PXD006118, and PXD013386. The datasets generated during the current study are available in the GitHub repository at https://github.com/Biocomputing-Research-Group/DeepFilter.