Abstract
Accurate and accessible nucleic acid diagnostics is critical to reducing the spread of COVID-19 and resuming socioeconomic activities. Here, we present an integrated platform for the direct detection of SARS-CoV-2 RNA targets near patients. Termed electrochemical system integrating reconfigurable enzyme-DNA nanostructures (eSIREN), the technology leverages responsive molecular nanostructures and automated microfluidics to seamlessly transduce target-induced molecular activation into an enhanced electrochemical signal. Through responsive enzyme-DNA nanostructures, the technology establishes a molecular circuitry that directly recognizes specific RNA targets and catalytically enhances signaling; only upon target hybridization, the molecular nanostructures activate to liberate strong enzymatic activity and initiate cascading reactions. Through automated microfluidics, the system coordinates and interfaces the molecular circuitry with embedded electronics; its pressure actuation and liquid-guiding structures improve not only analytical performance but also automated implementation. The developed platform establishes a detection limit of 7 copies of RNA target per μl, operates against the complex biological background of native patient samples, and is completed in <20 min at room temperature. When clinically evaluated, the technology demonstrates accurate detection in extracted RNA samples and direct swab lysates to diagnose COVID-19 patients.
Keywords: Molecular nanostructures, Automated microfluidics, Nucleic acid detection, Electrochemical sensor, COVID-19
1. Introduction
COVID-19 has caused an unprecedented and evolving pandemic (Bedford et al., 2020; Spinelli and Pellino, 2020; Singh et al., 2021; Zhou et al., 2021). Accessible and effective testing for SARS-CoV-2, the causal pathogen of COVID-19 (CSG, 2020), and its emerging variants (Volz et al., 2021; Madhi et al., 2021), is critical to not only controlling the disease spread but also resuming various socioeconomic activities (Bedford et al., 2020; Kerr et al., 2021). More than ever, accurate and easy-to-use molecular assays that can be performed near patients are urgently needed for distributed patient care (Laksanasopin et al., 2015; Nayak et al., 2016; Dong et al., 2014). To date, quantitative reverse transcription polymerase chain reaction (RT-qPCR) remains the clinical gold standard for COVID-19 detection (Kevadiya et al., 2021; Yüce et al., 2021). The nucleic acid detection uses dedicated primers to define SARS-CoV-2 target sequences and thermal cycling to replicate the target sequences before detection. Despite its sensitive performance, the approach requires exquisite primer design, extensive sample processing, specialized instrument and trained personnel (CDC, 2020); these assay characteristics not only pose challenges for versatile assay prototyping but also limit its large-scale implementation outside of centralized clinical laboratories (Esbin et al., 2020; Garg et al., 2019). To address these operation challenges, point-of-care testing platforms have been developed (Song et al., 2021; Valera et al., 2021; Mori et al., 2017; Issadore, 2015; Zhu et al., 2020; Yakoh et al., 2021) (e.g., rapid antigen tests); however, these commonly measure SARS-CoV-2 proteins, through direct antigen-antibody interactions, and have demonstrated low sensitivity and poor robustness.
Motivated by the sensitive performance of SARS-CoV-2 nucleic acid biomarkers and the ease of operation by direct protein detection, we propose to develop an integrated platform that marries the advantages of both to achieve sensitive and accessible detection of nucleic acid biomarkers. We have previously developed a molecular nanotechnology for the direct measurement of nucleic acids (Ho et al., 2018). The technology leverages catalytic molecular nanostructures – enzyme-DNA hybrid nanocomplexes – as programmable and specific transducers that activate upon target nucleic acid hybridization. In the absence of target nucleic acids, the nanocomplexes remain assembled and are catalytically inactivated; upon specific target hybridization, the nanocomplexes dissociate and liberate strong enzymatic activity to initiate downstream catalytic cascades. Importantly, the technology is highly programmable to accommodate various disease mutations; new assays can be readily developed by modifying a single sequence region in the nanostructure, without needing the complex design of multiple pairs of compatible PCR primers and dedicated probes (Ho et al., 2018). Nevertheless, to advance the technology for point-of-care COVID-19 detection, further developments are needed to improve the assay sensitivity and implementation.
To address these challenges, we develop an integrated platform of molecular nanotechnology and automated microfluidics to enable accurate and user-friendly detection of SARS-COV-2 nucleic acids. Termed electrochemical system integrating reconfigurable enzyme-DNA nanostructures (eSIREN), the technology incorporates multiple responsive molecular nanostructures to form a catalytic molecular circuitry, and automated microfluidics to organize and interface the molecular circuitry with embedded electronics. Specifically, the molecular circuitry comprises recognition and signaling molecular nanostructures to achieve target recognition and signal enhancement, respectively. Upon direct target binding, the recognition nanostructure is activated to initiate strong enzymatic activity; this enzyme activation catalyzes another signaling nanostructure to recruit other potent enzymes. The automated microfluidics incorporates pressure actuation and liquid guiders to coordinate the various molecular reactions, thereby seamlessly converting the target-induced molecular activation into an enhanced electrochemical signal. Such integration not only improves the assay's analytical performance but also optimizes the automated implementation (i.e., avoid carryover contamination by infectious samples).
Drawing on these synergistic advances, the developed technology demonstrates superior performance. As compared to conventional nucleic acid detection, instead of relying on target amplification, the technology catalyzes signal enhancement from target hybridization. This enables direct RNA measurement that bypasses all processing steps of conventional RT-qPCR, even against the complex biological background of native clinical samples. As compared to protein detection, the technology has a similar ease of operation and yet achieves sensitive nucleic acid measurement. Specifically, the eSIREN platform established a detection limit of ∼7 copies of RNA target per μl, which is 104-fold better than our previously developed technology. The entire assay is automated and completed in <20 min at room temperature. When applied for the clinical diagnostics of SARS-CoV-2, the technology demonstrated accurate detection using both direct swab lysates and extracted RNA samples.
2. Methods
2.1. Recognition nanostructure preparation
All oligonucleotide sequences were purchased from Integrated DNA Technologies (IDT) and can be found in Supplementary Table 1. The DNA oligonucleotide components (100 μм) were incubated at 95 °C for 5 min in a buffer containing 50 mM NaCl, 1.5 mM MgCl2 and 50 mM Tris–HCl (pH 8.5). This was followed by slow cooling to room temperature at 0.1 °C/s. Taq DNA polymerase (Promega) was subsequently added to form the recognition nanostructure. To characterize the response of recognition nanostructures to nucleic acid targets, synthetic oligonucleotides were used as matching target sequences. Titrations of target sequences were added to the recognition nanostructure solution and the resultant polymerase activity was measured through 5’ exonuclease degradation of a fluorescent probe.
2.2. Signaling nanostructure immobilization
Hairpin signaling oligonucleotide sequences were surface functionalized onto gold screen printed electrodes (4-mm diameter gold working electrode, with gold counter and silver pseudo-reference electrodes, Metrohm). Specifically, thiol-modified signaling oligonucleotides were introduced onto the electrodes, and incubated for 2 h at room temperature. After flushing with PBST to remove excess unbound oligonucleotides, the modified electrodes were blocked in 5% BSA solution for 1 h at room temperature and dried before assembly with the microfluidic device.
2.3. Microfluidic device fabrication
The microfluidic device included three layers and a functionalized screen-printed electrode. The cover layer and substrate layer were made from PMMA sheets and constructed using a tabletop CO2 laser engraver (Universal). The microchannel layer was fabricated through PDMS molding from a 3D-printed cast mold. Briefly, the cast mold was 3D printed using a stereolithographic 3D printer (ASIGA PICO2) with a photosensitive resin (SuperCAST v3). PDMS monomer and cross-linker were mixed at a ratio of 10:1 and cast on the 3D-printed molds, before overnight curing at 60 °C. After being peeled off from the cast mold, the microchannel layer was punched with hole punchers to form the reaction chamber and screw through-holes. To covalently bond the microchannel layer with the cover layer, the cover layer was surface activated with oxygen plasma, followed by immersing in APTES solution (5% v/v) for 20 min. Then the microchannel layer was surface activated with oxygen plasma and attached to the cover layer overnight at 70 °C. To form an enclosed reaction chamber, the screen-printed electrode was placed in the groove on the substrate layer and mechanically clamped to the microchannel layer by screw-fixing the cover layer to the substrate layer. Finally, the assembled device was vacuum-packed with a vacuum sealer (CHIGO ZG-06) after degassing in a vacuum chamber for 12 h.
2.4. Miniaturized pumping system
The miniaturized pumping system comprised six components: a pressure pump (RS Pro, D200), a manifold integrated with five solenoid valves (SMC, V100), a pressure sensor (SMC, PSE530), a USB relay controller (Huasing Electronics), a data acquisition device (NI, USB-6001) and a customized chip holder. The system was housed in a casing fabricated with a fused filament 3D printer (Ultimaker 3). The assembled system measured 17 cm (length) × 9 cm (width) × 7 cm (height) in dimension and was connected to a laptop installed with in-house developed controlling software.
2.5. Contact angle measurement
Contact angle measurements were conducted with a sessile-drop goniometer (Ramé-Hart 590) and recorded with commercial software (DROPimage Advanced v2.10) under ambient conditions (22 °C). A water droplet of 3 μl was deposited on the substrates and contact angles were measured within 5 s. The analysis of contact angles from recorded videos was made with the software ImageJ (Drop snake analysis).
2.6. Filling ratio measurement
To measure the ratio of the filled area in the reaction chamber, solution containing orange food dye was pumped into the microfluidic device using the miniaturized pumping system. The process of liquid filling was recorded with a camera (Canon 80D, EF 100 mm) and analyzed in ImageJ. The filling ratio was defined as the ratio of the filled area over the footprint of the reaction chamber.
2.7. Loading time measurement
A microfluidic device was taken out from its vacuum package before measurement. Reagents were applied to their respective inlets using disposable droppers. The outlet hole was sealed through pressure and immediately the reagents started to be drawn towards the reagent reservoirs by the vacuum loaders. This loading process was recorded with a camera (Canon 80D, EF 100 mm) and analyzed with a video player (VLC). The loading time was determined from the instance of outlet sealing to the moment that the reagent reservoirs were fully filled.
2.8. eSIREN workflow
For all eSIREN measurements, we first used disposable droppers to add reagents (30 μL) to their respective inlets of the microfluidic device, immediately after taking the device out from its vacuum package. Specifically, we applied a mixture containing sample, recognition nanostructure, and biotin-modified dNTPs (TriLink BioTechnologies) to Inlet 1; streptavidin-conjugated HRP (Thermo Scientific) in 0.5% BSA solution to Inlet 2; PBST washing buffer to Inlet 3; TMB substrate to Inlet 4. The outlet hole was then sealed until the reagent reservoirs were fully filled. After the microfluidic device was loaded into the miniaturized pumping system, an in-house developed LabVIEW interface was used to control reagent pumping and incubation in the reaction chamber in sequence. Briefly, it took ∼9 s to pump the reagents into the reaction chamber; the incubation of Inlets 1 and 2 solutions took 10 min and 5 min, respectively. During an incubation, the pump was turned on for 2 s, every 1 min, to bring fresh reagents to the reaction chamber. Before introducing new reagents, the reaction chamber was evacuated and then washed with new reagents/PBST washing buffer for 5 s. The electrochemical signal was recorded during the entire detection process using a miniaturized potentiostat (PalmSens, EmStat3). For each sample, sample-matched positive control (containing active polymerase) and negative control (scrambled recognition nanostructure) were run concurrently for data normalization.
2.9. RNA extraction and detection
Total RNA was isolated using RNeasy kit (Qiagen), according to the manufacturer's protocol. RNase inactivation reagent, included in the kit, was used to stabilize the extracted RNA. The quality and quantity of the extracted RNA were measured with a spectrophotometer (Thermo Scientific) and the product was stored at −80 °C before being used. To detect specific RNA targets through gold standard RT-qPCR analysis, extracted RNA was first reverse-transcribed to generate first-strand cDNA (MultiScribe Reverse Transcriptase, Thermo Scientific). For PCR analysis, we used Taqman Fast Advanced Master Mix (Thermo Scientific) and primer sets (Taqman gene expression assays, Thermo Scientific) as recommended by the manufacturer. Amplification conditions consisted of 1 cycle of 95 °C for 2 min, 45 cycles of 95 °C for 1 s and 60 °C for 20 s. All thermal cycling was performed on QuantStudio 5 real-time PCR system (Applied Biosystems).
2.10. Data normalization
The electrochemical signal was normalized with the equation:
| (1) |
where I norm is the normalized electrochemical signal; I target is the measured signal of the target sample; I positive and I control are the electrochemical signals of sample-matched positive and negative controls, respectively.
The RT-qPCR results were normalized with the equation:
| (2) |
where C norm is the normalized RT-qPCR signal; C T is the measured RT-qPCR results.
2.11. Clinical measurements
The study was approved by the National Healthcare Group Domain Specific Review Board (Ref: 2020/00106 and 2020/00120) and the NUS Institutional Review Board (H-19-001). SARS-CoV-2 positive clinical samples were handled according to the Singapore Ministry of Health Biosafety Branch and the NUS Institutional Biosafety Committee regulations in the Biosafety Level 3 (BSL-3) or Biosafety Level 2+ (BSL-2+) laboratories where appropriate. Subjects were recruited from multiple independent cohorts using Institutional Review Board-approved protocols.
A total of 21 samples consisting of extracted RNA and heat-inactivated swabs were evaluated in this study. To determine the diagnostic performance of the eSIREN assay, extracted RNA samples (positive, n = 9; negative, n = 6) and heat-inactivated swabs (positive, n = 4; negative, n = 2) were used directly on the eSIREN assay. Swabs were collected and heat-inactivated per published protocol (Smyrlaki et al., 2020). Briefly, swabs were lysed at 70 °C for 30 min, before introduction into the eSIREN microfluidic platform. The lysis/stabilization buffer contains the reaction buffer (50 mM NaCl, 1.5 mM MgCl2 and 50 mM Tris–HCl, pH 8.5) with 0.2 U/μl RNase inhibitor (Promega) to preserve RNA integrity in cell lysates. All SARS-CoV-2 clinical diagnoses were generated by commercial RT-qPCR assay (Fortitude Kit, MiRXES). PCR amplification conditions consisted of 1 cycle of 48 °C for 15 min, 1 cycle of 95 °C for 150 s, 42 cycles of 95 °C for 10 s and 59 °C for 42 s. CT value < 40 was determined as positive as per CDC guidelines (CDC, 2020). All measurements on clinical samples were performed in an anonymized and blinded fashion and finalized before comparison with clinical CT value.
2.12. Statistical analysis
Unless otherwise stated, all measurements were performed in triplicate, and the data displayed as mean ± standard deviation. For inter-sample comparisons, multiple pairs of samples were each tested via two-tailed Student's t-test, and the resulting P values were adjusted for multiple hypothesis testing using Bonferroni correction. Receiver operating characteristic (ROC) curves were generated from patient profiling data and constructed by plotting sensitivity versus (1 − specificity), and the values of area under the curve (AUC) were computed using the trapezoidal rule. The clinical reports were used as classifiers (true positives and true negatives). Statistical analyses were performed using MATLAB (2018a) and GraphPad Prism (v.8.0c).
3. Results and discussion
3.1. eSIREN platform
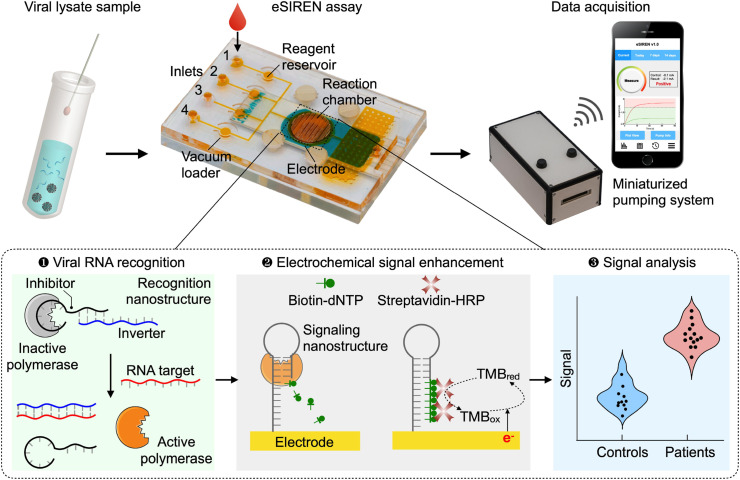
The eSIREN platform is designed for the direct detection of viral nucleic acids in a point-of-care setting. It leverages a molecular circuitry comprising responsive enzyme-DNA nanostructures to directly recognize target sequences and automated microfluidics to coordinate the molecular circuitry with embedded electrodes, so as to transduce the direct target recognition into an amplified electrical signal (Fig. 1 ). The platform is fully powered by a miniaturized pumping system to automate and streamline the assay operation. With respect to its molecular assay composition, the platform utilizes molecular DNA nanostructures to achieve three functional steps: viral RNA recognition, electrochemical signal enhancement and signal analysis. In the recognition step, clinical lysate samples containing SARS-CoV-2 (S gene) targets are mixed with a recognition nanostructure, before being introduced onto the cartridge. The recognition nanostructure is a hybrid molecular complex (Ho et al., 2018) that comprises DNA strands (i.e., inhibitor and inverter sequences) bound to a Taq DNA polymerase (Supplementary Fig. 1a). The inverter sequences are designed complementary to various SARS-CoV-2 RNA targets (Supplementary Table 1). To optimize the design, we determined the binding energy based on the Van't Hoff equation and compared the energy of the inhibitor and inverter sequence vs. that of the inverter and the target sequence, so as to balance polymerase inhibition against polymerase activation for highly specific detection. In the absence of target RNA, the nanostructure remains assembled as the DNA duplex binds strongly with the polymerase to inhibit the polymerase activity; in the presence of complementary target RNA, upon target hybridization with the inverter sequence, the nanostructure dissociates and liberates strong polymerase activity (Supplementary Fig. 1b). In the signal enhancement step, changes in the polymerase activity are measured through additional enzymatic recruitment and catalytic enhancement. Specifically, as the active polymerase elongates a self-primed hairpin signaling nanostructure, it incorporates biotin-modified deoxynucleotide triphosphates (biotin–dNTPs) into the signaling nanostructure, thereby recruiting streptavidin-conjugated horseradish peroxidase (HRP). As the signaling nanostructures are immobilized on gold electrodes, the HRP incorporation enhances redox cycling near the electrode surface for the development of strong electrochemical signals. Finally, during signal analysis, the resultant electrochemical current signals are correlated to viral RNA concentrations to identify SARS-CoV-2 infections.
Fig. 1.
Accessible SARS-CoV-2 detection through molecular nanostructures and automated microfluidics.
Schematics of the eSIREN platform. The platform incorporates molecular nanostructures, integrated microfluidics and a miniaturized pumping system for the direct and automated detection of viral RNA (S gene target of SARS-CoV-2). It leverages a molecular circuitry comprising catalytic enzyme-DNA nanostructures to directly recognize target RNA sequences and automated microfluidics to interface the molecular circuitry with the embedded electrodes, so as to transduce the direct target recognition into an amplified electrical signal. For the molecular circuitry, molecular nanostructures are organized to achieve three functional steps. In the target recognition step, the recognition nanostructure – a hybrid molecular complex comprises DNA strands (inhibitor and inverter sequences) that bind to and inhibit a DNA polymerase enzyme – is mixed with a clinical sample. Only in the presence of target RNA, upon target hybridization with the inverter sequence, the nanostructure dissociates to liberate strong polymerase activity. In the signal enhancement step, the activated polymerase elongates the signaling nanostructure – a self-primed DNA nanostructure immobilized onto the electrode – to incorporate biotin-modified deoxynucleotide triphosphates (biotin–dNTPs). This incorporation recruits streptavidin-conjugated horseradish peroxidase (HRP) near the electrode surface to enhance electrochemical signal amplification. The resultant electrochemical current changes are used to measure viral RNA targets. The entire assay is automated and can be completed in <20 min at room temperature, to achieve direct and user-friendly detection of SARS-CoV-2 infection near patients.
3.2. Characterization of eSIREN assay
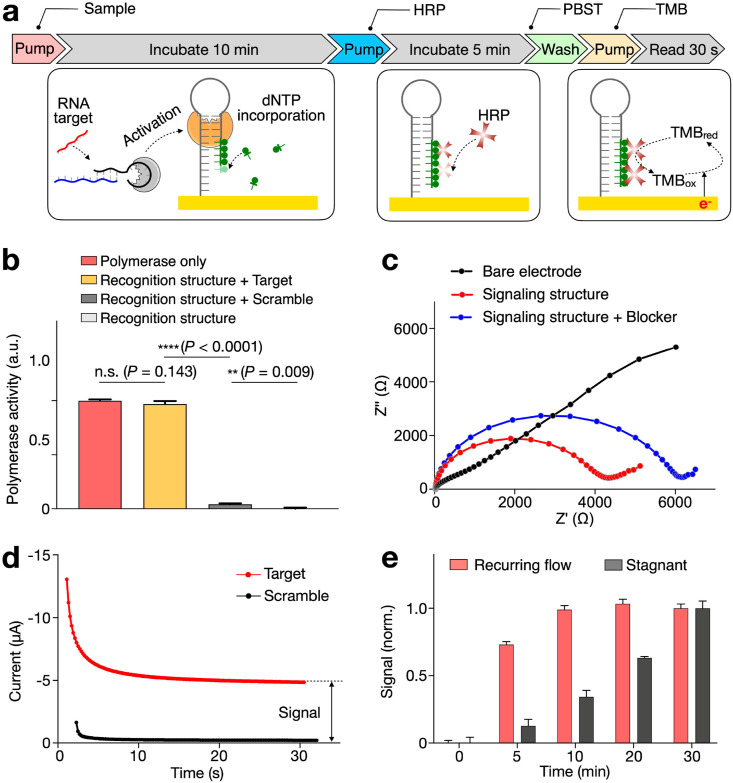
We first evaluated the molecular assay workflow and performed stepwise validation. Briefly, the eSIREN assay consists of a series of reagent introduction and incubation to activate the molecular circuitry for direct RNA detection (Fig. 2 a). All indicated conditions reflect the optimized eSIREN workflow. Specifically, a mixture containing the clinical sample, recognition nanostructures and biotin-modified dNTPs was introduced and incubated on the functionalized electrode for 10 min at room temperature. During the incubation, in the presence of specific RNA target, the polymerase is activated and elongates the immobilized signaling nanostructure with biotin-modified dNTPs. After a brief flushing, streptavidin-conjugated HRP was introduced for 5 min, to enable its binding onto the modified signaling nanostructures through streptavidin-biotin interaction. After another round of brief flushing, redox substrate (Tetramethylbenzidine, TMB) was introduced to start the electrochemical reaction. Continuous data acquisition was performed through a potentiostat to measure the induced electrochemical saturation current.
Fig. 2.
Characterization of the eSIREN assay.
(a) Workflow of the assay operation. Clinical sample, recognition nanostructure and biotin-dNTPs were first applied to the electrode surface. This was followed by a 10-min incubation, during which the recognition nanostructure was activated, in the presence target RNA, to elongate the signaling nanostructure. After a brief flushing, streptavidin-HRP was introduced for a 5-min incubation, to enable the binding of HRP onto the signaling nanostructures through streptavidin-biotin interaction. After washing with PBST buffer, redox substrate (Tetramethylbenzidine, TMB) was introduced to start the electrochemical reaction. Continuous electrochemical data acquisition was performed through a potentiostat. All assay reactions were completed at room temperature. (b) Characterization of the recognition nanostructures. Only in the presence of target RNA, the recognition nanostructures were activated. The resultant activity matched that of pure polymerase, suggesting the high specificity of the recognition nanostructures. (c) Characterization of the signaling nanostructures. Through electrochemical impedance spectroscopy, the increasing electrode resistance confirmed that the signaling nanostructures and protein blockers were successfully deposited in sequence onto the electrode surface. (d) An example of eSIREN measurements with target and control sequences. The complementary target generated a larger stabilized current, while the scrambled control generated negligible signal. (e) Incubation optimization with recurring flow. The incubation duration could be reduced by at least 3 × through the recurrent introduction of fresh reagents. All measurements were performed in triplicate and the data are presented as mean ± s.d in b, e.
To evaluate the step of target incubation, we characterized the performance of the recognition nanostructure (Fig. 2b). In comparison to pure polymerase only, the recognition nanostructure demonstrated low activity. In the presence of complementary RNA target, the polymerase activity fully recovered to attain a level comparable to that by pure polymerase; when scrambled sequences were introduced, the polymerase activity remained negligible. These results confirmed the high specificity of the engineered recognition nanostructure (Supplementary Fig. 1b). We next evaluated the signaling nanostructure functionalization on gold electrodes. All electrodes were functionalized with a layer of signaling nanostructures, before being blocked with proteins prior to assay reaction. We characterized the electrode preparation with electrochemical impedance spectroscopy (Fig. 2c). The increasing electrode resistance confirmed that the signaling nanostructure and protein blocker were successfully deposited in sequence onto the electrode. The addition of protein blocker not only reduced nonspecific binding, but also enhanced signal-to-noise ratio (Supplementary Fig. 2).
Finally, after verifying the recognition and signaling nanostructures, we evaluated the integrated assay process (Fig. 2d). When incubated with a complementary target, the assay generated a stabilized current (∼5 μA); this was significantly larger than the negligible current generated when the system was treated with scrambled control sequences. To optimize the eSIREN assay duration, we further investigated the condition of reagent incubation. Unlike in stagnant incubation (i.e., no flow during incubation), we created a recurring flow by introducing a small volume of reagent continuously during the incubation. By applying this flow setting (e.g., sample incubation with the recognition nanostructure), the incubation duration could be shortened by three-fold and demonstrated a comparable performance (Fig. 2e).
3.3. Microfluidic chip design
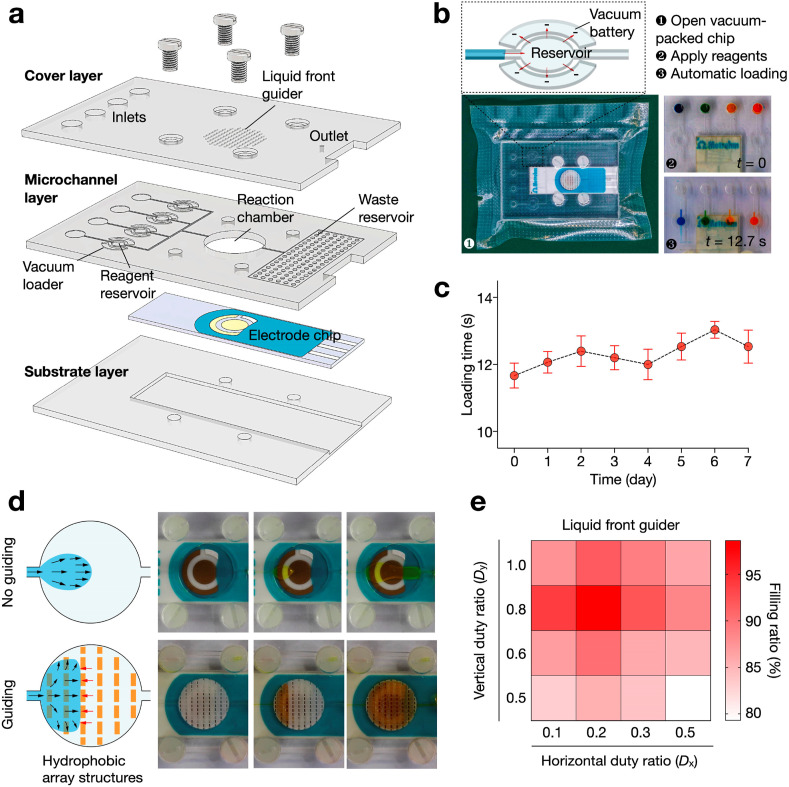
We next designed a microfluidic chip to implement and streamline the assay operation (Fig. 3 a). It comprised three in-house fabricated layers to embed the functionalized electrode. The cover and substrate layers were made from rigid PMMA and the microchannel layer from soft PDMS (Supplementary Fig. 3). The cover layer, microchannel layer and electrode chip were bonded to form an enclosed device through silane chemistry and screw-actuated clamping. To improve the robustness of the eSIREN platform, we incorporated two carefully-designed microfluidic components: the vacuum loader and liquid front guider. The vacuum loader is designed to automatically load assay reagents from the inlets to on-chip reservoirs (Fig. 3b). It comprised a reagent reservoir surrounded by two isolated chambers in negative pressure (i.e., vacuum battery, Fig. 3b, top). The vacuum battery generated negative pressure in the reservoir to automatically load the reagents in as little as 12.7 s (Fig. 3b, bottom). By storing the microfluidic cartridge in vacuum packing, the vacuum loader remained functional, even after 7 days of storage (Fig. 3c). As compared to housing the reagents in the inlets, this design eliminates possible interactions between reagents with external pumping systems; this feature was particularly designed to reduce any carryover contamination, in consideration of the system's COVID-19 application.
Fig. 3.
Microfluidic chip design.
(a) Exploded view of the chip. It consisted of two PMMA layers (cover and substrate layer) and a PDMS layer (microchannel layer) to embed the nanostructure-functionalized electrode. The layers were attached together with silane bonding and screw-actuated clamping. (b) Vacuum loader to automatically transfer reagents from inlets to the on-chip reagent reservoir. This design eliminates any possible contact between reagents and the pumping system, so as to minimize carryover contamination. (c) Time needed to load reagents into the reagent reservoirs (i.e., loading time) as a function of the storage time in vacuum packing. (d) Liquid front guider to improve fluid filling. Laser-engraved pillar array was implemented on the roof of the reaction chamber to form multi-layered perforated barriers perpendicular to the fluid flow. In the absence of the guider, when a liquid is filling a large chamber, it spreads naturally with a small divergence angle and leads to bubble retention. When the chamber is equipped with the guider, the liquid spreading is regulated to ensure complete filling. The liquid is guided to move between two pillar layers, due to a smaller resistance. Once the void between the two layers is completed filled, the liquid breaks the capillary resistance to fill the next interlayer space. (e) Filling ratio with different geometry of liquid front guider. The guider demonstrated the best performance when the horizontal (Dx) and vertical (Dy) duty ratios are 0.2 and 0.8, respectively. All measurements were performed in triplicate. In c, the data are presented as mean ± s.d; in e, the data are presented as mean.
The other microfluidic component – the liquid front guider – was designed to spread the fluid front towards sidewalls (Fig. 3d); this was designed to reduce bubble retention in the reaction chamber, a common problem particularly for liquid filling of large chambers (Wang et al., 2021; Zhao et al., 2020). The liquid front guider consisted of an array of laser-engraved pillars patterned on the roof of the reaction chamber. The pillars were arranged closely to form multi-layered perforated barriers perpendicular to the fluid flow (Supplementary Fig. 4a–c). As a result, it presented a significantly higher hydrophobicity (contact angle: 169.3°) than a bare PMMA surface (contact angle: 77.4°, Supplementary Fig. 5). In the absence of the guider, during liquid filling of a large chamber, the fluid spreads naturally with a small divergence angle (i.e., convex liquid front) and reaches the outlet before fully filling the chamber (Fig. 3d, top). This leads to bubble retention near the chamber sidewall and reduces reagent contact with the electrode surface; both of which result in aberrant eSIREN assay operation. In comparison, when the chamber is equipped with the guider, the liquid spreading is regulated to ensure complete filling of the reaction chamber (Fig. 3d, bottom and Supplementary Fig. 6). The liquid experiences a resistive capillary force, when encountering a layer of pillars; it is thus guided to move between the pillar layers, due to a smaller resistance, towards the chamber sidewall. Once the void between two pillar layers is completed filled, the liquid breaks the capillary resistance to fill the next interlayer space. To optimize the geometry of the liquid front guider, we measured the filling ratio of the chamber with different horizontal (D x) and vertical (D y) duty ratios (Supplementary Fig. 4d). The liquid front guider showed the best performance when D x and D y were 0.2 and 0.8, respectively.
3.4. Miniaturized pumping system
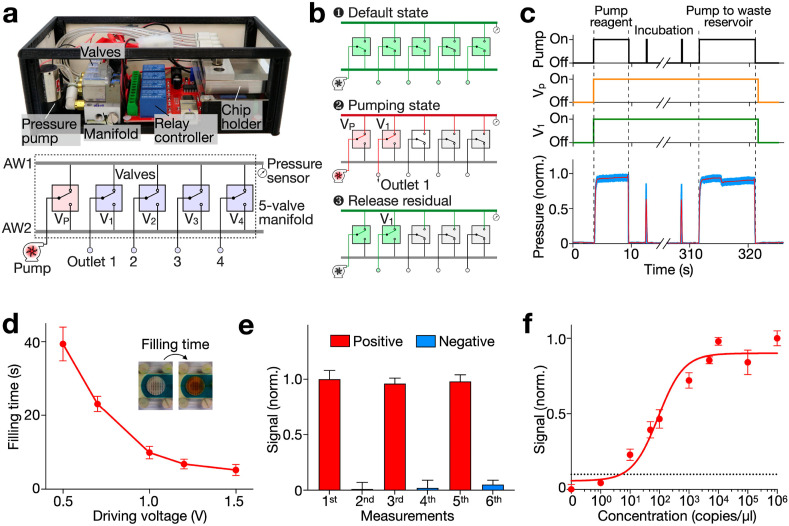
Next, we developed a miniaturized pumping system to automate the eSIREN operation. In view of its COVID-19 application, we designed the pumping system to be contact-free; its pressure-driven mechanism avoids any direct exposure to assay reagents to minimize potential carryover contamination. Specifically, the pumping system contained a pressure pump, a 5-valve manifold, a pressure sensor, a relay controller and a customized chip holder (Fig. 4 a). The positive pressure, generated by the pressure pump, is piloted to the target outlet by the manifold and relay controller, and then coupled to the microfluidic cartridge by the chip holder for liquid pumping. A single pumping event includes three steps (Fig. 4b). In the default state (Fig. 4b, top), all the outlets and the pressure pump are connected through Airway 2 (AW2) in the manifold. Here the pressure pump is switched off and thus functions as an outlet to the atmosphere. In the pumping state (Fig. 4b, middle), the pump and target outlet are connected through Airway 1 (AW1) in the manifold. In this state, the pressure pump is switched on to generate a positive pressure at the target outlet. Once pumping is done (Fig. 4b, bottom), the pressure pump is switched off to vent AW1 and the outlet. The pump and outlet are subsequently connected back to AW2 (i.e., to the default state).
Fig. 4.
Miniaturized pumping system and integration evaluation.
(a) Photograph and schematic of the pumping system. The system contained a pressure pump to generate positive pressure, a 5-valve manifold and relay controller to direct the positive pressure to target outlet, a pressure sensor, and a chip holder to couple the positive pressure to microfluidic chips. (b) Steps in a pumping event. (c) Operation of the pumping system in a typical reagent introducing procedure. (d) Time needed to full up the reaction chamber (filling time) as a function of the driving voltage of the pump. The driving voltage was optimized to 1.0
V for optimized filling efficiency and controllability. (e) System integration and performance evaluation. Positive and negative samples were introduced in a sequential manner. Weak signals were obtained with the negative sample after a positive run, suggesting negligible carryover from previous measurements. (f) Sensitivity of the eSIREN platform. The limit of detection (LOD) was determined by titrating known quantities of target and measuring their corresponding electrochemical signal. The automated eSIREN platform achieved a superior LOD of 7 copies of target per μl. The dashed line shows the LOD, defined as 3 × s.d. of eSIREN signal in a no-target control. Signals above the LOD are considered distinguishable with >99% confidence. All measurements were performed in triplicate and the data are presented as mean ± s.d in d–f.
To validate the pumping system, we performed a series of reagent introduction (Fig. 4c). First, a pathway was established between the pump and the target outlet by switching on their corresponding valves (i.e., Vp and V1). The pump was then switched on to direct reagents into the reaction chamber for a 10-min incubation. During the incubation, the pump was turned on for 2 s every minute to induce recurring flow for enhanced incubation. Finally, the pump was switched on to direct the reagents to the waste reservoir. Throughout the procedure, the system pressure was continuously monitored. Interestingly, we observed not only an elevated pressure during reagent pumping but also a slight pressure decreases when the reaction chamber was emptied. This suggests that the pressure signal could be further exploited to monitor the system operation. Furthermore, to optimize the assay operation, we measured the time needed to fill up the reaction chamber (filling time) as a function of the pump's driving voltage (Fig. 4d). The filling time decreased rapidly with increasing driving voltage and presented considerable variations at a low driving voltage, likely due to the weak pumping force compared to environmental fluctuations. We thus selected a driving voltage of 1.0 V for optimized filling efficiency and controllability.
Finally, we performed the entire eSIREN assay workflow via the miniaturized pumping system to evaluate the platform performance. As the system was designed to minimize potential carryover contamination, we introduced positive and negative samples in a sequential manner (Fig. 4e). The results showed strong signals for the positive runs and weak signals for the negative runs; weak signals were obtained with negative samples even after a prior positive run, suggesting negligible reagent carryover from previous measurements. Next, to validate the stability of the eSIREN platform in various conditions, we conducted eSIREN measurements with synthetic targets at different times of a day, in air-conditioned and ventilated rooms (Supplementary Fig. 7). The results show that despite environmental fluctuations in temperature and humidity, the eSIREN reports comparable results. We further evaluated the analytical performance in a titration experiment, where target sequences were serially diluted (Fig. 4f). The automated eSIREN platform achieved a superior limit of detection (LOD ∼ 7 copies of target per μl). The LOD showed >104-fold improvement to our previously reported colorimetric detection platform (Supplementary Table 2) (Ho et al., 2018).
3.5. eSIREN for clinical diagnosis
To evaluate the clinical utility of the eSIREN platform for SARS-CoV-2 detection, we finally conducted a feasibility study with patient samples. We aimed to determine: (1) if the eSIREN platform could be directly applied to detect clinical samples (i.e., extracted RNA of nasopharyngeal swab samples and inactivated swab lysates), and (2) the accuracy of eSIREN for COVID-19 diagnostics.
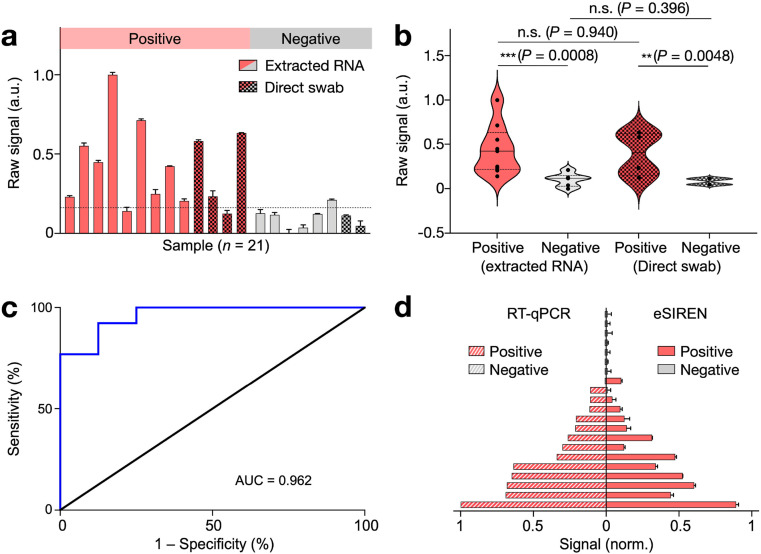
We conducted the eSIREN measurement with extracted RNA samples (n = 15) as well as heat-inactivated swab samples (n = 6, Fig. 5 a). Extracted RNA samples were processed through commercial kits. Heat-inactivated swab samples were heat lysed at 70 °C for 30 min, per published protocols (Smyrlaki et al., 2020). Out of the 21 clinical samples, 13 samples were determined as positive and 8 as negative for COVID-19 infection using gold-standard RT-qPCR assay. To further assess the eSIREN assay in different sample types, we performed multi-comparison analyses of positive and negative samples, respectively (Fig. 5b). Across both extracted RNA and direct swab samples, the positive samples showed elevated signals with respect to the negative samples. Importantly, the eSIREN results of the positive extracted RNA were comparable to that of the positive swab lysates. A similar comparison was observed between the different types of negative patient specimens, indicating technology compatibility with both types of clinical samples for point-of-care COVID-19 detection.
Fig. 5.
Clinical validation of eSIREN for COVID-19 diagnosis.
(a) eSIREN measurements were performed on extracted RNA samples (n = 15) as well as heat-inactivated swab samples (n = 6). Out of the 21 clinical samples, 13 samples were determined as positive and 8 as negative for COVID-19 infection using gold-standard RT-qPCR assay. (b) Statistical analysis of the eSIREN results. Across both extracted RNA and direct swab samples, the positive samples showed an elevated signal compared to the negative samples. (c) Receiver operator characteristic (ROC) curve of the eSIREN results. The platform demonstrated a high accuracy for SARS-CoV-2 detection (area under curve, AUC = 0.962). (d) eSIREN results for clinical classification. By applying an optimal assay threshold based on the Youden's index (the dotted line as indicated in a), the eSIREN platform showed sensitivity = 92.3%, specificity = 87.5% and overall accuracy = 90.5% across all tested clinical samples. The comparisons were performed against clinical RT-qPCR results of matched samples. All measurements were performed in triplicate. In a, c, the data are presented as mean ± s.d; in b, the data are presented as mean. (n.s., not significant, Student's t-test).. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To evaluate the assay performance, we next performed the receiver operating characteristic (ROC) analysis on the eSIREN clinical results (Fig. 5c). The platform showed superior performance (area under curve AUC = 0.962). Based on the Youden's index, we further determined an optimal assay threshold. Specifically, at the optimized assay threshold of 0.14, the eSIREN platform showed sensitivity of 92.3% and specificity of 87.5%. With this optimal assay thresholding for clinical classification, the eSIREN platform demonstrated an overall accuracy of 90.5% (19/21) across all tested clinical samples, as compared to clinical RT-qPCR results (Fig. 5d).
4. Conclusions
Aggressive testing for SARS-CoV-2 is critical to containing the unprecedented COVID-19 public health crisis. Current testing, however, relies primarily on RT-qPCR. Based on the conventional approach of target amplification, the detection requires extensive processing to convert SARS-CoV-2 viral RNA targets into DNA products (reverse transcription) and thermal cycling to replicate the products (PCR amplification) before detection. The approach is thus almost exclusively performed in centralized clinical laboratories due to the need for specialized instrument and trained personnel. This reliance has posed tremendous pressure on public health systems, leading to a significant shortage of supplies, delayed diagnoses and uncontrolled disease spread (Bedford et al., 2020; Kevadiya et al., 2021; Esbin et al., 2020). For prompt detection and efficient management, a user-friendly platform which can perform fast and accurate testing near patients is urgently needed, so as to relieve the pressure of frontline laboratories and facilitate massive pre-event testing for resuming socioeconomic activities (Kerr et al., 2021; Valera et al., 2021). Building upon our previously developed molecular nanotechnology (Ho et al., 2018), we designed the eSIREN platform. It leverages catalytic molecular nanostructures and automated microfluidics to form an integrated system: responsive enzyme-DNA nanostructures are organized as a molecular circuitry that interfaces seamlessly with the microfluidic electrochemical circuitry, thereby enabling direct and enhanced COVID-19 diagnostics near patients.
From the assay perspective, eSIREN uses various molecular nanostructures to form a cascading circuitry that bypasses all steps of conventional detection. It uses a programmable enzyme-DNA hybrid nanocomplex as the recognition nanostructure and a self-primed DNA as the signaling nanostructure. Only in the presence of target RNA, the recognition nanostructure is activated and dissociates upon target hybridization to liberate an active enzyme; the activated enzyme then elongates the signaling nanostructure, during which additional enzyme cascades (e.g., HRP) are recruited to catalyze strong signals. As compared with conventional PCR, eSIREN bypasses RNA extraction and target amplification; as compared with conventional electrochemical sensors based on direct target hybridization (Teengam et al., 2017), it incorporates cascading signal enhancement. Specifically, the eSIREN platform accurately detects RNA, even against the complex biological background of clinical swab lysates, thereby obviating any purification or extraction process. Furthermore, the assay reaction operates at room temperature; through cascading signal enhancement, the technology achieves responsive activation and highly sensitive detection. Notably, key reagents used in the assay (polymerase and dNTPs) are widely available in lyophilized condition, for long term storage (Yang and Wen, 2021), and are operationally stable at room temperature. The technology is thus uniquely designed to facilitate point-of-care detection.
From the implementation perspective, eSIREN uses automated microfluidics to organize and interface the molecular circuitry with electrochemical detection. This implementation not only coordinates the catalytic nanostructures into a potent network, but also interfaces the molecular nanotechnology with amplified electrochemistry to seamlessly convert the molecular activation into an electrical signal. In terms of operation, the automated microfluidics streamlines the assay workflow and improves the system robustness. Through the incorporation of vacuum loader and liquid front guider, the platform eliminates any direct contact between reagents, thereby substantially reducing the chance of carryover contamination. The liquid front guider flattens the fluid meniscus with hydrophobic pillar arrays; this effectively minimizes bubble trapping, even in large reaction chambers, to improve assay performance. The miniaturized pumping system further provides pressure-driven actuation to automate the assay workflow. Taken together, the integrated platform achieves direct and sensitive detection of SAR-CoV-2 targets. It demonstrates a LOD of ∼7 target copies per μl, which is > 104-fold improvement of our previously developed platform. The entire assay can be completed in <20 min at room temperature for rapid and accurate COVID-19 diagnostics.
The technology has the potential to be developed further. With the excellent programmability of molecular nanotechnologies (Li et al., 2017; Sundah et al., 2019), through multiplexing of recognition nanostructures, eSIREN could be readily expanded to identify different genetic loci of SARS-CoV-2. This improvement is likely to enhance the detection coverage and enable mutation identification. Likewise, further sensor incorporation (e.g., loading sensor) (Wang et al., 2016) could establish the platform with enhanced automation capabilities to identify and troubleshoot assay workflow, thereby facilitating diverse applications for at-home testing and self-testing. Finally, with the demonstrated robustness, beyond the current COVID-19 pandemic, eSIREN could be further expanded for accessible detection of other infectious diseases and improved patient stratification in resource-limited settings (Wu et al., 2020; Zhu et al., 2019). Technical improvements, through the integration of advanced microfluidics (Morikawa et al., 2020; Choi et al., 2019; Siedlik et al., 2021; Zhao et al., 2019) and arrayed sensor configuration (Lim et al., 2019; Jeong et al., 2019), could facilitate highly-parallel and large-scale testing.
CRediT authorship contribution statement
Haitao Zhao: Conceptualization, Methodology, Software, Validation, Investigation, Formal analysis, Writing – original draft, Writing – review & editing. Yan Zhang: Conceptualization, Methodology, Validation, Investigation, Formal analysis, Writing – original draft. Yuan Chen: Validation, Investigation. Nicholas R.Y. Ho: Validation, Investigation. Noah R. Sundah: Validation, Investigation. Auginia Natalia: Validation, Investigation. Yu Liu: Validation, Investigation. Qing Hao Miow: Investigation. Yu Wang: Investigation. Paul A. Tambyah: Resources. Catherine W.M. Ong: Resources. Huilin Shao: Conceptualization, Methodology, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank Y.J. Tan and J.M. Hong for their biosafety input, D.L. Beh, K.L. Chew, D. Chan, R. Jureen, B. Yan and C.K. Lee for facilitating clinical sample collection and transfer, P.M. Thong and X. Mao for assistance and access to BSL-2+ facilities.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113629.
Funding
This work was supported in part by funding from National University of Singapore (NUS), NUS Research Scholarship, National Medical Research Council, Ministry of Education, Institute for Health Innovation & Technology, IMCB Independent Fellowship, NUS Early Career Research Award, National University Health System Seed Grant and National Centre for Infectious Diseases Grant.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Bedford J., Enria D., Giesecke J., Heymann D.L., Ihekweazu C., Kobinger G., Lane H.C., Memish Z., Oh M.D., Sall A.A., Schuchat A., Ungchusak K., Wieler L.H., WHO Strategic and Technical Advisory Group for Infectious Hazards Lancet. 2020;395:1015–1018. doi: 10.1016/S0140-6736(20)30673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention CDC . 2020. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. [Google Scholar]
- Choi G., Murphy E., Guan W. ACS Sens. 2019;4:1957–1963. doi: 10.1021/acssensors.9b01067. [DOI] [PubMed] [Google Scholar]
- Coronaviridae Study Group (CSG) of the international committee on taxonomy of viruses. Nat. Microbiol. 2020;5:536–544. [Google Scholar]
- Dong Y., Xu Y., Liu Z., Fu Y., Ohashi T., Mawatari K., Kitamori T. Anal. Sci. 2014;30:359–363. doi: 10.2116/analsci.30.359. [DOI] [PubMed] [Google Scholar]
- Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. RNA. 2020;26:771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N., Boyle D., Randall A., Teng A., Pablo J., Liang X., Camerini D., Lee A.P. Lab Chip. 2019;19:1524–1533. doi: 10.1039/c8lc01303a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho N.R.Y., Lim G.S., Sundah N.R., Lim D., Loh T.P., Shao H. Nat. Commun. 2018;9:3238. doi: 10.1038/s41467-018-05733-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issadore D. Methods Mol. Biol. 2015;1256:123–137. doi: 10.1007/978-1-4939-2172-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H.H., Chen Z., Yadavali S., Xu J., Issadore D., Lee D. Lab Chip. 2019;19:665–673. doi: 10.1039/c8lc01267a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr C.C., Mistry D., Stuart R.M., Rosenfeld K., Hart G.R., Núñez R.C., Cohen J.A., Selvaraj P., Abeysuriya R.G., Jastrzębski M., George L., Hagedorn B., Panovska-Griffiths J., Fagalde M., Duchin J., Famulare M., Klein D.J. Nat. Commun. 2021;12:2993. doi: 10.1038/s41467-021-23276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevadiya B.D., Machhi J., Herskovitz J., Oleynikov M.D., Blomberg W.R., Bajwa N., Soni D., Das S., Hasan M., Patel M., Senan A.M., Gorantla S., McMillan J., Edagwa B., Eisenberg R., Gurumurthy C.B., Reid S.P.M., Punyadeera C., Chang L., Gendelman H.E. Nat. Mater. 2021;20:593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laksanasopin T., Guo T.W., Nayak S., Sridhara A.A., Xie S., Olowookere O.O., Cadinu P., Meng F., Chee N.H., Kim J., Chin C.D., Munyazesa E., Mugwaneza P., Rai A.J., Mugisha V., Castro A.R., Steinmiller D., Linder V., Justman J.E., Nsanzimana S., Sia S.K. Sci. Transl. Med. 2015;7:273re1. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- Li J., Green A.A., Yan H., Fan C. Nat. Chem. 2017;9:1056–1067. doi: 10.1038/nchem.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C.Z.J., Zhang Y., Chen Y., Zhao H., Stephenson M.C., Ho N.R.Y., Chen Y., Chung J., Reilhac A., Loh T.P., Chen C.L.H., Shao H. Nat. Commun. 2019;10:1144. doi: 10.1038/s41467-019-09030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E., Briner C., Kwatra G., Ahmed K., Aley P., Bhikha S., Bhiman J.N., Bhorat A.E., du Plessis J., Esmail A., Groenewald M., Horne E., Hwa S.H., Jose A., Lambe T., Laubscher M., Malahleha M., Masenya M., Masilela M., McKenzie S., Molapo K., Moultrie A., Oelofse S., Patel F., Pillay S., Rhead S., Rodel H., Rossouw L., Taoushanis C., Tegally H., Thombrayil A., van Eck S., Wibmer C.K., Durham N.M., Kelly E.J., Villafana T.L., Gilbert S., Pollard A.J., de Oliveira T., Moore P.L., Sigal A., Izu A. N. Engl. J. Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori E., Oohashi T., Imai H., Mawatari K., Kitamori T. Anal. Methods. 2017;9:2830–2834. [Google Scholar]
- Morikawa K., Kazoe Y., Takagi Y., Tsuyama Y., Pihosh Y., Tsukahara T., Kitamori T. Micromachines. 2020;11:995. doi: 10.3390/mi11110995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S., Sridhara A., Melo R., Richer L., Chee N.H., Kim J., Linder V., Steinmiller D., Sia S.K., Gomes-Solecki M. Sci. Rep. 2016;6:35069. doi: 10.1038/srep35069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedlik M.J., Yang Z., Kadam P.S., Eberwine J., Issadore D. Small. 2021;17 doi: 10.1002/smll.202005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Rahman S.A., Ehtesham N.Z., Hira S., Hasnain S.E. Nat. Med. 2021;27:1131–1133. doi: 10.1038/s41591-021-01397-4. [DOI] [PubMed] [Google Scholar]
- Smyrlaki I., Ekman M., Lentini A., Rufino de Sousa N., Papanicolaou N., Vondracek M., Aarum J., Safari H., Muradrasoli S., Rothfuchs A.G., Albert J., Högberg B., Reinius B. Nat. Commun. 2020;11:4812. doi: 10.1038/s41467-020-18611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q., Sun X., Dai Z., Gao Y., Gong X., Zhou B., Wu J., Wen W. Lab Chip. 2021;21:1634–1660. doi: 10.1039/d0lc01156h. [DOI] [PubMed] [Google Scholar]
- Spinelli A., Pellino G. Br. J. Surg. 2020;107:785–787. doi: 10.1002/bjs.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundah N.R., Ho N.R.Y., Lim G.S., Natalia A., Ding X., Liu Y., Seet J.E., Chan C.W., Loh T.P., Shao H. Nat. Biomed. Eng. 2019;3:684–694. doi: 10.1038/s41551-019-0417-0. [DOI] [PubMed] [Google Scholar]
- Teengam P., Siangproh W., Tuantranont A., Henry C.S., Vilaivan T., Chailapakul O. Anal. Chim. Acta. 2017;952:32–40. doi: 10.1016/j.aca.2016.11.071. [DOI] [PubMed] [Google Scholar]
- Valera E., Jankelow A., Lim J., Kindratenko V., Ganguli A., White K., Kumar J., Bashir R. ACS Nano. 2021;15:7899–7906. doi: 10.1021/acsnano.1c02981. [DOI] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O'Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F., Rey S.M., Nicholls S.M., Colquhoun R.M., da Silva Filipe A., Shepherd J., Pascall D.J., Shah R., Jesudason N., Li K., Jarrett R., Pacchiarini N., Bull M., Geidelberg L., Siveroni I., COG-UK C., Goodfellow I., Loman N.J., Pybus O.G., Robertson D.L., Thomson E.C., Rambaut A., Connor T.R. Cell. 2021;184:64–75.e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Zhu J., Hu X., Chen L., Zuo Y., Yang Y., Jiang F., Sun C., Zhao W., Han X. Lab Chip. 2021;21:1109–1117. doi: 10.1039/d0lc01057j. [DOI] [PubMed] [Google Scholar]
- Wang X., Phan D.T.T., Zhao D., George S.C., Hughes C.C.W., Lee A.P. Lab Chip. 2016;16:868–876. doi: 10.1039/c5lc01563d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhao H., Natalia A., Lim C.Z.J., Ho N.R.Y., Ong C.J., Teo M.C.C., So J.B.Y., Shao H. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Biosens. Bioelectron. 2021;176:112912. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Wen W. ACS Appl. Bio Mater. 2021;4:4354–4360. doi: 10.1021/acsabm.1c00131. [DOI] [PubMed] [Google Scholar]
- Yüce M., Filiztekin E., Özkaya K.G. Biosens. Bioelectron. 2021;172:112752. doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Zhang Y., Liu P.Y., Yap P.H., Ser W., Liu A.Q. Sensor. Actuator. B Chem. 2019;280:16–23. [Google Scholar]
- Zhao H., Chin L.K., Shi Y., Nguyen K.T., Liu P.Y., Zhang Y., Zhang M., Zhang J., Cai H., Yap E.P.H., Ser W., Liu A. Sensor. Actuator. B Chem. 2020;306:127562. [Google Scholar]
- Zhou B., Thao T.T.N., Hoffmann D., Taddeo A., Ebert N., Labroussaa F., Pohlmann A., King J., Steiner S., Kelly J.N., Portmann J., Halwe N.J., Ulrich L., Trüeb B.S., Fan X., Hoffmann B., Wang L., Thomann L., Lin X., Stalder H., Pozzi B., de Brot S., Jiang N., Cui D., Hossain J., Wilson M.M., Keller M.W., Stark T.J., Barnes J.R., Dijkman R., Jores J., Benarafa C., Wentworth D.E., Thiel V., Beer M. Nature. 2021;592:122–127. doi: 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
- Zhu J., Han G., Hu X., Zuo Y., Chen L., Wang F., Yang Y., Jiang F., Sun C., Zhao W., Han X. ACS Sens. 2020;5:1381–1388. doi: 10.1021/acssensors.0c00090. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Huang Z., Chen Q., Wu Q., Huang X., So P.K., Shao L., Yao Z., Jia Y., Li Z., Yu W., Yang Y., Jian A., Sang S., Zhang W., Zhang X. Nat. Commun. 2019;10:4049. doi: 10.1038/s41467-019-12089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.