Abstract
SARS-CoV-2 represents the causative agent of the current pandemic (COVID-19). The drug repurposing technique is used to search for possible drugs that can bind to SARS-CoV-2 proteins and inhibit viral replication. In this study, the FDA-approved antiplatelets are tested against the main protease and spike proteins of SARS-CoV-2 using in silico methods. Molecular docking and molecular dynamics simulation are used in the current study. The results suggest the effectiveness of vorapaxar, ticagrelor, cilostazol, cangrelor, and prasugrel in binding the main protease (Mpro) of SARS-CoV-2. At the same time, vorapaxar, ticagrelor, and cilostazol are the best binders of the spike protein. Therefore, these compounds could be successful candidates against COVID-19 that need to be tested experimentally.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11239-021-02558-5.
Keywords: SARS-CoV-2, COVID-19, Antiplatelet, Drug repurposing, Spike, Mpro
Introduction
The world is currently facing novel coronavirus disease 2019 (COVID-19), an unprecedented health crisis caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 is one of the families of coronaviruses (CoVs), which include the severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV), resulting in high mortality rates [1]. In December 2019, the first case of COVID-19 was discovered in Wuhan, Hubei, China, then rapidly spread worldwide [2]. The World Health Organization (WHO) in March 2020 announced that COVID-19 is a pandemic [3, 4]. Since the pandemic, about 200 million confirmed cases and over 4.2 million deaths globally around 220 countries, areas, or territories, with the majority in Europe and the Americas. The common symptoms of COVID-19 are fever, dry cough, nausea, shortness of breath, and respiratory complications. In many cases, olfactory sensory losses occur and sometimes leading to death [5].
SARS‐CoV‐2 was reported as an enveloped positive single-stranded RNA virus and categorized as a betacoronavirus family member [6]. SARS‐CoV‐2 consists of 16 non-structural proteins and four structural proteins, such as nucleocapsid (N), spike (S), envelope (E), and membrane (M) [7]. The ribonucleic acid (RNA) genome binds to N protein to form the viral nucleocapsid, whereas the S, E, and M proteins contribute to shaping the viral envelope. The glycoprotein spikes distinguish coronaviruses by their unique crown‐like appearance [8]. Cell internalization of SARS‐CoV‐2 utilizes the angiotensin-converting enzyme 2 (ACE2), expressed on many cell types in human tissues by interacting with spike protein [9]. After cell internalization, viral RNA is translated into two polyproteins (pp1a and pp1ab) encoding multiple critical non-structural proteins (nsPs), including two proteases; main protease (Mpro) or Chymotrypsin-like protease (3CLpro) and papain-like protease (PLpro), both of them processes the polypeptide pp1a and pp1ab in a specific way to generate 16 nsPs [9, 10]. The papain-like protease is responsible for the production of four nsPs. On the other hand, the rest of the vital nsPs are produced by Mpro, including methyltransferase, helicase, and RNA-dependent RNA polymerase (RdRp); all of them have a significant role in the viral infection cycle. Hence, the main protease and spike protein are considered prospective therapeutic targets to inhibit viral replication [9, 11].
Until now, there is no complete awareness of the pathogenic pathways of SARS-CoV-2. COVID-19 patients are suffering from severe acute respiratory syndrome and maybe in some cases leading to lung damage due to direct harmful viral effects on both types I and II pneumocytes and alveolar endothelial cells [12], resulting in uncontrolled severe inflammatory reaction and pulmonary microvascular thrombosis [13, 14]. Medical treatments have been proposed to avoid the symptoms of coagulopathy in COVID-19 patients [15]. Recent data showed that anticoagulant therapy has better results in moderate and severe COVID-19 patients with coagulopathy signs and those receiving mechanical ventilation [12, 16]. Additionally, the prophylactic impact of antithrombotic therapy on COVID-19 severity and mortality has been considered [17]. Moreover, it was reported that heparin could bind with the SARS-CoV-2 S1 receptor domain leading to a conformational change that hinders cell infection, which confirms the antiviral effect of heparin against COVID-19 [18]. No clinical findings have been reported concerning antiplatelet therapy's potential protective or therapeutic effects against COVID-19, unlike anticoagulant treatment [12]. Only one clinical trial (NCT04365309) has been started assuming aspirin's early usage is expected to reduce the incidence of COVID-19 due to its ability to inhibit viral proliferation, platelet aggregation, and lung injury.
So far, no specific drugs have shown promising therapeutic potential against COVID-19, while several attempts to find possible therapeutics are ongoing [19–22]. The Discovery of a new particular drug against SARS-CoV-2 will take more time utilizing the conventional method. So, drug repurposing has been launched as an inspiring approach, as recommended by WHO [23]. Our group recently revealed the promising role of antiplatelet FDA-approved drugs in the treatment of COVID-19 by inhibiting main protease (Mpro) and spike glycoprotein (S) [24].
This study will focus more intensely on screening the antiplatelet FDA-approved drugs against main protease and spike glycoprotein based on molecular dynamics (MD) simulation, which can confirm the stability of the interactions of the docked complexes.
Materials and methods
Structural retrieval and preparation
The structure of the ligands (FDA- approved Antiplatelets) are retrieved from the PubChem database and prepared for the docking study [24, 25]. Additionally, the SARS-CoV-2 Mpro and spike protein structures are retrieved from the Protein Data Bank (PDB ID: 6Y84 and 6YLA, respectively). The systems are prepared using PyMOL software, where water and ions are removed, and the missed H-atoms are added to the protein structures [26]. The protein structures are subjected to a 100 ns Molecular Dynamics Simulation (MDS) run. The CHARMM 36 force field is used to equilibrate the protein solvated in the TIP3P water model [27, 28]. The salt concentration is adjusted to the physiological conditions (154 mM NaCl and the pH value of 7.0) during the simulation. The temperature is adjusted to 310 K, and the system is simulated in NVT ensemble (constant number of atoms, volume, and temperature). NAno-scale Molecular Dynamics (NAMD) 2.13 and the Visualizing Molecular Dynamics (VMD) 1.9.3 software are utilized to run the simulation, input files preparation, and for data analysis along with in-house codes [29, 30]. The University of California San Francisco Chimera software is used to perform cluster analysis of the trajectories [31]. A representative conformation from each cluster is used in the docking experiments to test the antiplatelets' binding affinities against SARS-CoV-2 proteins. As shown below, twelve different conformations of the Mpro and seven conformations for the spike are used in the docking experiments.
Molecular docking
AutoDock Vina software is used to perform the docking experiments, while AutoDock Tools is used to prepare the input files and to set the grid box [32, 33]. Any missed H-atoms and charges are added to both the ligands and the proteins. The box sizes were set to 40 × 40 × 40 Å3 for both proteins, while the box center is set to be at the active site dyad (H41 and C145) in the case of Mpro and at the receptor-binding domain of the spike protein. In the Mpro the dyad residues are set to be flexible during the docking experiments. Average binding affinity values are plotted with their standard deviations (SD) for each drug. The docking complexes are examined using Protein–Ligand Interaction Profiler (PLIP) web server for detailed established interactions [34]. PyMOL software is used to represent selected complexes.
Results and discussion
The highly contagious COVID-19 pandemic affects the world leaving more than 4.2 M deaths and about 200 M infections. Drug repurposing is successfully helping COVID-19 patients by the reduction in the infection severity and the hospitalization period. [35]. Different in silico studies have suggested the previously approved drugs against the SARS-CoV-2. The suggestions are based on these drugs' binding ability against viral and host-cell proteins crucial for the infection [36–38].
The authors' recently published docking study suggests the possibility of antiplatelet drugs against COVID-19 [24]. To prove these drugs' effectiveness, we include molecular dynamics simulation for the SARS-CoV-2 protein targets Mpro and the spike protein for 100 ns (see the supplementary figure S1).
Figure 1 shows the average binding affinity for each drug of the antiplatelets against the SARS-CoV-2 spike (A) and Mpro (B). The spike protein was represented by seven different conformations representing the different clusters after MDS, while 12 conformations represented Mpro. The average binding affinity for nelfinavir is represented in the red columns, while the best compounds are in green. As reflected from the bar graphs, the best three compounds in their average binding affinity against both SARS-CoV-2 spike and Mpro are vorapaxar (-7.74 ± 0.35 and -8.08 ± 0.33 kcal/mol), ticagrelor (-7.03 ± 0.28 and -7.75 ± 0.35 kcal/mol), and cilostazol (-6.86 ± 0.43 and -7.27 ± 0.36 kcal/mol). Additionally, cangrelor (-6.78 ± 0.44 kcal/mol) and prasugrel (-6.76 ± 0.36 kcal/mol) show comparable average binding affinities against SARS-CoV-2 Mpro compared to nelfinavir (-6.92 ± 0.51 kcal/mol).
Fig. 1.
The average binding affinity (in kcal/mol) of the antiplatelet drugs against SARS-CoV-2 spike (A) and Mpro (B) proteins calculated by AutoDock Vina software. Error bars represent the standard deviation. Nelfinavir, the positive control, is shown in red columns
The interaction patterns' details of the antiplatelet drugs against SARS-CoV-2 proteins are summarized in Table 1 (spike) and Table 2 (Mpro). Two main types of interactions are established between the antiplatelet drugs and the SARS-CoV-2 proteins, which are H-bonding and hydrophobic contacts. The most-reported H-bonds are formed between the medicines and W436 and R509 of the spike, while residues F342 and W436 established hydrophobic contacts with the drugs. SARS-CoV-2 Mpro residues that form H-bonds with the medicines are G143, S144, C145 (one of the dyads), and E166. Additionally, L27 and E166 of the Mpro form hydrophobic contacts with the drugs. It is evident that the main interactions that stabilize the drug, icosapent ethyl, in the spike and Mpro are the hydrophobic contacts, while H-bonds are essential in stabilizing other drugs in the Mpro active site pocket (see Table 2).
Table 1.
The interaction pattern of the compounds having binding affinities against SARS-CoV-2 spike protein
| Compound | AutoDock Vina score (kcal/mol) | H-bonding | Hydrophobic interaction | ||
|---|---|---|---|---|---|
| Number | Interacting residues | Number | Interacting residues | ||
| Nelfinavir | −7.0 | 3 | T430, F515, and L517 | 1 | V382 |
| Anagrelide | −5.9 | 2 | T430(2) | 1 | F515 |
| Aspirin | −5.2 | 9 | R454(4), K458, S459, D467, and E471(2) | 2 | K458 and E471 |
| Cangrelor | −6.2 | 10 | D428, T430(2), G431, S514(2), F515(3), and L517 | ||
| Cilostazol | −7.1 | 5 | S373, W436, S438(2), and R509 | 4 | F342, V367, F374, and W436 |
| Clopidogrel | −5.7 | 2 | W436 and R509 | 3 | F342, F374, and W436 |
| Dipyridamole | −5.5 | 8 | Y396(2), D427, D428(2), D428, and E516(2) | 3 | P426, K462, and F464 |
| Epoprostenol | −5.9 | 2 | S375 and D405 | 5 | T376, V407, R408, V433, and V503 |
| Icosapent ethyl | −5.1 | 1 | T430 | 10 | V382, L387, L390, F392(2), P426, F464, F515(2), and L517 |
| Iloprost | −6.2 | 3 | W436, S438, and R509 | 4 | F342, V367, and L441(2) |
| Pentoxifylline | −5.7 | 2 | V382 and S514 | 7 | V382, L387, L390, F392(2), and F515(2) |
| Prasugrel | −6.4 | 3 | F338, G339, and N343 | 8 | F338, F342(2), V367, L368, F374, and W436(2) |
| Ticagrelor | −7.0 | 7 | F342, N343, A344, T345, W436, N437, and R509 | 2 | F374 and W436 |
| Ticlopidine | −5.5 | 1 | E516 | 4 | E516, L517(2), and L518 |
| Tirofiban | −6.1 | 4 | C336, G339, S373, and R509 | 9 | F342(3), V367(2), L368, F374, W436, and L441 |
| Vorapaxar | −7.6 | 2 | K462 and L517 | 4 | R355, P426, F429, and F464 |
The AutoDock Vina scores are listed among H-bonds and hydrophobic contacts and the residues that interact. Residue interacts with π-stacking is blue-colored, while the orange residue is that interacts with salt bridges. Bold residues are the most common residues
Table 2.
The interaction pattern of the compounds having better binding affinities than Nelfinavir against SARS-CoV-2 Mpro
| Compound | AutoDock Vina score (kcal/mol) | H-bonding | Hydrophobic interaction | ||
|---|---|---|---|---|---|
| Number | Interacting residues | Number | Interacting residues | ||
| Nelfinavir | −6.7 | 1 | E166 | 3 | T25, L27, and M165 |
| Anagrelide | −6.4 | 5 | N142, G143, S144, C145, and E166 | ||
| Aspirin | −5.2 | 4 | G143, S144, C145, and E166 | 1 | E166 |
| Cangrelor | −6.7 | 10 | N119, L141, N142, G143, S144, C145, E166, Q189(2), and D187 | ||
| Cilostazol | −7.2 | 5 | S144, E166, Q189, T190, and Q192 | 1 | Q189 |
| Clopidogrel | −5.7 | 2 | E166 and D187 | ||
| Dipyridamole | −6.2 | 8 | N28(2), N142, G143, C145(2), E166, and Q189 | 2 | T25 and L27 |
| Epoprostenol | −5.9 | 6 | N142, G143, S144, Q189, and Q192 | 1 | P168 |
| Icosapent ethyl | −4.8 | 0 | 7 | A70, V73(2), L75, T93, K97, and P122 | |
| Iloprost | −5.9 | 6 | R40, Y54(3), E55, and N84 | 2 | P52 and Y54 |
| Pentoxifylline | −5.7 | 5 | N142, G143, S144, C145, and E166 | 2 | T25 and L27 |
| Prasugrel | −7.0 | 7 | S46, L141, G143, S144(2), C145, and E166 | ||
| Ticagrelor | −7.8 | 7 | N142, G143, S144(3), C145, and E166 | 1 | T25 |
| Ticlopidine | −5.4 | 0 | 2 | M165 and Q192 | |
| Tirofiban | −5.9 | 5 | G143, S144, C145, and T190(2) | 2 | M165 and Q192 |
| Vorapaxar | −8.1 | 4 | T26, N119, N142, and E166 | 2 | E166 and R188 |
The AutoDock Vina scores are listed among H-bonds and hydrophobic contacts and the residues that interact. Residues that interact with π-stacking are blue-colored. Bold residues are the most common residues
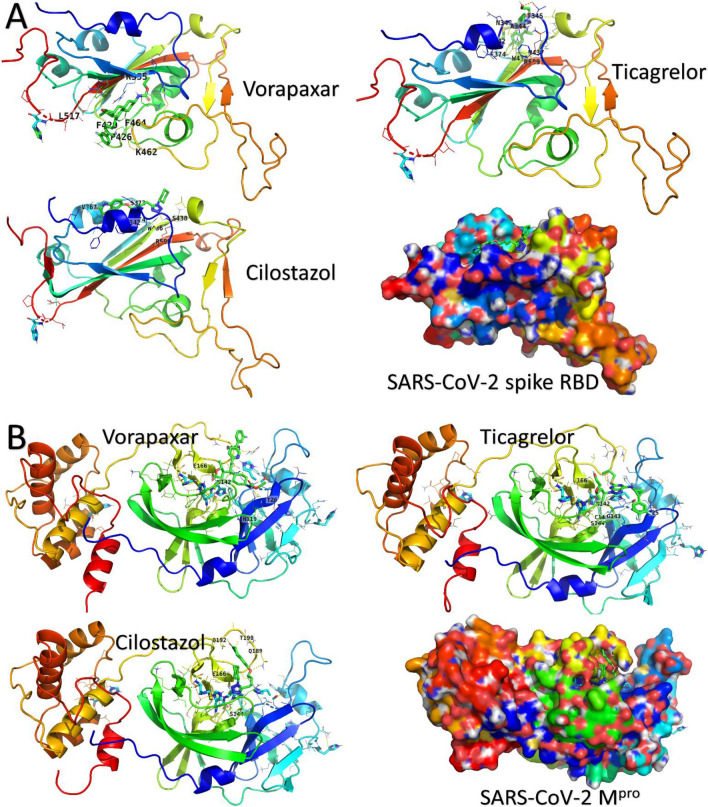
The best three drugs in binding SARS-CoV-2 spike and Mpro (vorapaxar, ticagrelor, and cilostazol) are represented in Figs. 2A and 2B. Proteins are expressed in the rainbow-colored cartoons and surface (N-blue and C-red). Interacting residues are labeled (one-letter) and shown in colored lines, while the drugs are in green sticks. Vorapaxar is the best binder drug to spike RBD and Mpro of SARS-CoV-2 with average binding affinity values of -7.74 ± 0.35 and -8.08 ± 0.33, respectively. Vorapaxar, ticagrelor, and cilostazol bind to the Mpro at the same pocket that contains the dyads (H41 and C145). Hence, these drugs could be potential candidates for SARS-CoV-2 inhibition by impairing Mpro function.
Fig. 2.
Docking complexes of the best three drugs against SARS-CoV-2 spike (A) and Mpro (B). Medicines are represented in green sticks while interacting residues are in lines and labeled with their one-letter code. The proteins are expressed in rainbow-colored (N-blue and C-red) cartoons. Surface representation of the proteins is found in the right-bottom corner for each figure
The current study suggests that vorapaxar, ticagrelor, and cilostazol are able to bind with both Mpro and SARS-CoV-2 spike RBD. Each one of them has an antithrombotic role besides the prospective antiviral effect. Vorapaxar is considered as protease-activated receptor 1 (PAR-1), is overexpressed by CD8 T lymphocytes after viral infection, working as an antagonist that inhibits thrombin-related platelet aggregation. It does not affect coagulation factors, ADP-mediated platelet aggregation, or bleeding time [39, 40]. Ticagrelor is a P2Y12 receptor antagonist which inhibits thrombosis and reduces the risk of myocardial infarction [41, 42]. Cilostazol is an antiplatelet agent and vasodilator used to treat intermittent claudication by inhibiting phosphodiesterase III and suppressing cyclic AMP (cAMP) degradation [43]. This study suggests the dual role of these three drugs against COVID-19 that needs to be tested experimentally.
We previously reported the enhanced binding contribution of the cell-surface Glucose Regulated Protein 78 (CS-GRP78), also termed Heat Shock Protein A5 (HSPA5), to the newly emerged variants SARS-CoV-2 (UK, South African, and Brazilian) spike protein compared to the wildtype variant [44, 45]. The predicted GRP78 recognition site of the spike is located in the C480-C488 region [46–48]. This recognition site is not affected by the binding of the antiplatelet drugs in our study. Additionally, antiplatelet drugs bind well with the spike protein apart from the binding surface to the main receptor for SARS-CoV-2, the Angiotensin-Converting Enzyme 2 (ACE2) (see Fig. 3). This makes the antiplatelet drugs bind to both wildtype and the new variant mutations of the spike.
Fig. 3.
The superposition of one of the complexes (Cilostazol against RBD of the spike) (surface) against the solved structure of SARS-CoV-2 spike and ACE2 (PDB ID: 6ACK) (magenta cartoon). The GRP78 recognition site is encircled
Even if not in the recognition surface, drug binding to the Receptor Binding Domain (RBD) of the spike protein can be of therapeutic importance since the spike is responsible for host cell entry [49]. Thus, any hindrance exerted by the drug bound firmly in the pockets of the RBD may affect the conformational changes that mediate cell entry. On the other hand, the spike's antiplatelet binding may not affect the host cell recognition to SARS-CoV-2.
Conclusion
Antiplatelet FDA-approved drugs are tested in this study against SARS-CoV-2 Mpro and spike proteins after molecular dynamics and trajectory clustering. The different conformations of Mpro (twelve) and the spike RBD (seven), gotten after clustering the MDS trajectories, support our previous finding that antiplatelet drugs could be successful candidates against COVID-19. Vorapaxar, ticagrelor, and cilostazol are able to bind firmly to both Mpro and the RBD of the SARS-CoV-2 spike. The drug-binding is not close to the mutations observed in the new variants of the spike (the UK, South African, and Brazilian); hence it could inhibit all variants of the spike. These in silico findings are yet to be verified by experimental activity assays and clinical trials.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The MDS calculations are performed over the Bibliotheca Alexandrina HPC, Alexandria, Egypt.
Data availability
The docking structures are available upon request from the corresponding author.
Declarations
Conflict of interest
All the authors declare that there is no competing interest in this work.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Paules CI, Marston HD, Fauci AS. Coronavirus infections-more than just the common cold. JAMA. 2020;323(8):707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 2.Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, Zhong W, Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramphul K, Mejias SG. Coronavirus disease: a review of a new threat to public health. Cureus. 2020;12(3):e7276. doi: 10.7759/cureus.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elfiky AA. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J Biomol Struct Dyn. 2021;39(9):3204–3212. doi: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui DS, E IA, Madani TA, Ntoumi F, Kock R, Dar O, Ippolito G, McHugh TD, Memish ZA, Drosten C, Zumla A, Petersen E The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infectious diseases : IJID : official Publication Int Society for Infectious Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salata C, Calistri A, Parolin C, Palu G. Coronaviruses: a paradigm of new emerging zoonotic diseases. Pathog Dis. 2019 doi: 10.1093/femspd/ftaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sohag AAM, Hannan MA, Rahman S, Hossain M, Hasan M, Khan MK, Khatun A, Dash R, Uddin MJ. Revisiting potential druggable targets against SARS-CoV-2 and repurposing therapeutics under preclinical study and clinical trials: a comprehensive review. Drug Dev Res. 2020 doi: 10.1002/ddr.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashour HM, Elkhatib WF, Rahman MM, Elshabrawy HA (2020) Insights into the Recent 2019 Novel Coronavirus (SARS-CoV-2) in Light of Past Human Coronavirus Outbreaks. Pathogens (Basel, Switzerland) 9 (3). doi:10.3390/pathogens9030186 [DOI] [PMC free article] [PubMed]
- 9.Kumar Y, Singh H, Patel CN. In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing. J Infect Public Health. 2020;13(9):1210–1223. doi: 10.1016/j.jiph.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281(18):4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mondal P, Natesh J, Abdul Salam AA, Thiyagarajan S, Meeran SM. Traditional medicinal plants against replication, maturation and transmission targets of SARS-CoV-2: computational investigation. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1842246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maldonado E, Tao D, Mackey K. Antithrombotic therapies in COVID-19 disease: a systematic review. J Gen Intern Med. 2020;35(9):2698–2706. doi: 10.1007/s11606-020-05906-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monteleone G, Sarzi-Puttini PC, Ardizzone S. Preventing COVID-19-induced pneumonia with anticytokine therapy. Lancet Rheumatol. 2020;2(5):e255–e256. doi: 10.1016/S2665-9913(20)30092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giani M, Seminati D, Lucchini A, Foti G, Pagni F. Exuberant plasmocytosis in bronchoalveolar lavage specimen of the first patient requiring extracorporeal membrane oxygenation for SARS-CoV-2 in Europe. J Thoracic Oncol: official Publication International Association for the Study of Lung Cancer. 2020;15(5):e65–e66. doi: 10.1016/j.jtho.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klok FA, Kruip M, van der Meer NJM, Arbous MS, Gommers D, Kant KM, Kaptein FHJ, van Paassen J, Stals MAM, Huisman MV, Endeman H. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res. 2020;191:145–147. doi: 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kow CS, Hasan SS. Use of antiplatelet drugs and the risk of mortality in patients with COVID-19: a meta-analysis. J Thromb Thrombolysis. 2021;52(1):124–129. doi: 10.1007/s11239-021-02436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corrochano M, Acosta-Isaac R, Mojal S, Miqueleiz S, Rodriguez D, Quijada-Manuitt MA, Fraga E, Castillo-Ocana M, Amaro-Hosey K, Albiol N, Soria JM, Antonijoan RM, Souto JC. Impact of pre-admission antithrombotic therapy on disease severity and mortality in patients hospitalized for COVID-19. J Thromb Thrombolysis. 2021 doi: 10.1007/s11239-021-02507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mycroft-West C, Su D, Elli S, Li Y, Guimond S, Miller G, Turnbull J, Yates E, Guerrini M, Fernig D, Lima M, Skidmore M. The 2019 coronavirus (SARS-CoV-2) surface protein (Spike) S1 Receptor Binding Domain undergoes conformational change upon heparin binding. BioRxiv. 2020 doi: 10.1101/2020.02.29.971093. [DOI] [Google Scholar]
- 19.Mahmud S, Elfiky AA, Amin A, Mohanto SC, Rahman ME, Acharjee UK, Saleh MA. Targeting SARS-CoV-2 nonstructural protein 15 endoribonuclease: an in silico perspective. Futur Virol. 2021;16(7):467–474. doi: 10.2217/fvl-2020-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gyebi GA, Elfiky AA, Ogunyemi OM, Ibrahim IM, Adegunloye AP, Adebayo JO, Olaiya CO, Ocheje JO, Fabusiwa MM. Structure-based virtual screening suggests inhibitors of 3-Chymotrypsin-Like Protease of SARS-CoV-2 from Vernonia amygdalina and Occinum gratissimum. Comput Biol Med. 2021;136:104671. doi: 10.1016/j.compbiomed.2021.104671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonousi A, Mahran HA, Ibrahim IM, Ibrahim MN, Elfiky AA, Elshemey WM. Novel adenosine derivatives against SARS-CoV-2 RNA-dependent RNA polymerase: an in silico perspective. Pharmacol Rep. 2021 doi: 10.1007/s43440-021-00300-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elfiky AA, Ibrahim IM, Amin FG, Ismail AM, Elshemey WM 2021 COVID-19 and Cell Stress. In: Rezaei N (ed) Coronavirus Disease - COVID-19. Springer International Publishing, Cham, pp 169–178. doi:10.1007/978-3-030-63761-3_10 [DOI] [PubMed]
- 23.Kupferschmidt K, Cohen J. Race to find COVID-19 treatments accelerates. Science. 2020;367(6485):1412–1413. doi: 10.1126/science.367.6485.1412. [DOI] [PubMed] [Google Scholar]
- 24.Abosheasha MA, El-Gowily AH. Superiority of cilostazol among antiplatelet FDA-approved drugs against COVID 19 M(pro) and spike protein: drug repurposing approach. Drug Dev Res. 2021;82(2):217–229. doi: 10.1002/ddr.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(D1):D1202–1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.2.4.1 V The PyMOL Molecular Graphics System, Version 2.4.1 Schrödinger, LLC.
- 27.Huang J, MacKerell AD., Jr CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J Comput Chem. 2013;34(25):2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mark P, Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J Phys Chem A. 2001;105(43):9954–9960. doi: 10.1021/jp003020w. [DOI] [Google Scholar]
- 29.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graphics. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 30.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 32.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Res. 2015;43(W1):W443–W447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khambholja K, Asudani D. Potential repurposing of Favipiravir in COVID-19 outbreak based on current evidence. Travel Med Infect Dis. 2020;35:101710. doi: 10.1016/j.tmaid.2020.101710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elfiky AA (2020) Corrigendum to "Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARSCoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study" [Life Sci. 253 (2020) 117592]. Life Sci 258:118350. doi:10.1016/j.lfs.2020.118350 [DOI] [PMC free article] [PubMed]
- 37.Elfiky AA. Reply to a letter to the editor. Life Sci. 2020;252:117715. doi: 10.1016/j.lfs.2020.117715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parvathaneni V, Gupta V. Utilizing drug repurposing against COVID-19 - Efficacy, limitations, and challenges. Life Sci. 2020;259:118275. doi: 10.1016/j.lfs.2020.118275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker NC, Lipinski MJ, Lhermusier T, Waksman R. Overview of the 2014 food and drug administration cardiovascular and renal drugs advisory committee meeting about vorapaxar. Circulation. 2014;130(15):1287–1294. doi: 10.1161/CIRCULATIONAHA.114.011471. [DOI] [PubMed] [Google Scholar]
- 40.group As, Vorapaxar for HIV-associated inflammation and coagulopathy (ADVICE): a randomised, double-blind, placebo-controlled trial. The lancet HIV. 2018;5(10):e553–e559. doi: 10.1016/S2352-3018(18)30214-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teng R, Oliver S, Hayes MA, Butler K. Absorption, distribution, metabolism, and excretion of ticagrelor in healthy subjects. Drug metabolism and disposition: the biological fate of chemicals. 2010;38(9):1514–1521. doi: 10.1124/dmd.110.032250. [DOI] [PubMed] [Google Scholar]
- 42.Dorsam RT, Kunapuli SP. Central role of the P2Y12 receptor in platelet activation. J Clin Invest. 2004;113(3):340–345. doi: 10.1172/JCI20986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Shakur Y, Yoshitake M, Kambayashi Ji J. Cilostazol (pletal): a dual inhibitor of cyclic nucleotide phosphodiesterase type 3 and adenosine uptake. Cardiovasc Drug Rev. 2001;19(4):369–386. doi: 10.1111/j.1527-3466.2001.tb00076.x. [DOI] [PubMed] [Google Scholar]
- 44.Elfiky AA, Ibrahim IM. Host-cell recognition through GRP78 is enhanced in the new UK variant of SARS-CoV-2, in silico. J Infect. 2021;82(5):186–230. doi: 10.1016/j.jinf.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ibrahim MI, Elfiky AA, Elgohary AM. Recognition through GRP78 is enhanced in the UK, South African, and Brazilian variants of SARS-CoV-2; An in silico perspective. Biochem Biophys Res Commun. 2021;562:89–93. doi: 10.1016/j.jinf.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ibrahim IM, Abdelmalek DH, Elshahat ME, Elfiky AA. COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect. 2020;80(5):554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elfiky AA. SARS-CoV-2 spike-heat shock protein A5 (GRP78) recognition may be related to the immersed human coronaviruses. Front Pharmacol. 2020;11:577467. doi: 10.3389/fphar.2020.577467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elfiky AA, Ibrahim IM, Ismail AM, Elshemey WM. A possible role for GRP78 in cross vaccination against COVID-19. J Infect. 2021;82(2):282–327. doi: 10.1016/j.jinf.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ismail AM, Elfiky AA. SARS-CoV-2 spike behavior in situ: a Cryo-EM images for a better understanding of the COVID-19 pandemic. Signal Transduct Target Ther. 2020;5(1):252. doi: 10.1038/s41392-020-00365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The docking structures are available upon request from the corresponding author.