Abstract
The novel coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread worldwide for almost 2 years. It starts from viral adherence to host cells through an interaction between spike glycoprotein 1 (S1) containing a receptor-binding domain (RBD) and human angiotensin-converting enzyme-2 (ACE2). One of the useful strategies to prevent SARS-CoV-2 infection is to inhibit the attachment of RBD to ACE2. Therefore, the current work proposed potent peptides against SARS-CoV-2 infection by carrying out MM-PBSA calculation based on the binding of 52 antiviral peptides (AVPs) to RBD. Considering the binding free energies of AVPs to RBD, cyanovirin-N (CV-N) showed the strongest RBD binding affinity among 52 AVPs. Upon structural analysis of RBD complex with CV-N, it was observed that 12 of the 13 key residues of RBD binding to ACE2 were hijacked by CV-N. CV-N bound to RBD at a smaller affinity of 14.9 nM than that of ACE2 and inhibited the recruitment of S1 to human alveolar epithelial cells. Further analysis revealed that CV-N suppressed SARS-CoV-2 S pseudovirion infection with a half-maximal inhibitory concentration (IC50) of 18.52 μg/mL. This study demonstrated a drug screening for AVPs against SARS-CoV-2 and discovered a peptide with inspiring antiviral properties, which provided a promising strategy for the COVID-19 therapeutic approach.
Graphic Abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s12539-021-00477-w.
Keywords: SARS-CoV-2, COVID-19, Antiviral peptides, Molecular simulation, Cyanovirin-N
Introduction
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is still seriously threatening public health and lives [1]. SARS-CoV-2 generally attacks the main human organs after viral entry [2], leading to respiratory failure, septicopyemia, cardiac failure, hemorrhage and kidney failure, which are the leading causes of the death of COVID-19 patients [3]. Due to the possible persistent infection of SARS-CoV-2 for years [4], it is urgent to develop effective antagonists to prevent the spread [5].
A ligand–receptor interaction established between SARS-CoV-2 spike glycoprotein 1 (S1) and host angiotensin-converting enzyme-2 (ACE2) is the initial step of viral infection [6]. The affinity of SARS-CoV-2 S1 containing a receptor-binding domain (RBD) binding to ACE2 is 15.2 nM [7]. X-ray crystal diffraction has resolved the detailed structure of SARS-CoV-2 RBD binding to ACE2 at 2.45 Å resolution, in which 18 residues of SARS-CoV-2 S1 constitute the receptor-binding motif (RBM) [8].
Drugs targeting the RBD are of interest for the development of therapeutics and diagnostics. There is an urgent need to identify novel approaches that could be used in the emergency phase. The screening and design of small molecule drugs to prevent the replication of SARS-CoV-2 have laid an important foundation for the rapid development of anti-SARS-CoV-2 drugs with clinical potential [9]. However, small molecule drugs are small in size and can only be applied to some small active pockets with partial hydrophobicity. Peptides rather than small molecules are powerfully to inhibit the recognition and binding of proteins since the protein–protein interaction usually has flat interface and large area.
Antiviral peptides (AVPs) naturally present in various species express a broad range of antiviral activities against several viruses with different antiviral mechanisms of action [10, 11]. They can either inhibit viral attachment by binding to viral targets on the host cell surface, such as HD5 binding to ACE2 [12], or target viral proteins as in the case of the envelope proteins; therefore, blocking viral fusion and entry into the host cell [13, 14]. Another mechanism of action is driven in cells, where spreading of the virus is inhibited by suppressing viral gene expression, inhibition of translation, or immune-modulatory activities [15].
Here we aim to repurpose natural substances as potential anti-SARS-CoV-2 agents since AVPs as potential drug candidates might block RBD and be used for curbing COVID-19 [16]. Herein, we created a drug library by selecting 52 AVPs followed by a successful drug screening and in vitro confirmation. Our analysis has revealed that cyanovirin-N (CV-N, PDB ID: 2EZM) has comparatively better inhibitory effect. Additionally, CV-N is the most favorable AVPs binding to RBD to block SARS-CoV-2 fusion and entry into the host cell. Previous reports have confirmed that CV-N is multi-target anti-viral peptide that can inhibit HIV-1 and HSV-2 [17, 18]. Besides, the current study unveils a novel function of CV-N, namely anti-SARS-CoV-2.
Materials and Methods
To search for a potent drug compound, a repository is needed to be scanned. Therefore, we created a drug library by selecting 52 AVPs from the antimicrobial peptide database [19] (APD, http://aps.unmc.edu/AP/). Structures of all the above AVPs were downloaded from RCSB protein data bank (http://www.rcsb.org). These AVPs have residues ranging from 13 to 133 amino acids. Their detailed information is listed in Table S1. The drug screening was performed using global molecular docking. The top AVPs as a result of global molecular docking were subjected to molecular dynamics, MM-PBSA calculation and structural analysis. For the sake of the visual inspection and rendering of biomolecular structures, the PyMOL software [20] was employed.
Molecular Docking of RBD-AVPs
The 3D structures of 52 AVPs were prepared by removing water molecules, adjusting the number of hydrogens, and fixing the hybridization state. The structures were protonated as well to avoid any misleading scores or poses as it is important for calculating the exact Coulombic interaction. An optimum pH, equal to 7 was maintained. The same procedure was repeated in the case of the RBD’s preparation. And since the RBD is located far from the N-glycosylation site [5], molecular docking was carried out without considering glycosylation. Just as our previous report on HD5 [12], the global molecular docking of RBD and AVPs was performed on the ZDOCK server [21] (http://zdock.umassmed.edu/). The ZDOCK algorithm is based on IFACE statistical potential, shape complementarity, and electrostatics [22], following the IRaPPA re-ranking [23]. Each global docking was repeated 1000 times without residue selection while only the top 10 docked conformations for each AVP were downloaded to analyze. All docked conformations were prioritized, and the best-docked conformations were chosen for molecular dynamics simulation to gauge their bonding stability.
Molecular Dynamic Simulation
All the RBD-AVP simulations were performed via GROMACS 2020.2 software package [24] using AMBER99SB-ildn force field [25] and TIP3P water model [26] with a time step of 2 fs. The initial periodic boundaries were placed ≥ 10 Å from all the atoms in the protein. Some counterions of Na+ or Cl− were added to neutralize the overall charge of the MD system to obtain a neutral system. First, 1000 steps of steepest descent minimization were carried out for obtaining a stable conformation. Then, for full relaxation, four 1 ns pre-equilibration simulations with restrained coordinates of the heavy atoms, main chain, backbone, and Cα, respectively, were performed step by step at 300 K. Finally, each equilibrium simulation with an isothermal–isobaric (NPT) ensemble at 1 atm and 300 K was performed for a duration of 100 ns. During all simulations, for the heavy atoms, the LINCS (LINear Constraint Solver) constraints [27] and the nonbonded pair list were updated every 10 steps. Electrostatic interactions were calculated using the particle mesh Ewald method [28]. The v-rescale temperature coupling method [29, 30] was used to maintain a constant temperature, and Berendsen coupling [31] was used to keep a constant pressure inside the box. The last 5 ns of each equilibrated trajectory was used for MM-PBSA calculation.
MM-PBSA Calculation
The binding free energies were calculated using the molecular mechanics energies combined with the Poisson–Boltzmann surface area continuum solvation (MM-PBSA) [32] as well as solvent accessible volume (SAV) model [33] in terms of g_mmpbsa procedure [33] for GROMACS based on 5000 snapshots sampled every 10 ps from total 5 ns equilibrated trajectory. During the calculation of MM-PBSA, the dielectric constant of water is set to 78 and the temperature is set as 300 K.
Identification of Non-covalent Interface Interactions
A PPCheck webserver for quantifying the strength of a protein–protein interface (http://caps.ncbs.res.in/ppcheck/index.html) [34] was used to identify the non-covalent interactions at the interface of RBD complexes with AVPs.
Bio-layer Interferometry (BLI)
The binding of CV-N (Guoping Pharmaceutical, Anhui Province, CHN) to biotinylated SARS-CoV-2 RBD (40592-V08B-B, Sino Biological, Beijing, CHN) was measured using Forte Bio’s “Octet Red 96” BLI (Sartorius, Göttingen, GER). The high-performance liquid chromatography and mass spectrum results of CV-N are shown in Fig. S1. RBD prepared in PBS was immobilized on SA biosensors at 15 μg/mL. CV-N were prepared in PBS with gradient concentrations. Association and disassociation were conducted at a shaking speed of 1000 rpm for 300 s. The data were processed using Fortebio Data Analysis 7.0 software. The equilibrium dissociation constant (KD) was obtained by a 1:1 fitting model.
Immunofluorescence Microscopy
Human alveolar epithelial A549 cells obtained from the cell bank of Chinese Academy of Sciences (CAS, Shanghai) and cultured in Dulbecco's modified Eagle medium (DMEM, Gibco, Thermo Fisher Scientific, Shanghai, CHN) containing 10% foetal bovine serum (FBS, Gibco) were seeded into a 12-well plate with sterile glass slides at a density of 2.5 × 105 cells/well. Cells cultured overnight and washed with sterile PBS were incubated with 10 μg/mL of S1 (40591-V08H, Sino Biological) pretreated with 20 μg/mL of CV-N at 37 °C for 15 min. Co-incubation was conducted at 4 °C for 1 h. The cells were then washed with PBS and fixed in 4% paraformaldehyde. A primary anti-spike rabbit monoclonal antibody (40150-R007, Sino Biological, 1:100) and a donkey anti-rabbit secondary antibody (Alexa Fluor 555, A0453, Beyotime, Shanghai, CHN) were employed to stain S1. A primary anti-ACE2 mouse monoclonal antibody (10108-MM36, Sino Biological, 1:100) and a goat anti-mouse secondary antibody (Alexa Fluor 488, A0428, Beyotime) were employed to stain ACE2. Nuclei were stained with 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI, C1002, Beyotime). The cells were observed using a Zeiss LSM 780 NLO confocal microscope.
Western Blot
A549 cell is a commonly used cell line for respiratory infections. Thus, we obtained A549 and human renal tubular epithelial HK-2 cells from the cell bank of CAS and cultured them in DMEM containing 10% FBS, and seeded them into a six-well plate at a density of 1 × 106 cells/well. The cells were incubated with 20 μg/mL of SARS-CoV-2 S1 pre-treated with different CV-N concentrations (1, 10, and 50 μg/mL) at 37 ℃ for 15 min. Co-incubation was performed at 4 ℃ for 1 h. After three times of wash with PBS, cells were collected and processed with RIPA lysis and extraction buffer (89900, Thermo Fisher Scientific, Shanghai, CHN). A primary anti-spike rabbit monoclonal antibody (40150-R007, Sino Biological, 1:500) and a goat anti-rabbit secondary antibody (A0208, Beyotime, 1:1000) were applied to detect S1. β-Actin determined by a mouse monoclonal antibody (AA128, Beyotime, 1:1000) was used as a reference. This experiment was repeated three times on different days to avoid counting any personal or environmental errors.
Cell Experiments with Pseudovirions
HEK-293T-hACE2 cells obtained from Prof. Ye and cultured in RPMI 1640 medium (Gibco, Thermo Fisher Scientific) containing 10% FBS were seeded into a 96-well plate at a density of 5 × 103 cells/well [35]. A total of 1 × 107 TU of SARS-CoV-2 S pseudovirions containing dual-luciferase reporter (Genewiz, Suzhou, CHN) were added to the culture medium after co-incubation with CV-N (100, 75, 50, 25, 12.5, 6.25, 3.13, and 1.56 μg/mL) at 37 ℃ for 30 min. The pseudovirions were removed after 12 h. The cells were cultured in RPMI 1640 medium containing 10% FBS for another 48 h. The infection of pseudovirions was assessed by determining the luciferase activity using a dual-luciferase reporter assay system (E1910, Promega, Beijing, CHN). This experiment was conducted in triplicate and repeated twice.
Results and Discussion
Peptide Screening by MM-PBSA Method
The final results derived from MM-PBSA calculation indicated that 23 AVPs with negative free binding energies could bind to RBD of SARS-CoV-2 while 7 AVPS have a comparatively stronger binding affinity towards RBD than the rest of the peptides. CV-N had the strongest RBD binding affinity among 52 AVPs (Fig. 1A, Table S2), indicating that CV-N may be the best inhibitor for RBD in the current research.
Fig. 1.
Screening histogram of AVPs (A), Venn diagram of key residues confirmed by current calculation of RBD-CV-N and experimental studies of RBD-ACE2 (B), BLI experimental curves of CV-N binding to SARS-CoV-2 RBD (C). Screening histogram indicated that CV-N had the strongest RBD-binding affinity among 52 AVPs. Venn diagram proved that 12 of the 13 residues of RBD binding to ACE2 were hijacked by CV-N. BLI experiment curves showed the affinity of CV-N binding to RBD was stronger than that of ACE2
Structural Analysis of RBD Complex with CV-N
According to the molecular simulation results, all residues that affected the binding between RBD and CV-N were shown in Fig. 1B. CV-N bound tightly to RBD, which formed 7 hydrogen bonds, 3 salt bridges, 3 favorable long-range electrostatic interaction, and 1 hydrophobic interaction. The detailed molecular interactions between RBD and CV-N were shown in Fig. 2 and Table 1. The frame backbone root mean square deviation (RMSD) analysis confirmed the complex’s stable nature, where minor fluctuation was observed in the first 10 ns from where it stabilized till the end of simulation ranging from 0.12 to 0.20 nm (Fig. S2). Also, CV-N could almost cover all key residues of RBD. For example, there were 17 residues of RBD within 5 Å of CV-N. According to the crystallography report of the 2019-nCoV RBD/ACE2-B0AT1 complex (PDB ID: 6M17) [36], the residues of RBD within 5 Å of ACE2 were 13 residues. 12 of the 13 key residues of RBD binding to ACE2 were hijacked by CV-N (Fig. 1B).
Fig. 2.
Exploring molecular interactions of RBD with CV-N. For clarity, RBD was colored in cyan and CV-N in red, and 11 hydrogen bonds were shown in yellow, 3 salt bridges were highlighted in orange, and 1 hydrophobic interaction was colored in purple. The molecular interactions were visualized through PyMOL
Table 1.
Intermolecular interactions of RBD complex with CV-N
| Hydrogen bonding | ||||
|---|---|---|---|---|
| No. | Residue of RBD | Hydrogen bond | Residue of CV-N | Distance (Å) |
| 1 | LYS417 | NZ—HZ2…OE2 | GLU68 | 1.65 |
| 2 | GLY446 | O…HH—OH | TYR9 | 1.72 |
| 3 | GLU484 | OE1…HZ2—NZ | LYS3 | 1.89 |
| 4 | GLU484 | OE2…HZ2—NZ | LYS3 | 2.04 |
| 5 | ASN487 | N—H…OD1 | ASP89 | 1.85 |
| 6 | TYR489 | OH—HH…OD2 | ASP89 | 1.51 |
| 7 | GLN493 | NE2—HE22…OD1 | ASP88 | 1.66 |
| 8 | GLN493 | OE1…HG—OG | SER5 | 1.72 |
| 9 | SER494 | OG…HE21—NE2 | GLN6 | 2.08 |
| 10 | GLY502 | N—H…OD1 | ASN10 | 1.96 |
| 11 | TYR505 | OH—HH…OG | SER66 | 2.01 |
| Salt bridges | ||||
|---|---|---|---|---|
| . | Residue of RBD | Cationic–anionic pair | Residue of CV-N | Distance (Å) |
| 1 | LYS417 | NZ…OE2 | GLU68 | 2.64 |
| 2 | GLU484 | OE1…NZ | LYS3 | 2.81 |
| 3 | GLU484 | OE2…NZ | LYS3 | 2.90 |
| Favorable electrostatic interactions | ||||
|---|---|---|---|---|
| No. | Residue of RBD | Electrostatic pairs | Residue of CV-N | Distance (Å) |
| 1 | LYS417 | CB…CB | GLU68 | 8.77 |
| 2 | LYS444 | CB…CB | GLU23 | 9.99 |
| 3 | GLU484 | CB…CB | LYS3 | 9.04 |
| Hydrophobic interactions | ||||
|---|---|---|---|---|
| No. | Residue of RBD | Hydrophobic groups | Residue of CV-N | Distance (Å) |
| 1 | TYR505 | CB…CB | TYR9 | 4.5 |
Previously, CV-N as lectin was supposed to target N-linked glycans of S glycoproteins. The binding free energy of CV-N with the S glycoprotein glycan [37] had been calculated as − 280.506 kJ/mol. Combined with our current research (− 356.979 kJ/mol), it can be concluded that CV-N is a multi-target peptide inhibitor, which can bind both glycan and RBD. The result proves that CV-N has the diversity of biological function and the potential medicinal prospect.
Bio-layer Interferometry (BLI) Determining the Binding Affinities
The recruitments of CV-N to RBD were measured with BLI experiment. The affinity of CV-N binding to RBD was 14.9 nM (Fig. 1C), smaller than that of RBD interacting with ACE2 (15.2 nM) [7], suggesting that CV-N was sufficient to inhibit the binding of RBD to ACE2.
Suppression of CV-N on S1 Recruitment and Pseudovirion Infection
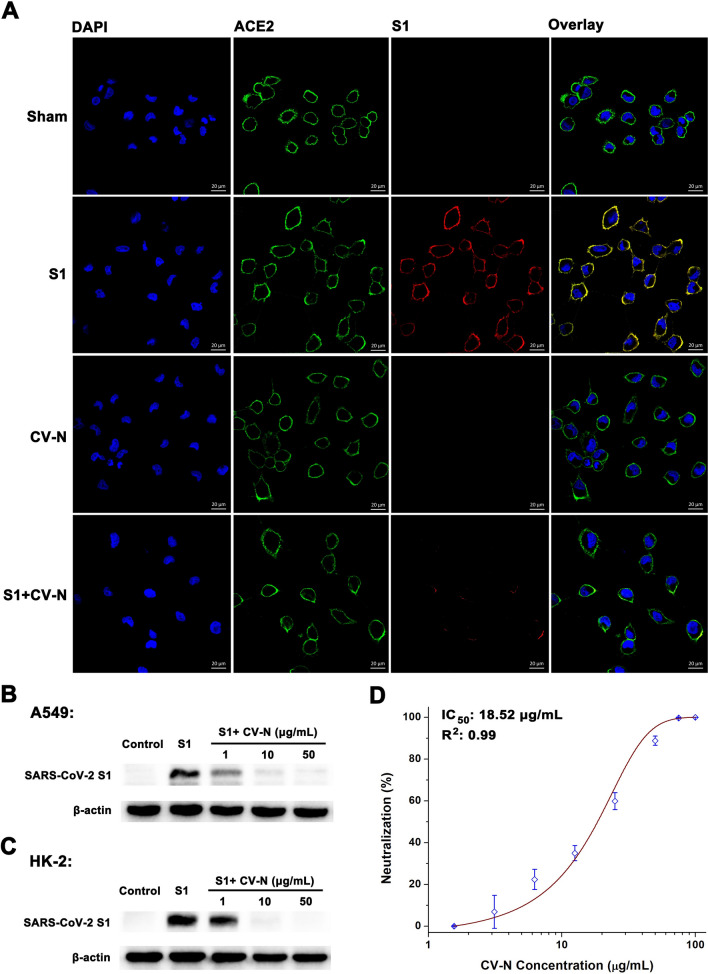
To evaluate the RBD-binding effect on S1 recruiting to ACE2, we conducted an experimental approach where the A549 cells were exposed to 10 μg/mL of S1 in the absence and presence of CV-N. Confocal microscopy showed that S1 adhered mainly to the cell surface in the lack of CV-N (Fig. 3A). Nevertheless, when cells were pretreated with 20 μg/mL of CV-N, it dramatically reduced the recruitment of S1. Western blot supported that CV-N dramatically decreased S1 adherence to A549 cells, and CV-N inhibition was in a dose-dependent manner (Fig. 3B). The inhibition of CV-N on S1 recruitment was also observed in human renal proximal tubular epithelial HK-2 cells abundant in ACE2 receptor (Fig. 3C) [38, 39]. SARS-CoV-2 S pseudovirions contain psPAX2 and luciferase reporter system, which were then employed to HEK-293T-hACE2 in the presence of increasing concentrations of peptides [40]. CV-N suppressed pseudovirion infection in a dose-dependent manner with a half-maximal inhibitory concentration (IC50) of 18.52 μg/mL (Fig. 3D), thus demonstrating that CV-N can inhibit SARS-CoV-2 invasion by blocking viral spike recruitment.
Fig. 3.
The inhibition of CV-N on SARS-CoV-2 S1 adherence and S pseudovirus infection. A Immunofluorescence microscopy revealing the adherence of S1 to A549 cells. In the sham group, cells were treated with normal rabbit IgG. Here, the scale bar indicated 20 μm. B Protein bands of S1 pretreated with increasing CV-N binding concentrations to A549 cells. C The inhibition of CV-N on S1 recruitment observed in human renal proximal tubular epithelial HK-2 cells abundant in ACE2 receptor. Here β-actin was provided as a reference. D IC50 determination. Results shown as mean ± standard deviation were processed by nonlinear curve fitting. The above results demonstrate that CV-N can inhibit SARS-CoV-2 invasion by blocking viral spike recruitment
Conclusions
To summarize the discovery of an effective inhibitor against SARS-CoV-2, the MM-PBSA was used to evaluate the binding of 52 AVPs to RBD, respectively. CV-N showed the strongest RBD binding affinity among 52 AVPs. Furthermore, the structural analysis of RBD complex with CV-N indicated that 12 of the 13 key residues of RBD binding to ACE2 were hijacked by CV-N. The BLI affinity of CV-N binding to RBD was 14.9 nM, smaller than that of RBD interacting with ACE2 (15.2 nM), which suggests that CV-N can inhibit effectively the binding of RBD to ACE2. Cell experiments and immunofluorescence microscopy also revealed the anti-SARS-CoV-2 virus effect of CV-N. Western blot supported that CV-N dramatically decreased S1 adherence to A549 cells, and the inhibition of CV-N was in a dose-dependent manner with an IC50 of 18.52 μg/mL. All of the above outcomes demonstrate that CV-N can inhibit SARS-CoV-2 invasion by blocking viral spike recruitment and prevent SARS-CoV-2 from adherence and entry into the host cell.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The heavy-duty computational operations were performed at the Peng Cheng Lab and the Center for High-Performance Computing, Shanghai Jiao Tong University. We owe our gratitude to Jingfei Qiu, Shaomeng Cao and Xuhui Shao from AI Research Center of Peng Cheng Laboratory, Shenzhen, Guangdong province, for their enormous technological support in computing resources and technology.
Abbreviations
- S1
Spike 1
- ACE2
Angiotensin-converting enzyme-2
- RBD
Receptor-binding domain
- LBD
Ligand-binding domain
- RBM
Receptor-binding motif
- COVID-19
Coronavirus disease 2019
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- AVP
Antiviral peptide
- NMR
Nuclear magnetic resonance
- CV-N
Cyanovirin-N
- HIV-1
Human immunodeficiency virus 1
- HSV-2
Herpes simplex virus 2
- MM-PBSA
Molecular mechanics energies combined with the Poisson–Boltzmann and surface area continuum solvation
- SAV
Solvent accessible volume
- BLI
Bio-layer interference
- IC50
Half maximal inhibitory concentration
- DMEM
Dulbecco’s modified Eagle medium
- PBS
Phosphate buffer saline
- FBS
Foetal bovine serum
- DAPI
2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride
- RIPA
Radio-Immunoprecipitation Assay
Author Contributions
DL and CW contributed equally to this paper. D-QW, JW and JZ conceived the study and designed the experiments. SW, JG, and XC performed the biological experiments and analyzed the data. DL, PC, JFQ and AM performed the computational experiments and analyzed the data. DL, CW, and AM wrote and edited the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 32070662, 61832019, 32030063 and 81725019), the Key Research Area Grant 2016YFA0501703 of the Ministry of Science and Technology of China, the Science and Technology Commission of Shanghai Municipality (Grant No. 19430750600), the Natural Science Foundation of Henan Province (Grant No. 162300410060), the program for scientific and technological innovation leader of Chongqing (Grant No. CQYC201903084), and the Natural Science Fund of Chongqing City (Grant No. cstc2019jcyj-msxmX0011). Partially Supported by Open Funding Project of State Key Laboratory of Microbial Metabolism (Grant No. MMLKF21-11) and Shanghai cryogenic biomedical technology professional service platform (Grant No. 18DZ2295700).
Data and Software Availability
The data and software for current research can be accessed from https://github.com/dxli75/SARS-CoV-II-RBD-AVPS-Screening.
Declarations
Conflict of interest
We have no competing financial interests to declare.
Footnotes
Daixi Li and Cheng Wang are the joint first author who contributed equally to this paper.
Contributor Information
Daixi Li, Email: dxli75@126.com.
Aamir Mehmood, Email: aamirmehmood@sjtu.edu.cn.
Junping Wang, Email: wangjunping@tmmu.edu.cn.
Dongqing Wei, Email: dqwei@sjtu.edu.cn.
References
- 1.The species Severe acute respiratory syndrome-related coronavirus classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020 doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wadman M, Couzin-Frankel J, Kaiser J, Matacic C. A rampage through the body. Sci (NY NY) 2020;368(6489):356–360. doi: 10.1126/science.368.6489.356. [DOI] [PubMed] [Google Scholar]
- 3.Zhang B, Zhou X, Qiu Y, Song Y, Feng F, Feng J, Song Q, Jia Q, Wang J. Clinical characteristics of 82 cases of death from COVID-19. PLoS ONE. 2020;15(7):e0235458. doi: 10.1371/journal.pone.0235458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kissler SM, Tedijanto C, Goldstein E, Grad YH, Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Sci (NY, NY) 2020;368(6493):860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369(6501):330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, Lu L, Jiang S, Yang Z, Wu Y, Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microb Infect. 2020;9(1):382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 9.Amin SA, Banerjee S, Ghosh K, Gayen S, Jha T. Protease targeted COVID-19 drug discovery and its challenges: Insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorg Med Chem. 2021;29:115860. doi: 10.1016/j.bmc.2020.115860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laosutthipong C, Kanthong N, Flegel TW. Novel, anionic, antiviral septapeptides from mosquito cells also protect monkey cells against dengue virus. Antiviral Res. 2013;98(3):449–456. doi: 10.1016/j.antiviral.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Mulder KC, Lima LA, Miranda VJ, Dias SC, Franco OL. Current scenario of peptide-based drugs: the key roles of cationic antitumor and antiviral peptides. Front Microbiol. 2013;4:321. doi: 10.3389/fmicb.2013.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C, Wang S, Li D, Wei DQ, Zhao J, Wang J. Human intestinal defensin 5 inhibits SARS-CoV-2 invasion by cloaking ACE2. Gastroenterology. 2020;159(3):1145–1147.e4. doi: 10.1053/j.gastro.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luteijn RD, Praest P, Thiele F, Sadasivam SM, Singethan K, Drijfhout JW, Bach C, de Boer SM, Lebbink RJ, Tao S, Helfer M, Bach NC, Protzer U, Costa AI, Killian JA, Drexler I, Wiertz E. A broad-spectrum antiviral peptide blocks infection of viruses by binding to phosphatidylserine in the viral envelope. Cells. 2020;9(9):1989. doi: 10.3390/cells9091989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Garrison JC, Poluektova LY, Bronich TK, Osna NA. Liver-targeted antiviral peptide nanocomplexes as potential anti-HCV therapeutics. Biomaterials. 2015;70:37–47. doi: 10.1016/j.biomaterials.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limthongkul J, Mapratiep N, Apichirapokey S, Suksatu A, Midoeng P, Ubol S. Insect anionic septapeptides suppress DENV replication by activating antiviral cytokines and miRNAs in primary human monocytes. Antiviral Res. 2019;168:1–8. doi: 10.1016/j.antiviral.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Vilas Boas LCP, Campos ML, Berlanda RLA, de Carvalho Neves N, Franco OL. Antiviral peptides as promising therapeutic drugs. Cell Mol Life Sci CMLS. 2019;76(18):3525–3542. doi: 10.1007/s00018-019-03138-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodrum BW, Maxwell J, Allen DM, Wilson J, Krumpe LR, Bobkov AA, Hill RB, Kibler KV, O’Keefe BR, Ghirlanda G. A designed, “nested” dimer of cyanovirin-N increases antiviral activity. Viruses. 2016;8(6):158. doi: 10.3390/v8060158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farr Zuend C, Nomellini JF, Smit J, Horwitz MS. Generation of a dual-target, safe, inexpensive microbicide that protects against HIV-1 and HSV-2 disease. Sci Rep. 2018;8(1):2786. doi: 10.1038/s41598-018-21134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44(D1):D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schrodinger, LLC, The PyMOL Molecular Graphics System, Version 1.8. 2015. http://www.schrodinger.com/products/pymol.
- 21.Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30(12):1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS ONE. 2011;6(9):e24657. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mintseris J, Pierce B, Wiehe K, Anderson R, Chen R, Weng Z. Integrating statistical pair potentials into protein complex prediction. Proteins. 2007;69(3):511–520. doi: 10.1002/prot.21502. [DOI] [PubMed] [Google Scholar]
- 24.Rakhshani H, Dehghanian E, Rahati A. Enhanced GROMACS: toward a better numerical simulation framework. J Mol Model. 2019;25(12):355. doi: 10.1007/s00894-019-4232-z. [DOI] [PubMed] [Google Scholar]
- 25.Pang YP. FF12MC: A revised AMBER forcefield and new protein simulation protocol. Proteins. 2016;84(10):1490–1516. doi: 10.1002/prot.25094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price DJ, Brooks CL., 3rd A modified TIP3P water potential for simulation with Ewald summation. J Chem Phys. 2004;121(20):10096–10103. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- 27.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 28.Abraham MJ, Gready JE. Optimization of parameters for molecular dynamics simulation using smooth particle-mesh Ewald in GROMACS 4.5. J Comput Chem. 2011;32(9):2031–2040. doi: 10.1002/jcc.21773. [DOI] [PubMed] [Google Scholar]
- 29.Bussi G, Zykova-Timan T, Parrinello M. Isothermal-isobaric molecular dynamics using stochastic velocity rescaling. J Chem Phys. 2009;130(7):074101. doi: 10.1063/1.3073889. [DOI] [PubMed] [Google Scholar]
- 30.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1):014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 31.Golo VL, Shaitan KV. Dynamic attractor for the Berendsen thermostat and the slow dynamics of biomacromolecules. Biofizika. 2002;47(4):611–617. [PubMed] [Google Scholar]
- 32.Aldeghi M, Bodkin MJ, Knapp S, Biggin PC. Statistical analysis on the performance of molecular mechanics Poisson-Boltzmann surface area versus absolute binding free energy calculations: bromodomains as a case study. J Chem Inf Model. 2017;57(9):2203–2221. doi: 10.1021/acs.jcim.7b00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumari R, Kumar R, Open Source Drug Discovery, C. Lynn A. g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 34.Sukhwal A, Sowdhamini R. Oligomerisation status and evolutionary conservation of interfaces of protein structural domain superfamilies. Mol Biosyst. 2013;9(7):1652–1661. doi: 10.1039/c3mb25484d. [DOI] [PubMed] [Google Scholar]
- 35.Chen X, Li R, Pan Z, Qian C, Yang Y, You R, Zhao J, Liu P, Gao L, Li Z, Huang Q, Xu L, Tang J, Tian Q, Yao W, Hu L, Yan X, Zhou X, Wu Y, Deng K, Zhang Z, Qian Z, Chen Y, Ye L. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol Immunol. 2020;17(6):647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan RH, Zhang YY, Li YN, Xia L, Guo YY, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lokhande KB, Apte GR, Shrivastava A, Singh A, Pal JK, Swamy KV, Gupta RK. Sensing the interactions between carbohydrate-binding agents and N-linked glycans of SARS-CoV-2 spike glycoprotein using molecular docking and simulation studies. J Biomol Struct Dyn. 2020;9:1–19. doi: 10.1080/07391102.2020.1851303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamming I, Timens W, Bulthuis M, Lely A, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du MCG, Chen F, Christiani DC, Zhang Z, Wang M. Multi-omics evaluation of gastrointestinal and other clinical characteristics of SARS-CoV-2 and COVID-19. Gastroenterology. 2020;158(8):2298–2301.e7. doi: 10.1053/j.gastro.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C, Wang S, Chen Y, Zhao J, Han S, Zhao G, Kang J, Liu Y, Wang L, Wang X, Xu Y, Wang S, Huang Y, Wang J, Zhao J. Membrane nanoparticles derived from ACE2-rich cells block SARS-CoV-2 infection. ACS Nano. 2021;15(4):6340–6351. doi: 10.1021/acsnano.0c06836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and software for current research can be accessed from https://github.com/dxli75/SARS-CoV-II-RBD-AVPS-Screening.