Abstract
Chronic hepatitis B virus (HBV) infection changes the composition of the extracellular matrix (ECM) and enables the onset and progression of hepatocellular carcinoma (HCC). The ensemble of ECM proteins and associated factors is a major component of the tumor microenvironment. Our aim was to unveil the matrisome genes from HBV‐related HCC. Transcriptomic and clinical profiles from 444 patients with HBV‐related HCC were retrieved from the Gene Expression Omnibus (GEO) and Cancer Genome Atlas (TCGA) repositories. Matrisome genes associated with HBV‐related hepatocarcinogenesis, matrisome gene modules, HCC subgroups, and liver‐specific matrisome genes were systematically analyzed, followed by identification of their biological function and clinical relevance. Eighty matrisome genes, functionally enriched in immune response, ECM remodeling, or cancer‐related pathways, were identified as associated with HBV‐related HCC, which could robustly discriminate HBV‐related HCC tumor from nontumor samples. Subsequently, four significant matrisome gene modules were identified as showing functional homogeneity linked to cell cycle activity. Two subgroups of patients with HBV‐related HCC were classified based on the highly correlated matrisome genes. The high‐expression subgroup (15.0% in the TCGA cohort and 17.9% in the GEO cohort) exhibited favorable clinical prognosis, activated metabolic activity, exhausted cell cycle, strong immune infiltration, and lower tumor purity. Four liver‐specific matrisome genes (F9, HPX [hemopexin], IGFALS [insulin‐like growth‐factor‐binding protein, acid labile subunit], and PLG [plasminogen]) were identified as involved in HBV‐related HCC progression and prognosis. Conclusion: This study identified the expression and function of matrisome genes from HBV‐related hepatocarcinogenesis, providing major insight to understand HBV‐related HCC and develop potential therapeutic opportunities.
Matrisome genes function as modules and sub‐classify HBV‐related HCC patients that exhibit distinct molecular patterns and clinical outcomes. Liver‐specific matrisome genes are involved in HBV‐related HCC, which could be ideal candidate therapeutic targets.
Abbreviations
- AFP
alpha‐fetoprotein
- AUC
area under the curve
- BCLC
Barcelona Clinic Liver Cancer
- CCL
chemokine (C‐C motif) ligand
- CXCL
chemokine (C‐X‐C motif) ligand
- ECM
extracellular matrix
- GEO
Gene Expression Omnibus
- GSEA
gene‐set enrichment analysis
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCL
hierarchical clustering
- HHMG
HBV‐related HCC‐associated matrisome gene
- IGFALS
insulin‐like growth‐factor‐binding protein, acid labile subunit
- HPX
hemopexin
- HSC
hepatic stellate cell
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MBL2
mannose binding lectin 2
- NES
normalized enrichment score
- NT
nontumor
- PC
principal component
- PCA
principal component analysis
- PI3K
phosphoinositide 3‐kinase
- PLG
plasminogen
- RNA‐seq
RNA sequencing
- ROC
receiver operating characteristic
- T
tumor
- TCGA
The Cancer Genome Atlas
- TNM
Tumor, Node, Metastasis
Chronic hepatitis B virus (HBV) infection accounts for approximately 80% of patients with virus‐related hepatocellular carcinoma (HCC), especially in Eastern Asian and most African countries, posing a serious threat to human health and quality of life.( 1 ) More than 250 million people worldwide will suffer from chronic HBV infection between 2015 and 2030, and about 5 million deaths will be attributed to HCC progression.( 1 ) Suppression of HBV reduces the risk of HCC( 2 ); yet, it is vital to understand the mechanisms underlying HBV‐related carcinogenesis to develop therapeutic options for HCC treatment.
To date, little progress has been made on the mechanisms driving carcinogenesis in HBV infection. A few well‐recognized events involved are (1) increased TERT or TP53 mutation( 1 ); (2) activation of Wnt, mammalian target of rapamycin (mTOR)/phosphoinositide 3‐kinase (PI3K)/Akt, and Ras/extracellular signal‐regulated kinase 1/2 signaling( 3 ); and (3) exhausted CD8+ T cells.( 4 ) Growing evidence also indicates that aberrant composition of the matrisome is involved in how the tumor microenvironment promotes HCC development, progression, and metastasis.( 5, 6 )
The matrisome comprises core extracellular matrix (ECM) molecules (collagens, glycoproteins, and proteoglycans) and ECM‐associated members (ECM regulators, ECM‐affiliated proteins, and secreted factors).( 7 ) The matrisome from two mouse models of HCC shows different composition,( 8 ) indicating that ECM remodeling during HCC onset is etiology‐specific. To date, the matrisome from human intrahepatic cholangiocarcinoma has been unveiled( 9 ); however, the matrisome from human HCC and, specifically from HBV‐related HCC, remains unknown.
Previous studies showed that the liver ECM is remodeled in HBV infection.( 10, 11 ) The hepatitis B e antigen activates hepatic stellate cells (HSCs), resulting in aberrant ECM production ( 10 ). The hepatitis B x protein (HBx) up‐regulates matrix metalloproteinases and increases HCC cell migration.( 11 ) Changes in the extracellular environment during HBV infection activate intracellular signaling pathways. For example, the oncogene collagen triple helix repeat containing‐1 facilitates progression of HBV‐related HCC, activating hypoxia inducible factor 1 alpha and vascular endothelial growth factor through the PI3K/Akt/mTOR pathway.( 12 ) Despite reports linking the matrisome with HBV‐related HCC, thorough identification and characterization are urgently needed.
In addition to anti‐HBV therapy and surgical resection for early HCC or first‐line treatment for advanced HCC, the ECM requires remodeling to return to its physiological state. Direct targeting of proteins from the ECM fails to be beneficial due to a stiff matrix barrier that limits drug delivery. Indeed, a clinical trial with a monoclonal antibody against lysyl oxidase like 2 (simtuzumab) to treat liver fibrosis failed.( 13 ) However, interventions focused on the matrisome genes could be helpful to prevent deposition or promote turnover of the tumor ECM. Therefore, our aim was to unveil the matrisome genes from HBV‐related HCC, which remain unknown.
Materials and Methods
Matrisome Genes
The matrisome genes included in this study were retrieved from the MatrisomeDB repository.( 7 ) They include 1,027 in silico–defined or experimentally confirmed ECM members from human samples. The matrisome genes are categorized into six subgroups: collagens, proteoglycans, ECM glycoproteins, ECM‐affiliated proteins, ECM regulators, and secreted factors.( 7 )
Patients and Samples
The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) repositories were explored for available transcriptomic profiles and/or clinical information from HBV‐related HCC or liver fibrosis (Supporting Table S1). Processed microarray data from patients with HBV‐related HCC (GSE121248 and GSE55092) were retrieved from the GEO for the identification of matrisome genes associated with HBV‐related carcinogenesis. Additional processed microarray data from patients with HBV‐related liver fibrosis (GSE84044) and RNA‐sequencing (RNA‐seq) data from patients with HBV‐related HCC (GSE65485, GSE94660, and GSE104310) were retrieved from the GEO to validate the identified matrisome genes of interest. Transcriptomic profiles from patients with HBV‐related HCC with available clinical information from the TCGA (n = 60) and GEO (GSE14520; n = 218) were included to further interpret the biological function or clinical relevance of the identified matrisome genes. In total, 152 nontumor and 470 tumor samples from 444 patients with HBV‐related HCC were used in this study (Supporting Table S1). Gene expression in the microarray data sets was normalized using the robust multichip average algorithm,( 14 ) and the average value of all corresponding probes was calculated for a given gene. Raw counts of gene expression in the RNA‐seq data sets were transformed into transcripts per kilobase million for subsequent analysis.
Flowchart and Analytical Approaches
The flowchart for data acquisition and analysis is shown in Supporting Fig. S1. Details on the analytical approaches are provided in the Supporting information.
Data Availability
The human matrisome gene list, processed GEO and TCGA data sets, and related R code have been deposited into the figshare platform (https://figshare.com/s/f7af736216ef73d7ee7d; https://doi.org/10.6084/m9.figshare.14069474).
Results
HBV‐Related HCC‐Associated Matrisome Genes Discriminate HBV‐Related HCC Tumor From Nontumor
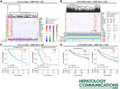
In HBV‐related carcinogenesis, the number of down‐regulated matrisome genes is 3 times higher than the up‐regulated ones in both the GSE55092 and GSE121248 microarray transcriptomic data sets (adjusted P < 0.05 and fold change >2 or <0.5; Fig. 1A). A total of 80 dysregulated matrisome genes were shared in the two data sets (P < 0.01), among which, 63 were significantly decreased and 17 increased (Supporting Fig. S2A,B and Supporting Table S2). These common abnormally expressed matrisome genes during HBV‐related carcinogenesis were named HHMGs. The primary cellular source for the identified HHMGs are hepatocytes (29 HHMGs), endothelial cells (39 HHMGs), HSCs (26 HHMGs), EPCAM+ cells and cholangiocytes (19 HHMGs), and Kupffer cells (12 HHMGs) (Supporting Fig. S3). They were significantly enriched in pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) and classified into three categories: immune response, ECM remodeling, and carcinogenesis (adjusted P < 0.05; Fig. 1B). Importantly, the identified HHMGs can be used as a combined signature to discriminate HBV‐related HCC tumor from nontumor samples. Indeed, samples from the merged GSE55092 and GSE121248 data sets were clustered as tumor or nontumor by hierarchical clustering (HCL) analysis (Fig. 1C). We further performed principal component analysis (PCA) on the 80 HHMGs, and the first two principal components (PCs), which were most informative, explained about 51.8% and 10.4%, respectively, of the total observed variances. As shown in Fig. 1D, the PCA plot further confirmed the classifying ability of the HCL analysis, because the first two PCs clearly distinguished tumor from nontumor samples in the merged two microarray transcriptomic data sets.
FIG. 1.
Abnormal matrisome gene expression during HBV‐related hepatocarcinogenesis. (A) Volcano plots of the HHMGs in the GSE55092 and GSE121248 data sets. Volcano plots were drawn using the ggplot2 R package (https://CRAN.R‐project.org/package=ggplot2). Blue indicates down‐regulation and red indicates up‐regulation. The most significant HHMGs are highlighted in green in the volcano plots under an extreme cutoff criterion (log10‐transformed [adjusted P] > 10 and fold change >4). (B) Significantly enriched KEGG pathways of the identified HHMGs. The size of the circle represents the gene number, and the color represents adjusted P value. An adjusted P < 0.05 was considered statistically significant. (C) Heatmap of the expression of the HHMGs in the merged liver samples from the GSE55092 and GSE121248 data sets. Log2‐transformed gene‐expression levels were scaled as a distribution with mean = 0 and SD = 1. The darker the blue, the lower the expression; the darker the red, the higher the expression. Both the row HHMGs and the column samples were clustered by HCL analysis with the average linkage method and “euclidean” as a distance metric. Tumor and nontumor samples from the GSE55092 and GSE121248 data sets are color‐coded. (D) PCA plot of all tumor and nontumor samples from the GSE55092 and GSE121248 data sets. Grouped samples are labeled with different colors. Abbreviations: NT, nontumor; T, tumor; TNF, tumor necrosis factor.
Diagnostic Ability of HHMGs Is Robust
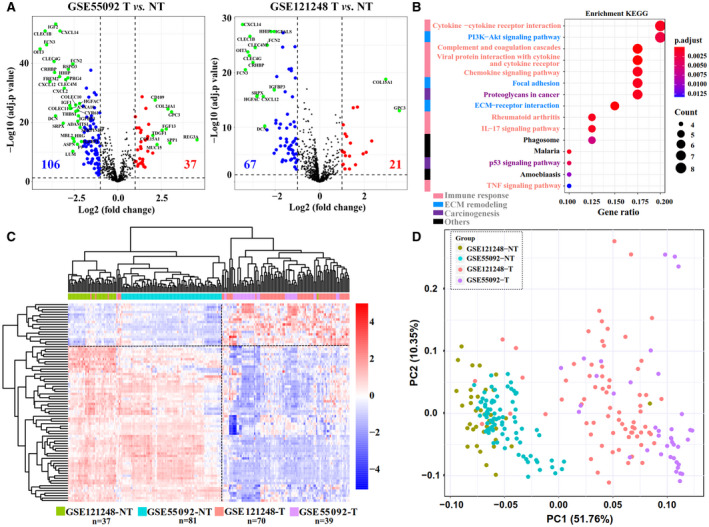
Given the heterogeneity of HCC tumor tissues, it is challenging to obtain signatures with stable diagnostic potential. To test the diagnostic performance of the identified HHMGs, we retrieved three independent RNA‐seq data sets of HBV‐related HCC from the GEO database. As expected, based on the expression of 80 HHMGs, HCL analysis clearly separated tumor from nontumor samples in all three RNA‐seq data sets (Fig. 2A, left). Interestingly, the total expression pattern of the HHMGs was not observed in HBV‐related liver fibrosis (Fig. 2A, right), further implying that the identified HHMGs were not associated with HBV‐related liver fibrosis but were HCC‐related. To eliminate the clustering method bias, PCA was carried out and achieved nearly the same results with HCL analysis (Fig. 2B). Receiver operating characteristic (ROC) curve analysis was done to measure the diagnostic ability of each HHMG. As shown in Fig. 2C, almost all HHMGs possess high and robust diagnostic ability to differentiate HBV‐related HCC tumor from nontumor samples, because the area under curve (AUC) value of most HHMGs is >0.7 in all five HBV‐related HCC data sets. Taken together, the diagnostic ability of the HHMGs is promising.
FIG. 2.
Robustness analysis of the identified HHMGs. (A) The gene‐expression pattern of the identified HHMGs was validated in the GSE65485, GSE94660, GSE104310, and GSE84044 data sets. Column samples were clustered by HCL analysis, with the average linkage method and “euclidean” as a distance metric. Log2‐transformed gene‐expression levels were scaled as a distribution with mean = 0 and SD = 1. The darker the blue, the lower the expression; the darker the red, the higher the expression. Tumor, nontumor, and fibrosis samples are color‐coded. (B) PCA plot of tumor, nontumor, or fibrosis samples in each data set, in which HHMGs were considered as observable variables. Grouped samples are labeled with different colors. (C) Heatmap of the AUC of each HHMG in each data set based on the ROC curve analysis. The darker the red, the higher the AUC value; the darker the blue, the lower the AUC value. Abbreviations: ABI3BP, ABI family, member 3 (NESH) binding protein; ADAMTS13, ADAM metallopeptidase with thrombospondin type 1 motif, 13; ADAMTSL2, ADAMTS‐like 2; ANGPTL1, angiopoietin‐like 1; ANGPTL6, angiopoietin‐like 6; ANXA10, annexin A10; ASPN, asporin; BGN, biglycan; BMPER, BMP binding endothelial regulator; CCBE1, collagen and calcium binding EGF domains 1; CCL14, chemokine (C‐C motif) ligand 14; CCL19, chemokine (C‐C motif) ligand 19; CCL2, chemokine (C‐C motif) ligand 2; CCL20, chemokine (C‐C motif) ligand 20; CCN1, cellular communication network factor 1; CD109, CD109 molecule; CLEC1B, C‐type lectin domain family 1, member B; CLEC4G, C‐type lectin domain family 4, member G; CLEC4M, C‐type lectin domain family 4, member M; COL15A1, collagen, type XV, alpha 1; COL4A1, collagen, type IV, alpha 1; COLEC10, collectin sub‐family member 10; COLEC11, collectin sub‐family member 11; CRHBP, corticotropin releasing hormone binding protein; CTHRC1, collagen triple helix repeat containing 1; CXCL12, chemokine (C‐X‐C motif) ligand 12; CXCL14, chemokine (C‐X‐C motif) ligand 14; CXCL2, chemokine (C‐X‐C motif) ligand 2; CXCL6, chemokine (C‐X‐C motif) ligand 6; DCN, decorin; DPT, dermatopontin; ECM1, extracellular matrix protein 1; ESM1, endothelial cell‐specific molecule 1; F0‐F4, Metavir scores; F9, coagulation factor IX; FBLN5, fibulin 5; FCN2, ficolin (collagen/fibrinogen domain containing lectin) 2; FCN3, ficolin (collagen/fibrinogen domain containing) 3; FGF13, fibroblast growth factor 13; FREM2, FRAS1 related extracellular matrix protein 2; GPC3, glypican 3; HABP2, hyaluronan binding protein 2; HGF, hepatocyte growth factor; HGFAC, HGF activator; HHIP, hedgehog interacting protein; IGF1, insulin‐like growth factor 1; IGF2, insulin‐like growth factor 2; IGFBP3, insulin‐like growth factor binding protein 3; INHBE, inhibin, beta E; LAMC1, laminin, gamma 1; LPA, lipoprotein, Lp(a); LUM, lumican; MASP1, mannan‐binding lectin serine peptidase 1 (C4/C2 activating component of Ra‐reactive factor); MMP12, matrix metallopeptidase 12; MXRA5, matrix‐remodelling associated 5; OIT3, oncoprotein induced transcript 3; PAMR1, peptidase domain containing associated with muscle regeneration 1; PRG4, P53‐responsive gene 4; PZP, pregnancy‐zone protein; REG3A, regenerating islet‐derived 3 alpha; RSPO3, R‐spondin 3 homolog; S100A8, S100 calcium binding protein A8; S100P, S100 calcium binding protein P; SERPINA4, serpin peptidase inhibitor, clade A (alpha‐1 antiproteinase, antitrypsin), member 4; SERPINA5, serpin peptidase inhibitor, clade A (alpha‐1 antiproteinase, antitrypsin), member 5; SERPINA7, serpin peptidase inhibitor, clade A (alpha‐1 antiproteinase, antitrypsin), member 7; SERPINB9, serpin peptidase inhibitor, clade B (ovalbumin), member 9; SERPINE1, serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1; SERPINF2, serpin peptidase inhibitor, clade F (alpha‐2 antiplasmin, pigment epithelium derived factor), member 2; SERPINI1, serpin peptidase inhibitor, clade I (neuroserpin), member 1; SPARCL1, SPARC‐like 1; SPP1, secreted phosphoprotein 1; SRPX, sushi‐repeat‐containing protein, X‐linked; SULF2, sulfatase 2; TDGF1, teratocarcinoma‐derived growth factor 1; THBS1, thrombospondin 1; THBS4, thrombospondin 4.
HHMGs Function as Modules During HBV‐Related Carcinogenesis
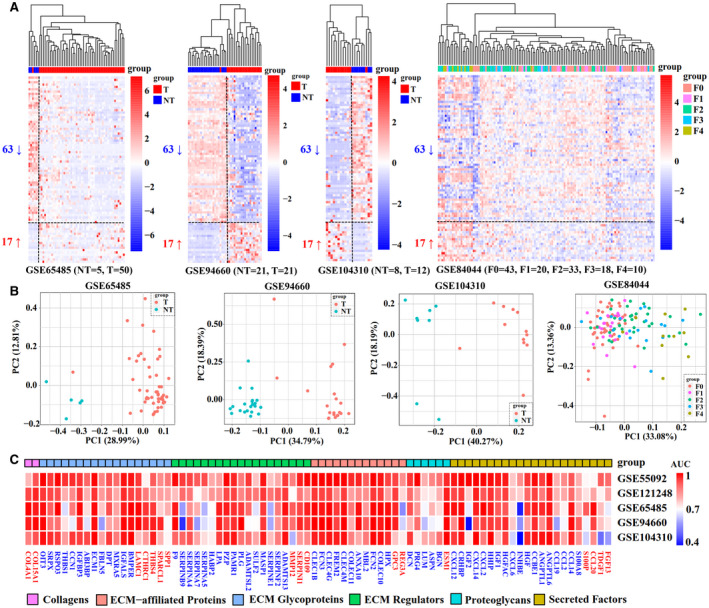
To further understand the role of the HHMGs involved in carcinogenesis in the setting of chronic HBV infection, we constructed the protein–protein interaction (PPI) network based on the identified HHMGs using the STRING database. As shown in Fig. 3A, 64 of 80 HHMGs are highly interconnected with 237 predicted interactions (enrichment P < 0.01). We then used the ClusterONE algorithm( 15 ) to mine the modules in which the matrisome genes showed a high extent of functional homogeneity. A total of four significant matrisome gene modules (P < 0.05) were found from the HHMG‐related PPI network (Fig. 3A). The matrisome genes in these modules constituted a combined signature, achieving almost perfect diagnostic potential for HBV‐related HCC tumors in the merged GSE55092 and GSE121248 data sets with AUC values of 98.9%, 98.6%, 100% and 99.5%, respectively (Fig. 3B). By HCL analysis, the four matrisome gene modules could independently separate all merged samples into two groups: Nontumor samples accounted for most in group 1, whereas tumor samples accounted for most in group 2 (Supporting Fig. S4). Gene‐set enrichment analysis (GSEA) analysis revealed that the cell cycle was activated in all of group 2, separated by the four identified matrisome gene modules (Fig. 3C and Supporting Table S3). Given that cell‐cycle progression plays a central role in promoting hepatocarcinogenesis,( 16 ) overall, it is possible that aberrant expression of these modular matrisome genes is associated with cell‐cycle dysregulation during HBV‐related carcinogenesis.
FIG. 3.
Matrisome gene modules and functional interpretation. (A) Abnormal matrisome genes’ regulatory network in HBV‐related HCC and functional modules. Significant matrisome gene modules are highlighted using different colors (P < 0.05). The node represents HHMGs, whose matrisome category is color‐coded. The edge between two nodes represents an interactive relationship. Down‐regulated HHMGs are marked using circles, and up‐regulated HHMGs are marked using diamonds. The size of the node represents the degree (i.e., the number of neighbors) of the indicated HHMG. The gene symbol of the nodes with a degree >15 is highlighted in red. (B) ROC curve analyses of four modules in the diagnosis of HBV‐related HCC from the merged samples in the GSE55092 and GSE121248 data sets. (C) GSEA analysis of the four matrisome modules. Merged samples in the GSE55092 and GSE121248 data sets were classified into two different subgroups, group 1 and group 2, based on the modular matrisome genes using HCL analysis. Adjusted P < 0.05 was considered statistically significant. Abbreviations: ADAMTS13, ADAM metallopeptidase with thrombospondin type 1 motif, 13; ADAMTSL2, ADAMTS‐like 2; ASPN, asporin; BGN, biglycan; CCL14, chemokine (C‐C motif) ligand 14; CCL19, chemokine (C‐C motif) ligand 19; CCL2, chemokine (C‐C motif) ligand 2; CCL20, chemokine (C‐C motif) ligand 20; CCN1, cellular communication network factor 1; CLEC1B, C‐type lectin domain family 1, member B; CLEC4G, C‐type lectin domain family 4, member G; COL15A1, collagen, type XV, alpha 1; COL4A1, collagen, type IV, alpha 1; COLEC10, collectin sub‐family member 10; COLEC11, collectin sub‐family member 11; CRHBP, corticotropin releasing hormone binding protein; CXCL12, chemokine (C‐X‐C motif) ligand 12; CXCL14, chemokine (C‐X‐C motif) ligand 14; CXCL2, chemokine (C‐X‐C motif) ligand 2; CXCL6, chemokine (C‐X‐C motif) ligand 6; DCN, decorin; DPT, dermatopontin; ECM1, extracellular matrix protein 1; F9, coagulation factor IX; FBLN5, fibulin 5; FCN2, ficolin (collagen/fibrinogen domain containing lectin) 2; FCN3, ficolin (collagen/fibrinogen domain containing) 3; FGF13, fibroblast growth factor 13; FREM2, FRAS1 related extracellular matrix protein 2; GPC3, glypican 3; HABP2, hyaluronan binding protein 2; HGF, hepatocyte growth factor; HGFAC, HGF activator; HHIP, hedgehog interacting protein; IGF1, insulin‐like growth factor 1; IGF2, insulin‐like growth factor 2; IGFBP3, insulin‐like growth factor binding protein 3; LAMC1, laminin, gamma 1; LPA, lipoprotein, Lp(a); LUM, lumican; MASP1, mannan‐binding lectin serine peptidase 1 (C4/C2 activating component of Ra‐reactive factor); MMP12, matrix metallopeptidase 12; MXRA5, matrix‐remodelling associated 5; NES, normalized enrichment score; PAMR1, peptidase domain containing associated with muscle regeneration 1; PRG4, P53‐responsive gene 4; PZP, pregnancy‐zone protein; RSPO3, R‐spondin 3 homolog; S100A8, S100 calcium binding protein A8; SERPINA4, serpin peptidase inhibitor, clade A (alpha‐1 antiproteinase, antitrypsin), member 4; SERPINA5, serpin peptidase inhibitor, clade A (alpha‐1 antiproteinase, antitrypsin), member 5; SERPINA7, serpin peptidase inhibitor, clade A (alpha‐1 antiproteinase, antitrypsin), member 7; SERPINE1, serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1; SERPINF2, serpin peptidase inhibitor, clade F (alpha‐2 antiplasmin, pigment epithelium derived factor), member 2; SERPINI1, serpin peptidase inhibitor, clade I (neuroserpin), member 1; SPARCL1, SPARC‐like 1; SPP1, secreted phosphoprotein 1; SRPX, sushi‐repeat‐containing protein, X‐linked; SULF2, sulfatase 2; THBS1, thrombospondin 1; THBS4, thrombospondin 4.
HHMGs Identify Molecular Distinct Subgroups of HBV‐Related HCC With Diverse Clinical Outcomes
We next explored the co‐expression relationship among the 80 HHMGs by Pearson correlation analysis based on the expression profiles in the TCGA cohort. As shown in Supporting Fig. S5, a consensus cluster with 19 HHMGs highly correlating among each other was identified following HCL clustering (r > 0.5; Supporting Table S4). The primary cellular source for the 19 HHMGs were endothelial cells (14 HHMGs) and HSCs (7 HHMGs) (Supporting Fig. S6). Patients with HBV‐related HCC in the TCGA cohort clustered into two subgroups according to the expression profile of these 19 HHMGs using HCL clustering (Fig. 4A). Subsequently, we performed independent analysis on another cohort of 218 patients with HBV‐related HCC (GSE14520) and identified two distinct molecular subgroups of HCC (ANGPTL6, CLEC4G, PAMR1, and OIT3 probes are absent in GSE14520) (Fig. 4B). To investigate whether the two subgroups represent clinically distinct patients, we compared age, gender, serum alanine aminotransferase (ALT) and alpha‐fetoprotein (AFP), multinodular status, tumor size, Tumor, Node, Metastasis (TNM) staging, and Barcelona Clinic Liver Cancer (BCLC) staging. There was almost no significant difference with respect to these parameters between the two subgroups, except that the high‐expression subgroup exhibited smaller tumor size than the low‐expression subgroup in the GEO cohort (P < 0.05; Fig. 4A,B). However, Kaplan‐Meier curves showed that the high‐expression subgroup exhibited not only notably prolonged overall survival, but recurrence‐free survival, in comparison with the low‐expression subgroup in both the TCGA and GEO cohorts (n = 60 and n = 218, respectively; P < 0.05; Fig. 4C,D). This indicates that patients with HBV‐related HCC with low expression of the 19 highly correlated HHMGs have unfavorable clinical outcome.
FIG. 4.
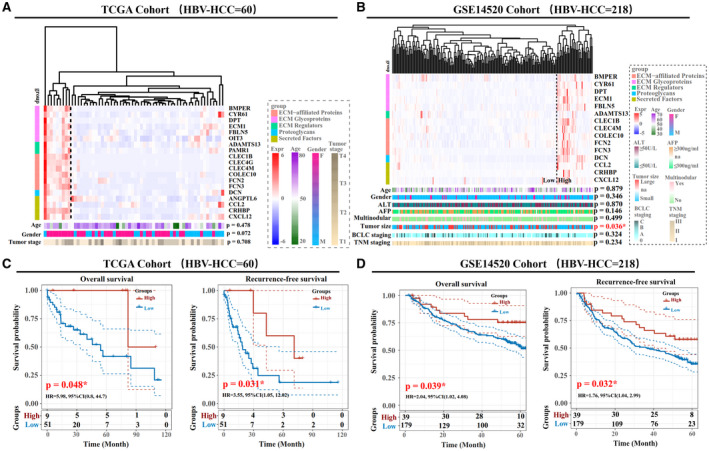
Clinical characteristics of HBV‐related HCC subgroups in the TCGA and GSE14520 cohorts. Heatmap of HBV‐related HCC subgroups in the TCGA (A) and GSE14520 (B) cohorts. The column patients were clustered by HCL analysis, with the average linkage method and “euclidean” as a distance metric. The matrisome category of HHMGs highly correlated with each other, and the clinical parameters of each patient with HCC are color‐coded. Log2‐transformed gene‐expression levels were scaled as a distribution with mean = 0 and SD = 1. The darker the blue, the lower the expression; the darker the red, the higher the expression. Overall survival and recurrence‐free survival analyses of the two subgroups (high and low expression) in the independent TCGA (C) and GSE14520 (D) cohorts. The statistical significance of the differences was determined by log‐rank test. *P < 0.05 was statistically significant. Abbreviations: ADAMTS13, ADAM metallopeptidase with thrombospondin type 1 motif, 13; ANGPTL6, angiopoietin‐like 6; BMPER, BMP binding endothelial regulator; CCL2, chemokine (C‐C motif) ligand 2; CLEC1B, C‐type lectin domain family 1, member B; CLEC4G, C‐type lectin domain family 4, member G; CLEC4M, C‐type lectin domain family 4, member M; COLEC10, collectin sub‐family member 10; CRHBP, corticotropin releasing hormone binding protein; CXCL12, chemokine (C‐X‐C motif) ligand 12; CYR61, cellular communication network factor 1 (CCN1); DCN, decorin; DPT, dermatopontin; ECM1, extracellular matrix protein 1; FBLN5, fibulin 5; FCN2, ficolin (collagen/fibrinogen domain containing lectin) 2; FCN3, ficolin (collagen/fibrinogen domain containing) 3; HR, hazard ratio; na, data lacking; OIT3, oncoprotein induced transcript 3; PAMR1, peptidase domain containing associated with muscle regeneration 1.
Subgroups of Patients With HBV‐Related HCC Show Discrete Molecular Functional Characteristics and Tumor Purity
We next explored the molecular functional characteristics in the two subgroups of patients. GSEA analysis showed that the high‐expression subgroup in both the TCGA and GEO cohorts was associated primarily with activation of immune‐related, metabolism‐related, and ECM‐related pathways (P < 0.05), whereas the low‐expression subgroup was associated with activation of carcinogenesis‐related pathways such as DNA replication and cell cycle (adjusted P < 0.05; Fig. 5A,B). Because tumor purity has been reported as linked with prognosis,( 17, 18, 19 ) we used the ESTIMATE algorithm( 20 ) to compare tumor purity between the two subgroups. As shown in Fig. 6A,B, the high‐expression subgroup in both the TCGA and GEO cohorts exhibited lower tumor purity, as the StromalScore, ImmuneScore, and ESTIMATEScore were significant higher when compared with the low‐expression subgroup (P < 0.05). We then used the MCPcounter algorithm( 21 ) to quantify the absolute abundance of nontumor cell populations between subgroups. Most nontumor cell types in the high‐expression subgroup were observed as more abundant than in the low‐expression subgroup; however, cytotoxic lymphocytes, B lineage, and natural killer cells were significant (P < 0.05) in the TCGA cohort (P < 0.05; Fig. 6A), whereas T cells, B‐cell lineage, myeloid dendritic cells, endothelial cells, and fibroblasts were significant in the GEO cohort (P < 0.05; Fig. 6B).
FIG. 5.
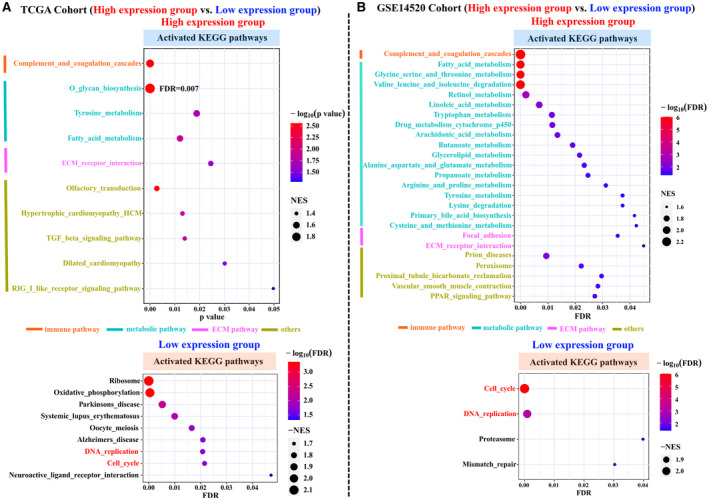
Functional comparison between two subgroups of patients with HBV‐related HCC. GSEA analysis of two HBV‐related HCC subgroups (high and low expression) in both the TCGA (A) and GSE14520 (B) cohorts. Significant activated KEGG pathways are shown in the dot plots. The size of the circle represents the NES, and the color indicates the P value or adjusted P value. The KEGG categories are color‐coded. Abbreviations: FDR, false discovery rate; NES, normalized enrichment score; PPAR, peroxisome proliferator‐activated receptor; TGF, transforming growth factor.
FIG. 6.
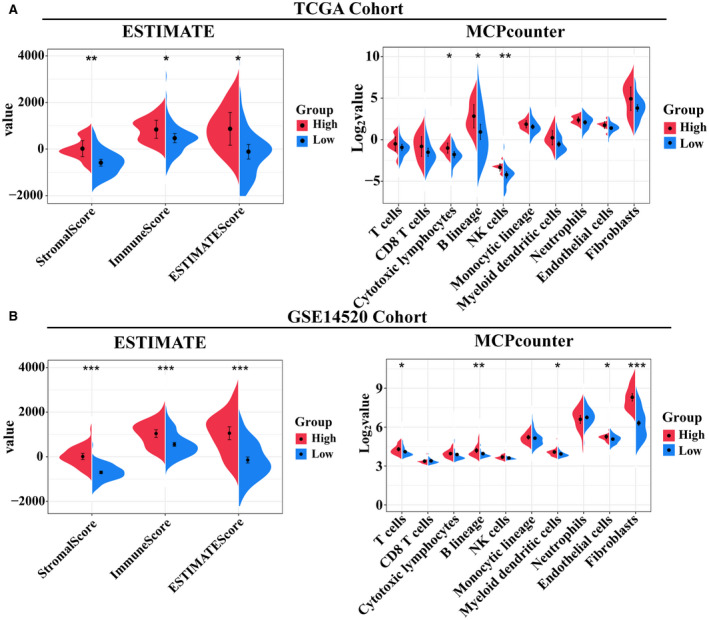
Comparison of the tumor purity between the two subgroups of patients with HBV‐related HCC. Violin plots of the StromalScore, ImmuneScore, ESTIMATEScore, and abundance of the immune or stromal cell populations in two subgroups of HBV‐related HCC from the TCGA (A) and GSE14520 (B) cohorts. The statistical difference was calculated with the Kruskal–Wallis test, and the P values are indicated above each violin plot with asterisks (*P < 0.05, **P < 0.01, and ****P < 0.001). Abbreviation: NK, natural killer cell.
Liver‐Specific HHMGs Are Implicated in HBV‐Related HCC
Screening the genotype‐tissue expression (GTEx) database, PLG [plasminogen], HABP2 [hyaluronan binding protein 2], HPX [hemopexin], IGFALS [insulin‐like growth‐factor‐binding protein, acid labile subunit], F9, MBL2 [mannose binding lectin 2], INHBE [inhibin subunit beta E], HGFAC [hepatocyte growth factor activator], SERPINF2 [serpin family F member 2], and SERPINA7 [serpin family A member 7] were found to be highly and specifically expressed in normal liver tissues (Supporting Fig. S7A). The result from the Human Protein Atlas database further confirmed the findings from the GTEx database (Supporting Fig. S7B). Single‐cell data analysis revealed that F9, HGFAC, INHBE, MBL2, and SERPINA7 are expressed only in hepatocytes, whereas HABP2, HPX, IGFALS, PLG, and SERPINF2 are expressed primarily in hepatocytes and cholangiocytes (Supporting Fig. S8). We next evaluated their potential as therapeutic targets by analyzing their clinical relevance and biological function using the GEO cohort. As indicated in Supporting Table S5 and Fig. 7A‐C, among all identified liver‐specific HHMGs, high expression of F9 was associated with low serum AFP activity, TNM, or BCLC staging (P < 0.05). Patients with high expression of HPX showed low AFP activity, small tumor size, and low TNM or BCLC staging (P < 0.05). High expression of IGFALS was a marker for small tumor size, low TNM staging, and prolonged overall or recurrence‐free survival (P < 0.05). Moreover, high expression of PLG could predict low‐serum ALT and AFP activities, less multinodular, low TNM or BCLC staging, and prolonged overall or recurrence‐free survival (P < 0.05). GSEA analysis revealed that low expression of F9, HPX, IGFALS, or PLG was negatively associated with activation of multiple metabolic pathways but positively associated with activation of cell cycle or DNA replication (adjusted P < 0.05; Supporting Table S6), suggesting that these liver‐specific HHMGs are tumor‐suppressor matrisome genes that may be potential antitumor targets for HBV‐related HCC.
FIG. 7.
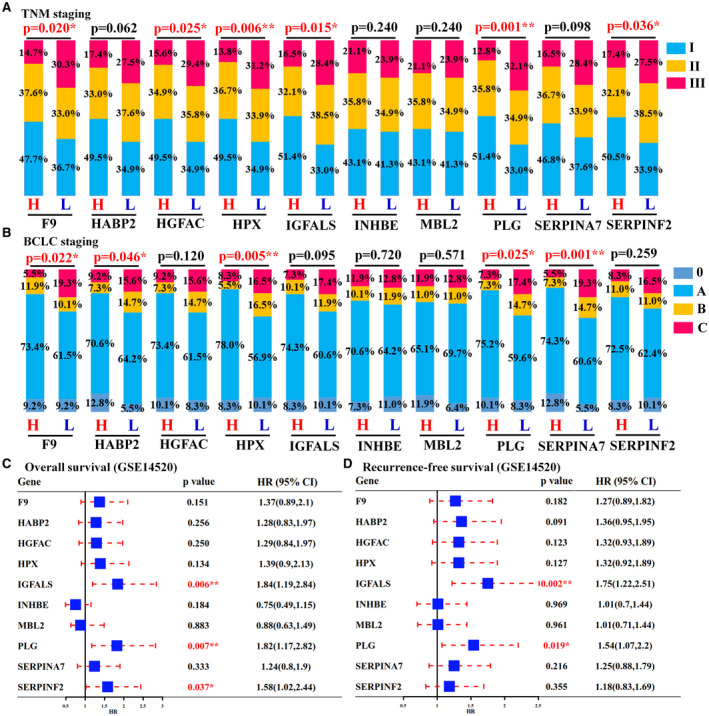
Liver‐specific HHMGs and their clinical relevance. TNM (A) and BCLC (B) staging comparisons between the high (H) and the low (L) expression groups. TNM or BCLC staging is shown as percentage. Statistical comparison was performed using chi‐square test (*P < 0.05 and **P < 0.01). Overall (C) and recurrence‐free (D) survival analyses of each liver‐specific HHMGs between the high‐expression and low‐expression groups. Comparisons between the two groups were performed using log‐rank test (*P < 0.05 and **P < 0.01). Abbreviation: HR, hazard ratio.
Discussion
HBV infection can affect the turnover of the liver matrisome.( 10, 11 ) A persistent low viral load of HBV in patients who received antiviral therapy shows higher rate of fibrosis progression,( 22 ) which could be explained by constant matrisome remodeling in HBV infection. Matrisome remodeling is a hallmark of liver fibrosis and also occurs during carcinogenesis, affecting cellular proliferation, migration or invasion, all involved in cancer progression, metastasis, and prognosis.( 23 ) The present study identified 80 matrisome genes changed during HBV‐related HCC. These matrisome genes are reliable and robust. First, they were identified from two independent microarray data sets with 118 nontumor and 109 tumor samples from 81 patients with HBV‐related HCC. Second, they were validated in three independent RNA‐seq data sets with 34 nontumor and 83 tumor samples from 85 patients with HBV‐related HCC. Third, these matrisome genes, as a single or combined signature, separate tumor from nontumor samples with chronic HBV infection using three different classification methods (HCL, PCA, and ROC curve analyses).
HBV‐related HCC often occurs in the absence of cirrhosis in Eastern Asia and most African countries( 1 ); specifically, cirrhosis is not an essential condition for HBV‐related carcinogenesis. Although the number of patients with a cirrhotic background enrolled in the studies from the GSE121248 and GSE55092 data sets were different (82% and 53.7%), the overlap of the differentially expressed matrisome genes shared by the two data sets was greater than what would be expected by chance (Fisher’s exact test; P < 0.01). In addition, the expression pattern of the identified 80 matrisome genes associated with HBV‐related HCC could not be verified in HBV‐related liver fibrosis, further implying that these dysregulated matrisome genes during HBV‐related HCC are, to some extent, cirrhosis‐independent. Notably, our results at the transcriptional level are not in line with findings at the posttranslational level from two HCC mouse models.( 8 ) Lai et al. found that most changes in core matrisome proteins between fibrotic tissues and tumor samples largely occurred between healthy and fibrotic tissues in two transgenic mouse models of liver cancer.( 8 ) This discordance can be partially explained by (1) different ECM components that may be assembled in each context( 8 ); (2) ECM deposition that occurs during cancer development, even though the corresponding genes are down‐regulated( 23 ); and (3) cross‐species variations.( 24 ) In addition, Lai et al. used global, rather than ECM‐based proteomics, which may not fully recapitulate the ECM microenvironment in HCC tumors.
Network modules are tensely clustered subnetworks with more internal connections than expected randomly in the entire network. Genes in the same module tend to have similar biological function. In this study, four significant matrisome gene modules were mined from the HBV‐related HCC‐related functional network. The merged samples in the GSE55092 and GSE121248 data sets could be separated into two groups based on the expression of modular members. The tumor‐biased group positively correlated with cell‐cycle progression. It is well known that HBx, which is essential for HBV replication in vivo, inhibits apoptosis and stimulates cell cycle,( 25, 26, 27 ) whereas cell‐cycle activation will elude growth suppressors, sustain proliferation, resist cell death, act on chromosome instability, and eventually trigger hepatocarcinogenesis.( 16 )
Among the four functional matrisome gene modules, all members in module 4 were chemokine genes including chemokine (C‐C motif) ligand (CCL) 2, CCL14, CCL19, CCL20, chemokine (C‐X‐C motif) ligand (CXCL) 2, CXCL6, CXCL12, and CXCL14. Chemokines are critical for attracting immune cells into the liver.( 28 ) For example, CCL2 is responsible for recruiting monocytes/macrophages; CCL19 is chemotactic for CD8+ T cells and dendritic cells; and CXCL2 and CXCL6 promote infiltration of immunosuppressive neutrophils and monocytes into the liver.( 28 ) In module 4, all chemokines except for CCL20 were significantly down‐regulated in HBV‐related HCC tumor samples, indicating weak and sparse immune infiltration. In agreement with this observation, Sia et al. found that only 25% of patients with HCC in a large cohort expressed markers of the inflammatory response, and two groups of patients with HCC were characterized by adaptive or exhausted immune responses.( 29 ) Immune cells fail to penetrate the tumor parenchyma and remain in the stroma surrounding tumor cell nests, probably attributable to (1) decreased migration of immune cells elicited by down‐regulation of chemokine genes and/or (2) obstruction of immune infiltration by stiff tumor ECM.( 30 ) Pan‐cancer analysis revealed that cancer tissues harboring lower expression of a core set of matrisome genes possess higher CD8+ T‐cell infiltration in multiple cancers,( 31 ) indicating that tumor stiffness exhibits an inverse relationship with immune activity.
The up‐regulated matrisome genes do not function as modules, indicating a weak intracellular biological significance; on the contrary, they are likely important for the extracellular tumor‐promoting microenvironment. COL4A1 (collagen type IV alpha 1 chain) and LAMC1 (laminin subunit gamma‐1) encode proteins that constitute the collagenous and noncollagenous components of the basement membrane; their overexpression is essential for HCC growth, metastasis, and survival.( 32, 33, 34 ) COL15A1 (collagen alpha‐1[XV] chain) encodes collagen XV and is a prominent histopathological component of sinusoidal capillarization in HCC.( 35 ) Because collagen XV is localized in the basement membrane, it may function to adhere it to the connective tissue stroma, which remains to be further investigated. Another interesting member is GPC3, which encodes Glypican‐3 and has been recognized as a better diagnostic marker of HCC, given its limited expression in normal and nontumoral livers but its high expression in HCC.( 36 ) Glypican‐3 acts as an ECM signal or “recruiter” in various signaling pathways, maintaining the concentration of extracellular ligands and promoting ligand–receptor interaction.( 36 ) Although other up‐regulated matrisome genes have been reported as associated with HCC development or prognosis, their extracellular tumor‐promoting role have not been unveiled in HCC, especially in HBV‐related HCC, and are worth studying.
Rapid evolution of genome‐wide transcriptomics technology is contributing to a more precise understanding of the correlations between clinicopathological characteristics and mediator molecules. By now, several laboratories have classified patients with HCC into proliferative and non‐proliferative subgroups, each representing 50% of patients with HCC( 37, 38, 39 ) and showing distinctions in metabolism and clinical outcome.( 40 ) A recent study based on integrated proteogenomic characterization reported that patients with HBV‐related HCC could be classified into three groups with obvious different molecular patterns and clinical prognosis.( 41 ) The present, prospective study—based on a core of matrisome genes that are highly correlated with each other—classified patients with HBV‐related HCC into two subgroups: high expression versus low expression. The high‐expression subgroup represented a small group of patients with HBV‐related HCC (15.0% in the TCGA cohort and 17.9% in the GEO cohort), but revealed significantly prolonged overall survival and recurrence‐free survival than the low‐expression group. This classification was validated in two independent cohorts with a total of 278 patients with HBV‐related HCC.
Our study also found a difference in the metabolic landscape between the two identified subgroups. Because cancer is usually viewed as a disease attributable to metabolic disorders,( 42 ) the exhausted metabolic activity in the low‐expression subgroup of patients with HBV‐related HCC may be one reason why this subgroup exhibited worse prognosis. The cell cycle was also activated in the low‐expression subgroup. As discussed previously,( 16 ) cell‐cycle progression could be another reason for unfavorable outcome in the low‐expression subgroup. Moreover, the low‐expression subgroup also exhibited higher tumor purity, as these samples presented low stromal and immune scores.( 20 ) Previous studies have revealed that low tumor purity is associated with unfavorable prognosis in colon cancer,( 17 ) gastric cancer,( 18 ) and glioma,( 19 ) whereas it is the opposite in HCC.( 43 ) The discordance is likely explained by the fact that HCC is not as desmoplastic as other cancers mentioned previously, in which a significant portion of matrisome is made by fibroblasts and other stromal cells. The cellular source of the identified HHMGs in this study is not only limited to immune cells and stromal cells, but to hepatocytes, EPCAM+ cells, and cholangiocytes (Supporting Fig. S3). Lower tumor purity shows higher proportion of mixed nonparenchymal cells including immune cells in the tumor region, whereas sparse infiltration of immune cells such as cytotoxic T cells into the tumor microenvironment has been widely reported as linked with favorable HCC prognosis.( 44, 45, 46 ) Understanding the purity or immunological characteristics of HBV‐related HCC may implement new approaches to personalized medicine.
Matrisome gene–based intervention will contribute to remodeling the tumor ECM to its physiological state. Liver‐specific matrisome genes can be ideal targets for HBV‐related HCC therapy, as they can avoid undesirable side effects. To conclude, this study identified 10 liver‐specific matrisome genes that are strictly expressed in hepatocytes and/or cholangiocytes, among which F9, HPX, IGFALS, and PLG have important clinical implications in HBV‐related HCC progression and prognosis. A previous bioinformatics analysis also confirmed that F9, IGFALS, and PLG were down‐regulated in HBV‐related HCC.( 47 ) PLG gene expression was also reported to be reduced in HCC tissue in an early publication.( 48 ) However, the specific role of these liver‐specific matrisome genes on the pathophysiology of HBV‐related HCC and the underlying mechanisms involved in their down‐regulation have not yet been reported, which are worth exploring.
Supporting information
Supplementary Material
Supported by a U.S. Public Health Service Grant from the National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK111677).
Potential conflict of interest: Nothing to report.
References
Author names in bold designate shared co‐first authorship.
- 1.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 2019;16:589‐604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su CW, Wu CY, Lin JT, Ho HJ, Wu JC. Nucleos(t)ide analogue continuous therapy associated with reduced adverse outcomes of chronic hepatitis B. J Chin Med Assoc 2020;83:125‐133. [DOI] [PubMed] [Google Scholar]
- 3.Torresi J, Tran BM, Christiansen D, Earnest‐Silveira L, Schwab RHM, Vincan E. HBV‐related hepatocarcinogenesis: the role of signalling pathways and innovative ex vivo research models. BMC Cancer 2019;19:707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Tian Z. HBV‐Induced Immune Imbalance in the development of HCC. Front Immunol 2019;10:2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong Y, Zheng Q, Wang Z, Lin X, You Y, Wu S, et al. Higher matrix stiffness as an independent initiator triggers epithelial‐mesenchymal transition and facilitates HCC metastasis. J Hematol Oncol 2019;12:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arteel GE, Naba A. The liver matrisome—looking beyond collagens. JHEP Rep 2020;2:100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao X, Taha IN, Clauser KR, Gao YT, Naba A. MatrisomeDB: the ECM‐protein knowledge database. Nucleic Acids Res 2020;48:D1136‐D1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai KKY, Shang S, Lohia N, Booth GC, Masse DJ, Fausto N, et al. Extracellular matrix dynamics in hepatocarcinogenesis: a comparative proteomics study of PDGFC transgenic and Pten null mouse models. PLoS Genet 2011;7:e1002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpino G, Overi D, Melandro F, Grimaldi A, Cardinale V, Di Matteo S, et al. Matrisome analysis of intrahepatic cholangiocarcinoma unveils a peculiar cancer‐associated extracellular matrix structure. Clin Proteomics 2019;16:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zan Y, Zhang Y, Tien P. Hepatitis B virus e antigen induces activation of rat hepatic stellate cells. Biochem Biophys Res Commun 2013;435:391‐396. [DOI] [PubMed] [Google Scholar]
- 11.Feng H, Li X, Niu D, Chen WN. Protein profile in HBx transfected cells: a comparative iTRAQ‐coupled 2D LC‐MS/MS analysis. J Proteomics 2010;73:1421‐1432. [DOI] [PubMed] [Google Scholar]
- 12.Zhang R, Cao Y, Bai L, Zhu C, Li R, He H, et al. The collagen triple helix repeat containing 1 facilitates hepatitis B virus‐associated hepatocellular carcinoma progression by regulating multiple cellular factors and signal cascades. Mol Carcinog 2015;54:1554‐1566. [DOI] [PubMed] [Google Scholar]
- 13.Chen W, Yang A, Jia J, Popov YV, Schuppan D, You H. Lysyl oxidase (LOX) family members: rationale and their potential as therapeutic targets for liver fibrosis. Hepatology 2020;72:729‐741. [DOI] [PubMed] [Google Scholar]
- 14.Han ES, Wu Y, McCarter R, Nelson JF, Richardson A, Hilsenbeck SG. Reproducibility, sources of variability, pooling, and sample size: important considerations for the design of high‐density oligonucleotide array experiments. J Gerontol A Biol Sci Med Sci 2004;59:306‐315. [DOI] [PubMed] [Google Scholar]
- 15.Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein‐protein interaction networks. Nat Methods 2012;9:471‐472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bisteau X, Caldez MJ, Kaldis P. The complex relationship between liver cancer and the cell cycle: a story of multiple regulations. Cancers (Basel) 2014;6:79‐111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao Y, Feng Q, Zheng P, Yang L, Liu T, Xu Y, et al. Low tumor purity is associated with poor prognosis, heavy mutation burden, and intense immune phenotype in colon cancer. Cancer Manag Res 2018;10:3569‐3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong Z, Zhang J, Guo W. Tumor purity as a prognosis and immunotherapy relevant feature in gastric cancer. Cancer Med 2020;9:9052‐9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C, Cheng W, Ren X, Wang Z, Liu X, Li G, et al. Tumor purity as an underlying key factor in Glioma. Clin Cancer Res 2017;23:6279‐6291. [DOI] [PubMed] [Google Scholar]
- 20.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres‐Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et al. Estimating the population abundance of tissue‐infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016;17:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Y, Wu X, Zhou J, Meng T, Wang B, Chen S, et al. Persistent low level of hepatitis B virus promotes fibrosis progression during therapy. Clin Gastroenterol Hepatol 2020;18:2582‐2591.e2586. [DOI] [PubMed] [Google Scholar]
- 23.Socovich AM, Naba A. The cancer matrisome: from comprehensive characterization to biomarker discovery. Semin Cell Dev Biol 2019;89:157‐166. [DOI] [PubMed] [Google Scholar]
- 24.Chen W, Xia X, Song N, Wang Y, Zhu H, Deng W, et al. Cross‐species analysis of gene expression and function in prefrontal cortex, hippocampus and striatum. PLoS One 2016;11:e0164295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong P, Zhang X, Zhang J, Zhang J, Luo H, Wang Z. Hepatitis B virus X protein in the proliferation of hepatocellular carcinoma cells. Front Biosci (Landmark Ed) 2013;18:1256‐1265. [DOI] [PubMed] [Google Scholar]
- 26.Motavaf M, Safari S, Saffari Jourshari M, Alavian SM. Hepatitis B virus‐induced hepatocellular carcinoma: the role of the virus x protein. Acta Virol 2013;57:389‐396. [DOI] [PubMed] [Google Scholar]
- 27.Ng SA, Lee C. Hepatitis B virus X gene and hepatocarcinogenesis. J Gastroenterol 2011;46:974‐990. [DOI] [PubMed] [Google Scholar]
- 28.Marra F, Tacke F. Roles for chemokines in liver disease. Gastroenterology 2014;147:577‐594.e1. [DOI] [PubMed] [Google Scholar]
- 29.Sia D, Jiao Y, Martinez‐Quetglas I, Kuchuk O, Villacorta‐Martin C, Castro de Moura M, et al. Identification of an immune‐specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology 2017;153:812‐826. [DOI] [PubMed] [Google Scholar]
- 30.Efthymiou G, Saint A, Ruff M, Rekad Z, Ciais D, Van Obberghen‐Schilling E. Shaping up the tumor microenvironment with cellular fibronectin. Front Oncol 2020;10:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bin Lim S, Chua MLK, Yeong JPS, Tan SJ, Lim WT, Lim CT. Pan‐cancer analysis connects tumor matrisome to immune response. NPJ Precis Oncol 2019;3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T, Jin H, Hu J, Li XI, Ruan H, Xu H, et al. COL4A1 promotes the growth and metastasis of hepatocellular carcinoma cells by activating FAK‐Src signaling. J Exp Clin Cancer Res 2020;39:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye G, Qin Y, Wang S, Pan D, Xu S, Wu C, et al. Lamc1 promotes the Warburg effect in hepatocellular carcinoma cells by regulating PKM2 expression through AKT pathway. Cancer Biol Ther 2019;20:711‐719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Désert R, Mebarki S, Desille M, Sicard M, Lavergne E, Renaud S, et al. “Fibrous nests” in human hepatocellular carcinoma express a Wnt‐related gene signature associated with poor clinical outcome. Int J Biochem Cell Biol 2016;81:195‐207. [DOI] [PubMed] [Google Scholar]
- 35.Kimura K, Nakayama M, Naito I, Komiyama T, Ichimura K, Asano H, et al. Human collagen XV is a prominent histopathological component of sinusoidal capillarization in hepatocellular carcinogenesis. Int J Clin Oncol 2016;21:302‐309. [DOI] [PubMed] [Google Scholar]
- 36.Guo M, Zhang H, Zheng J, Liu Y. Glypican‐3: a new target for diagnosis and treatment of hepatocellular carcinoma. J Cancer 2020;11:2008‐2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoshida Y, Toffanin S, Lachenmayer A, Villanueva A, Minguez B, Llovet JM. Molecular classification and novel targets in hepatocellular carcinoma: recent advancements. Semin Liver Dis 2010;30:35‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol 2015;12:408‐424. [DOI] [PubMed] [Google Scholar]
- 39.Zucman‐Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015;149:1226‐1239.e1224. [DOI] [PubMed] [Google Scholar]
- 40.Ng CKY, Piscuoglio S, Terracciano LM. Molecular classification of hepatocellular carcinoma: the view from metabolic zonation. Hepatology 2017;66:1377‐1380. [DOI] [PubMed] [Google Scholar]
- 41.Gao Q, Zhu H, Dong L, Shi W, Chen R, Song Z, et al. Integrated proteogenomic characterization of HBV‐related hepatocellular carcinoma. Cell 2019;179:561‐577.e522. [DOI] [PubMed] [Google Scholar]
- 42.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 2015;17:351‐359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ge PL, Li SF, Wang WW, Li CB, Fu YB, Feng ZK, et al. Prognostic values of immune scores and immune microenvironment‐related genes for hepatocellular carcinoma. Aging (Albany NY) 2020;12:5479‐5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foerster F, Hess M, Gerhold‐Ay A, Marquardt JU, Becker D, Galle PR, et al. The immune contexture of hepatocellular carcinoma predicts clinical outcome. Sci Rep 2018;8:5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parmiani G, Anichini A. T cell infiltration and prognosis in HCC patients. J Hepatol 2006;45:178‐181. [DOI] [PubMed] [Google Scholar]
- 46.Kurebayashi Y, Ojima H, Tsujikawa H, Kubota N, Maehara J, Abe Y, et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology 2018;68:1025‐1041. [DOI] [PubMed] [Google Scholar]
- 47.Xie S, Jiang X, Zhang J, Xie S, Hua Y, Wang R, et al. Identification of significant gene and pathways involved in HBV‐related hepatocellular carcinoma by bioinformatics analysis. PeerJ 2019;7:e7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinoshita M, Miyata M. Underexpression of mRNA in human hepatocellular carcinoma focusing on eight loci. Hepatology 2002;36:433‐438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
The human matrisome gene list, processed GEO and TCGA data sets, and related R code have been deposited into the figshare platform (https://figshare.com/s/f7af736216ef73d7ee7d; https://doi.org/10.6084/m9.figshare.14069474).


