Abstract
This study aimed to uncover underlying mechanisms and promising intervention targets of heart failure (HF)-related stroke. HF-related dataset GSE42955 and stroke-related dataset GSE58294 were obtained from the Gene Expression Omnibus (GEO) database. Weighted gene co-expression network analysis (WGCNA) was conducted to identify key modules and hub genes. Gene Ontology (GO) and pathway enrichment analyses were performed on genes in the key modules. Genes in HF- and stroke-related key modules were intersected to obtain common genes for HF-related stroke, which were further intersected with hub genes of stroke-related key modules to obtain key genes in HF-related stroke. Key genes were functionally annotated through GO in the Reactome and Cytoscape databases. Finally, key genes were validated in these two datasets and other datasets. HF- and stroke-related datasets each identified two key modules. Functional enrichment analysis indicated that protein ubiquitination, Wnt signaling, and exosomes were involved in both HF- and stroke-related key modules. Additionally, ten hub genes were identified in stroke-related key modules and 155 genes were identified as common genes in HF-related stroke. OTU deubiquitinase with linear linkage specificity(OTULIN) and nuclear factor interleukin 3-regulated(NFIL3) were determined to be the key genes in HF-related stroke. Through functional annotation, OTULIN was involved in protein ubiquitination and Wnt signaling, and NFIL3 was involved in DNA binding and transcription. Importantly, OTULIN and NFIL3 were also validated to be differentially expressed in all HF and stroke groups. Protein ubiquitination, Wnt signaling, and exosomes were involved in HF-related stroke. OTULIN and NFIL3 may play a key role in HF-related stroke through regulating these processes, and thus serve as promising intervention targets.
Keywords: Cardioembolic stroke, Heart failure, Bioinformatics, Weighted gene co-expression network analysis (WGCNA)
1. Introduction
Stroke is currently the second leading cause of death and a major cause of disability worldwide, costing approximately 3% to 4% of medical care expenditures in Western countries (Struijs et al., 2006; GBD 2015 Mortality and Causes of Death Collaborators, 2016; Ding et al., 2019). Cardioembolic stroke (CS), a subtype in which cardiogenic emboli cause cerebral thromboembolic ischemia, is estimated to account for 14% to 30% of ischemic stroke but has greater potential to lead to disability (Adams et al., 1993; Murtagh and Smalling, 2006). A growing number of cardiovascular diseases are recognized as causes of cardiogenic emboli, among which heart failure (HF) is responsible for 9% of all strokes (Pullicino and Homma, 2010). HF is a severe clinical syndrome derived from the progression of various heart diseases, with reduced cardiac pumping and filling functions. Previous studies showed that stroke occurred in 4%–5% of patients with severe HF, compared with 0.5% in those without HF (Mischie et al., 2013). Moreover, HF patients bear more than three times the risk of stroke compared with the general population (Kamel and Healey, 2017).
It is assumed that HF-related stroke may be related to the following pathophysiological mechanisms. First, the elevated or activated circulating markers like fibrin, pro-inflammatory cytokines, and platelets suggest a hypercoagulable state in HF patients (Thomas et al., 2016). Second, dyskinetic regions of the left ventricle where blood stasis happens create a hotbed of thrombosis, one of the potential sources of cardiogenic emboli (Schoner et al., 2015). Finally, atrial fibrillation (AF), which is well known as the chief precursor of CS, is a common complication of HF. HF accompanied by AF may present in approximately 2% of strokes (Bogiatzi et al., 2014; Kang et al., 2017; Gao et al., 2019). However, the underlying molecular biological processes of these pathophysiological mechanisms remain unclear.
Anticoagulation is the cornerstone of CS prevention and treatment (Aguilar et al., 2007). However, in contrast to the efficacy of anticoagulation in AF-related stroke, this therapeutic approach failed to reveal an overall clinical benefit for HF-related stroke regardless of hypercoagulability in HF patients. Two randomized controlled trials recruited HF patients treated with anticoagulant therapy and showed that no improvement in stroke episodes or other outcomes had taken place in this population (Cleland et al., 2004; Cokkinos et al., 2006). Also, the Warfarin and Antiplatelet Therapy in Chronic Heart Failure (WATCH) trial indicated that significantly increased bleeding risk counteracted the positive effects when HF patients received anticoagulants to prevent stroke (Massie et al., 2009). Thus, anticoagulation is less effective in preventing stroke for HF patients, and new intervention targets need to be identified.
Recent studies illustrated that genetic factors occupy an irreplaceable role in the pathogenesis of stroke (Malik and Dichgans, 2018; Dichgans et al., 2019; Zhou LY et al., 2019), and several gene expression profiling studies have been performed to reveal the underlying genes and biological processes during stroke occurrence and progression. Malik et al. (2018) identified 22 novel stroke risk loci and provided a framework for prioritization of stroke risk variants and genes. Zou et al. (2019) identified ZNF566, PDZK1IP1, ZFHX3, and PITX2 genes as significant biomarkers for AF-related stroke. Zhu WH et al. (2019) found that IL1α, IL1β, IL6, IL8, CXCL1, CXCL2, CXCL20, CCL4, ICAM1, and PTGS2 had a protective effect against stroke in females. However, no gene expression profile studies have yet focused on HF-related stroke, and the underlying genes and biological processes of HF-related stroke can be clearly understood by revealing its gene expression variations and patterns.
Weighted gene co-expression network analysis (WGCNA) is an advanced analytical method for discovering gene‒gene relationship and genetic network‒disease relationship, with the advantages of system-level insight and high sensitivity to genes with low abundance or small fold change (Wang et al., 2020). Recently, WGCNA has been increasingly used to construct co-expressed gene networks and aid in identifying underlying biological processes and key genes in disease-related networks (Raghow, 2016). In this study, we explored common biological processes from the co-expression networks of HF and CS so as to uncover the underlying biological processes of HF-related stroke. Even further, we identified key genes from the common genes of HF and CS to pinpoint promising intervention targets for HF-related stroke.
2. Materials and methods
2.1. Data sources
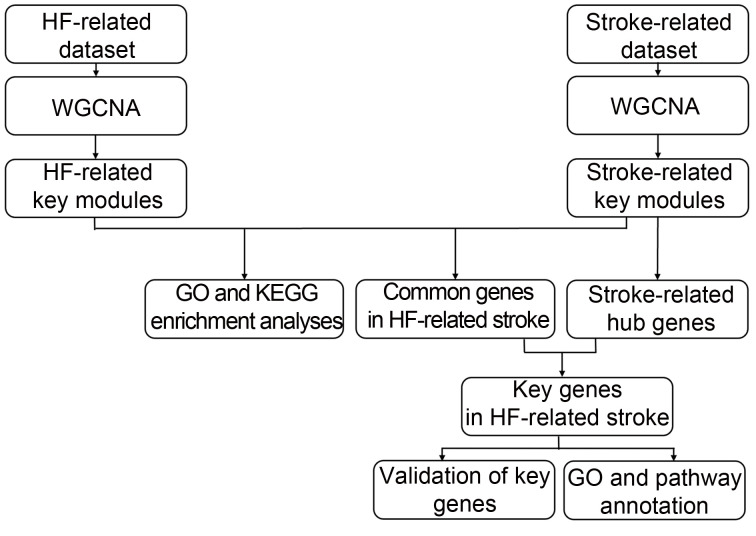
A flow chart for the present study is shown in Fig. 1. The HF-related microarray datasets (GSE42955 and GSE1145) and stroke-related datasets (GSE58294 and GSE22255) were obtained from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo). In dataset GSE42955, left ventricular (LV) samples from explanted hearts of HF patients (New York Heart Association (NYHA) functional classifications of III and IV) were regarded as HF group, and non-diseased donor hearts were regarded as control group. This dataset was verified using the GPL6244 platform (Affymetrix Human Gene 1.0 ST Array). In dataset GSE58294, whole blood from subjects with CS (stroke group) and healthy controls (control group) was collected. This dataset was verified using the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array). In dataset GSE1145, failing myocardial samples from explanted hearts of HF patients were regarded as HF group and non-diseased donor myocardial samples were used as control group. This dataset was verified using the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array). In dataset GSE22255, peripheral blood from subjects with stroke (stroke group) and healthy controls (control group) was collected. This dataset was verified using the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array). GSE1145 and GSE22255 were used as validation datasets.
Fig. 1. Flowchart of data analysis. HF: heart failure; WGCNA: weighted gene co-expression network analysis; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes.
2.2. Data preprocessing
The "Combat" function in the Surrogate Variable Analysis package (SVA, Version 3.13) was applied to remove batch effects among microarrays (Leek et al., 2012). Subsequently, all of the raw data were analyzed using bioinformatics methods, including background correction, quantile normalization, and log2-transformation of the expression values (Irizarry et al., 2003; Ritchie et al., 2015). The probes exhibiting the top 50% in high expression variance (and thus minimal loss of statistical information) were identified and retained with a genefilter package Version 1.64.0 (Gentleman et al., 2021) for subsequent analysis. Finally, the probe identifications (IDs) in the gene expression matrix were reannotated with gene symbols.
2.3. Construction of co-expressed gene modules
WGCNA was executed as follows: (1) outlier samples were detected by hierarchical clustering analysis; (2) an appropriate "soft" threshold (β) was selected by the "pickSoftThreshold" algorithm to obtain a biologically significant scale-free network (scale independence of >0.8); (3) the expression similarity between each gene pair m and n was defined as adjacency value: a mn=power(S mn, β)=|S mn|β(S mn was defined as the absolute value of the correlation coefficient between the profiles of genes m and n); (4) the adjacency matrix was transformed into a topological overlap matrix (TOM) by the "TOMsimilarity" algorithm to counter the effects of spurious or missing connections between genes m and n (Yip and Horvath, 2007); (5) modules were detected by hierarchical clustering analysis of TOM and the Dynamic Branch Cut method.
2.4. Identification of disease-related key modules
The analysis was conducted as follows: (1) module eigengene value was calculated by "moduleEigengenes" algorithm, which summarizes the gene expression profiles of a given module; (2) HF- or stroke-related key modules were identified through hierarchical clustering and Spearman correlation analysis of a module's module eigengene value and a sample's clinical traits; (3) the contribution of key modules to sample traits was assessed by principal component analysis (PCA).
2.5. GO and KEGG enrichment analyses of genes in disease-related key modules
Genes of HF-related key modules and stroke-related key modules were imported into the Metascape database ( https://metascape.org) to execute Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses (Zhou YY et al., 2019). GO describes the overrepresented biological functions among intramodular genes for three distinct aspects including molecular function, cellular component, and biological process (Ashburner et al., 2000; The Gene Ontology Consortium, 2019). KEGG describes the overrepresented pathways among intramodular genes (Kanehisa and Sato, 2020). A maximum P value of 0.01, minimum overlap of 3, and minimum enrichment of 1.5 were chosen as the threshold.
2.6. Identification of key genes in HF-related stroke
The common genes of HF-related stroke were obtained by taking the intersection of genes between HF-related key modules and stroke-related key modules. In addition, their protein–protein interaction (PPI) relationships were visualized with the STRING database (https://string-db.org). In stroke-related key modules, gene significance and module membership of each intramodular gene were calculated by the "geneTraitSignificance" and "geneModuleMembership" algorithms to find stroke-related hub genes. Key genes of HF-related stroke were obtained by taking the intersection of the common genes of HF-related stroke and stroke-related hub genes.
2.7. GO and pathway annotation of key genes in HF-related stroke
GO annotates the key genes of HF-related stroke for three distinct aspects including molecular function, cellular component, and biological process. The Reactome database ( https://reactome.org) is an open-source, open-access, manually curated, and peer-reviewed pathway database (Jassal et al., 2020). Here, we used the Reactome database to annotate and visualize key genes-involved pathways.
2.8. Predicting regulatory targets of transcription factor
Transcription factor was introduced into the Cytoscape software Version 3.8.2 (Shannon et al., 2003) and the iRegulon plug-in Version 1.3 (Janky et al., 2014) was used to predict the transcription factor targets.
2.9. Differential expression validation of key genes in HF- and stroke-related datasets
Differential expression analysis was performed with the limma package Version 3.38.3 (Ritchie et al., 2015) on two HF-related datasets and two stroke-related datasets. P-value of <0.05 was set as the threshold.
3. Results
3.1. Construction of co-expressed gene modules
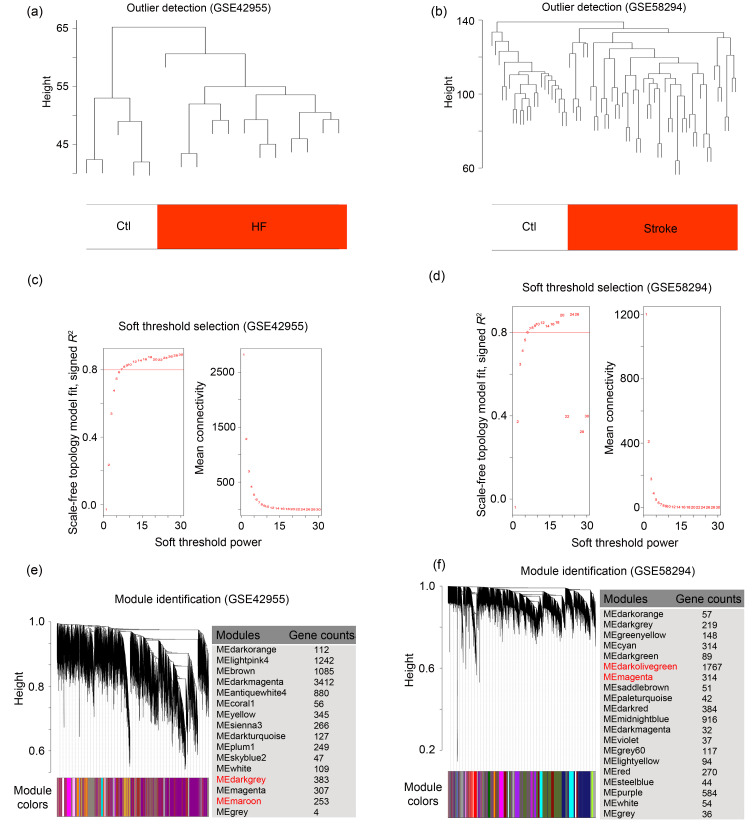
To identify co-expressed gene modules in HF-related and stroke-related datasets, we executed WGCNA. As shown in Figs. 2a and 2b, samples from both datasets were clustered into two clusters, exactly corresponding to the disease group (HF or stroke) and control group, with no outliers detected. Then, based on scale independence of >0.8, 8 and 6 were selected as the minimum β values for GSE42955 and GSE58294 respectively, to ensure biologically significant scale-free topology (Figs. 2c and 2d). As shown in Figs. 2e and 2f, genes in GSE42955 were clustered into 17 modules and genes in GSE58294 were clustered into 20 modules by hierarchical clustering analysis and the Dynamic Branch Cut methods for the gene dendrograms.
Fig. 2. Weighted co-expression network construction in HF- and stroke-related datasets. (a, b) Clustering dendrograms of samples to detect outliers based on their Euclidean distance. One branch represents a sample, and red marks distinguish the groups of samples. (c, d) Analysis of network topology for various soft thresholds (β). The left panel shows the scale-free fit index (scale independence, y-axis) as a function of the soft threshold power (x-axis); the right panel displays the mean connectivity (degree, y-axis) as a function of the soft threshold power (x-axis). (e, f) Gene dendrograms obtained by average linkage hierarchical clustering. The colored row underneath the dendrogram shows the module assignment determined by the Dynamic Tree Cut. Ctl: control; HF: heart failure; ME: module eigengene.
3.2. Identification of disease-related key modules
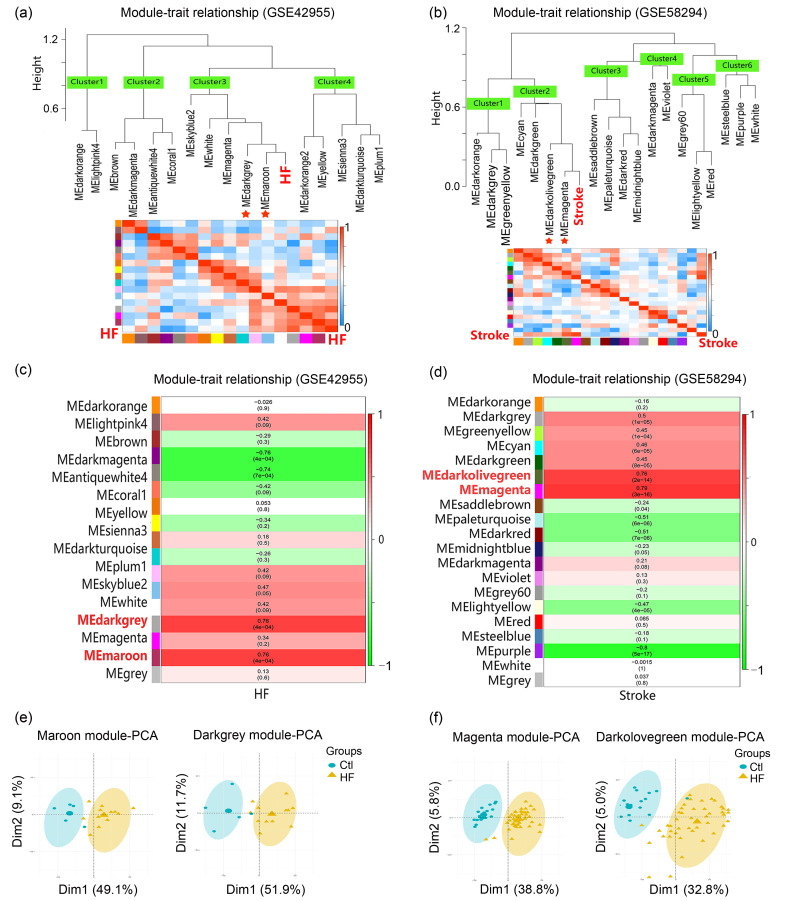
To identify HF- and stroke-related key modules, module eigengene values of modules were correlated with a sample's clinical traits by applying an average linkage hierarchical clustering algorithm and the Spearman correlation coefficient. As shown in Figs. 3a and 3c, hierarchical cluster dendrograms and correlation heatmaps of module eigengene values and clinical traits indicated that the Maroon and Darkgrey modules were highly correlated with HF (Maroon: correlation coefficient=0.76, correlation P-value=4×10-4; Darkgrey: correlation coefficient=0.76, correlation P-value=4×10-4). Similarly, the Magenta and Darkolivegreen modules were highly correlated with stroke (Magenta: correlation coefficient=0.79, correlation P-value=3×10-16; Darkolivegreen: correlation coefficient=0.76, correlation P-value=2×10-14) (Figs. 3b and 3d). Also, PCA results showed satisfactory distinguish ability for the Maroon and Darkgrey modules in response to HF, and for the Magenta and Darkolivegreen modules in response to stroke, indicating that genes in these four modules contributed to disease occurrence (Figs. 3e and 3f). Therefore, we identified the Maroon and Darkgrey modules as HF-related key modules and the Magenta and Darkolivegreen modules as stroke-related key modules.
Fig. 3. Identification of HF-related and stroke-related key modules. (a, b) Hierarchical cluster dendrograms and heatmaps of ME values and clinical traits. The adjacent branches indicate that corresponding modules are highly correlated. Red asterisks indicate HF- and stroke-related key modules. Each row in the heatmap corresponds to an ME (labeled by color), and each column to a clinical trait (HF or stroke). Red represents a positive correlation, and blue represents a negative correlation. (c, d) Module-trait associations. Each row in the heatmap corresponds to an ME, and each column to a clinical trait (HF or stroke). Each cell contains the corresponding correlation and P value. The table is color-coded by correlation according to the color legend. (e, f) PCA of genes within key modules in response to HF or stroke. HF: heart failure; ME: module eigengene; PCA: principal component analysis; Ctl: control; Dim: dimension.
3.3. Functional enrichment analysis of genes in disease-related key modules
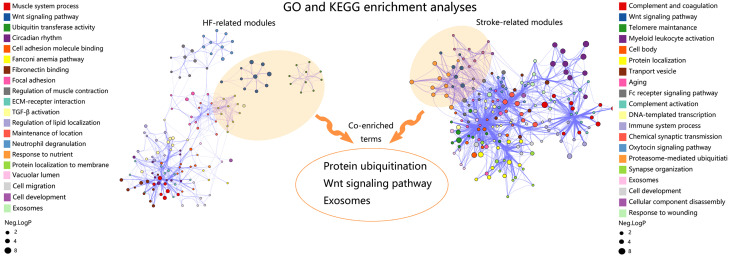
To reveal the underlying molecular biological processes in disease-related key modules, and more importantly, to find the common processes that link HF and CS, we performed GO and KEGG enrichment analyses. The top 20 GO and KEGG terms in HF-related and stroke-related key modules are shown in Fig. 4. In the HF-related key modules, the GO and KEGG terms were enriched in muscle system process, Wnt signaling, ubiquitin transferase activity, circadian rhythm, cell adhesion molecule binding, Fanconi anemia pathway, fibronectin binding, focal adhesion, extracellular matrix (ECM)-recepter interaction, transforming growth factor-β (TGF-β) activity, regulation of lipid localization, maintenance of location, response to nutrient availability, vacuolar lumen, cell migration, and exosomes. In stroke-related key modules, the GO and KEGG terms were enriched in complement and coagulation, Wnt signaling, telomere maintenance, myeloid leukocyte activation, cell body, transport vesicle, aging, Fc receptor signaling pathway, DNA-templated transcription, immune system process, chemical synaptic transmission, oxytocin signaling pathway, proteasome-mediated ubiquitination, synapse organization, exosomes, cellular component disassembly, and response to wounding. All the enriched GO and KEGG terms in HF-related and stroke-related key modules were shown in Table S1. Notably, genes in both HF-related key modules and stroke-related key modules were enriched in protein ubiquitination, Wnt signaling, and exosomes, which could prove to be biological processes for HF-related stroke.
Fig. 4. Identification of co-enriched biological processes of HF-related and stroke-related key modules. The sizes of the dots represent the negative lg(P-value). HF: heart failure; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes.
3.4. Identification of key genes in HF-related stroke
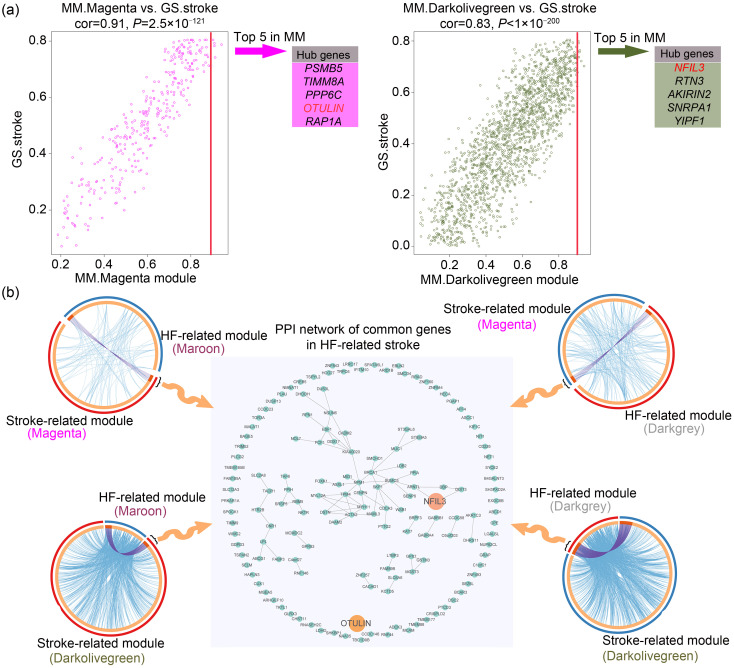
To identify key genes in HF-related stroke, the centrally located intramodular hub genes in stroke-related key modules were identified at first. Module membership summarized the intramodular centrality of each gene. Hence, the top five genes in terms of module membership were selected as hub genes in each stroke-related key module, and they were nuclear factor interleukin 3-regulated (NFIL3), RTN3, AKIRIN2, SNRPA1, YIPF1, PSMB5, TIMM8A, PPP6C, OTU deubiquitinase with linear linkage specificity(OTULIN), and RAP1A. Additionally, gene significance measure summarized the clinical significance of each gene. As shown in Fig. 5a, the high correlation between gene significance and module membership implied that hub genes in the Magenta and Darkolivegreen modules were also highly correlated with CS.
Fig. 5. Identification of key genes in HF-related stroke. (a) Identification of hub genes in stroke-related key modules. The high correlation between GS and MM implies that hub genes in the Magenta and Darkolivegreen modules are highly correlated with cardioembolic stroke. The genes with the top five MM values were selected as hub genes in each stroke-related key module. (b) Identification of common genes of HF-related stroke. The common genes between two HF-related modules and two stroke-related modules are shown in the four circos plots, where the inner circle represents gene lists while the color of the outer circle determines which module the genes in the inner circle belong to, and purple curves link identical genes between two modules. A PPI network constituted by common genes is presented at the center of the diagram. GS: gene significance; MM: module membership; HF: heart failure; PPI: protein‒protein interaction; cor: correlation.
Next, the common genes of HF-related and stroke-related modules were obtained by intersection. As shown in Fig. 5b, a total of 155 common genes were obtained and their PPI were revealed. After further intersecting these common genes with hub genes of stroke-related key modules, OTULIN and NFIL3 were obtained, indicating that the two genes played a more critical role in the pathogenesis of HF-related stroke than other common genes. Therefore, OTULIN and NFIL3 were identified as key genes of HF-related stroke.
3.5. Functional annotation on key genes of HF-related stroke
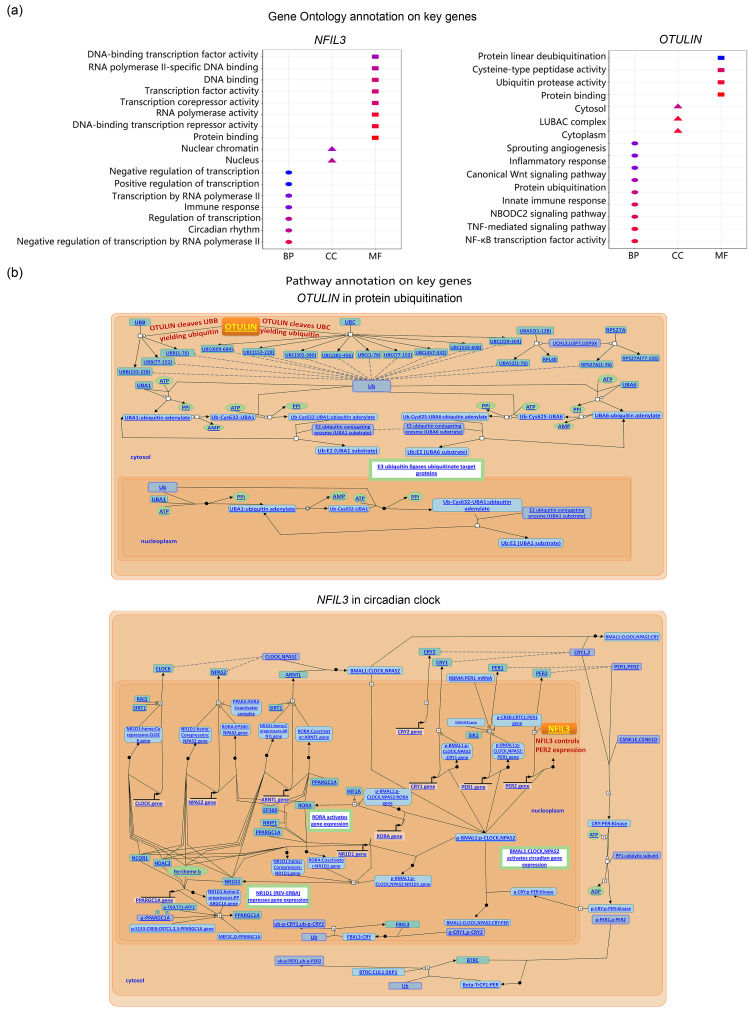
To reveal the functions of OTULIN and NFIL3 in HF-related stroke, we used GO and pathway annotation. As shown in Fig. 6, NFIL3 mainly functioned as a transcription factor in the nucleus. Notably, OTULIN was involved in protein ubiquitination and Wnt signaling in the cytoplasm. Since protein ubiquitination and Wnt signaling were identified above as potential biological processes in HF-related stroke, OTULIN might act in HF-related stroke by participating in these two processes.
Fig. 6. GO and pathway annotation for key genes of HF-related stroke. (a) GO terms involved by key genes. (b) Details of the roles of key genes in their involved pathways. GO: Gene Ontology; HF: heart failure; BP: biological process; CC: cellular component; MF: molecular function.
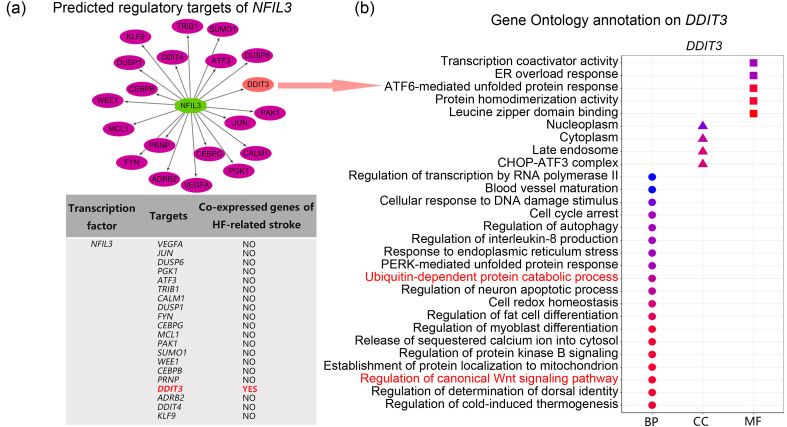
To further understanding how NFIL3 acts in HF-related stroke, we used the iRegulon plug-in (Version 1.3) in Cytoscape (Version 3.8.2) to predict its regulatory targets, and we observed that DDIT3, one of the common genes in HF-related stroke, was a potential target of NFIL3 (Fig. 7a). GO annotation on DDIT3 also revealed that NFIL3 was involved in regulation of protein ubiquitination and the Wnt signaling pathway (Fig. 7b). Thus, we believe that NFIL3 might control protein ubiquitination and Wnt signaling by regulating DDIT3 transcription in HF-related stroke.
Fig. 7. Association between NFIL3 and co-enriched processes. (a) Predicted regulatory targets of NFIL3. (b) GO annotation on DDIT3. GO: Gene Ontology; BP: biological process; CC: cellular component; MF: molecular function.
3.6. Differential expression validation in HF- and stroke-related datasets
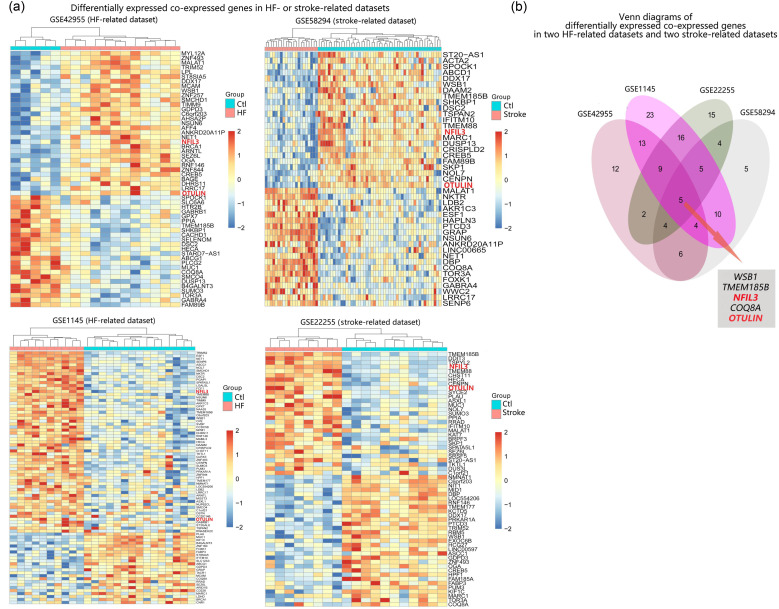
To validate the correlation between key genes and HF-related stroke in terms of fold change in expression, we performed differential expression analysis on the two datasets analyzed above (GSE42955 and GSE58294), and also on another HF-related dataset (GSE1145) and stroke-related dataset (GSE22255). As shown in Fig. 8a, with the threshold of P<0.05, 55 differentially expressed common genes were identified in GSE42955, 43 differentially expressed common genes were identified in GSE58294, 85 differentially expressed common genes were identified in GSE1145, and 60 differentially expressed common genes were identified in GSE22255. By further intersecting the differentially expressed common genes of the four datasets, we found that NFIL3 and OTULIN were among the significantly differentially expressed genes in all HF and stroke groups, in line with the above results showing that OTULIN and NFIL3 played key roles in HF-related stroke (Fig. 8b).
Fig. 8. Differential expression validation in HF-related and stroke-related datasets. (a) Heatmaps of the differential expressed co-expressed genes in HF- or stroke-related datasets. (b) Venn plot of differentially expressed common genes in the four datasets. HF: heart failure; Ctl: control.
4. Discussion
In this study, WGCNA was performed to identify two HF-related and two stroke-related key modules. Functional enrichment analysis of these key modules identified the underlying biological processes in HF-related stroke, including protein ubiquitination, Wnt signaling, and extracellular exosome. In addition, 155 genes were identified as common genes in both HF- and stroke-related key modules, and are thus the top possibilities for genes that play a biological role in HF-related stroke. Most importantly, among these common genes, OTULIN and NFIL3 were identified as the hub genes of stroke-related key modules, indicating their key roles in HF-related stroke. Functional annotation revealed that key genes acted in HF-related stroke by participating in or regulating the biological processes identified above. Finally, NFIL3 and OTULIN were validated to be up-regulated in HF and stroke groups, supporting the importance of OTULIN and NFIL3 in HF-related stroke.
Variations in gene expression and their patterns provide novel insight to the mechanism of HF-related stroke and assist in finding intervention targets. Jickling et al. (2010) found expression differences in HF-specific genes such as MAPK1, GNAQ, and MDM2 in peripheral blood of CS patients, and thus these genes might act in processes such as thrombopoietin signaling, nuclear factor-κB (NF-κB) activation, lymphocyte development, and inflammatory disorder. García-Berrocoso et al. (2020) described several dysregulated genes (e.g., CREM, CYBB, and PTEN) repeatedly identified in CS, which were associated with cardiomyopathy processes like cardiomyocyte apoptosis, ventricular abnormalities, and fibrosis. The current study is the first to perform gene co-expression pattern analysis (WGCNA) for CS. We applied WGCNA, a network-focused rather than individual gene/protein-focused analytic strategy, to evaluate expression similarity among genes under disease conditions and measure correlation between highly co-expressed gene sets (modules) and disease, which is a more effective technique for detecting gene expression with low abundance and variation compared to conventional differential expression analysis, as well as being less prone to information loss (Horvath and Dong, 2008; Pei et al., 2017). Quite a few studies have demonstrated its popularity in systematically identifying critical genes and relevant mechanisms of HF or stroke (Liu et al., 2019; Niu et al., 2019; Pu et al., 2020; Wang et al., 2020).
This study discovered three key biological processes in HF-related stroke by function enrichment analysis of HF-related and stroke-related key modules, including protein ubiquitination, Wnt signaling, and extracellular exosome. This finding indicated that the coagulation-anticoagulation pathway might not be a crucial mechanism for HF-related stroke, explaining the low effectiveness of anticoagulation therapy in stroke prevention for HF patients without AF. A previous study has shown that ubiquitin ligase blunts cardiomyocyte hypertrophy, the "knocking out" of which exacerbates cardiomyocyte hypertrophy in HF models (Barac et al., 2017). In an ischemic stroke model, on the other hand, the ubiquitin E3 ligase tumor necrosis factor receptor-associated factor 6 (TRAF6) aggravated ischemic stroke by ubiquitinating and activating Ras-related C3 botulinum toxin substrate 1 (Rac1) (Li et al., 2017). However, its effect on disease may be inversely depending on the target of ubiquitination because ubiquitination plays a central role in diverse cellular functions, so comprehensive and in-depth research is needed when studying its role in HF-related stroke (Wojcik and di Napoli, 2004; Barac et al., 2017). The Wnt signaling pathway has been proven to participate in cardiac maturation and it is generally believed that cardiac hypertrophy involves reactivation of fetal programs, meaning that Wnt signaling may be reactivated in HF (Blankesteijn et al., 2008). van de Schans et al. (2007) observed that knocking out a key member of Wnt signaling to block signal transduction inhibited the development of cardiac hypertrophy. In a mouse model of ischemic stroke, Wei et al. (2018) found a decrease in Wnt-3a (one of the canonical Wnt signaling ligands) and its downstream second messenger β-catenin, while Wnt-3a supplementation could alleviate stroke damage by its neuroprotective and regenerative actions. Changes in exosomes have been extensively reported in HF, and as one form of cell‒cell communication, exosomes exert various effects on cardiac hypertrophy and fibrosis depending on source cells, contents, and target cells (Bang et al., 2014; Yang, 2018; Ranjan et al., 2019). Exosomes may function throughout time-series processes from stroke occurrence to neurorestorative events after stroke. Stroke onset involves processes such as exosome-mediated thrombosis and neuroinflammation (Chen and Chopp, 2018). The neuroprotective effects of exosomes after stroke include pro-neurogenesis, pro-angiogenesis, and neuronal plasticity (Zhang and Chopp, 2016).
We identified 155 common genes, which might act in HF-related stroke. Among these common genes, OTULIN and NFIL3 were identified as the hub genes of CS, indicating that they played a critical role in HF-related stroke. OTULIN encodes enzymes and specifically recognizes and removes Met1-linked ubiquitin chains from protein substrates, giving it biological significance in angiogenesis, craniofacial and neural development, and regulation of Wnt signaling as well as the NF-κΒ pathway (Rivkin et al., 2013; Stangl et al., 2019). Lentivirus-mediated overexpression of OTULIN attenuates microglia activation, proinflammatory cytokines release, and neuroinflammation by depressing the NF-κB pathway in ischemic stroke models (Xu et al., 2018). OTULIN has not been reported to function in HF, but it is closely related to the immuno-inflammatory pathway and the Wnt signaling pathway, as we found by GO annotation, and both these pathways are known to be involved in HF. NFIL3 is well known as a basic leucine zipper transcription regulator, which activates interleukin 3-induced signaling pathways in T cells and hematopoietic cells, and also works in diverse cell processes such as development and survival of immune cells and circadian rhythm control (Tamai et al., 2014; Velmurugan et al., 2018). Studies have not confirmed its role in HF or stroke. However, a review was devoted to elucidating the potential of NFIL3 as a new therapeutic target for HF due to its impact on several receptor-mediated processes which participate in pathogenesis of HF: calcium signaling, autocrine signaling, and insulin-like growth factor II signaling (Velmurugan et al., 2018). In addition, NFIL3 appears to be neuroprotective owing to its pro-survival and anti-apoptotic effects on neurons (Tamai et al., 2014). Interestingly, we predicted that DDIT3 was a potential regulatory target of NFIL3, because it is reported to be involved in the regulation of protein ubiquitination and Wnt signaling. Therefore, based on these functions of OTULIN and NFIL3, we speculated that OTULIN was likely to act in HF-related stroke as an executor by participating in protein ubiquitination and Wnt signaling, while NFIL3 might control protein ubiquitination and Wnt signaling as a regulator by regulating DDIT3 transcription in HF-related stroke.
HF is a clinically, genetically, and pathophysiologically homogenous clinical entity. HF can be roughly classified into heart failure with reduced ejection fraction (HFrEF) and heart failure with preserved ejection fraction (HFpEF) (Simmonds et al., 2020). Although it had been found that there was no evident correlation between left ventricular ejection fraction (LVEF) and the risk of stroke in HF patients, HFrEF and HFpEF showed significant differences in genetics, pathophysiology, and outcomes (Kotecha et al., 2015; Chung et al., 2020). In HFpEF, genes enriched in mitochondrial adenosine triphosphate synthesis/electron transport are upregulated, which however are downregulated in HFrEF. HFpEF-specific altered genes are enriched in endoplasmic reticulum stress, autophagy, and angiogenesis (Hahn et al., 2021). In addition, HFrEF and HFpEF present with differences in inflammation and endothelial function, cardiomyocyte death and hypertrophy, alterations in the giant spring titin, and fibrosis (Simmonds et al., 2020). Moreover, incidences of death (all-cause, cardiovascular, and sudden), intracerebral hemorrhage, and HF hospitalization increase progressively in HFrEF patients compared with HFpEF patients (di Tullio et al., 2016). The patients included in our study were diagnosed as HFrEF with systolic dysfunction (LVEF of <40%); hence, our findings should be interpreted carefully and whether they can be applied to HFpEF warrants further investigation.
5. Conclusions
Based on our genetic findings, protein ubiquitination, Wnt signaling, and exosomes may be involved in HF-related stroke. A total of 155 common genes are likely to act in the mechanism of HF-related stroke, among which OTULIN and NFIL3 play a more critical role in HF-related stroke by participating in or regulating protein ubiquitination and Wnt signaling; they thus have the potential to become promising intervention targets.
Supplementary information
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81900387), the Guangdong Basic and Applied Basic Research Fund (No. 2019A1515011806), the Fundamental Research Funds for the Central Universities (No. 19ykpy97), and the Science and Technology Program of Guangzhou City of China (No. 201803040010).
Author contributions
Zhaoyu LIU and Jingfeng WANG directed the project. Haifeng ZHANG designed experiments. Chiyu LIU and Sixu CHEN performed experiments and data analysis. Chiyu LIU drafted the manuscript. Yangxin CHEN, Qingyuan GAO, and Zhiteng CHEN perfected the manuscript. All authors have read and approved the final manuscript and, therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines
Chiyu LIU, Sixu CHEN, Haifeng ZHANG, Yangxin CHEN, Qingyuan GAO, Zhiteng CHEN, Zhaoyu LIU and Jingfeng WANG declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Adams HP, Bendixen BH, Kappelle LJ, et al. , 1993. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke, 24(1): 35-41. 10.1161/01.str.24.1.35 [DOI] [PubMed] [Google Scholar]
- Aguilar MI, Hart R, Pearce LA, 2007. Oral anticoagulants versus antiplatelet therapy for preventing stroke in patients with non-valvular atrial fibrillation and no history of stroke or transient ischemic attacks. Cochrane Database Syst Rev, 3: CD006186. 10.1002/14651858.CD006186.pub2 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. , 2000. Gene ontology: tool for the unification of biology. Nat Genet, 25(1): 25-29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang C, Batkai S, Dangwal S, et al. , 2014. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest, 124(5): 2136-2146. 10.1172/jci70577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barac YD, Emrich F, Krutzwakd-Josefson E, et al. , 2017. The ubiquitin-proteasome system: a potential therapeutic target for heart failure. J Heart Lung Transplant, 36(7): 708-714. 10.1016/j.healun.2017.02.012 [DOI] [PubMed] [Google Scholar]
- Blankesteijn WM, van de Schans VAM, ter Horst P, et al. , 2008. The Wnt/frizzled/GSK-3β pathway: a novel therapeutic target for cardiac hypertrophy. Trends Pharmacol Sci, 29(4): 175-180. 10.1016/j.tips.2008.01.003 [DOI] [PubMed] [Google Scholar]
- Bogiatzi C, Hackam DG, McLeod AI, et al. , 2014. Secular trends in ischemic stroke subtypes and stroke risk factors. Stroke, 45(11): 3208-3213. 10.1161/strokeaha.114.006536 [DOI] [PubMed] [Google Scholar]
- Chen JL, Chopp M, 2018. Exosome therapy for stroke. Stroke, 49(5): 1083-1090. 10.1161/strokeaha.117.018292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Kim TH, Uhm JS, et al. , 2020. Stroke and systemic embolism and other adverse outcomes of heart failure with preserved and reduced ejection fraction in patients with atrial fibrillation (from the COmparison Study of Drugs for symptom control and complication prEvention of Atrial Fibrillation [CODE-AF]). Am J Cardiol, 125(1): 68-75. 10.1016/j.amjcard.2019.09.035 [DOI] [PubMed] [Google Scholar]
- Cleland JGF, Findlay I, Jafri S, et al. , 2004. The Warfarin/Aspirin study in heart failure (WASH): a randomized trial comparing antithrombotic strategies for patients with heart failure. Am Heart J, 148(1): 157-164. 10.1016/j.ahj.2004.03.010 [DOI] [PubMed] [Google Scholar]
- Cokkinos DV, Haralabopoulos GC, Kostis JB, et al. , 2006. Efficacy of antithrombotic therapy in chronic heart failure: the HELAS study. Eur J Heart Fail, 8(4): 428-432. 10.1016/j.ejheart.2006.02.012 [DOI] [PubMed] [Google Scholar]
- di Tullio MR, Qian M, Thompson JLP, et al. , 2016. Left ventricular ejection fraction and risk of stroke and cardiac events in heart failure: data from the warfarin versus aspirin in reduced ejection fraction trial. Stroke, 47(8): 2031-2037. 10.1161/strokeaha.116.013679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichgans M, Pulit SL, Rosand J, 2019. Stroke genetics: discovery, biology, and clinical applications. Lancet Neurol, 18(6): 587-599. 10.1016/s1474-4422(19)30043-2 [DOI] [PubMed] [Google Scholar]
- Ding HY, Xie YN, Dong Q, et al. , 2019. Roles of hyaluronan in cardiovascular and nervous system disorders. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 20(5): 428-436. 10.1631/jzus.B1900155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Gong YL, Xia L, et al. , 2019. Simulation of inter atrial block based on a human atrial model. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 20(4): 300-309. 10.1631/jzus.B1800420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Berrocoso T, Palà E, Consegal M, et al. , 2020. Cardioembolic ischemic stroke gene expression fingerprint in blood: a systematic review and verification analysis. Transl Stroke Res, 11(3): 326-336. 10.1007/s12975-019-00730-x [DOI] [PubMed] [Google Scholar]
- GBD 2015 Mortality and Causes of Death Collaborators , 2016. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the global burden of disease study 2015. Lancet, 388(10053): 1459-1544. 10.1016/s0140-6736(16)31012-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R, Carey V, Huber W, et al. , 2021. genefilter: methods for filtering genes from high-throughput experiments. Bioconductor version: Release (3.13). 10.18129/B9.bioc.genefilter [DOI] [Google Scholar]
- Hahn VS, Knutsdottir H, Luo X, et al. , 2021. Myocardial gene expression signatures in human heart failure with preserved ejection fraction. Circulation, 143(2): 120-134. 10.1161/circulationaha.120.050498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Dong J, 2008. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol, 4(8): e1000117. 10.1371/journal.pcbi.1000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, et al. , 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4(2): 249-264. 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- Janky R, Verfaillie A, Imrichová H, et al. , 2014. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol, 10(7): e1003731. 10.1371/journal.pcbi.1003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassal B, Matthews L, Viteri G, et al. , 2020. The reactome pathway knowledgebase. Nucleic Acids Res, 48(D1): D498-D503. 10.1093/nar/gkz1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jickling GC, Xu HC, Stamova B, et al. , 2010. Signatures of cardioembolic and large-vessel ischemic stroke. Ann Neurol, 68(5): 681-692. 10.1002/ana.22187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel H, Healey JS, 2017. Cardioembolic stroke. Circ Res, 120(3): 514-526. 10.1161/circresaha.116.308407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, 2020. KEGG mapper for inferring cellular functions from protein sequences. Protein Sci, 29(1): 28-35. 10.1002/pro.3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Kim J, Park JJ, et al. , 2017. Risk of stroke in congestive heart failure with and without atrial fibrillation. Int J Cardiol, 248: 182-187. 10.1016/j.ijcard.2017.07.056 [DOI] [PubMed] [Google Scholar]
- Kotecha D, Banerjee A, Lip GYH, 2015. Increased stroke risk in atrial fibrillation patients with heart failure: does ejection fraction matter?Stroke, 46(3): 608-609. 10.1161/strokeaha.114.008421 [DOI] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, et al. , 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics, 28(6): 882-883. 10.1093/bioinformatics/bts034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Qin JJ, Yang X, et al. , 2017. The ubiquitin E3 ligase TRAF6 exacerbates ischemic stroke by ubiquitinating and activating Rac1. J Neurosci, 37(50): 12123-12140. 10.1523/jneurosci.1751-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZH, Ma CG, Gu JH, et al. , 2019. Potential biomarkers of acute myocardial infarction based on weighted gene co-expression network analysis. BioMed Eng OnLine, 18: 9. 10.1186/s12938-019-0625-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik R, Dichgans M, 2018. Challenges and opportunities in stroke genetics. Cardiovasc Res, 114(9): 1226-1240. 10.1093/cvr/cvy068 [DOI] [PubMed] [Google Scholar]
- Malik R, Chauhan G, Traylor M, et al. , 2018. Multiancestry genome-wide association study of 52 0000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet, 50(4): 524-537. 10.1038/s41588-018-0058-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massie BM, Collins JF, Ammon SE, et al. , 2009. Randomized trial of warfarin, aspirin, and clopidogrel in patients with chronic heart failure: the Warfarin and Antiplatelet Therapy in Chronic Heart Failure (WATCH) trial. Circulation, 119(12): 1616-1624. 10.1161/circulationaha.108.801753 [DOI] [PubMed] [Google Scholar]
- Mischie AN, Chioncel V, Droc I, et al. , 2013. Anticoagulation in patients with dilated cardiomyopathy, low ejection fraction, and sinus rhythm: back to the drawing board. Cardiovasc Ther, 31(5): 298-302. 10.1111/1755-5922.12019 [DOI] [PubMed] [Google Scholar]
- Murtagh B, Smalling RW, 2006. Cardioembolic stroke. Curr Atheroscler Rep, 8(4): 310-316. 10.1007/s11883-006-0009-9 [DOI] [PubMed] [Google Scholar]
- Niu XW, Zhang JJ, Zhang LL, et al. , 2019. Weighted gene co-expression network analysis identifies critical genes in the development of heart failure after acute myocardial infarction. Front Genet, 10: 1214. 10.3389/fgene.2019.01214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei G, Chen L, Zhang W, 2017. WGCNA application to proteomic and metabolomic data analysis. Methods Enzymol, 585: 135-158. 10.1016/bs.mie.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Pu LY, Wang M, Li KX, et al. , 2020. Identification micro-RNAs functional modules and genes of ischemic stroke based on weighted gene co-expression network analysis (WGCNA). Genomics, 112(4): 2748-2754. 10.1016/j.ygeno.2020.03.011 [DOI] [PubMed] [Google Scholar]
- Pullicino P, Homma S, 2010. Stroke in heart failure: atrial fibrillation revisited?J Stroke Cerebrovasc Dis, 19(1): 1-2. 10.1016/j.jstrokecerebrovasdis.2009.09.002 [DOI] [PubMed] [Google Scholar]
- Raghow R, 2016. An ‘omics’ perspective on cardiomyopathies and heart failure. Trends Mol Med, 22(9): 813-827. 10.1016/j.molmed.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Ranjan P, Kumari R, Verma SK, 2019. Cardiac fibroblasts and cardiac fibrosis: precise role of exosomes. Front Cell Dev Biol, 7: 318. 10.3389/fcell.2019.00318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, et al. , 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res, 43(7): e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivkin E, Almeida SM, Ceccarelli DF, et al. , 2013. The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature, 498(7454): 318-324. 10.1038/nature12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoner A, Tyrrell C, Wu M, et al. , 2015. Endocardial endothelial dysfunction progressively disrupts initially anti then pro-thrombotic pathways in heart failure mice. PLoS ONE, 10(11): e0142940. 10.1371/journal.pone.0142940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, et al. , 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 13(11): 2498-2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds SJ, Cuijpers I, Heymans S, et al. , 2020. Cellular and molecular differences between HFpEF and HFrEF: a step ahead in an improved pathological understanding. Cells, 9(1): 242. 10.3390/cells9010242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangl A, Elliott PR, Pinto-Fernandez A, et al. , 2019. Regulation of the endosomal SNX27-retromer by OTULIN. Nat Commun, 10: 4320. 10.1038/s41467-019-12309-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struijs JN, van Genugten MLL, SMAA Evers, et al. , 2006. Future costs of stroke in the Netherlands: the impact of stroke services. Int J Technol Assess Health Care, 22(4): 518-524. 10.1017/s0266462306051464 [DOI] [PubMed] [Google Scholar]
- Tamai SI, Imaizumi K, Kurabayashi N, et al. , 2014. Neuroprotective role of the basic leucine zipper transcription factor NFIL3 in models of amyotrophic lateral sclerosis. J Biol Chem, 289(3): 1629-1638. 10.1074/jbc.M113.524389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium , 2019. The Gene Ontology Resource: 20 years and still going strong. Nucleic Acids Res, 47(D1): D330-D338. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas I, EncisoSilva J, Schlueter M, et al. , 2016. Anticoagulation therapy and NOACs in heart failure. In: Bauersachs J, Butler J, Sandner P (Eds.), Heart Failure. Handbook of Experimental Pharmacology, Vol. 243. Springer, Cham, p.515-535. 10.1007/164_2016_126 [DOI] [PubMed] [Google Scholar]
- van de Schans VAM, van den Borne SWM, Strzelecka AE, et al. , 2007. Interruption of Wnt signaling attenuates the onset of pressure overload-induced cardiac hypertrophy. Hypertension, 49(3): 473-480. 10.1161/01.Hyp.0000255946.55091.24 [DOI] [PubMed] [Google Scholar]
- Velmurugan BK, Chang RL, Marthandam Asokan S, et al. , 2018. A minireview of E4BP4/NFIL3 in heart failure. J Cell Physiol, 233(11): 8458-8466. 10.1002/jcp.26790 [DOI] [PubMed] [Google Scholar]
- Wang M, Wang LJ, Pu LY, et al. , 2020. LncRNAs related key pathways and genes in ischemic stroke by weighted gene co-expression network analysis (WGCNA). Genomics, 112(3): 2302-2308. 10.1016/j.ygeno.2020.01.001 [DOI] [PubMed] [Google Scholar]
- Wei ZZ, Zhang JY, Taylor TM, et al. , 2018. Neuroprotective and regenerative roles of intranasal Wnt-3a administration after focal ischemic stroke in mice. J Cereb Blood Flow Metab, 38(3): 404-421. 10.1177/0271678x17702669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik C, di Napoli M, 2004. Ubiquitin-proteasome system and proteasome inhibition: new strategies in stroke therapy. Stroke, 35(6): 1506-1518. 10.1161/01.STR.0000126891.93919.4e [DOI] [PubMed] [Google Scholar]
- Xu HB, Qin WY, Hu X, et al. , 2018. Lentivirus-mediated overexpression of OTULIN ameliorates microglia activation and neuroinflammation by depressing the activation of the NF-κB signaling pathway in cerebral ischemia/reperfusion rats. J Neuroinflammation, 15: 83. 10.1186/s12974-018-1117-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PC, 2018. Induced pluripotent stem cell (IPSC)-derived exosomes for precision medicine in heart failure. Circ Res, 122(5): 661-663. 10.1161/circresaha.118.312657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip AM, Horvath S, 2007. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics, 8: 22. 10.1186/1471-2105-8-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZG, Chopp M, 2016. Exosomes in stroke pathogenesis and therapy. J Clin Invest, 126(4): 1190-1197. 10.1172/jci81133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LY, Wang Y, Wang K, et al. , 2019. Potential therapeutic drugs for ischemic stroke based on bioinformatics analysis. Int J Neurosci, 129(11): 1098-1102. 10.1080/00207454.2019.1634072 [DOI] [PubMed] [Google Scholar]
- Zhou YY, Zhou B, Pache L, et al. , 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun, 10: 1523. 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu WH, Nan YN, Wang SQ, et al. , 2019. Bioinformatics analysis of gene expression profiles of sex differences in ischemic stroke. Biomed Res Int, 2019: 2478453. 10.1155/2019/2478453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou RJ, Zhang DE, Lv L, et al. , 2019. Bioinformatic gene analysis for potential biomarkers and therapeutic targets of atrial fibrillation-related stroke. J Transl Med, 17: 45. 10.1186/s12967-019-1790-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.