Abstract
The Myt1 protein kinase functions to negatively regulate Cdc2-cyclin B complexes by phosphorylating Cdc2 on threonine 14 and tyrosine 15. Throughout interphase, human Myt1 localizes to the endoplasmic reticulum and Golgi complex, whereas Cdc2-cyclin B1 complexes shuttle between the nucleus and the cytoplasm. Here we report that overproduction of either kinase-active or kinase-inactive forms of Myt1 blocked the nuclear-cytoplasmic shuttling of cyclin B1 and caused cells to delay in the G2 phase of the cell cycle. The COOH-terminal 63 amino acids of Myt1 were identified as a Cdc2-cyclin B1 interaction domain. Myt1 mutants lacking this domain no longer bound cyclin B1 and did not efficiently phosphorylate Cdc2-cyclin B1 complexes in vitro. In addition, cells overproducing mutant forms of Myt1 lacking the interaction domain exhibited normal trafficking of cyclin B1 and unperturbed cell cycle progression. These results suggest that the docking of Cdc2-cyclin B1 complexes to the COOH terminus of Myt1 facilitates the phosphorylation of Cdc2 by Myt1 and that overproduction of Myt1 perturbs cell cycle progression by sequestering Cdc2-cyclin B1 complexes in the cytoplasm.
The underlying goal of the cell division process is to accurately replicate the genetic material once during S phase and to ensure that identical chromosomal copies are segregated equally to the two daughter cells during mitosis. Cyclin-dependent protein kinases (Cdks) are key regulators of the eukaryotic cell division cycle. The Cdc2 kinase (Cdk1) is required for the onset of mitosis, and its activity is subject to multiple levels of regulation, including periodic association with the B-type cyclins and reversible phosphorylation (for reviews, see references 2 and 44). In higher eukaryotic organisms, phosphorylation of Cdc2 occurs on three regulatory sites, i.e., threonine 14, tyrosine 15, and threonine 161 (12, 13, 31, 45), whereas in fission yeast, Tyr 15 and Thr 167 are the major sites of phosphorylation (18, 19). Cdc2 is retained in an inactive state throughout the S and G2 phases of the cell cycle by Thr 14 and Tyr 15 phosphorylation (19, 31, 40, 48, 52, 62). In late G2, the Cdc25C phosphatase dephosphorylates Cdc2 on both Thr 14 and Tyr 15, which activates its kinase activity and promotes entry into mitosis (14, 17, 37, 61, 63).
In fission yeast, Tyr 15 phosphorylation is catalyzed by wee1+ and mik1+, which encode protein tyrosine kinases (15, 36, 41, 51, 52, 57). The human homologue of wee1+ encodes a tyrosine-specific protein kinase that phosphorylates Cdc2 exclusively on Tyr 15 (25, 26, 42, 43, 53, 54, 65). A second kinase, designated Myt1, has been identified in Xenopus and in humans (3, 9, 30, 40, 46). Myt1 is a dual-specificity protein kinase that phosphorylates Cdc2 on both Thr 14 and Tyr 15. In lysates prepared from Xenopus eggs, Myt1 accounts for the majority of the Thr 14 kinase activity (46). Both the phosphorylation and activity of Myt1 are cell cycle regulated. Activity is partially reduced during mitosis, and reduction in Myt1 kinase activity correlates with increased phosphorylation of Myt1 protein (9, 46). Two protein kinases have been shown to phosphorylate Myt1 in vitro. The ribosomal S6 protein kinase (p90rsk) phosphorylates Xenopus Myt1 (XeMyt1), thereby reducing the kinase activity of XeMyt1 (50). Human Myt1 is a substrate of Cdc2 in vitro, but Cdc2 phosphorylation does not appear to alter the enzymatic activity of Myt1 (9).
Although Myt1 and Wee1 have sequence similarities, the two kinases differ in several important ways. Wee1 is capable of phosphorylating Cdk2 complexed with either cyclin A or E in vitro (65). In contrast, Myt1 fails to recognize these complexes as substrates in vitro (9). Furthermore, human Myt1 is localized to the endoplasmic reticulum and Golgi complex (40), whereas Wee1 localizes to the nucleus (6, 43). These differences suggest that Wee1 and Myt1 may have distinct functions in regulating the cell cycle.
Previous studies reported that in interphase Cdc2-cyclin B1 complexes localize to the cytoplasm, whereas in prophase, just prior to nuclear envelope breakdown, Cdc2-cyclin B1 complexes translocate into the nucleus (5, 16, 49, 55, 56). Recent studies have demonstrated that throughout interphase, Cdc2-cyclin B1 complexes shuttle between the nucleus and the cytoplasm (21, 64, 66). The apparent cytoplasmic localization of Cdc2-cyclin B1 complexes is due to a nuclear export sequence in cyclin B1 which facilitates rapid export of Cdc2-cyclin B1 complexes from the nucleus. Phosphorylation of the nuclear export sequence in late prophase is proposed to block the nuclear export of cyclin B1 by interfering with the binding of the nuclear export receptor CRM1, leading to the nuclear accumulation of Cdc2-cyclin B1 complexes (39, 66). Cells engineered to express an active form of Cdc2 where Thr 14 and Tyr 15 are replaced by amino acids that cannot be phosphorylated (Cdc2AF) display only minor mitotic perturbations (8, 23, 28, 31). In contrast, coexpression of Cdc2AF with cyclin B1 fused to a nuclear localization sequence induces premature entry into mitosis in a significant number of cells (29). Thus, the initiation of mitosis in mammalian cells requires not only Cdc2 dephosphorylation but also the proper subcellular localization of the activated kinase.
Here we report that the overproduction of the human Myt1 protein kinase disrupts the intracellular trafficking of Cdc2-cyclin B1 and as a consequence cells delay in the G2 phase of the cell cycle. We have identified a Cdc2-cyclin B1 interaction domain in the COOH terminus of Myt1, and the integrity of this domain is required for overproduced Myt1 to sequester Cdc2-cyclin B1 complexes in the cytoplasm and to perturb cell cycle progression.
MATERIALS AND METHODS
Antibodies used for Western blotting.
Myt1 was detected with a c-Myc polyclonal antibody (A-14; Santa Cruz Biotechnology) or an affinity-purified peptide antibody (40). Cdc2 was detected with a monoclonal antibody (Cdc2 p34 [17]; Santa Cruz Biotechnology), and cyclin B1 was detected with an antibody raised against the C-terminal 12 amino acids of human cyclin B1. Secondary antibodies included horseradish peroxidase (HRP)–goat anti-mouse antibody (Cappel), HRP–goat anti-rabbit antibody (Zymed), and HRP-protein A (Amersham Life Science). Antibodies used for immunoprecipitation include anti-c-Myc (9E10) agarose conjugate (Santa Cruz Biotechnology) and anti-cyclin B1 polyclonal antibody (4).
Construction of plasmids.
pFASTBACHisMyt1 was made by subcloning the XbaI/XhoI fragment of pET15bMyt1 (40) into the XbaI and XhoI sites of the pFASTBAC baculovirus shuttle vector (Gibco-BRL). To make pFASTBACHisMyt1ΔC63, Myt1 cDNA clone 6-1 (40) was amplified by PCR with primers 5′-CCGGTACCTACCCTAGGCTGTCGTCAT-3′ and 5′-CTGGCCCATCTGCACAGC-3′. The PCR product was then digested with XmaI and KpnI and cloned into pFASTBACHisMyt1 that had been digested with the same enzymes. Kinase-inactive Myt1 was created by replacing Asn 238 (AAC) with Ala (GCC) by PCR mutagenesis. Specifically, cDNA clone 6-1 was amplified by PCR with primers 5′-CCTACTTCCGCCACGCAG-3′ and 5′-GGGGCCGGAGGAAGATGGCGGCAGGCTTGA-3′ (the underlined letters indicate a mutated codon). The PCR product was then digested with NcoI and cloned into pFASTBACHisMyt1 that had been digested with NcoI and SmaI. A Bsu36I-digested fragment was then isolated from the resulting construct and cloned into the Bsu36I site of pCDNAmycMyt1 to generate pCDNAmycMyt1N238A. To make pCDNAmycMyt1ΔC63, an AvrII/BamHI fragment containing the Myt1-coding sequence was isolated from pCDNAmycMyt1 and cloned into BamHI- and EcoRV-digested pCDNA3. An NheI linker (5′-CTAGCTAGCTAG-3′) was then inserted into the NotI site of the resulting plasmid to provide a stop codon immediately after the Myt1 open reading frame. To make pCDNAmycMyt1(3A), cDNA clone 6-1 was PCR amplified with primers 5′-CTGGCCCATCTGCACAGC-3′ and 5′-AACTCAGGTTGGGTCTAGGGTGTCCTCAAACAGGCTGAGGGCGGCGGCAGGCTCAAAGAAGGGGAA-3′ (the underlined sequence indicates the RNL-to-AAA mutations). The PCR product was then digested with SacII and SphI, and the 420-bp SacII/HpaI fragment was cloned into pCDNAmycMyt1 that had been digested with SacII and HpaI. Adenovirus shuttle vector pACCMVmycMyt1 was constructed by cloning the KpnI/XbaI fragment containing the myc-Myt1-coding region from pCDNAmyc-Myt1 into KpnI- and XbaI-digested pACCMV. pACCMVmycMyt1N238A was made in a similar way except that the insert was derived from pCDNAmycMyt1N238A. To generate adenoviruses with the pAdEasy system (22), pSmycMyt1ΔC63 and pSmycMyt1N238AΔC63 were constructed by subcloning the KpnI/XhoI fragments of pCDNAmycMyt1ΔC63 and pCDNAmycMyt1N238AΔC63 (40), respectively, into the KpnI and XhoI sites of the shuttle vector pShuttle-CMV.
Generation of recombinant viruses.
Recombinant baculoviruses were generated by using the BAC-TO-BAC Baculovirus Expression System (Gibco-BRL) and protocols suggested by the manufacturer. Adenoviruses expressing Myt1 and kinase-inactive Myt1 (Myt1N238A) were generated and propagated essentially as described previously (7). Specifically, 10 μg of pACCMVmycMyt1 or pACCMVmycMyt1N238A was cotransfected with 10 μg of pJM17 into low-passage 293 cells by the calcium phosphate method. Initial cell lysis was observed 3 to 4 weeks after transfection. Viral DNA was extracted from a fraction of viral particles to confirm the insertion of the Myt1-coding sequence by PCR or Southern blotting. Single plaques were then picked from an agarose overlay plate and used to infect 293 cells for two additional rounds to obtain a P2 lysate. For large-scale purification of recombinant adenoviruses, P2 lysates were used to infect 10 150-mm-diameter tissue culture plates of 293 cells. At 72 h after infection, the culture medium and cells were collected, and cells were lysed with 0.5% Nonidet P-40 (NP-40). Viral particles were precipitated with 10% polyethylene glycol 8000–1.25 M NaCl and subsequently purified by CsCl density gradient centrifugation. Purified virus was then dialyzed in 2 liters of phosphate-buffered saline (PBS) for 4 h at room temperature and stored in aliquots at −20°C in PBS containing 10 mM Tris (pH 8.0), 0.1% bovine serum albumin (BSA), and 50% glycerol. Viral titers were determined by using a PFU assay. Adenoviruses expressing mycMyt1ΔC63 and mycMyt1N238AΔC63 were generated by a recently published procedure (22). The pShuttle-CMV-based plasmids encoding Myt1 proteins and pAdEasy were cotransformed into Escherichia coli BJ5183 to achieve homologous recombination.
Cell synchronization and adenovirus infection.
HeLa cells were routinely grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 2 mM l-glutamine. To synchronize HeLa cells at the G1/S border, asynchronously growing cells were treated with 2 mM thymidine for 16 h. Cells were then released from the block by switching to complete growth medium containing 24 μM each thymidine and deoxycytidine. After 8 h, thymidine was added to the medium to a final concentration of 2 mM, and cells were cultured for another 16 h. The cells were then rinsed twice with PBS and cultured in complete growth medium. Samples were harvested at various times after the release. Synchronization of HeLa cells in mitosis was achieved by incubating asynchronously growing cells in medium containing 0.15 μg of nocodazole per ml for 18 h.
To study the effects of overexpression of Myt1 on cell cycle progression, HeLa cells were synchronized at the G1/S border by a double thymidine block-and-release protocol as described above. Three hours before release from the block, approximately 8 × 105 cells were infected for 45 to 60 min with adenoviruses encoding various Myt1 proteins at a multiplicity of infection of 50 in 0.5 ml of serum-free DMEM. Thymidine (2 mM) was included in the infection medium in order to maintain the G1/S synchronization. At the end of the infection, 4 ml of complete medium containing 2 mM thymidine was added, and cells were incubated for an additional 2 h. The cells were then washed twice with serum-free DMEM and cultured in complete growth medium. Cells were harvested at the desired times by trypsinization. Approximately one-third of the cells were lysed in mammalian cell lysis buffer (MCLB) (50 mM Tris [pH 8.0], 100 mM NaCl, 5 mM EDTA, and 0.5% NP-40) supplemented with 1 mM dithiothreitol, protease inhibitors (2 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin per ml, 20 μM leupeptin, and 5 μg of pepstatin per ml), and phosphatase inhibitors (1 mM Na3VO4 and 1 μM microcystin). One hundred micrograms of lysate was resolved on a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel and analyzed by Western blotting. The remaining cells were fixed in 90% ethanol and stained with 30 μg of propidium iodide per ml in PBS containing 1% BSA and 0.25 mg of RNase A per ml. Cell cycle profiles were determined by flow cytometry with a Becton-Dickinson FACScan, and data was analyzed with CELL QUEST software. In cases where indirect immunofluorescence staining was performed, adenovirus infections were done 2 h prior to release from the thymidine block.
For Myt1-Cdc2 coimmunoprecipitations, cells were harvested at 14 h following release from the synchrony-and-infection protocol described above. Cells were lysed in MCLB, and 2.5 mg of protein was incubated with 7.5 μl of Myc-agarose at 4°C for 3 h. The beads were then pelleted and washed three times with MCLB. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer was added, and the bead-bound proteins were resolved on an SDS–12% polyacrylamide gel. Additionally, 100 μg of lysate was resolved by SDS-PAGE to examine total cellular Cdc2. The proteins were transferred to nitrocellulose, and Cdc2 was visualized by immunoblotting.
Mitotic index measurements.
HeLa cells were subjected to G1/S synchronization and adenovirus infection as described above. Upon release of cells from the final thymidine block, 0.15 μg of nocodazole per ml was added to the medium to trap mitotic cells. Cells were harvested by trypsinization, washed once with PBS, and then resuspended in 75 mM KCl for 10 min (11). After centrifugation to remove the KCl, cells were treated with fixative (acetic acid-methanol [1:3, vol/vol]). The cells were then resuspended in fixative, spread onto slides, and allowed to air dry. Dried cells were then stained with 1 μg of DAPI (4′,6-diamidino-2-phenylindole) per ml for 5 min and then mounted and observed by fluorescence microscopy. A minimum of 600 nuclei were counted for each sample.
Indirect immunofluorescence.
HeLa cells infected with control (β-galactosidase [β-GAL]-encoding) or Myt1-encoding adenoviruses were fixed with 2% paraformaldehyde in PBS and then permeabilized in 2% Triton X-100 in PBS. Cells were incubated with 2% BSA to block nonspecific binding before being incubated with antibodies specific for cyclin B1 and/or Myc (for Myt1). Cyclin B1 monoclonal antibody (GNS1; Santa Cruz) was used at a 1:250 dilution, and the c-Myc polyclonal antibody (A-14; Santa Cruz) was used at a 1:1,000 dilution. Secondary antibodies included fluorescein isothiocyanate-conjugated goat anti-rabbit antibody (Cappel) and indocarbocyanine-conjugated donkey anti-mouse antibody (Jackson ImmunoResearch). Secondary antibodies were used at a 1:2,000 dilution. Both primary and secondary antibodies were diluted in PBS containing 2% BSA. Cellular DNA was stained with 0.1 μg of DAPI per ml for 2 min after secondary antibody incubation. Cells were observed with a conventional fluorescence microscope (model BX60; Olympus). In experiments where nuclear export of cyclin B was studied, cells were treated with 20 nM leptomycin B (LMB) for 2.5 h before fixation.
Transient transfections.
Transient transfections of HeLa cells were performed with SuperFect (Qiagen) according to the manufacturer’s protocol. Briefly, SuperFect-DNA complexes were prepared by mixing 5 μg of plasmid DNA with 30 μl of SuperFect in 150 μl of OptiMEM reduced-serum medium. HeLa cells grown on p60 tissue culture plates were then incubated with the SuperFect-DNA complexes in 1 ml of OptiMEM at 37°C for 2.5 h. At the end of the incubation, transfection medium was replaced with complete DMEM and cells were incubated at 37°C. Cells were typically harvested 24 h after the start of the transfection.
Cyclin B1-Myt1 coimmunoprecipitation.
Asynchronously growing HeLa cells and HeLa cells arrested in mitosis with nocodazole were lysed in MCLB. One microliter of cyclin B1 polyclonal antiserum was added to 1.5 mg each of asynchronous and mitotic cell lysates. The reaction mixtures were rocked for 2 h at 4°C. Protein A-Sepharose beads (20 μl) were then added, and the incubation was continued for an additional hour. Following precipitation, the beads were washed three times with MCLB. SDS-PAGE sample buffer was added to the reaction mixtures and to 150 μg of total cell lysate from asynchronous or M-phase cells. All proteins were resolved on an SDS–10% polyacrylamide gel. After transfer to nitrocellulose, cyclin B1 was detected by immunoblotting. This blot was stripped and reprobed to visualize coprecipitating Myt1.
Myt1 kinase assays.
Sf9 insect cells expressing HisMyt1 or HisMyt1ΔC63 were lysed with HIS lysis buffer (10 mM Tris [pH 7.5], 5 mM EGTA, 150 mM NaCl, 0.5% NP-40, 2 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin per ml, 20 μM leupeptin, 5 μg of pepstatin per ml). Recombinant Myt1 proteins were bound to 20 μl of Ni-nitrilotriacetic acid beads (Qiagen) by incubating the beads with lysate for 40 min at 4°C. The beads were washed twice with HIS lysis buffer followed by three times with incomplete kinase buffer (50 mM Tris [pH 8.0], 10 mM MgCl2). Kinase assays were then performed for 20 min at 30°C by incubating the beads in incomplete kinase buffer supplemented with 50 μM ATP, 10 μCi of [γ-32P]ATP, 1 mM dithiothreitol, and 0.5 μg of Cdc2 (K33R)-cyclin B1 complex that had been purified from Sf9 insect cells (40). Approximately 0.1 μg of HisMyt1 protein bound to beads was used in the kinase assay. Reactions were stopped by the addition of SDS-PAGE loading buffer, and proteins were resolved on an SDS–10% polyacrylamide gel.
RESULTS
Overproduction of kinase-active and -inactive forms of Myt1 perturbs cell cycle progression.
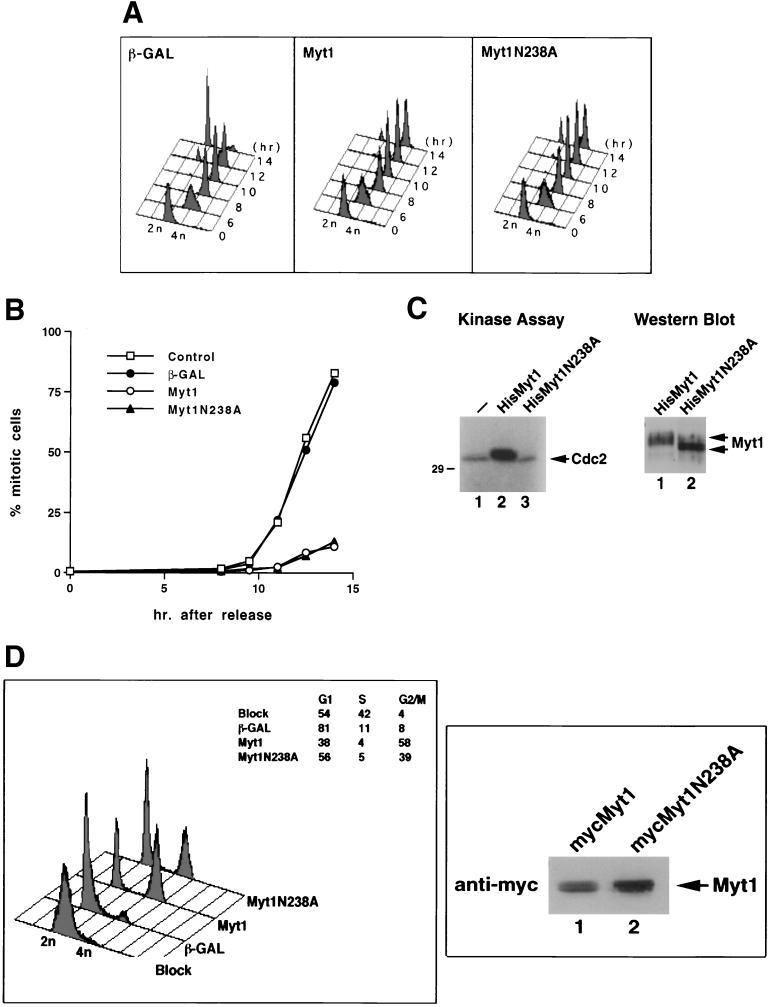
Human Myt1 phosphorylates Cdc2 on threonine 14 and tyrosine 15, thereby maintaining Cdc2-cyclin B complexes in an inactive state throughout interphase (40). Because Myt1 functions as a negative regulator of mitosis, overproduction of Myt1 might be expected to delay entry into mitosis. To test this prediction, recombinant adenovirus encoding human Myt1 was used to infect a population of HeLa cells that were synchronized at the G1/S border by a double thymidine block-and-release protocol (3). Flow cytometry was used to monitor the ability of cells to traverse the cell cycle after release from the block (Fig. 1A). Cells infected with a control adenovirus encoding β-GAL proceeded through the S, G2, and M phases of the cell cycle normally. By 14 h after the release, the vast majority of cells were in the G1 phase of the cell cycle, indicating normal progression through mitosis. In contrast, cells expressing wild-type Myt1 retained a 4N DNA content indicative of either a G2- or M-phase arrest.
FIG. 1.
Overexpression of Myt1 causes a G2 cell cycle delay. (A) HeLa cells synchronized at the G1/S border by a double thymidine block protocol were infected with recombinant adenoviruses encoding either β-GAL or the kinase-active (Myt1) or -inactive (Myt1N238A) form of Myt1 tagged with a Myc epitope. Cells were harvested at various times after release from the block, and the cellular DNA content was analyzed by flow cytometry. (B) Mitotic index measurements were made to assess whether cells were arresting in the G2 or M phase of the cell cycle. Cells were synchronized and infected with adenoviruses as described above. Nocodazole was added to trap cells in mitosis. At various times after release from the block, mitotic chromosome spreads were prepared and scored. At least 600 nuclei were counted in each experiment. (C) Kinase-active Myt1 (HisMyt1) or kinase-inactive Myt1 (HisMyt1N238A) was purified from Sf9 insect cells as a hexahistidine-tagged fusion protein. Glutathione S-transferase–cyclin B1-Cdc2 (K33R) complexes were also purified from insect cells as described previously (40). Kinase reactions were performed in vitro in the presence of substrate alone (left panel, lane 1) or substrate in the presence of either kinase-active (left panel, lane 2) or -inactive (left panel, lane 3) Myt1. Reaction mixtures were resolved by SDS-PAGE and subjected to autoradiography. Levels of kinase-active (right panel, lane 1) and kinase-inactive (right panel, lane 2) Myt1 in the reaction mixtures were assessed by Western blotting. (D) HeLa cells synchronized by a double thymidine block protocol were infected with recombinant adenoviruses encoding either β-GAL or the kinase-active (Myt1) or -inactive (Myt1N238A) form of Myt1 tagged with a Myc epitope. Cells were harvested at 14 h after release from the block, and the cellular DNA content was analyzed by flow cytometry (left panel). Levels of Myt1 and Myt1N238A were determined by immunoblotting (right panel).
Mitotic index measurements were made to determine whether cells overproducing Myt1 were arresting in the G2 or M phase of the cell cycle (Fig. 1B). There was little evidence of chromosome condensation or nuclear membrane disruption in Myt1-overproducing cells, even after treatment with the microtubule-disrupting agent nocodazole to stabilize any mitosis that may have occurred. These results demonstrate that overproduction of kinase-active Myt1 delayed cells in the G2 phase of the cell cycle with a nuclear DNA content of 4N.
Similar experiments were performed with a recombinant adenovirus encoding a kinase-inactive form of Myt1. Replacement of arginine at position 238 with alanine generated a kinase-inactive form of Myt1 as evidenced by the inability of this mutant to phosphorylate Cdc2 in vitro (Fig. 1C). Overproduction of kinase-inactive Myt1 (Myt1N238A) might be predicted to inhibit endogenous Myt1 function and thereby cause premature entry into mitosis (32). Alternatively, no discernible phenotype might be expected if the Wee1 tyrosine kinase was able to compensate under these circumstances. Surprisingly, overproduction of kinase-inactive Myt1 caused a G2 delay just as was observed upon overproduction of wild-type Myt1 (Fig. 1A and B).
Experiments were performed to determine whether the kinase-inactive mutant of Myt1 was as effective as wild-type Myt1 in inducing a G2 cell cycle delay (Fig. 1D). HeLa cells that were synchronized by a double thymidine block-and-release protocol were infected with recombinant adenoviruses under conditions where kinase-inactive Myt1 would be produced at higher levels than kinase-active Myt1 (Fig. 1D, right panel). Cells were harvested at 14 h after the release from the block, and flow cytometry was used to monitor the ability of cells to traverse the cell cycle (Fig. 1D, left panel). By 14 h after the release, 81% of cells that had been infected with the β-GAL-encoding virus were already in the G1 phase of the cell cycle, with only 8% remaining in G2/M. In contrast, cells expressing either wild-type Myt1 or kinase-inactive Myt1 exhibited a considerable G2 cell cycle delay; 58% of cells expressing wild-type Myt1 and 39% of cells expressing kinase-inactive Myt1 (Myt1N238A) were still in the G2 phase of the cell cycle at the 14-h time point. Given that kinase-inactive Myt1 was produced to higher levels than wild-type Myt1, these results indicate that wild-type Myt1 was more potent than kinase-inactive Myt1 at delaying cell cycle progression.
Interactions between Myt1 and Cdc2-cyclin B1 complexes.
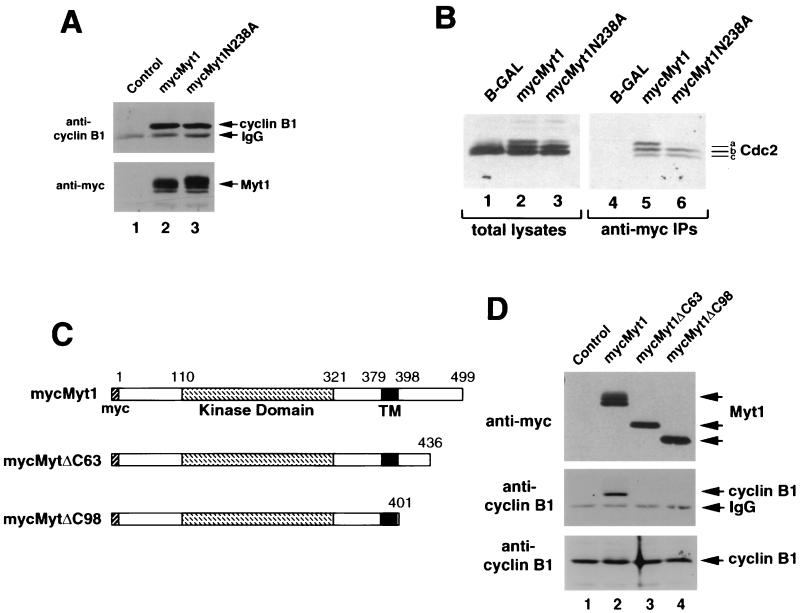
The finding that kinase-inactive Myt1 induced a G2 cell cycle delay was unexpected given that the mutant was incapable of inhibiting Cdc2 by phosphorylation. Because entry into mitosis requires both the activation of Cdc2 and the translocation of Cdc2-cyclin B1 complexes into the nucleus, we considered the possibility that kinase-inactive Myt1 prevented nuclear import of Cdc2 by binding to Cdc2-cyclin B1 complexes in the cytoplasm. To test if cyclin B1 could be found in a complex with Myt1, kinase-active and -inactive forms of Myt1 were transiently expressed in HeLa cells as Myc epitope-tagged proteins and were immunoprecipitated with an antibody specific for the Myc epitope. Immunoprecipitates were then examined for the presence of cyclin B1 by immunoblotting. As seen in Fig. 2A, cyclin B1 coimmunoprecipitated with both kinase-active (lane 2) and -inactive (lane 3) forms of Myt1.
FIG. 2.
The COOH terminus of Myt1 contains a Cdc2-cyclin B1 interaction domain. (A) Kinase-active (mycMyt1) (lane 2) and -inactive (mycMyt1N238A) (lane 3) forms of Myt1 were transiently expressed in HeLa cells as Myc epitope-tagged proteins and immunoprecipitated with antibody specific for the Myc epitope. Immunoprecipitates were then examined for the presence of cyclin B1 (top panel) and Myt1 (bottom panel) by immunoblotting. As a control, HeLa cells were transfected with vector lacking Myt1 (lane 1). IgG, immunoglobulin G. (B) HeLa cells synchronized at the G1/S border by a double thymidine block protocol were infected with recombinant adenoviruses encoding either β-GAL (lanes 1 and 4), kinase-active Myt1 (mycMyt1) (lanes 2 and 5), or kinase-inactive Myt1 (mycMyt1N238A) (lanes 3 and 6). Lysates were prepared when the control population of cells had reached mitosis (∼14 h after release from the block) and were resolved directly by SDS-PAGE (left panel), or Myt1 was immunoprecipitated with a monoclonal antibody against the Myc epitope tag prior to SDS-PAGE (right panel). The presence of Cdc2 was monitored by immunoblotting. Species a, b, and c are different electrophoretic forms of Cdc2 (see text for details). (C) Schematic representation of various deletion mutants of Myc epitope-tagged Myt1 used in the transient transfection assays in panel D. Numbers indicate amino acid residues of Myt1. The catalytic domain of Myt1 is indicated by a hatched box, and the putative membrane-targeting domain (TM) is represented by a filled box. The N-terminal Myc epitope tag is also shown. (D) HeLa cells were transiently transfected with vector alone (control) (lane 1) or with plasmids encoding either Myt1 (mycMyt1) (lane 2), Myt1 lacking its COOH-terminal 63 amino acids (mycMyt1ΔC63) (lane 3), or Myt1 lacking its COOH-terminal 98 amino acids (mycMyt1ΔC98) (lane 4). Cell lysates were prepared and either resolved by SDS-PAGE directly (bottom panel) or first incubated with antibody specific for the Myc epitope to precipitate Myt1. The precipitates were examined for the presence of Myt1 (top panel) and cyclin B1 (middle panel) by immunoblotting. To demonstrate that equal amounts of cyclin B1 were present in each sample, cellular lysates were immunoblotted with antibody specific for human cyclin B1 (bottom panel).
Experiments were also performed to determine if Cdc2 coimmunoprecipitated with Myt1 (Fig. 2B). HeLa cells synchronized at the G1/S border by a double thymidine block protocol were infected with recombinant adenoviruses encoding either β-GAL, kinase-active Myt1, or kinase-inactive Myt1. Following release from the second thymidine block, cell lysates were prepared when the control population of cells had completed mitosis (∼14 h after release). Lysates were resolved directly by SDS-PAGE (Fig. 2B, left panel), or Myt1 was immunoprecipitated with a monoclonal antibody against the Myc epitope tag prior to SDS-PAGE (right panel). The presence of Cdc2 was then monitored by immunoblotting, and the electrophoretic mobility of Cdc2 was used to assess whether it was phosphorylated on threonine 14, tyrosine 15, or both (3, 40). The slowest electrophoretic form of Cdc2 (species a) is phosphorylated on both Thr 14 and Tyr 15, whereas the intermediate form (species b) is phosphorylated on Thr 14 or Tyr 15, but not both. These two forms of Cdc2 have reduced kinase activity compared with Cdc2 that is not phosphorylated on Thr 14 and Tyr 15 (40). The fastest electrophoretic form of Cdc2 (species c) is not phosphorylated on either Thr 14 or Tyr 15 and represents either the active form of the kinase (phosphorylated on Thr 161 and bound to cyclin B) or Cdc2 that is not bound to cyclin (monomeric, inactive Cdc2). As expected, only the fastest electrophoretic form of Cdc2 was present in the control population of cells that had completed mitosis (Fig. 2B, lane 1). In contrast, all three electrophoretic forms were present in cells overproducing kinase-active Myt1 (lanes 2 and 5). Cells overproducing kinase-inactive Myt1 contained the fastest (species c) and intermediate (species b) electrophoretic forms of Cdc2 (lanes 3 and 6). This result suggests that endogenous Myt1 may still be capable of phosphorylating Cdc2 despite overproduction of kinase-inactive Myt1. However, this phosphorylation appears to be inefficient, as very little Cdc2 phosphorylated on both Thr 14 and Tyr 15 was detected in these cells (species a, lane 3). Cdc2 coimmunoprecipitated with both kinase-active and -inactive forms of Myt1 (Fig. 2B, right panel). The predominant forms of Cdc2 bound to Myt1 included either the doubly phosphorylated form (species a) or the singly phosphorylated form (species b). Although it was abundant in the extracts, very little of species c was found complexed to Myt1, suggesting that species c in these cells is monomeric Cdc2.
The COOH terminus of Myt1 contains a Cdc2-cyclin B1 interaction domain.
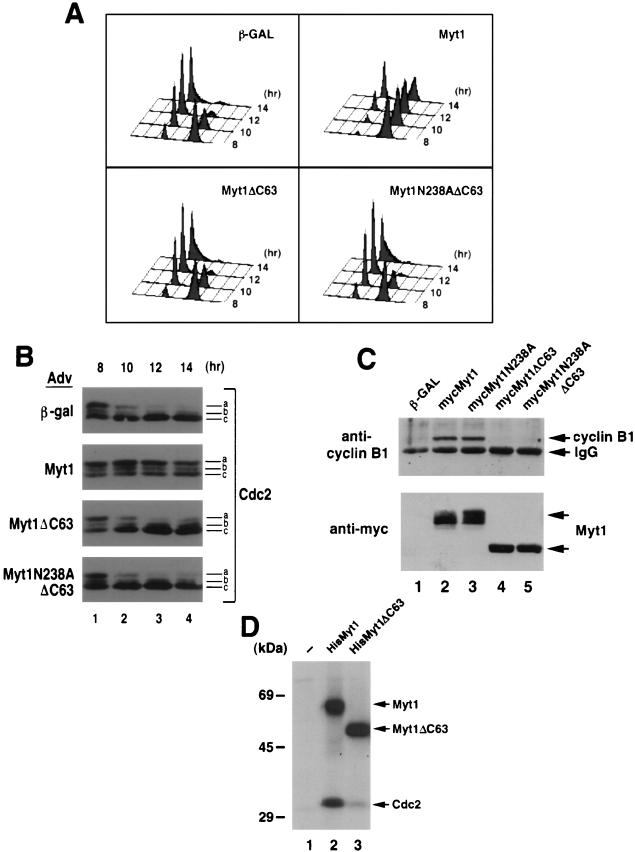
A deletion analysis was performed to identify regions of Myt1 that were critical for interactions with cyclin B1-Cdc2 (Fig. 2C and D). Removal of the COOH-terminal 63 amino acids of Myt1 completely abolished cyclin B1 binding (Fig. 2D, lane 3), thus identifying this region as a necessary component of the Cdc2-cyclin B1 interaction domain. To determine whether the G2 delay induced by kinase-active and -inactive forms of Myt1 could be eliminated if the Cdc2-cyclin B1 interaction domain was deleted, we tested the effects of overproducing forms of Myt1 lacking the COOH-terminal 63 amino acids. Recombinant adenoviruses encoding kinase-active (Myt1CΔ63) and -inactive (Myt1N238ACΔ63) forms of Myt1 lacking the binding domain were generated and used to infect HeLa cells that were synchronized at the G1/S border by the double thymidine block protocol. Flow cytometry was used to monitor the ability of cells to traverse the cell cycle after release from the block (Fig. 3A), and immunoblotting was performed to assess Cdc2 phosphorylation status (Fig. 3B). As was seen in Fig. 1, overproduction of wild-type Myt1 caused a significant G2 delay compared with overproduction of β-GAL. In contrast, there was no G2 delay observed in cells overproducing kinase-active or -inactive forms of Myt1 lacking the COOH-terminal 63 amino acids.
FIG. 3.
The G2 delay induced by overproduction of Myt1 requires the cyclin B-Cdc2 interaction domain. (A) HeLa cells synchronized at the G1/S border by a double thymidine block protocol were infected with recombinant adenoviruses encoding β-GAL, mycMyt1, mycMyt1ΔC63, or mycMyt1N238AΔC63 at a multiplicity of infection of 50. Cells were harvested at various times after release from the block, and the cellular DNA content was analyzed by flow cytometry. (B) Alternatively, cell lysates were prepared and resolved by SDS-PAGE, and Cdc2 was detected by immunoblotting. Species a, b, and c represent different electrophoretic forms of Cdc2 (see text for details). (C) HeLa cells were infected with adenoviruses encoding β-GAL (lane 1), mycMyt1 (lane 2), mycMyt1N238A (lane 3), mycMyt1ΔC63 (lane 4), or mycMyt1N238AΔC63 (lane 5). Myt1 proteins were immunoprecipitated with antibodies specific for the Myc epitope, and precipitates were analyzed for Myt1 (bottom panel) and cyclin B1 (top panel) by Western blotting. IgG, immunoglobulin G. (D) Full-length Myt1 (HisMyt1) (lane 2) or Myt1 lacking the C-terminal 63 amino acids (HisMyt1ΔC63) (lane 3) was purified from insect cells as a hexahistidine-tagged fusion protein. Kinase assays were performed in vitro in the presence of [γ-32P]ATP and purified glutathione S-transferase–cyclin B1-Cdc2 (K33R) complexes (40). Reaction mixtures were resolved by SDS-PAGE and subjected to autoradiography. Lane 1, substrate alone.
We next assessed the status of Cdc2 phosphorylation in each population of cells shown in Fig. 3A. In control cells overproducing β-GAL, Cdc2 was dephosphorylated on Thr 14 and Tyr 15 as cells moved from G2 (8-h time point) into mitosis, and this form of Cdc2 (species c) was maintained in the G1 population of cells (Fig. 3B). In contrast, the doubly phosphorylated form of Cdc2 (species a) was maintained throughout the entire time course in cells overproducing kinase-active Myt1 (Fig. 3B), consistent with the G2 delay observed in this population of cells (Fig. 3A). In cells overproducing forms of Myt1 lacking the COOH-terminal 63 amino acids, Cdc2 was dephosphorylated on both Thr 14 and Tyr 15, consistent with the failure of these forms of Myt1 to induce cell cycle delays. Immunoprecipitation of Myt1 from adenovirus-infected cells followed by immunoblotting confirmed that both kinase-active and -inactive forms of Myt1 require the COOH-terminal 63 amino acids for cyclin B1 binding (Fig. 3C). These results indicate that the binding of cyclin B1-Cdc2 to Myt1 is essential for the G2 delay induced upon overproduction of kinase-active and -inactive forms of Myt1.
The finding that kinase-active Myt1 lacking the Cdc2-cyclin B1 binding domain no longer induced a cell cycle delay upon overproduction suggested that binding to Myt1 might be critical for Cdc2 phosphorylation. Kinase assays were performed in vitro to test this possibility (Fig. 3D). Myt1 (lane 2) and the COOH-terminal deletion mutant of Myt1 (Myt1Δ63) (lane 3) were tested for their ability to phosphorylate purified catalytically inactive Cdc2-cyclin B1 complexes in vitro. Both Myt1 and Myt1Δ63 were active as judged by their ability to autophosphorylate. Strikingly, deletion of the COOH-terminal 63 amino acids of Myt1 impaired the ability of Myt1 to phosphorylate Cdc2 in vitro, suggesting that binding of Cdc2-cyclin B1 to Myt1 facilitates the phosphorylation of Cdc2.
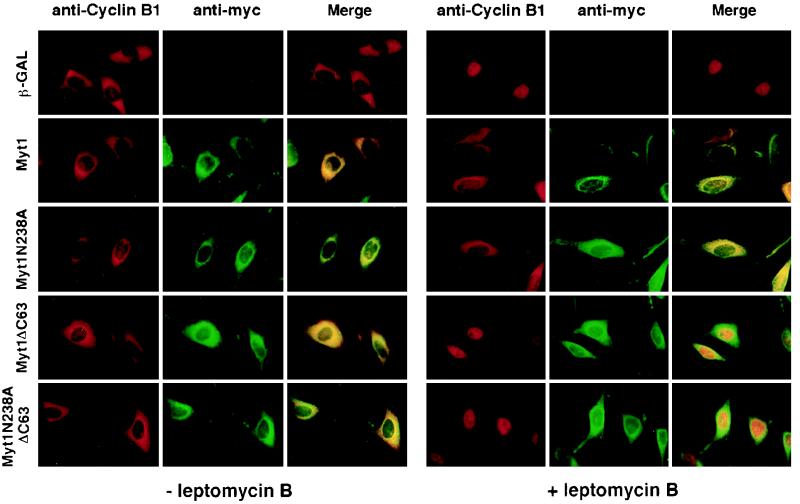
Overproduction of kinase-active and -inactive forms of Myt1 blocks trafficking of cyclin B1.
Throughout interphase, cyclin B1-Cdc2 complexes continuously shuttle between the nucleus and cytoplasm (21, 64, 66). Cyclin B1 appears to accumulate in the cytoplasm because of the rapidity with which it is exported from the nucleus. LMB, an inhibitor of the export factor exportin 1 (CRM1), blocks cyclin B1 nuclear export (21, 33, 47, 64, 66). We next examined the effects of Myt1 overproduction on the intracellular trafficking of cyclin B1 in vivo (Fig. 4). Indirect immunofluorescence was used to determine the localization of Myt1 and cyclin B1 in cells overproducing wild-type and mutant forms of Myt1. As seen in Fig. 4, kinase-active and -inactive forms of Myt1 localized to the cytoplasm, as did forms of Myt1 lacking the COOH-terminal 63 amino acids. This was true both in the presence (right panels) and absence (left panels) of LMB. In the absence of LMB, cyclin B1 was cytoplasmic under all experimental conditions, and the merged images indicate colocalization of cyclin B1 and overproduced Myt1. Nuclear accumulation of cyclin B1 was seen in LMB-treated cells overproducing either β-GAL or forms of Myt1 lacking the COOH-terminal 63 amino acids, indicating normal trafficking of cyclin B1 under these conditions. In contrast, cytoplasmic retention of cyclin B1 was seen in LMB-treated cells overproducing kinase-active and -inactive forms of Myt1 containing an intact COOH terminus. These results demonstrate that overproduction of active and inactive forms of Myt1 perturbs the trafficking of cyclin B1 in vivo and that the COOH terminus of Myt1 is required for this effect.
FIG. 4.
Overproduction of Myt1 blocks the intracellular trafficking of cyclin B1. HeLa cells synchronized at the G1/S border by a double thymidine block protocol were infected with recombinant adenoviruses encoding β-GAL or the indicated Myt1 proteins. Eight hours after release from the block, samples were either left untreated (− leptomycin B) or incubated with 20 μM LMB (+ leptomycin B). Cells were then fixed and subjected to indirect immunofluorescent staining with antibodies specific for cyclin B1 (anti-cyclin B1) or the Myc epitope to detect Myt1 (anti-myc). A merged image (Merge) is also shown for each sample.
The RXL motif in the COOH terminus of Myt1 is required for cyclin B1 binding.
The COOH-terminal 63 amino acids of Myt1 were examined for sequences that might contribute to cyclin B1 binding. It has been shown that cyclin A-Cdk2 binds to substrates containing an RXL motif, where R is arginine, X is any amino acid, and L is leucine (1, 10). The sequence RNL is contained within the COOH-terminal 63 amino acids of Myt1 at positions 486 to 488. We converted all of these amino acids to alanine and tested for the ability of the mutant protein [Myt1(3A)] to bind to cyclin B1 in vivo (Fig. 5). Myt1 and Myt1(3A) were transiently expressed in HeLa cells as Myc epitope-tagged proteins and immunoprecipitated with antibody specific for the Myc epitope. Immunoprecipitates were then examined for the presence of cyclin B1 by immunoblotting. As seen in Fig. 5, cyclin B1 coimmunoprecipitated with wild-type Myt1 (lane 2) but not the mutant form of Myt1 (lane 3), indicating that residues R, N, and L at positions 486, 487, and 488, respectively, are important for cyclin B1 binding.
FIG. 5.
The RXL motif in the COOH terminus of Myt1 is required for cyclin B1 binding. HeLa cells were transiently transfected with empty vector (control) (lane 1) or vectors encoding either Myc epitope-tagged Myt1 (mycMyt1) (lane 2) or Myc epitope-tagged Myt1 containing alanines in place of RNL at positions 486 to 488 [mycMyt1(3A)] (lane 3). Myt1 proteins were immunoprecipitated with antibody specific to the Myc epitope, and precipitates were analyzed for cyclin B1 (top panel) and Myt1 (bottom panel) by Western blotting. IgG, immunoglobulin G.
Loss of interactions between Myt1 and cyclin B1 during mitosis.
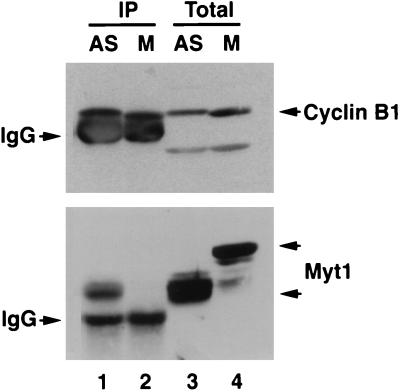
All of the experiments examining the interactions between Myt1 and cyclin B1 described above were done with cells transiently expressing exogenous Myt1. We wished to determine whether cyclin B1 bound to endogenous Myt1 (Fig. 6). To do this, lysates prepared from asynchronously growing cells or cells arrested in M phase with nocodozale were immunoprecipitated with cyclin B1-specific antibody. Immunoprecipitates were then examined for the presence of cyclin B1 (Fig. 6, top panel) and Myt1 (bottom panel) by immunoblotting. Interestingly, coimmunoprecipitation of Myt1 with cyclin B1 was observed in lysates prepared from asynchronously growing cells (lane 1, bottom panel) but not in those prepared from mitotic cells (lane 2, bottom panel). These results demonstrate that the hyperphosphorylated mitotic form of Myt1 does not associate with cyclin B-Cdc2 complexes.
FIG. 6.
Loss of Myt1-cyclin B1 complexes in mitosis. Lysates prepared from asynchronously growing HeLa cells (AS) (lanes 1 and 3) or HeLa cells arrested in mitosis with nocodazole (M) (lanes 2 and 4) were either analyzed directly by SDS-PAGE (Total) (lanes 3 and 4) or immunoprecipitated with polyclonal antibodies specific for cyclin B1 prior to SDS-PAGE (IP) (lanes 1 and 2). Cyclin B1 (top panel) and Myt1 (bottom panel) were detected by Western blotting. IgG, immunoglobulin G.
DISCUSSION
This study reports the identification of a novel functional domain in the COOH terminus of the human Myt1 kinase. This domain localizes to the COOH-terminal 63 amino acids (amino acids 436 to 499) of Myt1, with arginine, asparagine, and leucine (RNL) at positions 486, 487, and 488, respectively, serving as key residues within the domain. Disruption of this domain by deletion or by substitution of alanines for amino acids 486 to 488 eliminates the binding of Myt1 to cyclin B1-Cdc2 complexes. Thus, we assign this region of Myt1 as a cyclin B1-Cdc2 interaction domain. Residues just bordering and including amino acids 486 to 488 (PRNLL) in human Myt1 are conserved in XeMyt1, indicating that XeMyt1 may also bind cyclin B1-Cdc2 complexes. Between the protein kinase domain of Myt1 and the cyclin B1-Cdc2 interaction domain is another functional domain which is responsible for targeting Myt1 to membranes (40). Membrane targeting requires 20 amino acid residues bordered by arginine 378 and histidine 399. These residues are primarily hydrophobic or uncharged and are predicted to adopt an alpha-helical structure with the potential of spanning the lipid bilayer. It is unlikely that the membrane-targeting domain completely spans the lipid bilayer, as results from this study indicate that both the NH2-terminal kinase domain and COOH-terminal cyclin B1-Cdc2 interaction domain must be cytoplasmic.
Previous studies have reported a cyclin-Cdk recognition motif with the sequence ZRXL, where Z is basic or cysteine and X is either basic or nonpolar (1, 10). It has been proposed that this motif targets substrates (E2F1-3, p107, and p130), activators (Cdc25A), and cyclin-dependent kinase inhibitors (p21, p27, and p57) to cyclin-Cdk complexes (1, 10, 59, 60). The crystal structure of cyclin A-cdk2-p27 demonstrated that R and L of the ZRXL motif are of key importance. These residues are in the amino terminus of p27 and participate in hydrogen bonding and van der Waals contacts with cyclin A (58). p21 contains an additional ZRXL motif in its COOH terminus that is missing in the other cyclin-dependent kinase inhibitors and which targets p21 to PCNA (20). A hydrophobic patch on the surface of cyclin A has been shown to be essential for both binding to and phosphorylation of a RXL-containing substrates (60). Residues neighboring the RXL motif in Myt1 must be critical for conferring target specificity, as this domain in Myt1 facilitates the phosphorylation of Cdc2 when complexed with cyclin B1 but not that of Cdk2 when complexed with either cyclin E or A (9).
Overproduction of kinase-active Myt1 caused HeLa cells to delay in the G2 phase of the cell cycle. The mechanism of the G2 cell cycle delay is not obvious given that Myt1 can both inhibit Cdc2 by phosphorylation of Thr 14 and Tyr 15 and sequester Cdc2-cyclin B1 through a direct interaction. We observed that overproduction of kinase-active Myt1 prevented the nuclear-cytoplasmic shuttling of cyclin B1-Cdc2 complexes by sequestering the complex in the cytoplasm. Deletion of the cyclin B1-Cdc2 interaction domain in Myt1 eliminated binding of cyclin B1-Cdc2 complexes to Myt1 and restored the normal nuclear-cytoplasmic trafficking of cyclin B1-Cdc2 in vivo. Surprisingly, cell cycle progression was normal in cells overproducing kinase-active Myt1 lacking the cyclin B1-Cdc2 interaction domain even though Myt1 retained enzymatic activity. Kinase assays performed in vitro (Fig. 3D) and immunoblotting of endogenous Cdc2 (Fig. 3B) indicated that the integrity of the cyclin B1-Cdc2 interaction domain was also essential for the efficient phosphorylation of Cdc2 both in vitro and in vivo. Thus, deletion of the cyclin B1-Cdc2 interaction domain both restored the ability of cyclin B1 complexes to shuttle and reduced inhibitory phosphorylation of Cdc2, thereby allowing normal cell cycle progression.
Overproduction of kinase-inactive Myt1 also induced a G2 cell cycle delay, although not as efficiently as overproduction of wild-type Myt1. This is likely due to the fact that kinase-inactive Myt1 inhibits Cdc2 by sequestration but not by phosphorylation. Furthermore, if the physical interactions between Myt1 and Cdc2 are subject to negative regulation through phosphorylation of Myt1 by Cdc2, then kinase-inactive Myt1 would be expected to be more susceptible than kinase-active Myt1 to this form of negative regulation due to its inability to inactivate Cdc2 via phosphorylation. Two studies reported the presence of an inhibitor of Cdc2-cyclin B in Xenopus extracts (35, 38). In one case the inhibitor was shown to be titratable with excess Cdc2-cyclin B (35), and in the other case the inhibitor was shown to be membrane associated (38). In both cases the inhibitor was active against a Cdc2 mutant (Cdc2AF) lacking Thr 14 and Tyr 15, arguing that inhibition did not result from phosphorylation of Cdc2 on these sites. Although Myt1 was ruled out as a candidate because it would not be expected to inhibit the Cdc2AF mutant, this issue should be reconsidered given the results reported in this study. Human Myt1 has a Cdc2-cyclin B1 binding domain that would be capable of binding to Cdc2AF-cyclin B1 and functionally sequestering it. The sequence comprising the Cdc2-cyclin B1 binding domain is conserved in XeMyt1, leaving open the possibility that XeMyt1 might have been the inhibitor characterized in these two studies.
Our studies suggest a model whereby Cdc2-cyclin B1 complexes are targeted to the COOH terminus of Myt1 due to the presence of an RXL motif in Myt1. This facilitates phosphorylation of Cdc2 on Thr 14 and Tyr 15 by Myt1. The interaction between Myt1 and Cdc2-cyclin B1 complexes must be fairly transient under physiological conditions, as cyclin B1 complexes continuously shuttle between the nucleus and the cytoplasm throughout the S and G2 phases of the cell cycle. Thus, we speculate that upon dissociation from Myt1, Cdc2-cyclin B1 complexes enter the nucleus, where Wee1 might also contribute to Tyr 15 phosphorylation. The Cdc2-cyclin B1 complexes are then rapidly exported from the nucleus due to the presence of a nuclear export sequence in cyclin B1 (21, 64, 66). Overproduction of Myt1 can shift the equilibrium such that Cdc2-cyclin B1 complexes remain tethered to Myt1 and are prevented from shuttling between the nucleus and cytoplasm. In late G2, after cells have grown to the appropriate size and DNA replication is complete, Cdc2-cyclin B1 complexes begin to accumulate in the nucleus (5, 16, 49, 55, 56). This is presumably due to the phosphorylation of the cyclin B1 nuclear export sequence (39, 66). Concurrently, dephosphorylation of Cdc2 by the Cdc25C phosphatase results in the activation of Cdc2-cyclin B1 complexes, leading to phosphorylation of critical mitotic substrates. Myt1 becomes heavily phosphorylated during mitosis, and this is correlated with a modest, twofold reduction in Myt1 kinase activity (9). We found that the mitotic form of Myt1 no longer interacts with Cdc2-cyclin B1 complexes (Fig. 6), indicating that one function of Myt1 phosphorylation may be to reduce the affinity of Myt1 for Cdc2-Cyclin B1, rather than to affect the intrinsic activity of Myt1 per se. Interestingly, Palmer et al. (50) recently demonstrated that XeMyt1 is a substrate of p90rsk. Phosphorylation by p90rsk occurs in the C terminus of Myt1 and reduces the ability of Myt1 to phosphorylate Cdc2 in vitro. Palmer et al. (50) propose that Myt1 inhibition by p90rsk is a major regulatory step leading to the activation of Cdc2-cyclin B1 complexes during oocyte maturation. Perhaps p90rsk phosphorylates the COOH-terminal cyclin B1-Cdc2 interaction domain, thereby reducing the affinity of Myt1 for Cdc2-cyclin B1 complexes.
Several regulatory pathways interact to contribute to the abrupt transition observed as eukaryotic cells enter into mitosis. The Wee1 tyrosine kinase is inhibited by phosphorylation in late G2 and throughout mitosis and is an unstable protein whose levels decrease during mitosis (43, 54, 65). The intrinsic activity of the Cdc25C phosphatase is enhanced by phosphorylation in late G2 and throughout mitosis (24, 27, 34). Cyclin B1-Cdc2 complexes are prevented from exiting the nucleus due to phosphorylation of the cyclin B1 nuclear export sequence, thus leading to the nuclear accumulation of cyclin B1-Cdc2 complexes in late G2 (39, 66). Finally, the interactions between Myt1 and Cdc2-cyclin B1 complexes are disrupted during mitosis, possibly through the phosphorylation of the COOH terminus of Myt1. Further studies will be required to determine if the phosphorylation of Myt1 within its COOH terminus negatively regulates the interactions between Myt1 and Cdc2-cyclin B complexes.
ACKNOWLEDGMENTS
We thank M. Yoshida for providing LMB and H. Hermeking and B. Vogelstein for recombinant adenovirus encoding β-GAL. We thank Zhiqi Wu for technical assistance, Jeff Stanton for the human cyclin B1 antibody, and Julie Schwarz, David Crawford, and Paul Graves for helpful suggestions and comments.
This work was supported by the NIH and by NIH postdoctoral fellowships to F.L. and C.R.-O. H.P.-W. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Adams P D, Sellers W R, Sharma S K, Wu A D, Nalin C M, Kaelin W G. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atherton-Fessler S, Hannig G, Piwnica-Worms H. Reversible tyrosine phosphorylation and cell cycle control. Semin Cell Biol. 1993;4:433–442. doi: 10.1006/scel.1993.1051. [DOI] [PubMed] [Google Scholar]
- 3.Atherton-Fessler S, Liu F, Gabrielli B, Lee M S, Peng C-Y, Piwnica-Worms H. Cell cycle regulation of the p34cdc2 inhibitory kinases. Mol Biol Cell. 1994;5:989–1001. doi: 10.1091/mbc.5.9.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atherton-Fessler S, Parker L L, Geahlen R L, Piwnica-Worms H. Mechanism of p34cdc2 regulation. Mol Cell Biol. 1993;13:1675–1685. doi: 10.1128/mcb.13.3.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailly E, Pines J, Hunter T, Bornens M. Cytoplasmic accumulation of cyclin B1 in human cells: association with a detergent-resistent compartment and with the centrosome. J Cell Sci. 1992;101:529–545. doi: 10.1242/jcs.101.3.529. [DOI] [PubMed] [Google Scholar]
- 6.Baldin V, Ducommun B. Subcellular localisation of human wee1 kinase is regulated during the cell cycle. J Cell Sci. 1995;108:2425–2432. doi: 10.1242/jcs.108.6.2425. [DOI] [PubMed] [Google Scholar]
- 7.Becker T C, Noel R J, Coats W S, Gomez-Foix A M, Alam T, Gerard R D, Newgard C B. Use of recombinant adenovirus for metabolic engineering of mammalian cells. In: Roth M G, editor. Methods in cell biology. New York, N.Y: Academic Press, Inc.; 1994. pp. 161–189. [DOI] [PubMed] [Google Scholar]
- 8.Blasina A, Paegle E S, McGowan C H. The role of inhibitory phosphorylation of Cdc2 following DNA replication block and radiation-induced damage in human cells. Mol Biol Cell. 1997;8:1013–1023. doi: 10.1091/mbc.8.6.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Booher R N, Holman P S, Fattaey A. Human Myt1 is a cell cycle regulated kinase that inhibits Cdc2 but not Cdk2 activity. J Biol Chem. 1997;272:22300–22306. doi: 10.1074/jbc.272.35.22300. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Saha P, Kornbluth S, Dynlacht B D, Dutta A. Cyclin-binding motifs are essential for the function of p21Cip1. Mol Cell Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cliby W A, Roberts C J, Cimprich K A, Stringer C M, Lamb J R, Schreiber S L, Friend S H. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draetta G, Beach D. Activation of cdc2 protein kinase during mitosis in human cells: cell cycle-dependent phosphorylation and subunit rearrangement. Cell. 1988;54:17–26. doi: 10.1016/0092-8674(88)90175-4. [DOI] [PubMed] [Google Scholar]
- 13.Draetta G, Piwnica-Worms H, Morrison D, Druker B, Roberts T, Beach D. Human cdc2 protein kinase is a major cell-cycle regulated tyrosine kinase substrate. Nature. 1988;336:738–744. doi: 10.1038/336738a0. [DOI] [PubMed] [Google Scholar]
- 14.Dunphy W G, Kumagai A. The cdc25 protein contains an intrinsic phosphatase activity. Cell. 1991;67:189–196. doi: 10.1016/0092-8674(91)90582-j. [DOI] [PubMed] [Google Scholar]
- 15.Featherstone C, Russell P. Fission yeast p107wee1 mitotic inhibitor is a tyrosine/serine kinase. Nature. 1991;349:808–811. doi: 10.1038/349808a0. [DOI] [PubMed] [Google Scholar]
- 16.Gallant P, Nigg E A. Cyclin B2 undergoes cell cycle-dependent nuclear translocation and, when expressed as a non-destructible mutant, causes mitotic arrest in HeLa cells. J Cell Biol. 1992;117:213–224. doi: 10.1083/jcb.117.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gautier J, Solomon M J, Booher R N, Bazan J F, Kirschner M W. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 1991;67:197–211. doi: 10.1016/0092-8674(91)90583-k. [DOI] [PubMed] [Google Scholar]
- 18.Gould K L, Moreno S, Owen D J, Sazer S, Nurse P. Phosphorylation at Thr 167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 1991;10:3297–3309. doi: 10.1002/j.1460-2075.1991.tb04894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gould K L, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- 20.Gulbis J M, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 21.Hagting A, Karlsson C, Clute P, Jackman M, Pines J. MPF localization is controlled by nuclear export. EMBO J. 1998;17:4127–4138. doi: 10.1093/emboj/17.14.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He T-C, Zhou S, DaCosta L T, Yu J, Kinzler K W, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heald R, McLoughlin M, McKeon F. Human Wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated Cdc2 kinase. Cell. 1993;74:463–474. doi: 10.1016/0092-8674(93)80048-j. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann I, Clarke P R, Marcote M J, Karsenti E, Draetta G. Phosphorylation and activation of human cdc25-C by cdc2-cyclin B and its involvement in the self-amplification of MPF at mitosis. EMBO J. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honda R, Ohba Y, Yasuda H. The cell cycle regulator, human p50wee1, is a tyrosine kinase and not a serine/tyrosine kinase. Biochem Biophys Res Commun. 1992;186:1333–1338. doi: 10.1016/s0006-291x(05)81552-9. [DOI] [PubMed] [Google Scholar]
- 26.Igarashi M, Nagata A, Jinno S, Suto K, Okayama H. Wee1+-like gene in human cells. Nature. 1991;353:80–83. doi: 10.1038/353080a0. [DOI] [PubMed] [Google Scholar]
- 27.Izumi T, Walker D H, Maller J M. Periodic changes in phosphorylation of the Xenopus cdc25 phosphatase regulate its activity. Mol Biol Cell. 1992;3:927–939. doi: 10.1091/mbc.3.8.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin P, Gu Y, Morgan D O. Role of inhibitory Cdc2 phosphorylation in radiation-induced G2 arrest in human cells. J Cell Biol. 1996;134:963–970. doi: 10.1083/jcb.134.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin P, Hardy S, Morgan D O. Nuclear localization of cyclin B1 controls mitotic entry after DNA damage. J Cell Biol. 1998;141:875–885. doi: 10.1083/jcb.141.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kornbluth S, Sebastian B, Hunter T, Newport J. Membrane localization of the kinase which phosphorylates p34cdc2 on threonine 14. Mol Biol Cell. 1994;5:273–282. doi: 10.1091/mbc.5.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krek W, Nigg E A. Differential phosphorylation of vertebrate p34cdc2 kinase at the G1/S and G2/M transitions of the cell cycle: identification of major phosphorylation sites. EMBO J. 1991;10:305–316. doi: 10.1002/j.1460-2075.1991.tb07951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krek W, Nigg E A. Mutations of p34cdc2 phosphorylation sites induce premature mitotic events in HeLa cells: evidence for a double block to p34cdc2 kinase activation in vertebrates. EMBO J. 1991;10:3331–3341. doi: 10.1002/j.1460-2075.1991.tb04897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J Biol Chem. 1997;272:29742–29751. doi: 10.1074/jbc.272.47.29742. [DOI] [PubMed] [Google Scholar]
- 34.Kumagai A, Dunphy W G. Regulation of the cdc25 protein during the cell cycle in Xenopus extracts. Cell. 1992;70:139–151. doi: 10.1016/0092-8674(92)90540-s. [DOI] [PubMed] [Google Scholar]
- 35.Kumagai A, Dunphy W G. Control of the cdc2/cyclin B complex in Xenopus egg extracts arrested at a G2/M checkpoint with DNA synthesis inhibitors. Mol Biol Cell. 1995;6:199–213. doi: 10.1091/mbc.6.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee M S, Enoch T, Piwnica-Worms H. Mik1+ encodes a tyrosine kinase that phosphorylates p34cdc2 on tyrosine 15. J Biol Chem. 1994;269:30530–30537. [PubMed] [Google Scholar]
- 37.Lee M S, Ogg S, Xu M, Parker L L, Donoghue D J, Maller J L, Piwnica-Worms H. cdc25+ encodes a protein phosphatase that dephosphorylates p34cdc2. Mol Biol Cell. 1992;3:73–84. doi: 10.1091/mbc.3.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee T H, Kirschner M W. An inhibitor of p34cdc2/cyclin B that regulates the G2/M transition in Xenopus extracts. Proc Natl Acad Sci USA. 1996;93:352–356. doi: 10.1073/pnas.93.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Meyer A N, Donoghue D J. Requirement for phosphorylation of cyclin B1 for Xenopus oocyte maturation. Mol Biol Cell. 1995;6:1111–1124. doi: 10.1091/mbc.6.9.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu F, Stanton J J, Wu Z, Piwnica-Worms H. The human Myt1 kinase preferentially phosphorylates Cdc2 on threonine 14 and localizes to the endoplasmic reticulum and Golgi complex. Mol Cell Biol. 1997;17:571–583. doi: 10.1128/mcb.17.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lundgren K, Walworth N, Booher R, Dembski M, Kirschner M, Beach D. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell. 1991;64:1111–1122. doi: 10.1016/0092-8674(91)90266-2. [DOI] [PubMed] [Google Scholar]
- 42.McGowan C H, Russell P. Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr 15. EMBO J. 1993;12:75–85. doi: 10.1002/j.1460-2075.1993.tb05633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGowan C H, Russell P. Cell cycle regulation of human WEE1. EMBO J. 1995;14:2166–2175. doi: 10.1002/j.1460-2075.1995.tb07210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgan D O. Cyclin dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Deve Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 45.Morla A O, Draetta G, Beach D, Wang J Y J. Reversible tyrosine phosphorylation of cdc2: dephosphorylation accompanies activation during entry into mitosis. Cell. 1989;58:193–203. doi: 10.1016/0092-8674(89)90415-7. [DOI] [PubMed] [Google Scholar]
- 46.Mueller P R, Coleman T R, Kumagai A, Dunphy W G. Myt1: a membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- 47.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade or crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;9:6320–6324. [PubMed] [Google Scholar]
- 48.Norbury C, Blow J, Nurse P. Regulatory phosphorylation of the p34cdc2 protein kinase in vertebrates. EMBO J. 1991;10:3321–3329. doi: 10.1002/j.1460-2075.1991.tb04896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ookata K, Hisanaga S-I, Okano T, Tachibana K, Kishimoto T. Relocation and distinct subcellular localization of p34cdc2-cyclin B complex at meiosis reinitiation in starfish oocytes. EMBO J. 1992;11:1763–1772. doi: 10.1002/j.1460-2075.1992.tb05228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmer A, Gavin A, Nebreda A. A link between MAP kinase and p34cdc2/cyclin B during oocyte maturation: p90rsk phosphorylates and inactivates the p34cdc2 inhibitory kinase Myt1. EMBO J. 1998;17:5037–5047. doi: 10.1093/emboj/17.17.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parker L L, Atherton-Fessler S, Lee M S, Ogg S, Falk F L, Swenson K I, Piwnica-Worms H. Cyclin promotes the tyrosine phosphorylation of p34cdc2 in a wee1+ dependent manner. EMBO J. 1991;10:1255–1263. doi: 10.1002/j.1460-2075.1991.tb08067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parker L L, Atherton-Fessler S, Piwnica-Worms H. p107wee1 is a dual specificity kinase that phosphorylates p34cdc2 on tyrosine 15. Proc Natl Acad Sci USA. 1992;89:2917–2921. doi: 10.1073/pnas.89.7.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parker L L, Piwnica-Worms H. Inactivation of the p34cdc2-cyclin B complex by the human wee1 tyrosine kinase. Science. 1992;257:1955–1957. doi: 10.1126/science.1384126. [DOI] [PubMed] [Google Scholar]
- 54.Parker L L, Sylvestre P J, Byrnes III M J, Liu F, Piwnica-Worms H. Identification of a 95-kDa WEE1-like tyrosine kinase in HeLa cells. Proc Natl Acad Sci USA. 1995;92:9638–9642. doi: 10.1073/pnas.92.21.9638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pines J, Hunter T. Human cyclin A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pines J, Hunter T. The differential localization of human cyclin A and B is due to a cytoplasmic retention signal in cyclin B. EMBO J. 1994;13:3772–3781. doi: 10.1002/j.1460-2075.1994.tb06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- 58.Russo A A, Jeffrey P D, Patten A K, Massague J, Pavletich N P. Crystal structure of the p27kip1 cyclin dependent kinase inhibitor bound to the cyclin A-cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 59.Saha P, Eichbaum Q, Silberman E D, Mayer B J, Dutta A. p21CIP1 and Cdc25A: competition between an inhibitor and an activator of cyclin-dependent kinases. Mol Cell Biol. 1997;17:4338–4345. doi: 10.1128/mcb.17.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schulman B A, Lindstrom D L, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci USA. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sebastian B, Kakizuka A, Hunter T. Cdc25M2 activation of cyclin-dependent kinases by dephosphorylation of threonine-14 and tyrosine-15. Proc Natl Acad Sci USA. 1993;90:3521–3524. doi: 10.1073/pnas.90.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Solomon M J, Lee T, Kirschner M W. Role of phosphorylation in p34cdc2 activation: identification of an activating kinase. Mol Biol Cell. 1992;3:13–27. doi: 10.1091/mbc.3.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strausfeld U, Labbe J C, Fesquet D, Cavadore J C, Picard A, Sadhu K, Russell P, Doree M. Dephosphorylation and activation of a p34cdc2/cyclin B complex in vitro by human cdc25 protein. Nature. 1991;351:242–245. doi: 10.1038/351242a0. [DOI] [PubMed] [Google Scholar]
- 64.Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watanabe N, Broome M, Hunter T. Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the cell cycle. EMBO J. 1995;14:1878–1891. doi: 10.1002/j.1460-2075.1995.tb07180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang J, Bardes E S G, Moore J D, Brennan J, Powers M A, Kornbluth S. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]