Abstract
Activation of poly(ADP-ribose) polymerase (PARP) by DNA breaks catalyzes poly(ADP-ribosyl)ation and results in depletion of NAD+ and ATP, which is thought to induce necrosis. Proteolytic cleavage of PARP by caspases is a hallmark of apoptosis. To investigate whether PARP cleavage plays a role in apoptosis and in the decision of cells to undergo apoptosis or necrosis, we introduced a point mutation into the cleavage site (DEVD) of PARP that renders the protein resistant to caspase cleavage in vitro and in vivo. Here, we show that after treatment with tumor necrosis factor alpha, fibroblasts expressing this caspase-resistant PARP exhibited an accelerated cell death. This enhanced cell death is attributable to the induction of necrosis and an increased apoptosis and was coupled with depletion of NAD+ and ATP that occurred only in cells expressing caspase-resistant PARP. The PARP inhibitor 3-aminobenzamide prevented the NAD+ drop and concomitantly inhibited necrosis and the elevated apoptosis. These data indicate that this accelerated cell death is due to NAD+ depletion, a mechanism known to kill various cell types, caused by activation of uncleaved PARP after DNA fragmentation. The present study demonstrates that PARP cleavage prevents induction of necrosis during apoptosis and ensures appropriate execution of caspase-mediated programmed cell death.
Apoptosis and necrosis are two forms of cell death, with distinct morphological and biochemical features. While apoptosis accounts for most physiological cell deaths, necrosis is usually induced in pathological situations by accidental, acute damage to cells (2, 61). Apoptosis, or programmed cell death, is a tightly regulated process under normal conditions. This process is controlled by a hierarchical set of cell death molecules identified originally in Caenorhabditis elegans (22) and later in mammalian cells (10).
For many cell types, the apoptotic cascade has been well studied with a model involving tumor necrosis factor (TNF-α) receptor and Fas (APO-1/CD95) mediation (39). After death ligands bind to and activate TNF-α and Fas receptors, several prominent events occur, including activation of caspases, which are critical players in execution of apoptosis (41), and of the DNA fragmentation factor (DFF/ICAD) (15, 35), which degrades chromatin DNA, a hallmark of apoptosis.
Another prominent event during apoptosis is the selective cleavage of poly(ADP-ribose) polymerase (PARP) by several caspases, especially by caspase-3 (26, 31). Following treatment of cells with TNF-α or anti-Fas antibodies, caspase-3 cleaves the 113-kDa PARP at the DEVD site between Asp214 and Gly215, to generate 89- and 24-kDa polypeptides (40, 54). PARP cleavage is an universal phenomenon observed during programmed cell death induced by a variety of apoptotic stimuli. However, the significance of this event in the apoptotic cascade is not clear. Recent studies demonstrate that PARP−/− cells exhibit a normal apoptotic response to various stimuli, including TNF-α and anti-Fas treatment, suggesting that PARP itself is dispensable in various apoptotic pathways (32, 59).
PARP is an abundant, chromatin-associated enzyme which, in response to DNA damage, binds rapidly to DNA strand breaks and undergoes automodification by forming long, branched poly(ADP-ribose) polymers by using NAD+ (34). Continued attempts by cells to resynthesize NAD+ lead to depletion of the ATP pool (51), which has been proposed as a mechanism for DNA damage-induced cell death in many cell types (4, 17, 53). Recent studies have demonstrated that the intracellular ATP levels influence the mode of cell death. Whereas high levels of ATP enable cells to undergo apoptosis, low ATP levels shift cells from apoptosis to necrosis (13, 33). Necrosis is characterized by swelling of the cells, disruption of the cell membrane, and lysis, leading to release of the cell contents, which may result in an inflammatory response (19, 28). We reasoned that the cleavage of PARP at early phases of apoptosis has a significant function in this process and is a critical event for cells to ensure normal apoptosis by preventing NAD+ and ATP depletion and thereby to inhibit unwanted necrosis. To test this hypothesis, we have generated cell lines stably expressing a cleavage-resistant mutant PARP in a PARP-null background and have studied their responsiveness to apoptotic stimuli.
MATERIALS AND METHODS
Site-directed mutagenesis.
A eukaryotic-prokaryotic expression vector, pSG9M (kindly provided by G. Lamm, Research Institute of Molecular Pathology, Vienna, Austria), containing full-length human PARP cDNA under the control of simian virus 40 (SV40) and T7 promoters (PARP/WT), was used to generate caspase-resistant PARP. A point mutation (G→A at nucleotide 640 of the human gene) (7) was introduced into the DEVD box (codons 211 to 214) of PARP/WT by use of a site-directed mutagenesis system (Stratagene, La Jolla, Calif.). Mutagenesis was performed according to the manufacturer’s recommendation with primers (G→A is underlined) GGCGATGAGGTGAATGGAGTGGATG and CATCCACTCCATTCACCTCATCGCC. The resulting plasmid was designated D214N. Introduction of the mutation was confirmed by sequencing.
In vitro transcription-translation and cleavage assay.
PARP/WT and D214N were transcribed and translated in vitro by using a T7 promoter and T7 polymerase-directed reticulocyte lysate system (Promega, Madison, Wis.) to generate [35S]methionine-labeled protein. A 1.5-μl portion of the translated product was incubated with 4 U of purified human recombinant caspase-3 (a kind gift of D. Nicholson, Merck Frosst, Pointe Claire-Dorval, Canada) for 1 h at 30°C in a buffer containing 50 mM HEPES-KOH (pH 7), 10% (wt/vol) sucrose, 2 mM EDTA, 0.1% (wt/vol) 3-[(3-cholamidoproplyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and 5 mM dithiothreitol. The reaction mixtures were boiled for 5 min in a standard reducing buffer and subjected to sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE). Following fixation and amplification (Amplify; Amersham, Buckinghamshire, United Kingdom), the gel was dried under a vacuum and subjected to autoradiography.
Cell transfection and induction of apoptosis.
Cells were routinely maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum at 37°C in 5% CO2. To establish stable cell lines expressing wild-type or mutant PARP, immortalized fibroblasts (A11) derived from a PARP−/− embryo (58) were used. Semiconfluent cultures grown in six-well plates were transfected with 1.5 μg of linearized PARP/WT or D214N plasmid by use of Lipofectamine according to the instructions of the manufacturer (Life Technologies, Gaithersburg, Md.). For positive selection, all cultures were cotransfected with 0.15 μg of pPGK-Hygro (kindly provided by E. F. Wagner, Research Institute of Molecular Pathology) and selected with 600 μg of hygromycin per ml. Resistant clones were isolated and characterized by Southern blot analysis for transgene integration. For induction of apoptosis in all experiments, unless otherwise stated, cells were treated with 40 ng of recombinant TNF-α (W. Fiers, University of Ghent, Ghent, Belgium) per ml in medium containing 1 μg of actinomycin D (Calbiochem, La Jolla, Calif.) per ml.
Western blot analysis of PARP expression and cleavage in transfected cells.
Cells were lysed in a modified radioimmunoprecipitation assay buffer containing 0.5 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin per ml, 0.5 μg of leupeptin per ml, and 1 μM pepstatin. Fifty micrograms of protein per lane was subjected to SDS–10% PAGE analysis, followed by transfer to nitrocellulose membrane (Bio-Rad, Hercules, Calif.) and hybridization with rabbit polyclonal Vic-5 antiserum against PARP (Boehringer Mannheim, Mannheim, Germany). Proteins were visualized with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) (Sigma, St. Louis, Mo.) followed by use of the ECL chemiluminescence system (Amersham, Little Chalfont, United Kingdom).
Cellular localization and enzymatic activity of PARP.
To detect the subcellular localization of mutant PARP, cells were cultured on coverslips and fixed in ice-cold pure methanol at −20°C for 30 min. After being washed, the cells were incubated with rabbit polyclonal Vic-5 anti-PARP antibody for 60 min. Cultures were then washed with phosphate-buffered saline (PBS), incubated with fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG (Boehringer Mannheim), and examined under a fluorescence microscope. Poly(ADP-ribose) polymer formation was detected as described previously (58). Briefly, cells were cultured on coverslips and treated with a 200 μM concentration of the alkylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) at 37°C for 30 min. The cells were then immediately washed and fixed in ice-cold 10% trichloroacetic acid in PBS, followed by dehydration through graded dilutions of ethanol. Samples were incubated with the monoclonal antibody 10H raised against poly(ADP-ribose) polymers (27) and FITC-labeled goat anti-mouse IgG (Southern Biotechnology Associates, Birmingham, Ala.). Signal was visualized by fluorescence microscopy.
Caspase-3 activity assay.
Apoptosis was induced by treatment of cells with 20 ng of TNF-α per ml for 12 h. Cell lysates were incubated with a specific substrate, DEVD-AFC (50 μM) (Clontech, Palo Alto, Calif.), and caspase-3 activity was measured fluorometrically with a fluorometer (Perkin-Elmer, Norwalk, Conn.) equipped with a 400-nm excitation filter and a 505-nm emission filter. As a control for caspase-3 specificity, lysates were preincubated with 10 μM DEVD-CHO (Clontech), a caspase-3 inhibitor, at 37°C for 30 min.
Flow cytometry.
Cells were cultured in six-well plates and treated with TNF-α for 12 h. Apoptosis was quantified by flow cytometry after staining with FITC-conjugated annexin V (Clontech), which binds to externalized phosphatidylserine on the surface of apoptotic cells, and/or propidium iodide (PI), which stains cells that have lost membrane integrity. Fluorescence-activated cell sorter (FACS) analyses were carried out on a FACScalibur (Becton Dickinson, San Jose, Calif.) with CellQuest software.
TUNEL analysis.
DNA fragmentation in apoptotic cells was visualized by terminal deoxyribonucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) (18). Cells were grown on coverslips and treated with TNF-α for 12 h. Samples were air dried and fixed for 30 min with 4% paraformaldehyde in PBS, followed by permeabilization with 0.1% Triton X-100 and 0.1% sodium citrate for 2 min on ice. Cells were treated with the fluorescein in situ cell death detection mixture according to the instructions of the manufacturer (Boehringer Mannheim). After the final washing steps, coverslips were mounted with Vectashield mounting medium containing PI (Vector Laboratories, Burlingame, Calif.) and examined under a fluorescence microscope. For detection of poly(ADP-ribose) polymer formation in apoptotic cells, a protocol combining immunochemical staining of polymers and TUNEL assay was employed. Briefly, after incubation with the fluorescein in situ cell detection mixture, cells were rinsed with PBS containing 3% bovine serum albumin and incubated with monoclonal anti-poly(ADP-ribose) polymer antibody 10H for 1 h. Following washing with PBS, samples were incubated with Cy3–anti-mouse IgG (Sigma) for 30 min at 37°C and mounted with antifade medium containing DAPI (4′,6-diamidino-2-phenylindole) (Vector Laboratories) before examination under a fluorescence microscope.
MTT assay.
MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenytetrazolium bromide] detection of cell viability was performed as described previously (47). Briefly, cells were plated in 96-well plates and cultured overnight. Apoptosis was induced with TNF-α for various times in time course experiments. Cultures were incubated with staining mixtures, and absorbance was read at 600 nm in a spectrophotometer.
Morphological examination of necrotic cell death.
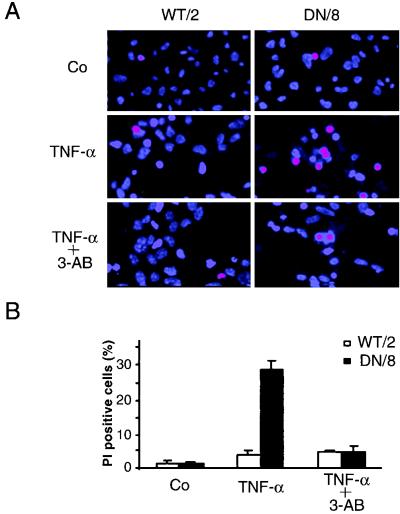
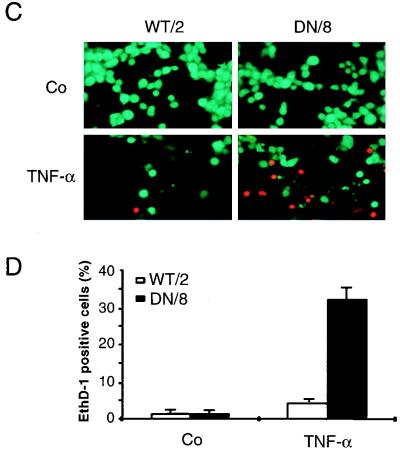
The procedure to score necrosis was basically that described previously (33, 48). Cells were incubated either with 10 μM Hoechst 33342 (Ho-342) (Molecular Probes, Eugene, Oreg.) and 10 μM PI (Clontech) for 10 min or with 2 μM calcein-acetoxymethyl ester (CAM) (Molecular Probes) and 4 μM ethidium homodimer (EthD-1) (Molecular Probes) for 45 min and then analyzed under a fluorescence microscope. Necrotic cell death was quantified by counting cells taking up the vital dye, i.e., those with pink double-stained nuclei (Ho-342–PI staining) or red-stained nuclei (CAM–EthD-1 staining).
LDH release assay.
In order to reduce background absorbance, cells were grown in Dulbecco modified Eagle medium without phenol red (Gibco BRL) supplemented with 5% fetal calf serum in 96-well plates. Following cell death induction, released lactate dehydrogenase (LDH) in the supernatant was measured with a coupled enzymatic assay (CytoTox 96 assay; Promega). Fifty microliters of sample was mixed with 50 μl of substrate mix, and following 30 min of incubation in the dark, absorbance was recorded at 492 nm in a spectrophotometer.
Measurements of NAD+ and ATP levels.
For measurement of cellular NAD+, an enzymatic cycling techniques with alcohol dehydrogenase from Saccharomyces cerevisiae, adapted for 96-well plates, was used (23). For ATP measurement, we used a method described previously (36, 52). Briefly, 106 cells were suspended in 50 μl of dilution buffer (100 mM Tris-HCl [pH 7.75], 4 mM EDTA), 450 μl of boiling dilution buffer was added, and cells were incubated for 2 min at 100°C. Samples were spun at 10,000 × g for 1 min, 20 μl of supernatant was added to 50 μl of luciferase reagent (Bioluminescence Assay Kit CLS II; Boehringer Mannheim), and luciferase activity was determined in a luminometer and directly converted to the ATP concentration.
RESULTS
Mutation of the DEVD site renders PARP protein resistant to caspase cleavage.
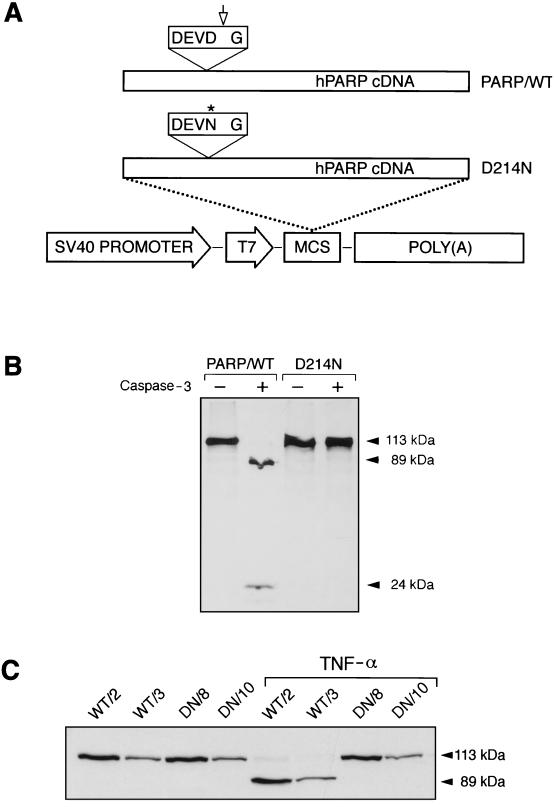
To generate a cleavage-resistant PARP, a G-to-A point mutation was introduced at nucleotide 640 of the human cDNA, resulting in an Asp (D)-to-Asn (N) transition at codon 214 in the caspase-3 recognition sequence DEVD (codons 211 to 214). The resultant D214N mutation was confirmed by sequencing and used to construct an expression vector under the control of the T7 and SV40 promoters (Fig. 1A). [35S]methionine-labeled wild-type and mutant PARP proteins were synthesized in a cell-free system by in vitro transcription-translation. Both wild-type (PARP/WT) and mutant (D214N) PARP proteins were incubated with recombinant caspase-3 and then subjected to SDS-PAGE analysis. As expected, products from both PARP/WT and D214N constructs gave rise to proteins of 113 kDa (Fig. 1B). After incubation with caspase-3, wild-type PARP protein was cleaved into two fragments with molecular masses of 89 and 24 kDa, whereas mutant PARP protein remained intact and yielded a 113-kDa band (Fig. 1B). This result demonstrates that the mutation at codon 214 renders PARP resistant to caspase cleavage.
FIG. 1.
Abolition of caspase-3 cleavage of PARP by mutation at the cleavage site (DEVD). (A) The full-length human PARP (hPARP) cDNA was under control of the T7 and SV40 promoters (PARP/WT). An Asp (D)→Asn (N) transition (codon 214, asterisk) was introduced into the DEVD site, resulting in a mutant construct designated D214N. The polyadenylation signal [(poly (A)] was from SV40. MCS, multiple cloning sites. The arrow indicates the caspase cleavage site. (B) [35S]methionine-labeled PARP was generated by an in vitro coupled transcription-translation system. Translated products from PARP/WT and D214N vectors were incubated either with (+) or without (−) purified human recombinant caspase-3. (C) Western blot analysis of PARP expression and cleavage in transfected fibroblasts. Stably transfected clones expressing wild-type PARP (WT/2 and WT/3) and those expressing mutant PARP (DN/8 and DN/10) were either treated or not treated with TNF-α (20 ng/ml); this was followed by Western blot analysis with polyclonal antiserum Vic-5. Full-length PARP (113 kDa) and two cleaved fragments (89 and 24 kDa) are indicated by arrowheads.
Establishment of cell lines exclusively expressing wild-type or caspase-resistant PARP.
To study the effect of noncleavable PARP during apoptosis, an SV40 promoter-driven D214N mutant PARP vector was transfected into immortalized mouse fibroblasts (A11) originally isolated from a PARP−/− embryo (58), the use of which avoids the influence of endogenous PARP. To control the transfection and experimental procedures, and to better compare the effects of cleavage and noncleavage of PARP on apoptosis, an SV40-driven wild-type (PARP/WT) vector was also transfected into A11 cells. Several cell clones were isolated, and these cell lines were similar to each other and to the parental A11 cells in terms of proliferation properties (data not shown). Western blot analyses of cell lysates from these clones revealed various expression levels of PARP (Fig. 1C). For further studies, we chose two pairs of cell lines based on the expression levels of the transgenes: WT/2 and DN/8 (high expressors; expression ca. three- to fourfold higher than that in wild-type embryonic fibroblasts) and WT/3 and DN/10 (low expressors; expression similar to that in wild-type cells). The PARP expression levels within each group were comparable (Fig. 1C).
To determine cleavability of the mutant PARP by caspases, these four clones were treated with TNF-α and analyzed by Western blotting. While the wild-type proteins in WT/2 and WT/3 cells were completely cleaved into 89- and 24-kDa peptides, the mutant PARP in DN/8 and DN/10 cells remained intact under the same conditions, demonstrating complete resistance of mutant PARP to caspase cleavage in vivo (Fig. 1C). These results confirm the findings obtained with the cell-free system and indicate that the Asp residue at the P1 position in the P4DEVDP1 sequence is required for efficient cleavage by caspase-3-like proteases in cells undergoing apoptosis.
PARP with a mutation in DEVD retains normal enzymatic function and does not inhibit caspase-3 activity.
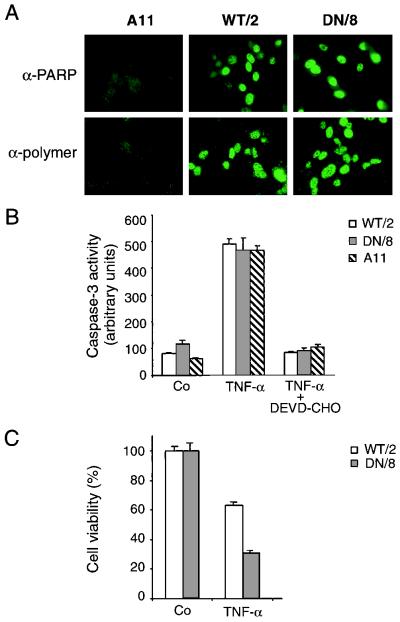
Since DEVD lies within the bilateral motifs of the nuclear localization signal, we determined if the mutation at the cleavage site affects subcellular localization of PARP. Indirect immunofluorescence staining with an anti-PARP antiserum revealed nuclear staining in both wild-type PARP- and mutant PARP-expressing cells (Fig. 2A), indicating that the D214N mutation did not affect nuclear targeting of PARP.
FIG. 2.
Characterization of wild-type and caspase-resistant PARP in transfected mouse fibroblast cells. (A) Upper panels, immunofluorescence analysis detects PARP expression in nuclei of WT/2 cells and DN/8 cells with a polyclonal anti-PARP antiserum, Vic-5. Lower panels, following treatment with MNNG, poly(ADP-ribose) polymer formation in WT/2 and DN/8 cells was detected by immunofluorescence after staining with a mouse monoclonal antibody, 10H. A11 cells (PARP−/−) were used as a control. (B) Analysis of caspase-3 activity in WT/2, DN/8, and A11 cells which were either treated or not treated with 20 ng of TNF-α per ml for 12 h. Cell lysates were preincubated with or without the caspase-3 inhibitor DEVD-CHO, and caspase-3 activity was measured by using a fluorescent substrate, DEVD-AFC, in a fluorometer. Data are from one of three independent experiments and are expressed as means ± standard deviations for duplicate samples. (C) Expression of caspase-resistant PARP sensitizes fibroblasts to TNF-α-induced cell death. WT/2 and DN/8 cells were either treated or not treated with TNF-α for 12 h, and cell viability was analyzed by the MTT assay. The results are shown as means ± standard deviations for quadruplicate samples. Data are from one of three independent experiments.
To test if mutation at the cleavage site affects PARP enzymatic function, namely, poly(ADP-ribosyl)ation following DNA damage, we used an indirect immunofluorescence assay to detect poly(ADP-ribose) polymer formation in these transfectants. While PARP activity was not detectable in untreated cells (neither in WT/2 nor in DN/8 cells) (data not shown), after treatment with the alkylating agent MNNG, fluorescent signals were readily detected in the nuclei of both WT/2 and DN/8 cells by staining with an antibody against poly(ADP-ribose) polymers (Fig. 2A). These results demonstrate that wild-type PARP and D214N mutant PARP can be activated by DNA breaks to catalyze poly(ADP-ribose) polymer formation.
To rule out the possibility that caspase-resistant PARP might act as an inhibitor of caspase-3-like proteases, the activity of caspase-3 in cells expressing uncleavable PARP was measured with a fluorometer following the cleavage of DEVD-AFC, a caspase substrate conjugated with a fluorescence tag. Figure 2B shows that similar levels of caspase-3 activity was observed in DN/8 and WT/2 as well as A11 clones, whereas caspase-3 activity was completely inhibited by DEVD-CHO, a specific caspase-3 inhibitor. These results indicate that the prevention of PARP cleavage by caspases is not due to the mutation at codon 214 of the protein having an inhibitory effect on caspase-3-like proteases and that caspase activity in PARP−/− cells is not altered.
Caspase-resistant PARP sensitizes fibroblasts to TNF-α-induced death.
To define an optimal stage to analyze the death response and molecular changes in these cells, we first performed a time course experiment by using the MTT assay. After TNF-α treatment, cells start to die after 2 h; about 40 to 50% die after around 10 to 12 h, and all cells die after 24 h (data not shown). We decided to analyze the death response during 12 h from the beginning of apoptotic treatment. At 12 h after TNF-α treatment, more DN/8 cells expressing cleavage-resistant PARP (63%) than WT/2 cells expressing wild-type protein (∼30%) had died (Fig. 2C). To further characterize the mode of cell death, we applied various measurements to detect apoptosis versus necrosis.
Cells expressing caspase-resistant PARP exhibit increased apoptosis.
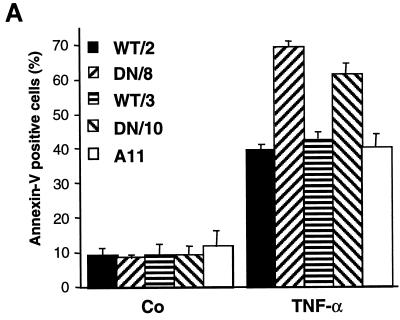
Since annexin V binds to externalized phosphatidylserine on membranes of early apoptotic cells (37), we used flow cytometry (FACS) analysis after staining cells with FITC-conjugated annexin V and PI. A 12-h treatment with TNF-α resulted in a higher percentage of apoptotic cells (annexin V positive and PI negative) in both mutant clones DN/8 and DN/10 than in their WT/2 and WT/3 counterparts as well as untransfected A11 cells (Fig. 3A). While about 70 and 60% of the DN/8 and DN/10 cells, respectively, were positive for annexin V staining, approximately 40% of the WT/2 and WT/3, as well as A11, cells were apoptotic (Fig. 3A). The enhanced apoptotic response in cells expressing caspase-resistant PARP was also observed after staining of cells by the TUNEL technique, which detects DNA fragmentation occurring in apoptosis. After treatment with TNF-α, there was a higher proportion of TUNEL-positive DN/8 cells (∼60%) than of TUNEL-positive WT/2 cells (34%) (Fig. 3B and C). Together, these experiments show that uncleavable PARP sensitizes cells to TNF-α-induced apoptosis.
FIG. 3.
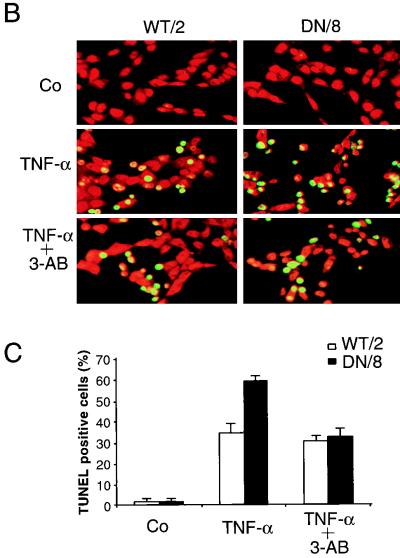
Enhanced apoptosis in cells expressing caspase-resistant PARP. (A) Flow cytometry analysis of two cell clones expressing wild-type PARP (WT/2 and WT/3) and two clones expressing mutant PARP (DN/8 and DN/10) as well as nontransfected cells (A11) after treatment with TNF-α and staining with FITC-annexin V and PI. The results are given as the means ± standard deviations for triplicate samples. Data are from one of three independent experiments. Co, control, not treated. (B) Fluorescence analysis of apoptosis in wild-type and caspase-resistant PARP-expressing cells by TUNEL staining. WT/2 or DN/8 cells were either not treated (control [Co]) or treated with TNF-α or TNF-α together with 3-AB for 12 h and then stained by the TUNEL technique and with PI. TUNEL-positive cells show bright yellow fluorescent nuclei (middle panels), whereas PI staining visualizes nuclei of all cells (red). The increased apoptosis in DN/8 cells was repressed by 3-AB to the level in WT/2 cells, as judged by the reduced number of TUNEL-positive cells (lower panels). (C) The percentage of TUNEL-positive cells was quantified by scoring yellow nuclei (see above). At least 100 cells were evaluated in each sample. Data show means ± standard deviations for triplicate samples from two independent experiments.
Necrosis is induced in cells expressing caspase-resistant PARP after TNF-α treatment.
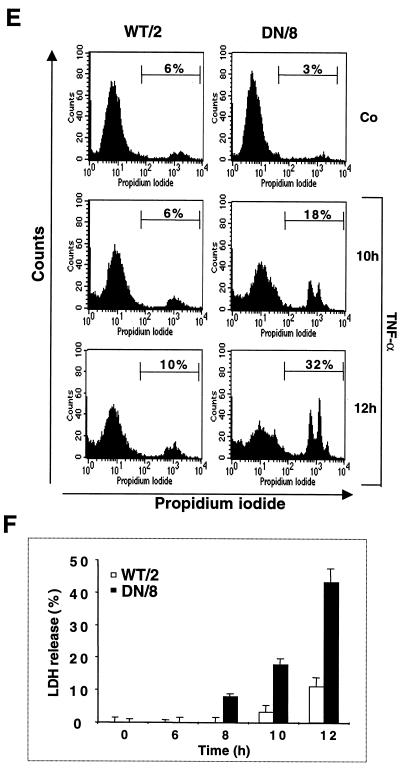
We next examined necrotic cell death in WT/2 and DN/8 cells after TNF-α treatment. After 12 h of treatment with TNF-α, cells were stained with Ho-342 and PI or with CAM and EthD-1, followed by fluorescence microscopy analysis. After Ho-342 and PI staining, the necrotic cells lost their membrane integrity as shown by uptake of PI, resulting in pink nuclear staining. A larger proportion of DN/8 cells (27%) than of WT/2 cells (∼5%) was stained positive for this dye (Fig. 4A and B). Similar results were also obtained by using CAM and EthD-1 staining. While ∼30% EthD-1-positive cells were observed in the DN/8 population, only a small proportion of necrotic cells was observed in the WT/2 population (Fig. 4C and D). FACS analysis confirmed this observation, because TNF-α treatment resulted in 32% of DN/8 cells taking up PI, whereas only 10% of WT/2 cells could take up the dye, an increase from 6% for the untreated group (Fig. 4E). All three assays were consistent and revealed similar levels (about 30%) of necrotic death in DN/8 cells. The disruption of the necrotic cell membrane was further tested by examining LDH release from cultures of these cells. As shown in Fig. 4F, a dramatic increase of LDH was observed in the supernatant of DN/8 cell cultures in a time-dependent manner, whereas only a small increase was found in that of WT/2 cells. This increased cell lysis in DN/8 cells was also seen by the trypan blue exclusion assay, which measures the necrotic cells as they lose their membrane integrity and fail to exclude the dye (data not shown). Taken together, these results show that cells expressing uncleavable PARP after apoptotic induction exhibit necrotic characteristics.
FIG. 4.
Induction of necrosis in cells expressing caspase-resistant PARP. (A to D) Fluorescence microscopy analyses of necrosis in WT/2 or DN/8 cells. Cells were either not treated (control [Co]) or treated with TNF-α or TNF-α together with 3-AB for 12 h and then stained with either Ho-342 and PI (A and B) or CAM and EthD-1 (C and D) (see Materials and Methods). Necrotic cells were quantified by scoring pink nuclei (A and B) or red nuclei (C and D). At least 100 cells were analyzed in each sample. Data show means ± standard deviations for triplicate samples from two independent experiments. (E) Analysis of necrotic cells by flow cytometry. Cells were treated with TNF-α and stained with PI at the indicated time points. The percentage of necrotic cells, seen as a shift to stronger PI fluorescence, was quantified by using CellQuest software. (F) After TNF-α induction, the extent of necrotic cell death was assessed by LDH release measurement at the indicated time points, and the LDH contents in supernatants of WT/2 and DN/8 cultures were corrected to that without TNF-α treatment. The data are means ± standard deviations for quadruplicate samples and are representative of two independent experiments.
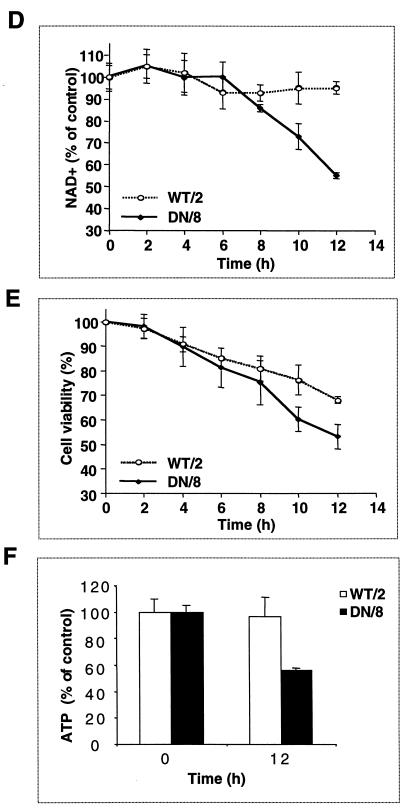
NAD+ and ATP pools are depleted in apoptotic cells expressing uncleavable PARP.
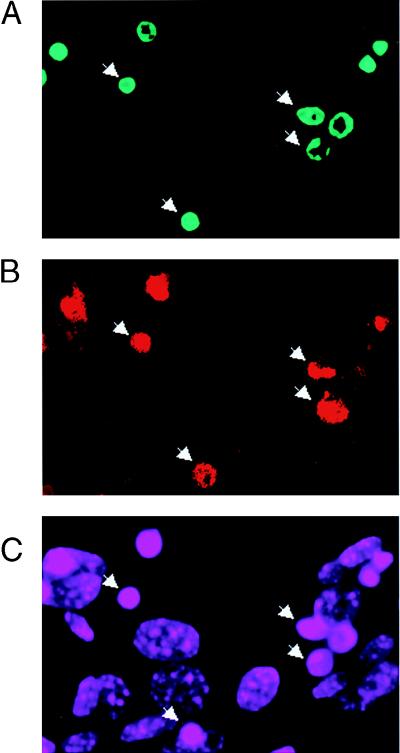
Since activation of PARP is dependent solely on DNA breaks, which lead to poly(ADP-ribose) polymer formation and subsequent depletion of intracellular NAD+, we hypothesized that noncleavable PARP could be activated by apoptotic fragmented DNA in TUNEL-positive cells and NAD+ would be consumed. First, we examined the enzymatic function of PARP in these cells by double staining with TUNEL and the antibody against poly(ADP-ribose) polymers. After TNF-α induction, polymer formation can be detected concomitantly in DN/8 cells that are TUNEL positive, although some TUNEL-positive cells were negative for polymer formation (Fig. 5A to C), indicating polymer synthesis by uncleaved PARP in cells with fragmented DNA. Next, a time course study of NAD+ consumption in correlation with the cell death profile was performed after incubation with TNF-α. Figure 5D shows that the NAD+ levels in DN/8 cells were unchanged for the first 6 h of TNF-α treatment and then decreased, falling to 55% of the levels in untreated cells at 12 h. In contrast, the levels of NAD+ in TNF-α-treated WT/2 cells were slightly decreased but remained significantly higher than those in DN/8 cells at the 12-h point (Fig. 5D), suggesting that wild-type PARP is inactive, most likely due to its cleavage by caspases. In parallel, cell survival following TNF-α treatment was measured at the same time points by using the MTT assay. The results show that the proportion of dead cells in DN/8 cultures was significantly higher than that in WT/2 cultures after 8 h (Fig. 5E).
FIG. 5.
Enzymatic activity of uncleavable PARP in a late stage of apoptosis. (A to C) Immunofluorescence analysis of DN/8 cells by triple staining with TUNEL, antibody against poly(ADP-ribose) polymers, and DAPI. After 10 h of TNF-α treatment, TUNEL-positive cells show bright green staining (FITC) (A), and polymer formation was visualized as red (rhodamine) (B). Polymer formation was detected in most but not all of the TUNEL-positive cells (arrows in A and B). DAPI staining visualizes all cell nuclei (C). (D and E) Time course study of intracellular NAD+ levels and cell viability during apoptosis. Cells were treated with TNF-α and analyzed for NAD+ content at the indicated time points. The total NAD+ content in the control (100%) was 1.43 ± 0.037 pmol for 103 WT/2 cells and 1.25 ± 0.047 pmol for 103 DN/8 cells (D). In parallel, cell viability was measured by the MTT assay at the same time points (E). (F) Intracellular ATP content of WT/2 and DN/8 cells. Cells were either treated or not treated with TNF-α for 12 h, and the intracellular ATP level was determined by using the luciferase assay. Data are means ± standard deviations for at least triplicate samples and are representative of two independent experiments.
To test whether the reduction of NAD+ could also deplete the ATP pool (51), we measured the intracellular ATP levels in both cell types. At 12 h after treatment with TNF-α, cell lysates from DN/8 cells contained less than 60% of the control ATP content (Fig. 5F), which correlates well with the similar level of NAD+ in these cells (Fig. 5D). In contrast, the ATP level in the WT/2 population was not changed after TNF-α treatment, which is consistent with unchanged NAD+ levels (Fig. 5D and F). Since polymer formation by uncleaved PARP was detected in TUNEL-positive cells (Fig. 5A and B), the partial reduction of the NAD+ and ATP pool (about 45%) most likely resulted from a subpopulation of DN/8 cells containing apoptotic DNA breaks and thus corresponded to more than 90% depletion of the intracellular NAD+ and ATP pool in polymer-forming cells. These results suggest that apoptosis-induced chromatin fragmentation activates uncleaved PARP, resulting in NAD+ and ATP depletion.
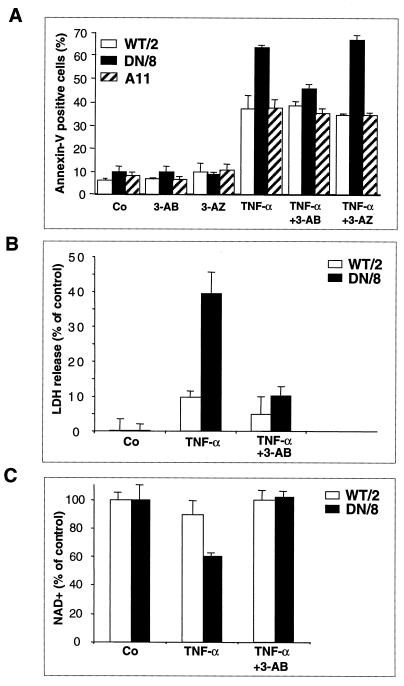
PARP inhibition counteracts the enhanced apoptotic response and abolishes necrosis of cells expressing caspase-resistant PARP.
To further investigate the finding that the increased death response is caused by NAD+ depletion due to the activation of uncleaved PARP induced by DNA breaks, DN/8 and WT/2 cells were preincubated with 3-aminobenzamide (3-AB), a widely used PARP inhibitor, or its noninhibitory analog, 3-aminobenzoic acid (3-AZ). FACS analysis after annexin V staining revealed that the levels of TNF-α-induced apoptosis in DN/8 cells were reduced from 65 to 45%, approximately the levels in WT/2 cells (Fig. 6A). However, 3-AB did not inhibit apoptosis in WT/2 cells, suggesting that this substance does not possess a general antiapoptotic property. The inhibition of increased magnitudes of apoptosis by 3-AB seems to be specific for the inhibition of PARP function, as 3-AZ did not affect the apoptotic response in either DN/8 or WT/2 cells, compared to the groups without 3-AB treatment (Fig. 6A). Moreover, preincubation with 3-AB inhibited the increased proportion of TUNEL-positive DN/8 cells to a level similar to that for WT/2 cells (Fig. 3C), indicating that the enhanced apoptosis is due to the activity of uncleaved PARP.
FIG. 6.
The PARP inhibitor 3-AB prevents NAD+ depletion and counteracts both necrosis and enhanced apoptosis in cells expressing noncleavable PARP. (A) WT/2 and DN/8 cells were preincubated for 30 min with 3 mM 3-AB or its noninhibitory analog 3-AZ. After treatment with TNF-α, samples were stained with FITC-annexin V and PI and analyzed by flow cytometry as described for Fig. 3. (B) Cells were preincubated with 3 mM 3-AB for 90 min. After treatment with TNF-α, LDH release in the supernatants of the indicated clones was measured. Data are means ± standard deviations for triplicate samples and are from one of three independent experiments. (C) After treatment with 3-AB and TNF-α, NAD+ levels were measured as described for Fig. 5D. Data are means ± standard deviations and are representative of two experiments.
We have also studied the effect of 3-AB on the induction of necrotic cell death after TNF-α treatment and found that the inhibitor reduced PI-positive fractions of Ho-342- and PI-stained DN/8 cells to a level similar to that for WT/2 cells (Fig. 4A and B) and that LDH release in DN/8 cells is also markedly prevented (Fig. 6B). Furthermore, 3-AB treatment preserves the level of NAD+ content in DN/8 cells during TNF-α-induced cell death (Fig. 6C). These results suggest that the NAD+ drop is a consequence of activity of uncleavable PARP and indicate that this is coupled with the enhanced cell death.
DISCUSSION
Failure of PARP cleavage leads to increased cell death induced by TNF-α.
A characteristic event of apoptosis is the proteolytic cleavage of PARP, suggesting a role for this protein in apoptosis. However, PARP−/− lymphocytes, fibroblasts, and hepatocytes, as well as neuronal cells, undergo normal apoptosis induced by various stimuli, including TNF-α and anti-Fas, indicating that PARP is dispensable in the apoptotic cascade (32, 59). Perhaps surprisingly, the present study using mouse fibroblast cells expressing caspase-resistant PARP demonstrates that prevention of PARP cleavage induces necrosis and also enhanced apoptosis after treatment with TNF-α, indicating that PARP cleavage by caspases is an important event in response to apoptotic stimuli.
We can rule out the possibility that the additional cell death is attributable to the toxicity of poly(ADP-ribose) polymers as is the case with yeast, where overexpression of PARP results in inhibition of proliferation due to interference of poly(ADP-ribosyl)ated proteins with chromatin replication (3, 9, 25). Since cells after caspase activation and DNA fragmentation are no longer able to replicate their DNA, this mechanism is not applicable in the observed cell death.
The ability of PARP to synthesize poly(ADP-ribose) polymers by using NAD+ is absolutely dependent on DNA strand breaks (34). The enhanced cell death in cells expressing uncleavable PARP is most likely caused by the activation of caspase-resistant PARP in response to DNA fragmentation and the consequent depletion (more than 90%) of intracellular NAD+ and ATP in the apoptotic (TUNEL-positive) fraction. The present study provides several lines of evidence to support this conclusion. (i) Polymer formation was found in TUNEL-positive cells that contain uncleavable PARP, suggesting that uncleavable PARP is activated by chromatin DNA breaks. (ii) The PARP inhibitor 3-AB prevents the NAD+ drop and concomitantly blocks the elevated cell death response. (iii) NAD+ levels in wild-type-PARP-expressing cells were not changed, indicating that wild-type PARP is inactivated by caspase cleavage prior to DNA fragmentation. (iv) Finally, inhibition of caspases by specific tetrapeptide also blocks the increased death (data not shown), suggesting that this event is dependent on caspase-mediated downstream events, e.g., DNA fragmentation.
Failure of PARP cleavage leads to NAD+ depletion-induced necrosis.
It has been proposed that the cleavage (i.e., the inactivation) of PARP by caspases inhibits poly(ADP-ribosyl)ation and prevents depletion of NAD+ and ATP pools, which would otherwise cause necrosis leading to a pathological inflammatory response (12). Our data are in agreement with this hypothesis, since NAD+ and ATP depletion in cells expressing caspase-resistant PARP induces necrosis, as determined by several criteria, including fluorescence microscopy after staining of cells with Ho-234 and PI or with CAM and EthD-1, FACS analysis of PI uptake, and LDH release measurement. This was further substantiated by using the PARP inhibitor 3-AB, which can preserve the NAD+ level and concomitantly abolish necrosis.
The necrotic cells most likely originated from apoptotic cells, i.e., TUNEL-positive cells. Since the TUNEL-positive fraction of DN/8 cells displays polymer formation, it is reasonable to speculate that the drop in NAD+ and ATP occurs in the same cells, which can be readily detected by necrotic criteria. However, due to technical limitations, we were not able to confirm whether TUNEL-positive cells also simultaneously exhibit features of necrosis at the single-cell level. This necrosis seems to occur specifically and more rapidly in cells containing caspase-resistant PARP than in cells expressing wild-type PARP, although the latter would eventually undergo secondary necrosis at a late stage of apoptosis, as judged by LDH release (Fig. 4F and data not shown). While we cannot rule out the possibility that the necrosis seen in DN/8 cells is secondary to accelerated apoptosis, many of the PI- and EthD-positive cells exhibit no obvious breakage of nuclei and lack of dye conversion in their cytoplasm (Fig. 4A and C), suggesting a necrotic death disrupting the apoptotic program. Nevertheless, this result is consistent with the theory known as the suicide hypothesis, originally proposed by Berger (4), in which PARP-mediated NAD+ depletion can cause reduction in glycolysis, disturbed purine nucleotide metabolism, and impaired synthesis of ATP and macromolecules. This mechanism has been demonstrated to be responsible for the death of several cell types after DNA damage, such as neuronal cells (14, 16, 62), pancreatic islet cells (20), and muscle cells (55). However, the exact molecular mechanism by which NAD+ or ATP depletion kills cells is presently unknown.
It has been shown that various apoptosis-inducing stimuli can trigger both apoptotic and necrotic pathways which can be regulated by mitochondrial permeability transition, caspase activation, and ATP supply (13, 21, 33, 57). Other factors, such as intensity and duration of treatment, may also affect the determination of these two distinct pathways (5, 37). In contrast to these events controlling the mode of cell death, activation of uncleavable PARP is a rather late event to induce necrosis, after apoptotic DNA fragmentation.
PARP cleavage plays a regulatory role in the process of apoptosis.
Although the cleavage of a few proteins, such as DFF/ICAD (15, 35) as well as Bcl-2 (6) and Bcl-xL (8), has been shown to be involved in the initiation and execution of the apoptotic cascade, the significance of caspase cleavage of most cellular substrates has not been explored. Our findings that failure of PARP cleavage sensitizes cells to apoptosis indicate that PARP cleavage is an important step in the apoptotic process. The fact that uncleavable PARP sensitizes cells to apoptotic treatment is unexpected in the light of the previously proposed hypotheses. (i) PARP is an enzyme involved in DNA repair, and its cleavage may inactivate their DNA repair system, which otherwise might impede progression of apoptosis (44, 59). (ii) Inactivation of PARP is suggested to be required for deribosylation of some endonucleases in order to facilitate DNA fragmentation in apoptosis (49). (iii) The cleavage of cellular proteins, such as PARP, may be just one unrelated event during apoptosis (41, 44).
However, the cause of accelerated apoptosis by uncleavable PARP is not clear. One possibility is that NAD+ depletion and compromised energy metabolism may alter mitochondrial permeability transition (21), which causes disturbance of the mitochondrial transmembrane potential leading to the release of proapoptotic factors and may affect one or several of the self-amplifying feedback loops (29, 45). Another explanation is that since necrosis is an uncontrolled cell death in response to massive cell damage, it may passively contribute to an already-started apoptotic cascade. Finally, poly(ADP-ribosyl)ation and PARP activity were suggested to positively correlate with the apoptotic response (60).
The result that uncleavable PARP accelerates apoptosis is consistent with earlier inhibitor studies, where inhibition of PARP activity delayed apoptosis (1, 30, 38, 42). Although the mechanism underlying the results of the inhibitor studies is not clear, it is possible that chemical inhibition of PARP prevents its release from DNA ends, which may impede subsequent DNA fragmentation during apoptosis. The increased apoptosis seems to be a consequence of PARP activity, because the PARP inhibitor 3-AB counteracts only an elevated fraction of apoptosis in mutant PARP-expressing cells and does not inhibit apoptosis in WT/2 cells. This result also suggests that PARP activity is not essential for induction of apoptosis, which is consistent with previous observations that cells lacking PARP exhibit normal apoptosis (32, 59). However, these findings appear to be in contrast to the recent observations that PARP is transiently activated in the early phase of apoptosis (50) and that inactivation of PARP by a dominant-negative mutation or gene targeting at the fourth exon facilitates DNA fragmentation induced by alkylating agents and gamma radiation (11, 46), although death pathways induced by these agents are complex and have not yet been defined. This result is also in contradiction to the recent findings by Oliver et al. that transient transfection of uncleavable PARP renders fibroblasts resistant to Fas antibody-induced apoptosis (43). The discrepancy in these data is probably due to the nature of the genetic modification introduced into the cells used. While the cells in the present study were originally derived from mutant mice generated by gene targeting to the second exon, which destroys the PARP function, Oliver et al. (43) used cells in which the PARP gene was disrupted at the fourth exon, which might maintain its DNA binding activity (24). In addition, perhaps due to the use of the transient-transfection technique, apoptosis was scored only by morphological criteria, such as flat cells (alive) versus rounded cells (dead) or nuclear condensation (43).
In conclusion, the present study, using fibroblasts which express caspase-resistant PARP, demonstrates that although PARP per se is dispensable in apoptosis, its cleavage has evolved as a mechanism to ensure the normal speed and order of apoptotic events and to protect cells from necrotic death. Inappropriate apoptosis can lead to several degenerative diseases, autoimmune processes, and carcinogenesis (56). Although the significance of this event is obscure in cell culture systems, abnormal accelerated apoptosis and induction of unwanted necrosis, with release of cell content in vivo, may result in the disturbance of biological processes, such as tissue homeostasis, or an inflammatory response, as well as tumor and disease development.
ACKNOWLEDGMENTS
We thank B. Auer for providing human PARP cDNA, W. Fiers for TNF-α, D. Nicholson for recombinant caspase-3, and A. Bürkle for 10H antibody. We also thank R. Kurzbauer and S. Aigner for technical assistance. We are grateful to G. Mollon for preparation of photographs. We are also grateful to A. E. Grigoriadis, J. Hall, C. Morrison, K. Schulze-Osthoff, K. Sabapathy, and E. F. Wagner for critical comments and discussions.
Z.H. is in receipt of an IARC Special Training Award. This project was initiated at the Research Institute of Molecular Pathology, Vienna, Austria.
REFERENCES
- 1.Agarwal S, Drysdale B E, Shin H S. Tumor necrosis factor-mediated cytotoxicity involves ADP-ribosylation. J Immunol. 1988;140:4187–4192. [PubMed] [Google Scholar]
- 2.Arends M J, Wyllie A H. Apoptosis: mechanisms and roles in pathology. Int Rev Exp Pathol. 1991;32:223–254. doi: 10.1016/b978-0-12-364932-4.50010-1. [DOI] [PubMed] [Google Scholar]
- 3.Avila M A, Velasco J A, Smulson M E, Dritschilo A, Castro R, Notario V. Functional expression of human poly(ADP-ribose) polymerase in Schizosaccharomyces pombe results in mitotic delay at G1, increased mutation rate, and sensitization to radiation. Yeast. 1994;10:1003–1017. doi: 10.1002/yea.320100803. [DOI] [PubMed] [Google Scholar]
- 4.Berger N A. Poly(ADP-ribose) in the cellular response to DNA damage. Radiat Res. 1985;101:4–15. [PubMed] [Google Scholar]
- 5.Bonfoco E, Krainc D M, Ankarcrona, Nicotera P, Lipton S A. Apoptosis and necrosis: two distinct events induced, respectively, by mild and intense insults with N-methyl-d-aspartate or nitric oxide/superoxide in cortical cell culture. Proc Natl Acad Sci USA. 1995;92:7162–7166. doi: 10.1073/pnas.92.16.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng E H, Kirsch D G, Clem R J, Ravi R, Kastan M B, Bedi A, Ueno K, Hardwick J M. Conversion of Bcl-2 to a Bax-like death effector by caspases. Science. 1997;278:1966–1968. doi: 10.1126/science.278.5345.1966. [DOI] [PubMed] [Google Scholar]
- 7.Cherney B W, McBride O W, Chen D, Alkhatib H, Bhatia K, Hensley P, Smulson M E. cDNA sequence, protein structure, and chromosomal location of the human gene for poly(ADP-ribose) polymerase. Proc Natl Acad Sci USA. 1987;84:8370–8374. doi: 10.1073/pnas.84.23.8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clem R J, Cheng E H, Karp C L, Kirsch D G, Ueno K, Takahashi A, Kastan M B, Griffin D E, Earnshaw W C, Veliuona M A, Hardwick J M. Modulation of cell death by Bcl-XL through caspase interaction. Proc Natl Acad Sci USA. 1998;95:554–559. doi: 10.1073/pnas.95.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collinge M A, Althaus F R. Expression of human poly(ADP-ribose) polymerase in Saccharomyces cerevisiae. Mol Gen Genet. 1994;245:686–693. doi: 10.1007/BF00297275. [DOI] [PubMed] [Google Scholar]
- 10.Cryns V, Yuan J. Proteases to die for. Genes Dev. 1998;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- 11.de Murcia J M, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver F J, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earnshaw W C. Apoptosis: lessions from in vitro systems. Trends Cell Biol. 1995;5:217–220. doi: 10.1016/s0962-8924(00)89006-6. [DOI] [PubMed] [Google Scholar]
- 13.Eguchi Y, Shimizu S, Tsujimoto Y. Intracellular ATP levels determine cell death fate by apoptosis or necrosis. Cancer Res. 1997;57:1835–1840. [PubMed] [Google Scholar]
- 14.Eliasson M J, Sampei K, Mandir A S, Hurn P D, Traystman R J, Bao J, Pieper A, Wang Z-Q, Dawson T D, Snyder S H, Dawson V L. Poly (ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 15.Enari M, Sahahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 16.Endres M, Wang Z-Q, Namura S, Waeber C, Moskowitz M A. Ischemic brain injury is mediated by the activation of poly(ADP-ribose) polymerase. J Cereb Blood Flow Metab. 1997;17:1143–1151. doi: 10.1097/00004647-199711000-00002. [DOI] [PubMed] [Google Scholar]
- 17.Gaal J C, Smith K R, Pearson C K. Cellular euthanasia mediated by a nuclear enzyme: a central role for nuclear ADP-ribosylation in cellular metabolism. Trends Biochem Sci. 1987;12:129–130. [Google Scholar]
- 18.Gavrieli Y, Sherman Y, Ben-Sasson S A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grooten J, Goossens V, Vanhaesebroeck B, Fiers W. Cell membrane permeabilization and cellular collapse, followed by loss of dehydrogenase activity: early events in tumour necrosis factor-induced cytotoxicity. Cytokine. 1993;5:546–555. doi: 10.1016/s1043-4666(05)80003-1. [DOI] [PubMed] [Google Scholar]
- 20.Heller B, Wang Z-Q, Wagner E F, Radons J, Bürkle A, Fehsel K, Burkart V, Kolb H. Inactivation of the poly(ADP-ribose) polymerase gene affects oxygen radical and nitric oxide toxicity in islet cells. J Biol Chem. 1995;270:11176–11180. doi: 10.1074/jbc.270.19.11176. [DOI] [PubMed] [Google Scholar]
- 21.Hirsch T, Marchetti P, Susin S A, Dallaporta B, Zamzami N, Marzo I, Geuskens M, Kroemer G. The apoptosis-necrosis paradox. Apoptogenic proteases activated after death. Oncogene. 1997;15:1573–1581. doi: 10.1038/sj.onc.1201324. [DOI] [PubMed] [Google Scholar]
- 22.Horvitz H R, Shaham S, Hengartner M O. The genetics of programmed cell death in the nematode Caenorhabditis elegans. Cold Spring Harbor Symp Quant Biol. 1994;59:377–385. doi: 10.1101/sqb.1994.059.01.042. [DOI] [PubMed] [Google Scholar]
- 23.Jacobson E L, Jacobson M K. Tissue NAD as a biochemical measure of niacin status in humans. Methods Enzymol. 1997;280:221–230. doi: 10.1016/s0076-6879(97)80113-9. [DOI] [PubMed] [Google Scholar]
- 24.Jeggo P A. PARP—another guardian angel? Curr Biol. 1998;8:R49–R51. doi: 10.1016/s0960-9822(98)70032-6. [DOI] [PubMed] [Google Scholar]
- 25.Kaiser P, Auer B, Schweiger M. Inhibition of cell proliferation in Saccharomyces cerevisiae by expression of human NAD+ ADP-ribosyltransferase requires the DNA binding domain (“zinc fingers”) Mol Gen Genet. 1992;232:231–239. doi: 10.1007/BF00280001. [DOI] [PubMed] [Google Scholar]
- 26.Kaufmann S H, Desnoyers S, Ottaviano Y, Davidson N E, Poirier G G. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 27.Kawamitsu H, Hoshino H, Okada H, Miwa M, Momoi H, Sugimura T. Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry. 1984;23:3771–3777. doi: 10.1021/bi00311a032. [DOI] [PubMed] [Google Scholar]
- 28.Kerr J F, Winterford C M, Harmon B V. Apoptosis. Its significance in cancer and cancer therapy. Cancer. 1994;73:2013–2026. doi: 10.1002/1097-0142(19940415)73:8<2013::aid-cncr2820730802>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 29.Kroemer G. The proto-oncogene Bcl-2 and its role in regulating apoptosis. Nat Med. 1997;3:614–620. doi: 10.1038/nm0697-614. [DOI] [PubMed] [Google Scholar]
- 30.Kuo M L, Chau Y P, Wang J H, Shiah S G. Inhibitors of poly(ADP-ribose) polymerase block nitric oxide-induced apoptosis but not differentiation in human leukemia HL-60 cells. Biochem Biophys Res Commun. 1996;219:502–508. doi: 10.1006/bbrc.1996.0263. [DOI] [PubMed] [Google Scholar]
- 31.Lazebnik Y A, Kaufmann S H, Desnoyers S, Poirier G G, Earnshaw W C. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 32.Leist M, Single B, Kunstle G, Volbracht C, Hentze H, Nicotera P. Apoptosis in the absence of poly-(ADP-ribose) polymerase. Biochem Biophys Res Commun. 1997;233:518–522. doi: 10.1006/bbrc.1997.6491. [DOI] [PubMed] [Google Scholar]
- 33.Leist M, Single B, Castoldi A F, Kuhnle S, Nicotera P. Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. J Exp Med. 1997;185:1481–1486. doi: 10.1084/jem.185.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindahl T, Satoh M S, Poirier G G, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Zou H, Slaughter C, Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 36.Lundin A, Hasenson M, Persson J, Pousette A. Estimation of biomass in growing cell lines by adenosine triphosphate assay. Methods Enzymol. 1986;133:27–42. doi: 10.1016/0076-6879(86)33053-2. [DOI] [PubMed] [Google Scholar]
- 37.Martin S J, Reutelingsperger C P, McGahon A J, Rader J A, van Schie R C, LaFace D M, Green D R. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J Exp Med. 1995;182:1545–1556. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monti D, Cossarizza A, Salvioli S, Franceschi C, Rainaldi G, Straface E, Rivabene R, Malorni W. Cell death protection by 3-aminobenzamide and other poly(ADP-ribose)polymerase inhibitors: different effects on human natural killer and lymphokine activated killer cell activities. Biochem Biophys Res Commun. 1994;199:525–530. doi: 10.1006/bbrc.1994.1260. [DOI] [PubMed] [Google Scholar]
- 39.Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 40.Nicholson D W, Ali A, Thornberry N A, Vaillancourt J P, Ding C K, Gallant M, Gareau Y, Griffin P R, Labelle M, Lazebnik Y A, Munday N A, Raju S M, Smulson M E, Yamin T-T, Yu V L, Miller D K. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 41.Nicholson D W, Thornberry N A. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 42.Nosseri C, Coppola S, Ghibelli L. Possible involvement of poly(ADP-ribosyl) polymerase in triggering stress-induced apoptosis. Exp Cell Res. 1994;212:367–373. doi: 10.1006/excr.1994.1156. [DOI] [PubMed] [Google Scholar]
- 43.Oliver F J, de la Rubia G, Rolli V, Ruiz-Ruiz M C, de Murcia G, Murcia J M. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. J Biol Chem. 1998;273:33533–33539. doi: 10.1074/jbc.273.50.33533. [DOI] [PubMed] [Google Scholar]
- 44.Patel T, Gores G J, Kaufmann S H. The role of proteases during apoptosis. FASEB J. 1996;10:587–597. doi: 10.1096/fasebj.10.5.8621058. [DOI] [PubMed] [Google Scholar]
- 45.Reed J C. Cytochrome c: can’t live with it—can’t live without it. Cell. 1997;91:559–562. doi: 10.1016/s0092-8674(00)80442-0. [DOI] [PubMed] [Google Scholar]
- 46.Schreiber V, Hunting D, Trucco C, Gowans B, Grunwald D, de Murcia G, Menissier de Murcia J. A dominant-negative mutant of human poly(ADP-ribose) polymerase affects cell recovery, apoptosis, and sister chromatid exchange following DNA damage. Proc Natl Acad Sci USA. 1995;92:4753–4757. doi: 10.1073/pnas.92.11.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulze-Osthoff K, Krammer P H, Droge W S. Divergent signalling via APO-1/Fas and the TNF receptor, two homologous molecules involved in physiological cell death. EMBO J. 1994;13:4587–4596. doi: 10.1002/j.1460-2075.1994.tb06780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu S, Eguchi Y, Kamiike W, Waguri S, Uchiyama Y, Matsuda H, Tsujimoto Y. Bcl-2 blocks loss of mitochondrial membrane potential while ICE inhibitors act at a different step during inhibition of death induced by respiratory chain inhibitors. Oncogene. 1996;13:21–29. [PubMed] [Google Scholar]
- 49.Shiokawa D, Ohyama H, Yamada T, Takahashi K, Tanuma S. Identification of an endonuclease responsible for apoptosis in rat thymocytes. Eur J Biochem. 1994;226:23–30. doi: 10.1111/j.1432-1033.1994.tb20022.x. [DOI] [PubMed] [Google Scholar]
- 50.Simbulan-Rosenthal C M, Rosenthal D S, Iyer S, Boulares A H, Smulson M E. Transient poly(ADP-ribosyl)ation of nuclear proteins and role of poly(ADP-ribose) polymerase in the early stages of apoptosis. J Biol Chem. 1998;273:13703–13712. doi: 10.1074/jbc.273.22.13703. [DOI] [PubMed] [Google Scholar]
- 51.Sims J L, Berger S J, Berger N A. Poly(ADP-ribose) polymerase inhibitors preserve nicotinamide adenine dinucleotide and adenosine 5′-triphosphate pools in DNA-damaged cells: mechanism of stimulation of unscheduled DNA synthesis. Biochemistry. 1983;22:5188–5194. doi: 10.1021/bi00291a019. [DOI] [PubMed] [Google Scholar]
- 52.Stanley P E. Extraction of adenosine triphosphate from microbial and somatic cells. Methods Enzymol. 1986;133:14–22. doi: 10.1016/0076-6879(86)33051-9. [DOI] [PubMed] [Google Scholar]
- 53.Szabo C, Dawson V L. Role of poly(ADP-ribose) synthetase in inflammation and ischaemia-reperfusion. Trends Pharmacol Sci. 1998;19:287–298. doi: 10.1016/s0165-6147(98)01193-6. [DOI] [PubMed] [Google Scholar]
- 54.Tewari M, Quan L T, O’Rourke K, Desnoyers S, Zeng Z, Beidler D R, Poirier G G, Salvesen G S, Dixit V M. Yama/CPP32B, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 55.Thiemermann C, Bowes J, Myint F P, Vane J R. Inhibition of the activity of poly(ADP ribose) synthetase reduces ischemia-reperfusion injury in the heart and skeletal muscle. Proc Natl Acad Sci USA. 1997;94:679–683. doi: 10.1073/pnas.94.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson C B. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 57.Vercammen D, Beyaert R, Denecker G, Goossens V, Van Loo G, Declercq W, Grooten J, Fiers W, Vandenabeele P. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J Exp Med. 1998;187:1477–1485. doi: 10.1084/jem.187.9.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z-Q, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner E F. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 59.Wang Z-Q, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner E F. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright S C, Wei Q S, Kinder D H, Larrick J W. Biochemical pathways of apoptosis: nicotinamide adenine dinucleotide-deficient cells are resistant to tumor necrosis factor or ultraviolet light activation of the 24-kD apoptotic protease and DNA fragmentation. J Exp Med. 1996;183:463–471. doi: 10.1084/jem.183.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wyllie A H, Kerr J F, Currie A R. Cell death: the significance of apoptosis. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 62.Zhang J, Dawson V L, Dawson T M, Snyder S H. Nitric oxide activation of poly(ADP-ribose) synthetase in neurotoxicity. Science. 1994;263:687–689. doi: 10.1126/science.8080500. [DOI] [PubMed] [Google Scholar]