Abstract
Increasing genome data are coming out. Genome size estimation plays an essential role in guiding genome assembly. Several months ago, other researchers were the first to publish a draft genome of the red gromwell (i.e. Lithospermum erythrorhizon). However, we considered that the genome size they estimated and assembled was incorrect. This study meticulously estimated the L. erythrorhizon genome size to should be ∼708.74 Mb and further provided a reliable genome version (size ≈ 693.34 Mb; contigN50 length ≈ 238.08 Kb) to support our objection. Furthermore, according to our genome, we identified a gene family of the alkannin/shikonin O-acyltransferases (i.e. AAT/SAT) that catalysed enantiomer-specific acylations in the alkannin/shikonin biosynthesis (a characteristic metabolic pathway in L. erythrorhizon’s roots) and further explored its evolutionary process. The results indicated that the existing AAT/SAT were not generated from only one round of gene duplication but three rounds; after different rounds of gene duplication, the existing AAT/SAT and their recent ancestors were under positive selection at different amino acid sites. These suggested that a combined power from gene duplication plus positive selection plausibly propelled AAT/SAT’s functional differentiation in evolution.
Keywords: Lithospermum erythrorhizon genome, alkannin/shikonin O-acyltransferases, gene duplication, positive selection
1. Introduction
Red gromwell (Fig. 1A), i.e. Lithospermum erythrorhizon Siebold & Zucc., is a traditional Chinese medicine plant [former name: L. officinale var. erythrorhizon (Siebold & Zucc.) Maxim.1; No. of chromosomes: all records 2n = 282]. In the past, L. erythrorhizon was recognized as a variant of L. officinale L. (former name: L. officinale var. stewartii Kazmi1; No. of chromosomes: most records 2n = 282), although they are now separated species. Besides, based on current molecular evidence, L. erythrorhizon is still determined as the closest species of L. officinale.3
Figure 1.
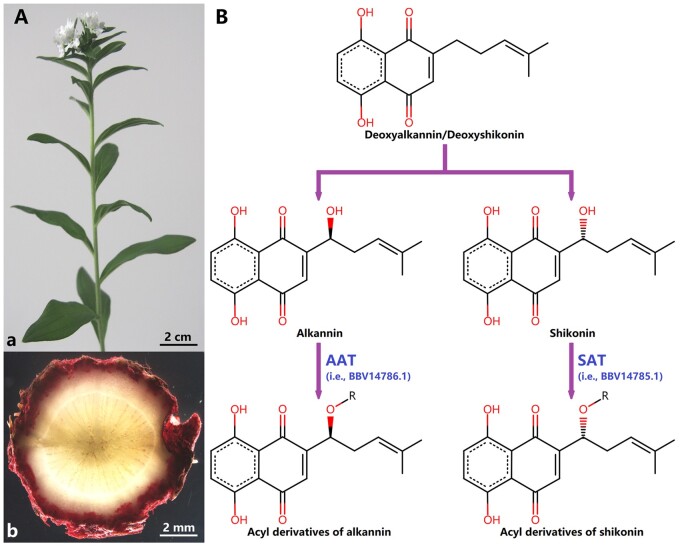
Lithospermum erythrorhizon and alkannin/shikonin’s acylation reactions. (A) An individual L. erythrorhizon in our greenhouse. a. Leaves, stems, and flowers; b. Roots’ cross-section. (B) Alkannin/shikonin’s enantiomer-specific O-acylations. AAT: alkannin O-acyltransferase (NCBI ID: BBV14786.1); SAT: shikonin O-acyltransferase (NCBI ID: BBV14785.1).
Lately, Auber et al.4 published a hybrid assembled genome of L. erythrorhizon (estimated genome size ≈ 369.34 Mb; assembled genome size ≈ 367.41 Mb) using our short Illumina data (NCBI ID: SRX2882373; Supplementary Table S1) plus their long ONT data (NCBI IDs: SRX7432848–SRX7432852; Supplementary Table S1). However, Pustahija et al.5 reported that L. officinale’s genome size was ∼743 Mb (1C ≈ 0.76 pg), significantly greater than the L. erythrorhizon’s genome size estimated and assembled by Auber et al.4 Since the chromosome numbers between L. erythrorhizon and L. officinale are almost identical,2 we consider that this significant difference is not due to polyploidization but to Auber et al.’s misestimation and misassembly.4 Therefore, in this study, we carried out a rigorous genome size estimation and further provided a new version of the L. erythrorhizon genome to support our objection.
Alkannin/shikonin and their acyl derivatives are main secondary-metabolites in L. erythrorhizon’s root periderm (Fig. 1A).4,6 Recently, Oshikiri et al.7 verified that two enzymes (NCBI IDs: BBV14785.1 and BBV14786.1) were enantiomer-specific alkannin/shikonin O-acyltransferases (i.e. AAT/SAT; Fig. 1B) in L. erythrorhizon. However, AAT/SAT family members in L. erythrorhizon and their evolutionary process were still indistinct. Therefore, we detailedly identified AAT/SAT-like superfamily members in the genomes of L. erythrorhizon plus other nine representative species (Supplementary Table S2),8–16 and further conducted a series of analyses to illuminate AAT/SAT’s evolutionary process.
2. Materials and methods
2.1. Plant materials
The seeds of L. erythrorhizon were purchased from ShiJie Seed Breeding Company (https://cfsjzy.1688.com/), located in Chifeng, Inner Mongolia Autonomous Region, China. The healthy seeds were germinated in several pots and then cultured in a greenhouse. Young leaves from flowering individuals were applied for genome sequencing.
2.2. Genome sequencing
Genomic DNA was extracted using a Magnetic Plant Genomic DNA Kit (Cat. no: 4992407; Tiangen, China). After quality control, a paired-end library (insert size ∼170 bp) was constructed using a TIANSeq Fast DNA Library Kit (Cat. no: 4992261; Tiangen, China) and then was sequenced by an Illumina HiSeq 2000 sequenator (Illumina, USA). Subsequently, a SMRTbell library (∼20 Kb) was constructed using a SMRTbell Express Template Prep Kit (PN: 100-938-900; PacBio, USA) and then was sequenced by a PacBio Sequel sequenator (PacBio, USA).
2.3. Data processing
Trimmomatic v0.3617 and FastUniq v1.118 filtered Illumina raw data to remove adapters, low-quality reads, poly-N reads, and PCR-duplicated reads. SMRT Link v6.0 (https://www.pacb.com/support/software-downloads) filtered PacBio raw data to remove adapters and too-short reads (i.e. length < 1 Kb). Furthermore, NanoFilt v2.5.019 filtered ONT raw data that Auber et al. published (NCBI IDs: SRX7432848–SRX7432852; Supplementary Table S1)4 to remove too-short reads (i.e. length < 1 Kb) and low-quality reads (i.e. RQ < 7.0).
2.4. Genome size estimation
Illumina clean data were applied to estimate genome sizes: (i) kmers were counted, and then were exported to histogram files using Jellyfish v2.2.1020 (key parameter: jellyfish histo-h Max_count); (ii) GenomeScope v1.0,21 GenomeScope v2.0,22 and GCE v1.0.223 with the corresponding key parameters (Supplementary Table S3) were applied to calculate genome sizes, respectively. Furthermore, the chloroplast reads were removed in total Illumina clean data via BWA v0.7.1724 (key parameter: bwa mem) and SAMtools v1.1025 (key parameter: samtools view -bF 4) based on three Lithospermeae chloroplast genomes downloaded from NCBI (NCBI IDs: MT975394.1, MT975393.1, and NC_049569.1; Supplementary Table S4).26,27 Subsequently, the cpclean data (i.e. chloroplast-filtered) also were used to estimate genome sizes according to the identical steps mentioned above.
2.5. Genome assembly and annotation
PacBio and ONT clean data were first corrected via NextDenovo v2.3.1 (key parameter: read_cutoff = 1,000; seed_cutoff = 10,000) (https://github.com/Nextomics/NextDenovo), separately. The total corrected data were then applied for genome assembly using NextDenovo v2.3.1 (key parameter: nextgraph_options = –a 1), SmartDenovo v1.0.028 (key parameter: –J 5000; –k 16), Flye v2.8.129 (key parameter: –i 1), and Wtdbg v2.530 (key parameter: –L 5000; –k 15; –p 0; –S 2), independently. Finally, the NextDenovo-assembled version was further polished three rounds via Pilon v1.2331 based on Illumina clean data. In addition, BUSCO v2.0.132 was applied to evaluate genome completeness.
Repetitive sequences were identified via RepeatMasker v4.1.1 (http://www.repeatmasker.org) based on a combined database including curated Dfam v3.2,33 RepBase (RepeatMasker Edition-20181026),34 plus a custom L. erythrorhizon library constructed via RepeatModeler v2.0.1 (key parameter: -LTRStruct) (http://www.repeatmasker.org/RepeatModeler). Subsequently, protein-coding genes were predicted as the following process: (i) repetitive sequences were masked first; (ii) AUGUSTUS v3.3.3,35 GlimmerHMM v3.0.4,36 and SNAP37 were used for ab initio prediction; (iii) GeMoMa v1.6.438 was applied for homology prediction based on four published genome data (Supplementary Table S2)8,10,12,16; (iv) PASA v2.4.139 and TransDecoder v5.5.0 (https://github.com/TransDecoder/TransDecoder) were used to identify transcripts based on transcriptome data that we published previously (NCBI IDs: SRX3978407–SRX3978409; Supplementary Table S1)6; (v) total results were finally integrated into a union set without overlap using EVidenceModeler v1.1.1.40
2.6. Identification of AAT/SAT-like superfamily
The identification process was as follows: (i) with AAT/SAT’s amino acid sequences (i.e. BBV14785.1 and BBV14786.1) as the queries and 10 genomes (i.e. L. erythrorhizon plus nine representative species; Supplementary Table S2) as a database, similarity searches were severally performed using DIAMOND v2.0.541 (key parameter: -f 6 –more-sensitive -e 1e-5 -k 1000); (ii) the redundant sequences were first removed in the results; (iii) then, unusual sequences (i.e. containing abnormal bases, lacking initiation codon and/or termination codon) were filtered out; (iv) batch CD-Search42 was used to further identify protein domains (key parameter: e-value: 1e-5; database: Pfam v32.043); since AAT/SAT contained only one complete domain (i.e. PF02458|Transferase) as their characteristic structure (Supplementary Table S11),44–46 the sequences containing redundant domains and/or incomplete PF02458 domain are deleted; (v) finally, MEME v5.2.047 was applied to search and identify protein motifs (key parameter: –mod oops –nmotifs 20 –minw 5 –maxw 100; e-value for search: 1e-1000; e-value for identification: 1e-5); since AAT/SAT contained two characteristic motifs (i.e. HXXXD and DFGWG; the DFGWG motif is not absolutely conservative; Supplementary Table S12),44–46 the sequences containing both these two motifs were retained as AAT/SAT-like superfamily members. Furthermore, according to the Swiss-Prot database, we applied DIAMOND v2.0.541 (key parameter: -f 6 -e 1e-100 –id 99 -k 3) to confirm which members had been functionally verified by previous studies.
2.7. Phylogenetic analysis
The amino acid sequences of the identified AAT/SAT-like superfamily members were aligned via MUSCLE v3.8.31.48 Subsequently, the preliminary alignment was trimmed using trimAl v1.4.149 (key parameter: –gt 0.50). The trimmed alignment was used to construct a phylogenetic tree via IQ-TREE v2.0.350 according to the maximum likelihood (i.e., ML) method (best-fit model: VT + F + R10; key parameter: –seqtype AA -m MFP –alrt 1000 -B 1000). Furthermore, we distinguished the AAT/SAT-like family according to the tree structure.
Based on the codon model, the L. erythrorhizon’s AAT/SAT-like family members’ nucleotide sequences were aligned via PRANK v17042751 (key parameter: -F -codon). Then, the preliminary alignment was trimmed by trimAl v1.4.149 (key parameter: –gt 0.50). The trimmed alignment was transformed back to amino acid sequences, and this amino acid alignment was used to construct a phylogenetic tree via MEGA-X52 based on the ML method (best-fit model: JTT + G4; bootstrap replications: 1,000). Besides, this tree and its trimmed codon alignment were used for the following selection pressure analysis.
2.8. Ks calculation
Total 12 L. erythrorhizon’s AAT/SAT-like family members combined to produce 66 gene pairs C122. Each gene pair was aligned via MUSCLE v3.8.3148 based on the corresponding amino acid sequences, and each alignment was transformed back to nucleotide sequences. Ks values for each gene pair were calculated via KaKs_Calculator v2.053 (key parameter: –m NG).
2.9. Gene duplication identification
Our L. erythrorhizon gene set was applied for all-vs.-all similarity searches via DIAMOND v2.0.541 (key parameter: -f 6 –more-sensitive -e 1e-30 -k 6). The results plus the corresponding gff file of the gene set were further input into the ‘duplicate_gene_classifier’ module in MCScanX54 to identify duplication types for each gene (priority: WGD/Segmental > Tandem > Proximal > Dispersed > Singleton).
2.10. Selection pressure analysis
According to the branch-site models, the CodeML module in PAML v4.9j55 was used to detect positive sites on foreground branches: (i) first, a target foreground branch was labelled in the corresponding tree; (ii) an alternative model (i.e. Model A) was set to that sites were under positive selection on the labelled foreground branch (key parameter: model = 2, NSsites = 2, fix_omega = 0, omega = 1.5); (iii) a null model (i.e. Model A null) was then set to that sites were under neutral selection on the labelled foreground branch (key parameter: model = 2, NSsites = 2, fix_omega = 1, omega = 1); (iv) the likelihood ratio test (i.e. LRT)56 was then applied to determine which model was accepted [threshold: when P < 0.05, the alternative model (i.e. Model A) was accepted], (v) furthermore, the bayes empirical bayes test (i.e., BEB)57 was used to determine which site was under positive selection (threshold: when posterior probabilities > 0.90, that site probably was under positive selection).
3. Results and discussion
3.1. Lithospermum Erythrorhizon genome
Based on our Illumina data [NCBI ID: SRX2882373 (SRR5644206); Supplementary Table S1], Auber et al.4 estimated L. erythrorhizon’s genome size to be ∼369.34 Mb using GenomeScope v1.021 with default parameters (i.e. parameter ‘Kmer length’ = 21 and parameter ‘Max kmer coverage’ = 1e + 03). We repeated their calculation and obtained an identical result (Supplementary Table S3 and Fig. S1). The original intention of setting parameter ‘Max kmer coverage’ = 1e + 03 was to avoid interference from high-frequency non-nuclear reads (e.g. organelle reads and contamination reads).21 However, the practice had proven that this obsolete default parameter (i.e. ‘Max kmer coverage’ = 1e + 03) was improper (https://github.com/schatzlab/genomescope/issues/22; https://github.com/schatzlab/genomescope/issues/28). Thus, software developers suggested this parameter to be set to 1e + 06 (https://github.com/schatzlab/genomescope/issues/30), and further changed this default from 1e + 03 to all (i.e. ‘Max kmer coverage’ = –1) in the GenomeScope latest version (i.e. v2.0).22
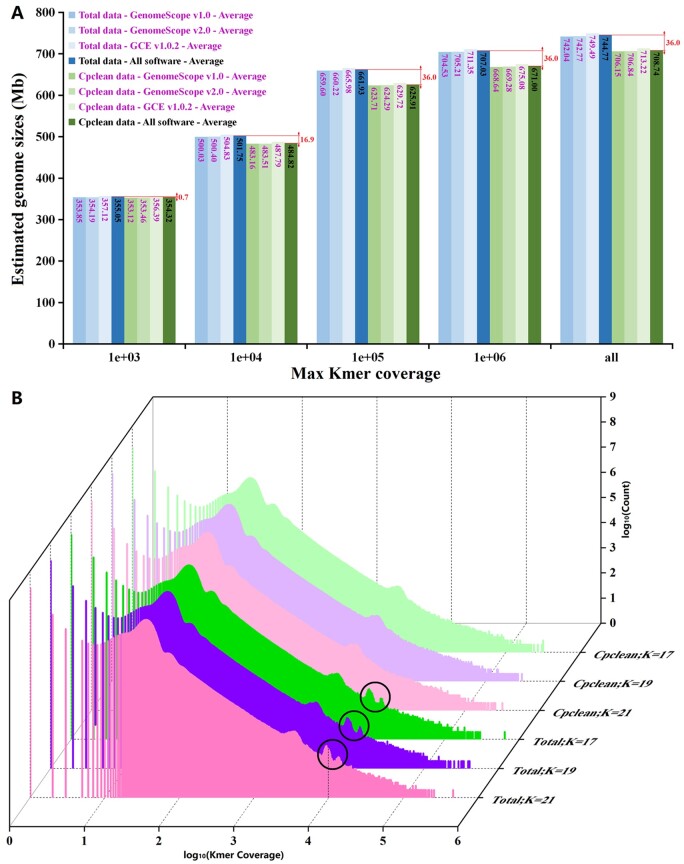
For Spermatophyta, high-frequency non-nuclear reads primarily come from chloroplast because current materials used for genome sequencing are generally green leaves rather than etiolated leaves. Accordingly, through applying GenomeScope v1.0,21 GenomeScope v2.0,22 and GCE v1.0.2,23 we calculated the L. erythrorhizon’s genome size at five thresholds of parameter ‘Max kmer coverage’ (i.e. 1e + 03, 1e + 04, 1e + 05, 1e + 06, and all) with three levels of parameter ‘Kmer length’ (i.e. 17, 19, and 21), based on total Illumina data and corresponding chloroplast-filtered data (i.e. cpclean data; Supplementary Table S4). The results (Fig. 2A and Supplementary Table S3) showed that: (i) different software (or versions) and parameter ‘Kmer length’ had little effect on genome size estimation when parameter ‘Max kmer coverage’ was fixed; (ii) the estimated genome sizes continued to increase as ‘Max kmer coverage’ became large; (iii) when ‘Max kmer coverage’ ≥ 1e + 04, the genome sizes estimated by total data were significantly greater than that estimated by cpclean data; but, the corresponding differences remained almost constant (∼36.0 Mb; Fig. 2A) when ‘Max kmer coverage’ ≥ 1e + 05; these suggested that chloroplast reads significantly skewed the estimated genome size, and these reads mainly concentrated at around ‘Kmer coverage’ ≈ 1e + 04, consistent with the kmer distribution (Fig. 2B) and previous study21; (iv) coincidently, the genome size (∼707.03 Mb) estimated by total data at ‘Max kmer coverage’ = 1e + 06 was approximately equal to the size (∼708.74 Mb) estimated by cpclean data at ‘Max kmer coverage’ = all, due to the increased size caused by chloroplast reads exactly offset the decreased size caused by a lack of high-kmer reads (i.e. the reads at ‘Max kmer coverage’ > 1e + 6); this probably was why developers first suggested parameter ‘Max kmer coverage’ to be set to 1e + 06 and further changed it to all; in other words, to make the calculation more accurate, parameter ‘Max kmer coverage’ was recommended to be set to all when high-frequency non-nuclear reads can be filtered out, whereas this parameter was suggested to be set to 1e + 06 as an empirical value when high-frequency non-nuclear reads cannot be filtered out due to a lack of reference databases (e.g. a chloroplast genome).22 Therefore, we believed that the actual L. erythrorhizon’s genome size should be ∼708.74 Mb, which approached the L. officinale’s genome size (∼743 Mb) as Pustahija et al. reported5 rather than ∼369.34 Mb as Auber et al. estimated.4
Figure 2.
Lithospermum erythrorhizon’s estimated genome sizes and kmer frequency histogram. Total: total clean Illumina data; Cpclean: chloroplast-filtered data. (A) Estimated genome sizes at five thresholds of parameter ‘Max kmer coverage’ (i.e. 1e + 03, 1e + 04, 1e + 05, 1e + 06, and all) via using GenomeScope v1.0, GenomeScope v2.0 and GCE v1.0.2. (B) Kmer frequency histograms at three thresholds of parameter ‘Kmer length’ (i.e. K = 17, 19, and 21). Black circles: the peaks of the chloroplast reads.
Additionally, we noticed Auber et al.’s descriptions about our Illumina data [NCBI ID: SRX2882373 (SRR5644206); https://www.ncbi.nlm.nih.gov/sra/SRX2882373; Supplementary Table S1]4 were incorrect. Auber et al. wrote in their article:
To create a reference genome, we combined L. erythrorhizon ONT genomic DNA (gDNA) reads generated in-house from Siebold & Zucc. Plants with publicly available Illumina gDNA reads sequenced by Nanjing University in 2018 from an unknown accession (SRR5644206). The Illumina data consisted of ∼21.7 Gb Illumina HiSeq paired-end short reads (150 bp) with an estimated heterozygosity of 0.39% and projected genome size of 369.34 Mb.4
In fact, these Illumina data were not an unknown accession. We can know that the submitter was Dr. Chengyi Tang by inquiring about the corresponding BioSample ID (NCBI ID: SAMN06972300; https://www.ncbi.nlm.nih.gov/biosample/SAMN06972300). And, these data should contain a total of 173,693,157 × 2 paired-end reads. The corresponding total length should be ∼34.7 Gb (Gigabases) (Supplementary Table S1). The ‘∼21.7 Gb (Gigabytes)’ Auber et al. wrote4 was just a computer file size. The reads length should be 100 bp rather than 150 bp (i.e. SpotLen = 200 = 100 × 2). All these corresponding statistical information had already been published in NCBI on 2018-04-15 (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRX2882373&o=acc_s%3Aa). In view of Auber et al.’s thoughtless attitude for reference data, we have no faith in Auber et al.’s ability to acquire an actual L. erythrorhizon genome size via our data.
Furthermore, we assembled the L. erythrorhizon genome. The total long reads used for assembling were 31.5 Gb (Supplementary Table S1). The sizes of the preliminary assembled genomes were 680.91–745.64 Mb (Supplementary Table S5). Again, this proved that the actual genome size should be ∼708.74 Mb as estimated above rather than ∼369.34 Mb as Auber et al. reported.4 If the estimated genome size was ∼369.34 Mb, the data coverage could reach up to 85.29× (i.e. 31.5 Gb/369.34 Mb ≈ 85.29); with such sufficient coverage, it is impossible that the estimated size (∼369.34 Mb) was significantly lesser than the assembled size (680.91∼745.64 Mb). The NextDenovo-assembled version was then selected for an error correction because its size and continuity were better than others (Supplementary Table S5). The final genome size was ∼693.34 Mb, and the contigN50 length was ∼238.08 Kb (Supplementary Table S5). BUSCO evaluation showed that ∼88.68% of complete BUSCOs from Embryophyta.odb932 could map to our genome (i.e. 1,277/1,440; Supplementary Table S6), indicating that our genome completeness was acceptable and better than Auber et al.’s (their mapped ratio was only ∼79.31%,4 i.e. 1,141/1,440). Subsequently, we predicted that our genome contained ∼480.93 Mb (∼69.36%) repetitive sequences in which tandem repeats were ∼4.76 Mb and interspersed repeats were ∼472.45 Mb (Supplementary Table S7); and, our genome also contained 35,932 protein-coding genes, in which 28,995 genes (∼80.69%) were supported by transcriptome data (Supplementary Table S8 and Fig. S2).
3.2. AAT/SAT-like family
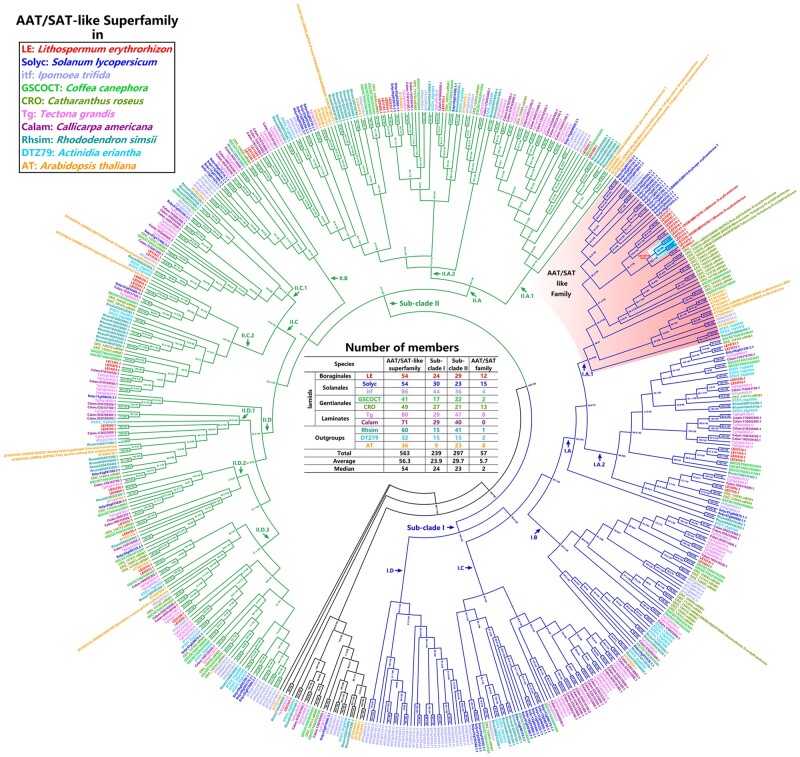
Lithospermum erythrorhizon belongs to Boraginales; and, Boraginales, together with three other orders (i.e. Solanales, Gentianales, and Laminates), are the four core groups in the lamiids clade.6,58 Therefore, through sequence similarity search, we identified 1,233 AAT/SAT-like genes (Supplementary Table S9) in L. erythrorhizon, six other lamiids species (i.e. two Solanales species: Solanum lycopersicum, Ipomoea trifida; two Gentianales species: Coffea canephora, Catharanthus roseus; two Lamiales species: Tectona grandis, Callicarpa americana), and three outgroup species (i.e. Rhododendron simsii, Actinidia eriantha, and Arabidopsis thaliana) (Supplementary Table S2). As expected, we found that the AAT’s equivalent was LE32265.1, and the SAT’s equivalent was LE01141.1 (Supplementary Table S9). After removing redundant sequences and unusual encoding sequences, a total of 674 genes were retained (Supplementary Table S10). Since AAT/SAT contained one characteristic domain (i.e. PF02458|Transferase) and two characteristic motifs (i.e. HXXXD and DFGWG) (Supplementary Tables S11 and S12),44–46 the sequences containing abnormal domains and motifs were further filtered. Finally, a total of 563 genes (Fig. 3 and Supplementary Table S12) were retained as AAT/SAT-like superfamily members, in which at least 18 members had been verified by previous studies [i.e. 2 (i.e. AAT/SAT) + 16 from the Swiss-Prot database (Supplementary Table S13)]. According to the above structural and functional information, the AAT/SAT-like superfamily should be the BAHD superfamily (i.e. benzylalcohol O-acetyltransferase, anthocyanin O-hydroxycinnamoyltransferase, N-hydroxycinnamoyl anthranilate benzoyltransferase, and deacetylvindoline O-acetyltransferase superfamily), which catalysed various acylation reactions in plant metabolism (e.g. lignins, anthocyanins, terpenoids, and various esters).44–46
Figure 3.
An ML phylogenetic tree of the AAT/SAT-like superfamily across 10 representative species (i.e. seven lamiids species plus three outgroup species; Supplementary Table S1). */* is SH-aLRT support/ultrafast bootstrap support. Two main sub-clades: Sub-clade I (blue branches) and Sub-clade II (green branches). The Sub-clade I contains five sub-categories, i.e. I.A.1∼2, I.B, I.C, and I.D; the Sub-clade II contains eight sub-categories, i.e. II.A.1∼2, II.B, II.C.1∼2, and II.D.1∼3. The sub-category I.A.1 is named as ‘AAT/SAT-like family’ in this study.
To further classify the AAT/SAT-like superfamily, we constructed a phylogenetic tree. The results (Fig. 3) showed that: (i) this superfamily was roughly divided into three sections, i.e. Sub-clade I, Sub-clade II, and some oddments; furthermore, the Sub-clade I was roughly divided into four broad categories and five sub-categories (i.e. I.A.1∼2, I.B, I.C, and I.D), and the Sub-clade II was roughly divided into four broad categories and eight sub-categories (i.e. II.A.1∼2, II.B, II.C.1∼2, and II.D.1∼3); (ii) AAT/SAT belonged to the sub-category I.A.1; thus, we named this sub-category as ‘AAT/SAT-like family’ in this study; in addition, this AAT/SAT-like family (i.e. the sub-category I.A.1) contained a total of eight verified members, in which CRO_120021.mRNA1 (i.e. Swiss-Prot ID: Q9ZTK5|deacetylvindoline O-acetyltransferase from C. roseus; Supplementary Table S13) was used to name the BAHD superfamily in previous studies44–46; (iii) although S. lycopersicum and I. trifida belonged to Solanales, the numbers of the AAT/SAT-like family members they each contained were significantly different (i.e. S. lycopersicum: 15 vs. I. trifida: 4; Fig. 3), and a similar numerical difference was also found in two Gentianales species (i.e. C. canephora: 2 vs. C. roseus: 13; Fig. 3); in addition, two Lamiales species (i.e. T. grandis and C. americana) did not contain any members in this AAT/SAT-like family, although they owned abundant members in the Sub-clade I and the whole superfamily (Fig. 3); therefore, all these indicated that the number of AAT/SAT-like family members significantly expanded or contracted in different species, which might be related to the species-specific properties.
3.3. AAT/SAT’s evolutionary process
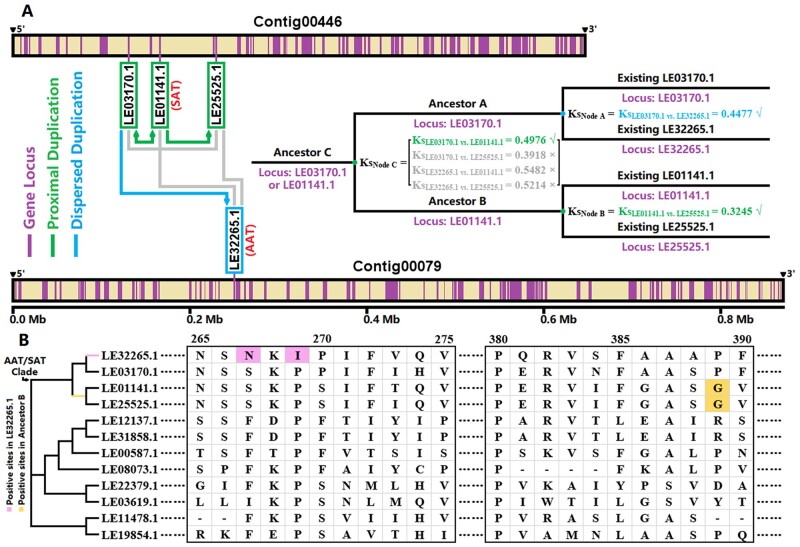
In the AAT/SAT-like family (i.e. the sub-category I.A.1), AAT/SAT (i.e. LE32265.1 and LE01141.1) plus two other members (i.e. LE03170.1 and LE25525.1) seem to converge into a common clade (Fig. 3). Therefore, we named this clade as ‘AAT/SAT clade’ in this study. These four members should be real gene loci because the evidence collectively supports them on three fronts (i.e. Ab initio + Homology + Transcriptome; Supplementary Table S8). Based on the tree reconstructed only using 12 L. erythrorhizon members (Supplementary Fig. S3) and the corresponding Ks values (Supplementary Table S14), we further reconfirmed that these four members should be descended from a recent common ancestor (i.e. a common clade) due to Ksclade I (i.e.AAT/SAT clade) (≈ 0.46) ≪ Ksclade I vs. clade II (≈ 1.97) and Ksclade I ≪ Ks
clade I vs. clade III (≈ 2.52).
Subsequently, we identified duplication types for each gene in the L. erythrorhizon genome. The results indicated that three members (i.e. LE01141.1, LE03170.1, and LE25525) came from ‘proximal duplications’, whereas another one (i.e. LE32265.1) came from a ‘dispersed duplication’ (Supplementary Table S15). These were consistent with their loci information in the genome: LE01141.1, LE03170.1, and LE25525.1 were closely located in the Contig00446, whereas LE32265.1 was located in the Contig00079 alone (Fig. 4A). The phylogenetic relationship in the AAT/SAT clade had been confirmed as ((LE32265.1, LE03170.1)Node A, (LE01141.1, LE25525.1)Node B)Node C (Fig. 3 and Supplementary Fig. S3), and the Ks values between these four members were also known (Supplementary Table S14). Therefore, (i) one round of dispersed duplication should occur in Node A at KsNode A = KsLE03170.1 vs. LE32265.1 = 0.4477 because only LE32265.1 was identified as ‘dispersed duplication’; (ii) one round of proximal duplication should occur in Node B at KsNode B = KsLE01141.1 vs. LE25525.1 = 0.3245 because both LE01141.1 and LE25525.1 were identified as ‘proximal duplications’; (iii) and, another round of proximal duplication should occur in Node C at KsNode C = KsLE03170.1 vs. LE01141.1 = 0.4976 because there was only one round of dispersed duplication in the AAT/SAT clade (thus, KsLE32265.1 vs. LE01141.1 and KsLE32265.1 vs. LE25525.1 were excluded), and KsNode C must be greater than KsNode A and KsNode B (thus, KsLE03170.1 vs. LE25525.1 were excluded) (Fig. 4A). To sum up, we inferred that the AAT/SAT’s evolutionary process probable underwent three rounds of gene duplication (Fig. 4A): (i) first, one round of proximal duplication occurred in Node C at Ks = 0.4976 and made ancestor C produce ancestor A and ancestor B; ancestor A probable located on the existing LE03170.1 locus, and ancestor B probable located on the existing LE01141.1 locus, due to KsLE03170.1 vs. LE01141.1 was assigned to KsNode C; (ii) subsequently, one round of dispersed duplication occurred in Node A at Ks = 0.4477 and made ancestor A produce the existing LE03170.1 and LE32265.1 (i.e. AAT); (iii) finally, another round of proximal duplication occurred in Node B at Ks = 0.3245 and made ancestor B produce the existing LE01141.1 (i.e. SAT) and LE25525.1.
Figure 4.
AAT/SAT’s evolutionary process in L. erythrorhizon. (A) AAT/SAT and two other members’ locus information and duplication process. ‘√’: these Ks values can be accepted; ‘×’: these Ks values cannot be accepted. The Ks values are based on Supplementary Table S14; and, the duplication types are based on Supplementary Table S15. (B) Potential positive selection sites inside the AAT/SAT clade. The complete amino acid alignment (after trimAl) is exhibited in Supplementary Fig. S4.
Furthermore, we detected whether positive selection sites existed on each branch inside the AAT/SAT clade. The results showed two potential positive sites (i.e. sites 267 and 269) were on the branch LE32265.1 and one potential positive site (i.e. site 389) was on the branch ancestor B (Fig. 4B and Supplementary Table S16). In other words, (i) after the proximal duplication in Node C, ancestor B was possibly subjected to positive selection, while ancestor A was not; (ii) after the dispersed duplication in Node A, LE32265.1 was possibly subjected to positive selection, while LE03170.1 was not; (iii) after the proximal duplication in Node B, both LE01141.1 and LE25525.1 were not under positive selection. To sum up, the above evidence suggested that gene duplication and positive selection collectively propelled AAT/SAT’s functional differentiation in evolution.
Supplementary data
Supplementary data are available at DNARES online.
Supplementary Material
Acknowledgements
Thanks for some financial supports from the Natural Science Foundation of Jiangsu Province (Grant Numbers: BK20180332).
Accession numbers
The genome sequencing data of L. erythrorhizon were stored in the NCBI, and their accession IDs were SRX2882373 and SRX9956727 (Supplementary Tables S1). The L. erythrorhizon genome we assembled was stored in the NCBI (accession ID: JAIEZA000000000), the EMBL (accession ID: CAJVUH010000000) and the CNGBdb (accession ID: CNA0029378), respectively. In addition, the corresponding gene annotation files (i.e. gff/cds/pep) were stored in the CNGBdb, and their shared accession ID was CNA0029378. Other data and genomes downloaded from previous studies were listed in Supplementary Tables S1, S2, and S4.
Conflict of interest
The authors declare that they have no competing interests.
Author contributions
C.T. completed this whole project and wrote this manuscript.
References
- 1. The Plant List 2013, Version 1.1. Published on the internet. http://www.theplantlist.org
- 2. Rice A., Glick L., Abadi S., et al. 2015, The chromosome counts database (CCDB) – a community resource of plant chromosome numbers, New Phytol., 206, 19–26. [DOI] [PubMed] [Google Scholar]
- 3. Weigend M., Gottschling M., Selvi F., Hilger H.H.. 2009, Marbleseeds are gromwells – systematics and evolution of Lithospermum and allies (Boraginaceae tribe Lithospermeae) based on molecular and morphological data, Mol. Phylogenet. Evol., 52, 755–68. [DOI] [PubMed] [Google Scholar]
- 4. Auber R.P., Suttiyut T., McCoy R.M., et al. 2020, Hybrid de novo genome assembly of red gromwell (Lithospermum erythrorhizon) reveals evolutionary insight into shikonin biosynthesis, Hortic. Res., 7, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pustahija F., Brown S.C., Bogunić F., et al. 2013, Small genomes dominate in plants growing on serpentine soils in West Balkans, an exhaustive study of 8 habitats covering 308 taxa, Plant Soil., 373, 427–53. [Google Scholar]
- 6. Tang C.Y., Li S., Wang Y.T., Wang X.. 2020, Comparative genome/transcriptome analysis probes Boraginales’ phylogenetic position, WGDs in Boraginales, and key enzyme genes in the alkannin/shikonin core pathway, Mol. Ecol. Resour., 20, 228–41. [DOI] [PubMed] [Google Scholar]
- 7. Oshikiri H., Watanabe B., Yamamoto H., Yazaki K., Takanashi K.. 2020, Two BAHD acyltransferases catalyze the last step in the shikonin/alkannin biosynthetic pathway, Plant Physiol., 184, 753–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tomato Genome Consortium. 2012, The tomato genome sequence provides insights into fleshy fruit evolution, Nature, 485, 635–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu S., Lau K.H., Cao Q., et al. 2018, Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement, Nat. Commun., 9, 4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Denoeud F., Carretero-Paulet L., Dereeper A., et al. 2014, The coffee genome provides insight into the convergent evolution of caffeine biosynthesis, Science., 345, 1181–4. [DOI] [PubMed] [Google Scholar]
- 11. Franke J., Kim J., Hamilton J.P., et al. 2019, Gene discovery in Gelsemium highlights conserved gene clusters in monoterpene indole alkaloid biosynthesis, Chembiochem., 20, 83–7. [DOI] [PubMed] [Google Scholar]
- 12. Zhao D., Hamilton J.P., Bhat W.W., et al. 2019, A chromosomal-scale genome assembly of Tectona grandis reveals the importance of tandem gene duplication and enables discovery of genes in natural product biosynthetic pathways, Gigascience., 8, giz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hamilton J.P., Godden G.T., Lanier E., et al. 2020, Generation of a chromosome-scale genome assembly of the insect-repellent terpenoid-producing Lamiaceae species, Callicarpa americana, Gigascience., 9, giaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang F.S., Nie S., Liu H., et al. 2020, Chromosome-level genome assembly of a parent species of widely cultivated azaleas, Nat. Commun., 11, 5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang W., Sun X., Yue J., et al. 2019, Chromosome-scale genome assembly of kiwifruit Actinidia eriantha with single-molecule sequencing and chromatin interaction mapping, Gigascience., 8, giz027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lamesch P., Berardini T.Z., Li D., et al. 2012, The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools, Nucleic Acids Res., 40, D1202–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bolger A.M., Lohse M., Usadel B.. 2014, Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics., 30, 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu H., Luo X., Qian J., et al. 2012, FastUniq: a fast de novo duplicates removal tool for paired short reads, PLoS One., 7, e52249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Coster W., D’Hert S., Schultz D.T., Cruts M., Van Broeckhoven C.. 2018, NanoPack: visualizing and processing long-read sequencing data, Bioinformatics., 34, 2666–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marçais G., Kingsford C.. 2011, A fast, lock-free approach for efficient parallel counting of occurrences of k-mers, Bioinformatics., 27, 764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vurture G.W., Sedlazeck F.J., Nattestad M., Underwood C.J., Fang H., Gurtowski J., Schatz M.C.. 2017, GenomeScope: fast reference-free genome profiling from short reads, Bioinformatics., 33, 2202–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ranallo-Benavidez T.R., Jaron K.S., Schatz M.C.. 2020, GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes, Nat. Commun., 11, 1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu B., Shi Y., Yuan J.. 2013, Estimation of genomic characteristics by analyzing K-mer frequency in de novo genome projects. arXiv., 1308.2012v2.
- 24. Li H. 2013, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv., 1303.3997v2.
- 25. Li H., Handsaker B., Wysoker A., et al. ; 1000 Genome Project Data Processing Subgroup. 2009, The sequence alignment/map format and SAMtools, Bioinformatics., 25, 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park I., Yang S., Song J.H., Moon B.C.. 2020, Dissection for floral micromorphology and plastid genome of valuable medicinal borages Arnebia and Lithospermum (Boraginaceae), Front. Plant Sci., 11, 606463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen Q., Zhang D.. 2019, The complete chloroplast genome sequence of Onosma paniculatum Bur. et Franch. (Boraginaceae), a medicinal plant in Yunnan and its adjacent regions, Mitochondrial DNA B Resour., 4, 3330–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu H., Wu S., Li A., Ruan J.. 2020, SMARTdenovo: a de novo assembler using long noisy reads, Preprints., 2020090207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kolmogorov M., Yuan J., Lin Y., Pevzner P.A.. 2019, Assembly of long, error-prone reads using repeat graphs, Nat. Biotechnol., 37, 540–6. [DOI] [PubMed] [Google Scholar]
- 30. Ruan J., Li H.. 2020, Fast and accurate long-read assembly with wtdbg2, Nat. Methods., 17, 155–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walker B.J., Abeel T., Shea T., et al. 2014, Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement, PLoS One., 9, e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M.. 2015, BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs, Bioinformatics., 31, 3210–152. [DOI] [PubMed] [Google Scholar]
- 33. Storer J., Hubley R., Rosen J., Wheeler T.J., Smit A.F.. 2021, The Dfam community resource of transposable element families, sequence models, and genome annotations, Mob. DNA., 12, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bao W., Kojima K.K., Kohany O.. 2015, Repbase update, a database of repetitive elements in eukaryotic genomes, Mob. DNA., 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stanke M., Steinkamp R., Waack S., Morgenstern B.. 2004, AUGUSTUS: a web server for gene finding in eukaryotes, Nucleic Acids Res., 32, W309–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Majoros W.H., Pertea M., Salzberg S.L.. 2004, TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders, Bioinformatics., 20, 2878–9. [DOI] [PubMed] [Google Scholar]
- 37. Korf I. 2004, Gene finding in novel genomes, BMC Bioinformatics., 5, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keilwagen J., Wenk M., Erickson J.L., Schattat M.H., Grau J., Hartung F.. 2016, Using intron position conservation for homology-based gene prediction, Nucleic Acids Res., 44, e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haas B.J., Delcher A.L., Mount S.M., et al. 2003, Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies, Nucleic Acids Res., 31, 5654–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haas B.J., Salzberg S.L., Zhu W., et al. 2008, Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments, Genome Biol., 9, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Buchfink B., Xie C., Huson D.H.. 2015, Fast and sensitive protein alignment using DIAMOND, Nat. Methods., 12, 59–60. [DOI] [PubMed] [Google Scholar]
- 42. Marchler-Bauer A., Bryant S.H.. 2004, CD-Search: protein domain annotations on the fly, Nucleic Acids Res., 32, W327–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. El-Gebali S., Mistry J., Bateman A., et al. 2019, The Pfam protein families database in 2019, Nucleic Acids Res., 47, D427–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. D’Auria J.C. 2006, Acyltransferases in plants: a good time to be BAHD, Curr Opin Plant Biol., 9, 331–40. [DOI] [PubMed] [Google Scholar]
- 45. Tuominen L.K., Johnson V.E., Tsai C.J.. 2011, Differential phylogenetic expansions in BAHD acyltransferases across five angiosperm taxa and evidence of divergent expression among Populus paralogues, BMC Genomics., 12, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang T., Huo T., Ding A., et al. 2019, Genome-wide identification, characterization, expression and enzyme activity analysis of coniferyl alcohol acetyltransferase genes involved in eugenol biosynthesis in Prunus mume, PLoS One., 14, e0223974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bailey T.L., Boden M., Buske F.A., et al. 2009, MEME SUITE: tools for motif discovery and searching, Nucleic Acids Res., 37, W202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Edgar R.C. 2004, MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Res., 32, 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T.. 2009, trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses, Bioinformatics., 25, 1972–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q.. 2015, IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies, Mol. Biol. Evol., 32, 268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Löytynoja A. 2014, Phylogeny-aware alignment with PRANK, Methods Mol. Biol., 1079, 155–70. [DOI] [PubMed] [Google Scholar]
- 52. Kumar S., Stecher G., Li M., Knyaz C., Tamura K.. 2018, MEGA X: molecular evolutionary genetics analysis across computing platforms, Mol. Biol. Evol., 35, 1547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang D., Zhang Y., Zhang Z., Zhu J., Yu J.. 2010, KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies, Genomics. Proteomics Bioinformatics., 8, 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Y., Tang H., Debarry J.D., et al. 2012, MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity, Nucleic Acids Res., 40, e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang Z. 2007, PAML 4: phylogenetic analysis by maximum likelihood, Mol Biol Evol., 24, 1586–91. [DOI] [PubMed] [Google Scholar]
- 56. Anisimova M., Bielawski J.P., Yang Z.. 2001, The accuracy and power of likelihood ratio tests to detect positive selection at amino acid sites, Mol Biol Evol., 18, 1585–92. [DOI] [PubMed] [Google Scholar]
- 57. Yang Z., Wong W.S., Nielsen R.. 2005, Bayes empirical Bayes inference of amino acid sites under positive selection, Mol Biol Evol., 22, 1107–18. [DOI] [PubMed] [Google Scholar]
- 58. The Angiosperm Phylogeny Group. 2016, An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV, Bot J Linn Soc., 181, 1–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.