Abstract
Small open reading frames (small ORFs/sORFs/smORFs) are potentially coding sequences smaller than 100 codons that have historically been considered junk DNA by gene prediction software and in annotation screening; however, the advent of next-generation sequencing has contributed to the deeper investigation of junk DNA regions and their transcription products, resulting in the emergence of smORFs as a new focus of interest in systems biology. Several smORF peptides were recently reported in non-canonical mRNAs as new players in numerous biological contexts; however, their relevance is still overlooked in coding potential analysis. Hence, this review proposes a smORF classification based on transcriptional features, discussing the most promising approaches to investigate smORFs based on their different characteristics. First, smORFs were divided into non-expressed (intergenic) and expressed (genic) smORFs. Second, genic smORFs were classified as smORFs located in non-coding RNAs (ncRNAs) or canonical mRNAs. Finally, smORFs in ncRNAs were further subdivided into sequences located in small or long RNAs, whereas smORFs located in canonical mRNAs were subdivided into several specific classes depending on their localization along the gene. We hope that this review provides new insights into large-scale annotations and reinforces the role of smORFs as essential components of a hidden coding DNA world.
Keywords: genome annotation, smORF peptides, long non-coding RNA, dual functional RNA, alternative ORFs
1. Introduction
The big data era promoted by next-generation sequencing (NGS) has irreversibly changed genetics and systems biology. The amount of biological data available to scientists has been rapidly increasing during the last 15 years,1 which makes the intersection between experimental sciences and computational biology even more imperative to overcome new scientific barriers.
One of the most important achievements in NGS technology has been the development of deep transcriptome sequencing approaches, such as RNA-seq and ribosome profiling, which have greatly improved proteomic, peptidomic and phenotypic analyses.2–4 New molecular components have thus been discovered,5 especially in hidden proteomes expressed from coding small open reading frames (small ORFs/sORFs/smORFs).6
Generally, open reading frames (ORFs) are defined as nucleotide sequences between a translation start codon and the nearest in-frame stop codon (Fig. 1).7 SmORFs differ from other ORFs in size, which typically range from the lower theoretical limit of two codons8 to 100 codons9; however, upper thresholds from 150 to 250 codons have rarely been proposed in the literature.8,10,11 In prokaryotes, a 50-codon size limit is normally accepted.12
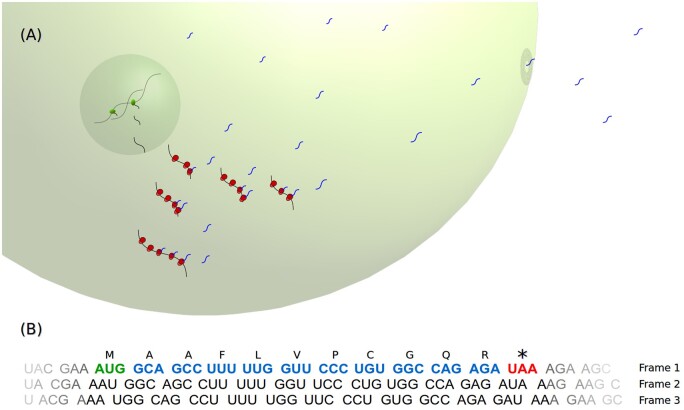
Figure 1.
smORF peptide biosynthesis. (A) smORF transcription, translation and cellular/extracellular trafficking. smORF peptide biosynthesis occurs directly via ribosome translation after smORF gene transcription. smORF peptides can play several roles inside and outside the cell. RNA polymerase in the nucleus is shown in green; ribosomes in the cytoplasm are shown in red; and smORF peptides in the cytoplasm are shown as blue winding lines. (B) Schematic representation of a hypothetical ORF. The illustrated ORF is a smORF within the first of the three RNA frames (Frame 1). The smORF is highlighted in bold font; the start codon is shown in green; the stop codon is shown in red; and the remaining codons are shown in blue. Above the smORF codons are their corresponding one-letter-code amino acids, encoding a hypothetical 11 amino acid smORF peptide.
SmORFs have historically been dismissed in annotation screenings due to methodological challenges. Pioneering smORF studies performed before the NGS era (e.g., Refs13–17) were even more challenging without deep sequencing data, but greatly contributed to the acceptance of the field when new sequencing technologies were developed. Currently, gene prediction algorithms and genome annotations usually overlook smORFs owing to their low statistical coding potential,18,19 which has been justified by the fact that millions of non-functional smORFs occur stochastically in genomes due to their small size. Thus, the computational prediction and experimental analysis of coding smORFs are challenging tasks, akin to searching for a needle in the haystack10,20; however, several biologically relevant smORF peptides have been discovered throughout the three domains of life (study in Archaea21; Eukarya review22; Bacteria review12) and viruses.23 SmORFs are also significant targets of biomedical studies.24–31 Moreover, recent discussions have revealed that smORFs are important precursors of de novo protein-coding gene birth.32,33 Thus, although smORFs were previously hidden among the so-called ‘junk DNA’, their study is now an emerging field.
Our new classification scheme comprises 12 smORF classes and subclasses based on transcription features. Some of the classes proposed have been covered in previous reviews and screening reports during recent years, under different perspectives.34–39 Here, we contribute to this knowledge by offering a ‘divide and conquer’ perspective to discuss the most promising approaches to study each smORF class. By providing a detailed explanation of the smORF classes and organizing a comprehensive scheme, we note how the features of each smORF group influence the coding smORF detection. We also discuss the molecular findings that support the coding potential of each class and the future prospects for the smORF field. We expect that this classification scheme may direct further studies and discoveries of new coding smORFs by offering a comprehensive landscape of smORF diversity and their detection challenges.
First, smORFs are divided into two general groups, namely expressed and non-expressed. Non-expressed smORFs are also known as intergenic smORFs (Fig. 2A).34 Second, the expressed smORFs are subdivided into two categories: smORFs located in canonical mRNAs or smORFs located in transcripts with non-coding characteristics (Fig. 2A); in the second case, transcripts may evolve dual functional roles. Finally, we subdivided the two expressed smORF groups into several other classes according to their transcriptional features (Fig. 2B). Importantly, all proposed classes may contain random and non-coding smORFs, with the exception of the group where the smORFs are the reference CDSs (coding DNA sequences) along mRNAs. Throughout the text we focus on the study of the coding smORFs of each class.
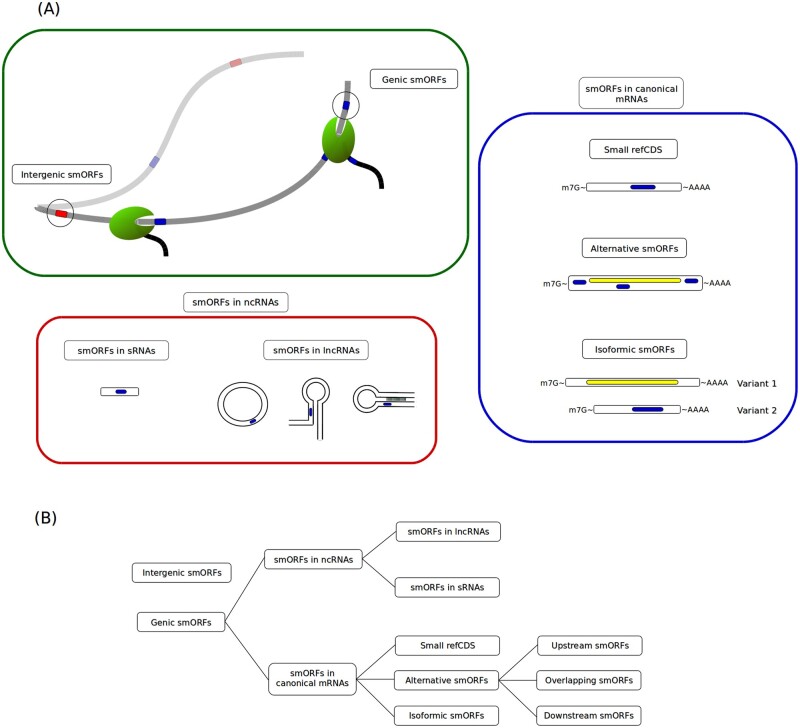
Figure 2.
Proposed smORF classification. (A) smORF classes and their representative locations. Hundreds of thousands of smORFs in the genome are non-expressed and are therefore classified as intergenic smORFs (green box). Expressed smORFs are classified as genic smORFs (green box) and are subdivided into smORFs located in non-coding RNAs (ncRNAs) (red box) and smORFs located in canonical mRNAs (blue box). Different types of ncRNAs and canonical mRNAs with their respective classes of smORFs are represented in red and blue boxes. Red tracks represent intergenic smORFs; blue tracks represent genic smORFs; yellow tracks represent large ORFs. (B) smORF classification chain. SmORF classes can be organized into groups and subgroups defined by transcriptional features.
2. Intergenic smORFs
Intergenic smORFs are not generally functional or transcribed; they constitute the great majority of smORFs in the genome and originate randomly via the simple arrangement and rearrangement of nucleotides, causing them to be classified as junk DNA.34
2.1. Strategic insights into the investigation of intergenic smORFs
Analysis based on sequence similarity indicated that thousands of theoretically non-expressed smORFs are evolutionarily conserved, which suggests coding potential.40 Importantly, most coding smORFs are overlooked in genome annotation screenings and are usually classified as intergenic DNA stretches. Thus, strategies to uncover new intergenic smORFs must include smORF detection in intergenic regions using bioinformatic tools, such as the getORF program provided by EMBOSS,41 followed by filters of evolutionary conservation and comparison to RNA sequence databases to verify if the sequence is expressed (a similar approach was performed in Ladoukakis et al.42). Then, an analysis of differences in expression under stress conditions is a useful approach since recent evidence from bacteria suggests that intergenic smORFs can be expressed under osmotic stress, lower temperatures and specific growth conditions, indicating that intergenic smORFs are a potential source of peptides with condition-dependent expression.43
3. smORFs in small RNAs
Small RNAs are transcripts smaller than ∼200 − 300 nucleotides.44–46 The regulatory roles of small RNAs are widely studied in all domains of life,47–49 as small RNAs are involved in cellular responses to biotic and abiotic stresses50,51; however, the coding capacity of small RNAs has been neglected, and reports of functional smORFs within this type of transcript exist but are scarce.
3.1. Strategic insights into the investigation of smORFs in small RNAs
The most promising source of smORFs in small RNAs are prokaryotic dual-function transcripts, as previously reported.52–54 Strictly coding small RNAs are an undescribed phenomenon, but their existence is theoretically possible. The coding capacity of eukaryotic small RNAs is still unexplored and requires investigation.
In prokaryotes, the widespread distribution of potentially coding small RNAs has been confirmed by computational predictive approaches. For instance, it was recently shown that at least 0.5% of all small RNAs in 14 bacterial species contain smORFs under purifying selection, and the proportion reaches ∼20% in some taxa.55 Moreover, mass spectrometry and ribosome profiling database analyses have confirmed that dozens of these smORFs in small RNAs are in fact translated.55 Thus, a bioinformatic approach integrating small RNA identification56,57 followed by smORF detection pipelines based on evolutionary conservation and ribosome profiling footprints58–61 must be considered to detect new coding small RNAs in large-scale screening. In addition, the expression under different stress conditions is certainly an important factor to be analysed, because at least 40 putative dual-function small RNAs encode smORFs smaller than 30 codons in Methanosarcina mazei (Archaea) and their expression is modulated by different levels of nitrogen availability.62 Importantly, the experimental detection of small RNAs is highly dependent on the RNA isolation method because small RNAs can be lost during precipitation steps due to their size. Column-based or acrylamide gel isolation approaches should be preferentially applied for the isolation of potential smORF-containing small RNAs.
Because evidence suggests that most coding small RNAs are dual-functional (examples in the section below), another interesting strategy is the search for smORF sequences in known regulatory small RNAs using bioinformatic tools, such as the Expasy Translate Tool.63 Then, each hypothetical smORF peptide identified should be submitted to tBLASTn analysis64 against related species databases applying non-stringent parameters for smORF detection.65 Finally, putative smORFs could be indicated by evolutionary conservation. The use of hypothetical peptide sequences in BLAST searches avoids false negative results caused by synonymous modifications in small coding sequences. This strategy can be adapted for any smORF class.
3.2. Examples of smORF peptides from small RNAs
One of the first described dual-function small RNAs was SgrS, which is transcribed under glucose-phosphate stress in Escherichia coli.66 Interestingly, both the SgrS transcript and its smORF peptide play roles in glucose flux, but in different pathways. The SgrS transcript downregulates an important glucose transporter by base pairing with its mRNA, and SgrS also encodes a 43 amino acid peptide, sgrT, that modulates the influx of glucose by inhibiting its transporters.66
The functional analysis of another small RNA, SR1, in Bacillus subtilis showed that the coding and non-coding activities of this small RNA are distinct.67 The regulatory role of SR1 involves the inhibition of the translation of an important transcription activator via nucleotide pairing. The coding activity of SR1 is mediated by a 39 amino acid peptide, SR1P, encoded by the transcript. SR1P binds to the glycolytic enzyme GapA, triggering the formation of a complex with RNAse J1 and thereby increasing the affinity of RNAse for its substrates.67 In addition, SR1P-GapA binding promotes the stabilization of gapA operon mRNA.67
In Staphylococcus aureus, the RNAIII transcript regulates the translation and/or stability of virulence factors, cell wall metabolism enzymes and transcription factor mRNAs (reviewed in Bronesky et al.52). Moreover, RNAIII encodes the smORF peptide δ-haemolysin,68 also known as hld (26 amino acids length), which can lyse red blood cells, trigger membrane disorders, and exert antimicrobial activities (reviewed in Verdon et al.69).
Mass spectrometry analysis of the archaeal species M. mazei undergoing cell growth under different stress conditions identified three smORF peptides between 23 and 61 amino acids in length.21 The identified smORF peptides exhibit high conservation among Methanosarcina species as well as highly conserved secondary structures of their small RNAs. Based on the concentration of the peptides during nitrogen restriction stress, oligopeptide 36 (61 amino acids) might modulate an essential protein associated with nitrogen metabolism.21
4. smORFs in long non-coding RNAs
By definition, long non-coding RNAs (lncRNAs) are untranslated RNA molecules longer than ∼200 nucleotides.35 Typical lncRNAs are expressed at low levels and are non-conserved. On the other hand, lncRNA expression is associated with biological processes such as differentiation, proliferation, embryonic development, cancer, apoptosis and stress responses (reviewed in Chekulaeva and Rajewsky9 and Perry and Ulitsky70).
The coding potential of lncRNAs is still disregarded owing to certain lncRNA characteristics, including (i) the absence of major ORFs71; (ii) degeneration frequency similar to that of introns and much higher than that of exons, which disfavours new coding ORF fixation72,73 and (iii) when translated, the instability and rapid degradation of lncRNA peptides, suggesting that the translation of these peptides is a random and neutral process.74 In contrast, the effective translation of smORFs located in lncRNAs has been described, and these translated smORFs exhibit sequence, structural and functional conservation.24,31,75,76 Currently, smORF peptides translated from lncRNAs are considered to represent a new frontier in biomedical studies focussed on new biomarkers and molecular targets in cancer.29
Our classification scheme indicates that smORFs discovered in strictly ‘coding lncRNAs’ should be classified as small reference coding sequences (small refCDSs) when their transcripts are reannotated as mRNAs (Fig. 3). Therefore, small refCDS functions will be discussed in its corresponding section. In the case of coding circular RNAs (circRNAs) lacking non-coding functional annotation, we still classify their coding smORFs as smORFs in lncRNAs, because the field is recent and the molecular niche of circular transcripts is poorly known, but recent evidence points to regulatory effects without translation.77–79
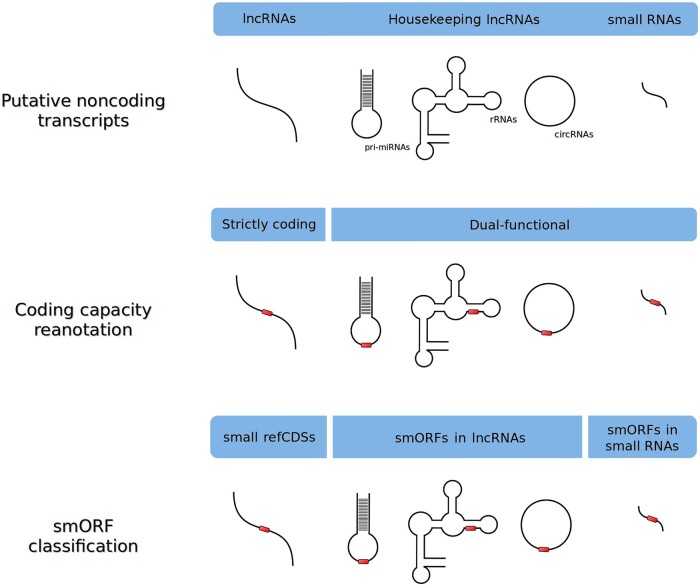
Figure 3.
General scheme of the smORF classification of misannotated non-coding transcripts. Strictly coding lncRNAs are reclassified as smORF located in mRNAs, while ncRNAs showing both coding and regulatory roles are reannotated as dual-function transcripts. Three smORF classes can be identified in ncRNAs: small refCDSs, smORFs in lncRNAs (dual functional) and smORFs in small RNAs (generally dual functional). Red tracks represent coding smORFs.
4.1. Strategic insights into the investigation of smORFs in lncRNAs
Among all smORF classes, lncRNAs are the most promising source of smORF discoveries. For instance, ∼98% of annotated lncRNAs in Metazoa contain at least one unannotated smORF.34 Studies in Arabidopsis thaliana show that thousands of smORFs in lncRNAs display evidence of translation, as confirmed by Ribo-Seq analysis.80 Another clue is provided by the wide gene coverage of smORFs, because ∼70% of human genes are expressed as lncRNAs.81 These data show that a plethora of ‘coding lncRNAs’ are still misannotated or even overlooked.
In this context, some features should be considered in annotation screenings searching for smORFs in lncRNAs. For instance, the amino acid usage of theoretical unannotated smORF peptides in lncRNAs differs from that of canonical proteins, although it is not random,34 suggesting a new biological pattern that should not be dismissed. Another important feature is that misannotated lncRNAs can exhibit typical mRNA structures or non-canonical characteristics.82,83 In the second case, in addition to the absence of polyadenylation sites, Kozak consensus sequences and 5′ caps,84,85 in some cases these transcripts exhibit a circular structure.86,87 Furthermore, lncRNAs are usually non-conserved, although their smORFs can still show signs of positive selection at amino acid sequences.61 However, even in the absence of orthologues, species-specific smORFs can be functional via orphan gene generation.88
Bioinformatic prediction approaches are a promising strategy to detect putative smORFs based on the aforementioned features, examples include machine learning programs89 and codon usage comparisons between random and functional smORF peptides. Importantly, the diversity of lncRNA types enables several approaches to investigate their coding potential. For instance, some types of housekeeping lncRNAs can also encode smORF peptides (Fig. 3), making them dual-function transcripts, as described for rRNAs,90 and pri-miRNAs.91 The translation of smORFs in lncRNAs might even include antisense and intronic transcripts.92 Thus, smORF detection performed on known functional lncRNAs followed by comparative analysis using BLAST is also an interesting strategy.
After the selection of potentially coding smORFs, a promising experimental approach is the development of precise CRISPR-Cas9 genome editing, such as precise deletions or point mutations, on conserved smORFs in lncRNAs differentially expressed in important biological contexts, such as cancer. Recent yeast and fruit fly studies used similar strategies.93,94
4.2. Examples of smORF peptides from lncRNAs
Several smORF peptides have been recently discovered in housekeeping lncRNAs. For example, the pri-miR171b and pri-miR165a transcripts, which are miRNA precursors, encode two peptides of 18 and 21 amino acids, respectively. Both peptides exhibit cis-acting regulatory functions by increasing the accumulation of their correlated miRNAs, thereby indirectly promoting the negative modulation of their targets associated with root development in Medicago truncatula and A. thaliana.91
Another example of a dual-function transcript is mammalian mitochondrial 12S rRNA, which encodes the smORF peptide MOTS-c (16 amino acids), a regulator of metabolic homeostasis in the nucleus.90,95 In mice, MOTS-c treatment prevents insulin resistance and diet-induced obesity and reverses age-related insulin resistance in muscles.90,95 Additionally, MOTC-c is involved in cold stress defence by increasing adipose thermogenesis.96
The smORF peptide humanin (24 amino acids), encoded by human mitochondrial 16S rRNA,13 was identified as a new cDNA involved in Alzheimer’s disease. Humanin interacts with the apoptosis regulator Bax (BCL-2-associated X protein), preventing its activation and, thus, cell death via apoptosis.13
For many years, circRNAs have been considered atypical products in cells; however, it was recently reported that these molecules are stable and are generated via a mechanism known as back-splicing.77,97 circRNAs might act as miRNA and protein sequesterers and may be involved in the splicing regulation of RNA polymerase II-mediated transcription (reviewed in Refs77–79) Moreover, circRNAs have been shown to be involved in several pathologies, such as diabetes, neurological diseases and cancer.78 Interestingly, circRNAs lack optimal translation sequences, such as the 5′ end 7-methylguanosine (m7G) cap structure and 3′ poly(A) tail.97 This new class of regulatory RNAs can also be translated into conserved smORF peptides.85–87,98,99
An example of a coding circRNA is circMbl1, which encodes a peptide of ∼10 kDa that moves to synapses in response to starvation and FOXO expression, suggesting that smORF peptides encoded by circRNAs may be involved in the mechanisms of neuronal communication.85 Another example of a coding circRNA is the circular form of (Long Intergenic Noncoding RNA p53-Induced Transcript (LINC-PINT), a glioblastoma suppressor,87 which encodes an 87 amino acid smORF peptide that directly interacts with Polymerase Associated Factor complex (PAF1c), thereby inhibiting transcription elongation of multiple oncogenes. Interestingly, its expression is lower in glioblastoma cells than in normal tissues.87 Finally, the circPPP1R12A circRNA gene triggers tumour pathogenesis and metastasis in colon cancer by activating the Hippo-YAP signalling pathway. Interestingly, this circRNA gene contains a smORF encoding a 73 amino acid peptide.99 All of these findings suggest that circRNAs are the newest reservoir of smORFs to be explored.
5. smORFs as reference CDSs
A reference CDS is the main or unique coding ORF of an mRNA, which can be flanked by translatable alternative ORFs.100 All reference CDSs smaller than 100 codons are defined as small refCDSs in this classification. A strong criterion for the annotation of the coding capacity of a transcript is related to ORF size.101 Thus, size restriction during automatic annotation was probably one of the reasons for the mis-annotation of important small refCDS transcripts as long non-coding RNAs in past years.14,24,25,30,31,91,102–108
5.1. Strategic insights into the investigation of smORFs as reference CDSs
Studies based on ribosome profiling and mass spectrometry are promising approaches because they have indicated the widespread translation of potential small refCDSs, but often at low expression rates,37,58,60,109–111 which possibly indicates the coding potential immaturity of the transcripts.33 Moreover, some eukaryotic mRNAs exhibit more than one small refCDS, constituting polycistronic transcripts (Fig. 4), which might even contain conserved and duplicated smORFs.24,65,112 Thus, a strategy based on a crosslink between ribosome profiling and mass spectrometry can indicate which smORFs are translated, even in polycistronic transcripts. Additionally, analysis of conservation at the amino acid level among different species coupled to transcript and peptide localization techniques (e.g. in situ hybridization and antibody staining, respectively) are promising approaches to identify biologically relevant small refCDSs. Knockdown experiments using RNA interference (RNAi) or loss-of-function via CRISPR-Cas9 are also important techniques if available in selected species; however, RNAi results are not conclusive when applied to polycistronic transcripts because they do not reveal which smORF is responsible for the studied phenotype.
Figure 4.
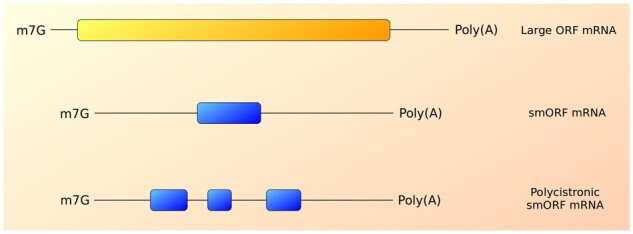
Comparison between large ORF mRNAs and smORF transcripts, which can occur in polycistronic arrangements. The yellow track represents large reference CDS; blue tracks represent small reference CDSs (coding smORFs). The lower panel represents a polycistronic mRNA containing three smORFs (blue tracks).
5.2. Examples of small refCDS peptides
One of the most emblematic examples of misannotated small refCDS transcripts is toddler/apela/ELABELA/ende, whose coding potential has been widely discussed (reviewed in Pauli et al.113) The toddler/apela/ELABELA/ende is a 55 amino acid peptide highly conserved among vertebrates that acts as an embryonic signal, binds to apelin receptors and triggers mesendodermal cell migration during gastrulation.76
Another classic example is the polycistronic gene pri/mlpt/tal (polished-rice/mille-pattes/tarsal-less, respectively), first described in T. castaneum as a gap gene. Pri/mlpt/tal is highly conserved among Pancrustacea and encodes two to five duplicated peptides (10 − 30 amino acids) containing the same LDPTGXY motif.65,112Pri/mlpt/tal is a well-known smORF gene, and its characterization has strongly contributed to the acceptance of smORFs as new developmental players in animals.22 Mutations and knockdown of pri/mlpt/tal promote lethal embryonic phenotypes that differ between species and among biological processes, such as the modification of epidermal structures,106,114 changes in the number of locomotor appendages 65,115,116 and tarsal deformations.75,117 The main mechanism of action of pri/mlpt/tal smORF peptides is to trigger the truncation of the transcription factor Ovo/Shavenbaby N-terminus, thereby converting it from a repressor to an activator.118
In the Hemiptera order (bedbugs and aphids), our group identified a new smORF within the polycistronic gene pri/mlpt/tal, named smHemiptera.116 SmHemiptera consists of ∼80 codons in Rhodnius prolixus and exhibits a GHR(Y/N)WMTHLPLSRP region shared among all derived hemipterans whose pri/mlpt/tal sequence is available.116 In addition, smHemiptera contains two large introns,116 which is an uncommon pattern in genes encoding small proteins.119
Two transmembrane smORF peptides, sarcolamban A and B (28 and 29 amino acids, respectively), were discovered in a misannotated non-coding transcript of Drosophila melanogaster.24 Sarcolamban peptides evolved 550 million years ago and contain orthologues with conserved roles in humans (phospholamban and sarcolipin).24 These peptides are involved in calcium transport regulation in the sarcoplasmic reticulum and act specifically during myocardial contraction; as a result, several studies on cardiac arrhythmia target sarcolamban peptides and their orthologues.24 Moreover, two smORF peptides from the same family, myoregulin (46 amino acids)30 and its antagonist DWORF (34 amino acids),31 are involved in calcium regulation in skeletal muscles. Myoregulin and DWORF were also discovered in misannotated lncRNAs in rats and are important targets of muscle performance studies.30,31
The polycistronic gene Enod40 exhibits two highly conserved smORFs in plants (12 and 24 amino acids in soybeans), which are expressed during early stages of root nodule organogenesis.14 Interestingly, these smORFs overlap, and their peptides bind to nodulin-100, a sucrose synthase subunit, suggesting their involvement in sucrose uptake control in nitrogen-fixing nodules.14
The transmembrane smORF peptide hemotin (88 amino acids) was identified in D. melanogaster. This peptide acts by regulating endosome maturation during phagocytosis, allowing the phagocytic digestion of microorganisms.120 Hemotin deletion results in the accumulation of undigested material within endolysosomes, decreased resistance against infections and reduced lifespan.120 Interestingly, the same study identified a homologous relationship between hemotin and stannin,120 a vertebrate smORF peptide (88 amino acids in humans).121 Both smORF peptides exhibit sequence, structural and functional conservation.120
Recently, the mitochondrial smORF peptide brawnin (71 amino acids length) was identified in vertebrates as an essential player in respiratory complex III assembly. In zebrafish, brawnin deletion causes complete complex III loss, which promotes early death.122
6. smORFs as isoforms of major ORFs (isoformic smORFs)
Isoformic smORFs are small variants of major ORFs. The main mechanism of isoformic smORF generation is alternative splicing (Fig. 5),10 which is a widespread regulatory process throughout multicellular eukaryotes due to their greater numbers of introns.123 Furthermore, other regulatory mechanisms, such as alternative transcription initiation (Fig. 5B), alternative polyadenylation (Fig. 5C) and alternative refCDS translation (Fig. 5D), could theoretically generate isoformic smORFs (reviewed in de Klerk and 't Hoen124 and Touriol et al.125) Even though smORF peptides translated from pseudogenes are not precise isoforms of their reference proteins, they may play a similar role as smaller forms of large CDS variants (Fig. 5E). Thus, isoform generation is an impressive mechanism underlying genetic variability,126 and thousands of variants could fall within the smORF size thresholds.
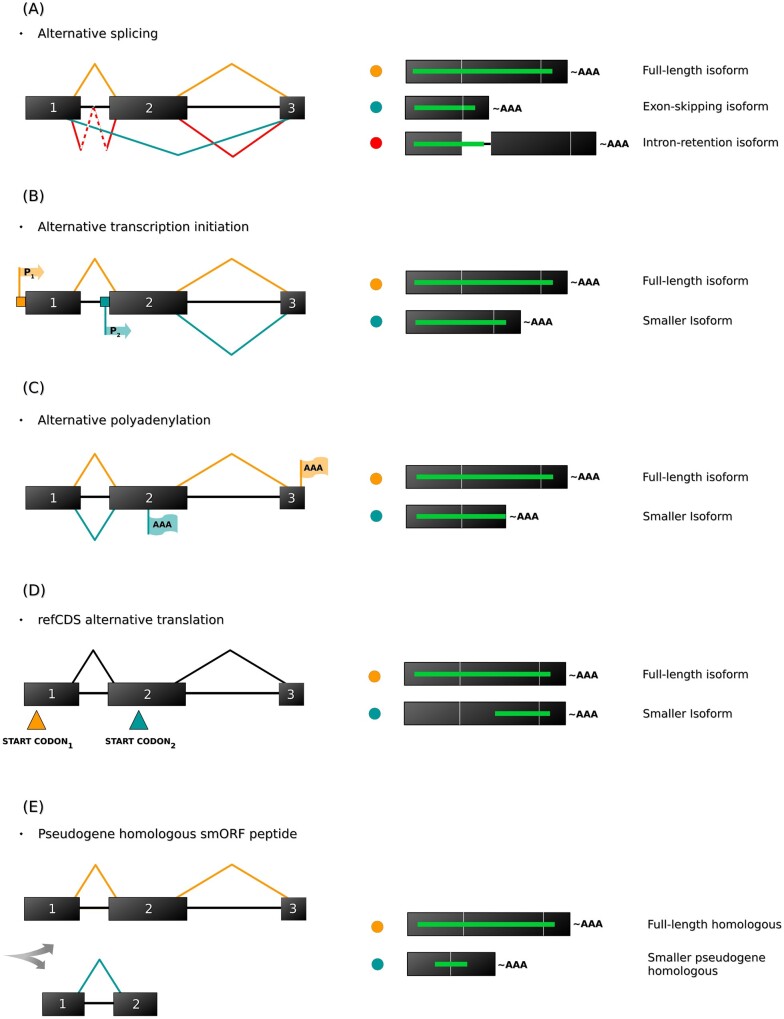
Figure 5.
Mechanisms of isoformic smORF biosynthesis. Isoformic smORFs are generated via large ORF transcriptional editing, which fragments large CDSs into smaller variants that can fall within the smORF length limits. (A) Alternative splicing (AS) is the best described mechanism of isoformic smORF biosynthesis; however, other molecular processes, such as (B) alternative transcription initiation via alternative promoters, (C) alternative polyadenylation cleavage, (D) alternative refCDS translation mediated by downstream start codons and (E) the fragmentation of homologous pseudogenes, could theoretically generate isoformic smORFs. Black tracks represent exons; black lines represent introns; green lines represent ORF variants; red, yellow and blue circles indicate the respective processes on the left.
Isoformic smORF generation via alternative splicing is possible via different pathways, such as exon deletion, which promotes major ORF truncation to generate small fragment(s)127 (Fig. 5A), and intron retention128 (Fig. 5A), which theoretically inserts stop and/or start codons within a major ORF. These processes lead to the generation of smaller isoforms with different carboxy and amino termini, although premature termination codon (PTC) insertion can also trigger the nonsense-mediated mRNA decay pathway.128 Moreover, more than one isoformic smORF can be generated by the same canonical RNA depending on the number of non-inactivating truncations produced.129,130
6.1. Strategic insights into the investigation of isoformic smORFs
Vertebrates are potentially the best models for the discovery of isoformic smORFs regulated by alternative splicing in animals. Vertebrate species are thought to exhibit a higher occurrence of alternative splicing than representatives of non-chordate species.130,131 Considering that the number of coding genes does not differ between invertebrates and vertebrates (e.g. ∼20,000 genes in humans <www.ncbi.nlm.nih.gov/genome/guide/human/> versus ∼20,000 genes in Caenorhabditis elegans132) evolution, differentiation and complexity may not be precisely linked to the number of genes in a taxon but are instead linked to the diversity of variants produced.123 Thus, many isoformic smORFs might be overlooked in vertebrates because alternative splicing (among other mechanisms) allows the number of transcripts to reach up to 10 times the number of precursor genes.133 Importantly, the experimental prediction of alternative splicing variants is as challenging as the prediction of smORFs itself because splicing complexity has not been fully elucidated, especially in the case of deleterious mutants (reviewed in Blakeley et al.134) however, a large-scale analysis in Physcomitrella patens (moss) identified 6,092 smORFs regulated by alternative splicing in 4,389 different genes.39 The same study reported that isoformic smORF peptides tend to follow the activity pathways of their precursors as well as their amino acid usage,39 which are important leads for functional investigations.
The bioinformatic prediction of isoformic smORFs performed only on transcriptome sequencing data is limited by de novo assemblies that could not discriminate alternative gene products without a reference genome. If genome sequences are available, the use of computational predictors of splicing sites,135 splicing variants136 or even alternative transcription initiation and polyadenylation sites137 prior to smORF detection is advantageous, because these software can generate alternative transcription data as reference for transcriptome assemblers. On the other hand, the functional investigation of isoformic smORFs at the DNA level is particularly difficult due to the overlap between smORFs and their major variants. Thus, post-transcriptional and post-translational approaches, such as RNAi, in situ hybridization and antibody staining, as well as comparisons to ribosome profiling and shotgun proteomic data, are more suitable.
Pseudogene homologous analysis requires pseudogene prediction pipelines prior to smORF detection.138 Then, an interesting strategy is the evaluation of evolutionary constraints on pseudogene homologous smORFs to detect coding potential for further investigation. Consistent with this strategy, 50 pseudogenes encoding homologous smORF peptides of canonical proteins were reported to undergo translation and the resulting peptides are under evolutionary constraints (Ka/Ks ratio < 0.3), which suggests potential functional roles.139
smORF functional analysis of pseudogenes using CRISPR-Cas9 is also challenging. For instance, if the target pseudogene region is similar or identical to the parental gene, the specificity of CRISPR-Cas9 will be problematic because the guide RNA might misdirect the Cas9 nuclease to non-target regions.
6.2. Examples of isoformic smORF peptides
The most representative examples of isoformic smORFs encode interference peptides (small interfering peptides (siPEPs) or microproteins (miPs)) akin to microRNAs.34 siPEPs/miPs consist of a single protein−protein interaction domain, which allows them to bind to larger proteins, typically transcription factors, generating non-functional complexes.140,141 Moreover, siPEPs/miPs can control their targets via substrate competition.140,141 siPEP/miP generation occurs via single-gene-unit expression or alternative splicing; in the second case, they can regulate their own alternative splicing transcript variants.140–143
The tumour suppressor TEL (also known as ETV6) belongs to the ETS transcription factor family.17 Five alternative splicing variants of TEL may be generated by exon deletion.17 Two isoforms encode the smORF peptides TEL-c and TEL-d (both ranging from 20 to 30 amino acids), which lack the activity of their full-length variant suppressors but exhibit considerable expression in individuals with myelodysplastic syndrome-derived leukaemia.17 Interestingly, TEL-c and TEL-d exhibit lower expression in myelodysplastic syndrome before cancer progression, suggesting a link to smORF expression and leukaemia development.17
Members of the Phytochrome Interacting Factor (PIF) transcription factor family are important players in seed dormancy and germination control in response to environmental stimuli in A. thaliana.144 A previous analysis showed that PIF6 contains an alternative splicing isoformic peptide of ∼180 amino acids known as PIF6-β, which is generated by the insertion of a premature stop codon and the excision of the DNA ligand domain due to the deletion of exon 3.144 PIF6-β is highly expressed in seed development; however, PIF6 gene silencing, which includes full-length variant knockdown, increases primary seed dormancy. On the other hand, the overexpression of PIF6-β alone promotes dormancy reduction.144 Even though the length of PIF6-β does not correspond to the most accepted smORF threshold of 100 codons, it is an interesting example of how alternative splicing can generate small variants with important biological effects. Unfortunately, annotation screenings have neglected the roles of alternative splicing smORF variants across species, possibly as a consequence of methodological obstacles in smORF detection.
7. smORFs as alternative CDSs in mRNAs
Alternative smORFs are alternatively or pervasively translated from the same mRNAs as canonical large CDSs. In contrast to isoformic smORFs, alternative smORFs are not variants of large CDSs generated via RNA editing but rather are different CDSs located in different parts of the mature mRNA.
In early genomics research, the ‘one gene, one protein’ dogma145 was confronted by the capacity of mRNAs to encode more than one protein from alternative ORFs. In this context, the classic configuration of a eukaryotic mRNA was described as a monocistronic transcript divided into three regions: a CDS, usually consisting of a major ORF, flanked by 5′ and 3′ untranslated regions (UTRs) (Fig. 6A).146
Figure 6.
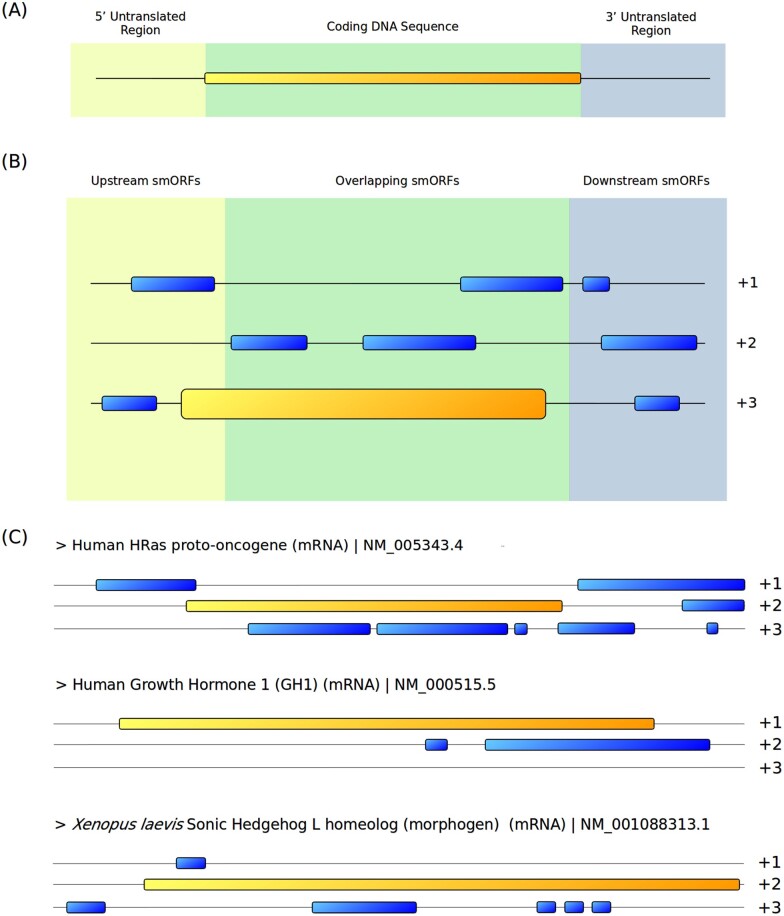
Location and distribution of alternative smORFs. (A) Canonical monocistronic mRNA paradigm comprising a unique large CDS between untranslated regions (UTRs). (B) Alternative smORF division and distribution within an mRNA. Upstream smORFs are located in the 5′UTR, and their stop codons may extend across the reference CDS region. The start codons of overlapping smORFs strictly overlap with the reference CDS region, but their sequences may extend to the 3′UTR. Downstream smORFs are located totally within the 3′UTR. (C) Examples of well-known representative mRNAs exhibiting several alternative smORFs (sequence analysis by the authors). Alternative smORFs are commonly encountered in mRNAs, and their coding potential is still underappreciated. Yellow tracks represent reference CDSs (large ORFs); blue tracks represent alternative smORFs.
Mechanisms such as alternative splicing126 and mature RNA editing147,148 are well-known processes underlying alternative protein evolution from mRNAs; however, emerging evidence suggests that alternative ORF translation is also an important biological pathway contributing to genetic variability, either from overlapping genes149–151 or alternative ORFs in unique transcripts.152 In the latter case, translation may involve post-transcriptional regulation mechanisms, such as the reinitiation of translation,153 ribosomal frame shifting154 and stop codon read-through.155
Although de novo protein generation via alternative ORFs is atypical, especially in eukaryotes,156,157 the distribution of coding alternative ORFs is speculated to be underestimated due to detection difficulties.158 Proteomic and transcriptomic approaches corroborate the existence of putative alternative smORFs in the flanking regions of or overlapping with refCDSs in different frames,38,58,100,159 by definition comprising new polycistronic mRNAs.160 For instance, recent data suggest that 15% of alternative smORFs in humans are preceded by Kozak consensus sequences,38 suggesting that smORFs can show efficiency in ribosome recognition.161,162 Importantly, hundreds of alternative smORFs with evidence of translation in humans are highly conserved in distant taxa, such as basal vertebrates, invertebrates and yeast.100 Another study identified 149 alternative smORFs that are conserved among humans and rats. Dozens of these smORFs overlap with reference CDSs but do not show evolutionary sequence constraints on their codon composition.159 Recently, a large-scale approach using cross-linked mass spectrometry followed by shotgun proteomics revealed that alternative ORFs act as regulators in NCH82 human glioma cell reprogramming via protein kinase A activation.163
Alternative smORFs are an emerging frontier in the study of mechanisms of genetic variability36; however, RNA-based phenotype analysis techniques such as RNAi and in situ hybridization cannot distinguish the roles of refCDSs and alternative smORFs since mRNAs are identical in these two cases. Thus, new tools such as CRISPR-Cas9 have arisen as a promising approach for studies on this topic, although modifications in overlapping smORFs still pose the challenge of refCDS constraints, which requires careful experimental planning.
Alternative smORFs present peculiarities inherent to their positions in transcripts. Alternative ORFs that are located in 5′UTRs, overlap with refCDSs or are located in 3′UTRs are referred to as upstream ORFs, overlapping smORFs and downstream smORFs, respectively 8,35 (Fig. 6B). Alternative smORFs are frequently present in all types of mRNAs (some examples are presented in Fig. 6C), even though functions may be lacking. Thus, discovering which smORFs are biologically relevant is particularly challenging. The characteristics of each subgroup of alternative smORFs are detailed below.
7.1. smORFs as alternative CDSs in mRNAs: upstream ORFs
Upstream ORFs (uORFs) are smORFs located in 5′UTRs and have been reported in 20 − 50% of eukaryotic mRNAs.164 uORFs play roles in several post-transcriptional control mechanisms, particularly by modulating ribosomal access to downstream refCDSs.165 This mechanism promotes the regulation of translation efficiency and triggers mRNA degradation.165 Post-transcriptional regulation promoted by uORFs is highly important to prevent the overexpression of central proteins of cellular networks; in this context, mRNAs of regulatory proteins such as transcription factors tend to evolve uORFs,161,166 and mRNAs without uORFs encode more abundant proteins.167,168 uORFs modulate the translation of several disease-related mRNAs (reviewed in Zhang et al.169) including transcripts modulated during cancer.170
uORF peptides can function by interacting with refCDS proteins or by binding to other molecules to trigger ribosomal stalling.171–173 Cis-acting regulatory uORF peptides are known as peptoswitches, analogous to riboswitches, which are modulators of RNA transcription and translation.174 On the other hand, the translation of uORFs can generate non-functional peptides, in which translation itself is the regulatory event.113 For instance, uORF stop codons can be recognized as PTCs, thereby triggering the nonsense-mediated mRNA decay (NMD) pathway, which promotes transcript degradation.175 Another interesting regulatory mechanism occurs when uORF stop codons overlap with the initial portion of refCDSs (known as overlapping uORFs, or oORFs); in these cases, translational control is achieved via direct competition between the oORF and the refCDS for ribosomal coverage.176
Even though uORFs perform several cis-regulatory roles, a significant portion may encode trans-acting peptides.15,177–180
7.1.1. Strategic insights into the investigation of uORF peptides
Reannotation of mRNAs focussing on uORF discovery is a promising approach. For instance, in zebrafish, a computational analysis showed that over 60% of all protein-coding genes contain uORFs.181 This number can reportedly reach 50% in mammals166,182,183 and 30% in plants184; however, a small portion of uORFs show coding potential in terms of amino acid sequence conservation. Only 1.1% (87) of the uORFs found in mice, 1.7% (149) in humans, 0.3% (27) in zebrafish181 and 0.5% (44) in Arabidopsis185 appear to be under selective pressure to maintain their encoded amino acid sequences. In zebrafish, over 60% of conserved uORFs show evidence of translation, as indicated by ribosome profiling.181 Interestingly, ∼20% of uORFs exhibit start codons in Kozak consensus optimal regions in humans.186 In addition, some uORF peptides that are conserved between humans and mice are under stabilizing selection.139 Based on the aforementioned data, we suggest that a conservative coding uORF annotation approach focuses on amino acid sequence conservation, the presence of Kozak consensus and stabilizing selection. In addition, the amino acid usage of uORFs is an important hint because it differs from that of canonical proteins,34 and alternative start codons are common.187,188 Considering that most functional uORFs perform cis-regulatory roles, sequences that do not follow these parameters are potentially non-coding or pervasively translated.
7.1.2. Examples of uORF peptides
uORF peptides have well-established roles in cis-acting regulatory pathways. For instance, the transcript of human S-adenosylmethionine decarboxylase, an enzyme involved in the responses to changes in polyamine levels, displays one of the smallest smORF peptides described to date (six amino acids) whose translation triggers ribosomal stalling and modulates the translation of its downstream refCDS.189
Another interesting example comes from the model legume Medicago truncatula, in which the 62 amino acid peptide uORF1p is generated via partial intron retention in the mRNA of the transcription factor MtHAP2-1. uORF1p also regulates the translation of its refCDS via a trans-acting regulatory mechanism.190 Once translated, uORF1p binds to the 5′ leader sequence of the MtHAP2-1 transcript, thereby decreasing MtHAP2-1 translation.190
Another uORF peptide that acts via trans-acting mechanisms occurs in the 1A glucocorticoid receptor transcript.15 Receptor synthesis is completely inhibited by the deletion of the second (uORF-2) of five uORFs in its mRNA. Interestingly, uORF-2 translation results in a 93 amino acid peptide that also modulates the expression of the 1A glucocorticoid receptor via an unknown mechanism. Therefore, the uORF-2 peptide may be involved in the translation of this receptor, probably by interacting with other molecules.15
uORF peptides can also play roles that are wholly independent of the corresponding refCDS regulation. For instance, the gene encoding the MKKS protein, associated with McKusick−Kaufman syndrome, contains three regulatory uORFs. Two of these uORFs encode highly conserved peptides (63 and 45 amino acids in humans) that can be observed in the mitochondrial membrane, while the MKKS protein remains in the cytoplasm. These data suggest that uORFs can act independently from their respective refCDSs.177
The 5′UTR of the c-akt proto-oncogene transcript displays an uORF encoding a 10 amino acid tumour rejection antigen (pRL1), which is recognized by cytotoxic T lymphocytes in BALB/c radiation-induced leukaemia RL♂1 cells.191 Interestingly, sequence analysis showed that pRL1 amino acid residues are identical to the 269 − 278 stretch of the protein expressed by the c-akt viral homologue v-akt.191 These findings demonstrate that uORFs can also play important immunological roles, although overlapping smORFs have been more frequently identified as antigen precursors.192
7.2. smORFs as alternative CDSs in mRNAs: overlapping smORFs
Overlapping smORFs are smORFs that overlap with the refCDS in another frame. The start codons of overlapping ORFs strictly overlap with refCDSs and can extend to the 3′UTR,193 unlike oORFs, as mentioned in the previous section. The great majority of overlapping ORFs are smORFs.38,100 Several overlapping smORF peptides have been described, and their widespread expression suggests many different roles.194–196
7.2.1. Strategic insights into the investigation of overlapping smORFs
Overlapping smORFs are abundant in mRNAs but were initially considered to represent a strategy for increasing genetic variability in size-restricted species genomes, such as those of prokaryotes156,159,197; however, overlapping ORFs have also been discovered in complex eukaryotes.100,193,198,199 For instance, in humans, ∼41% of mRNAs contain at least one unannotated overlapping ORF, and most of these ORFs encode peptides smaller than 90 amino acids.100 In another study, 217 overlapping ORFs containing Kozak consensus sequences were shown to be conserved in rodents and humans according to RefSeq transcript analysis.193 To distinguish random sequences from coding stretches, some features must be considered. For example, overlapping smORFs suffer from the sequence constraints imposed by refCDSs because evolutionary pressures on one frame affect the others, thereby challenging phenotypic analysis; however, previous studies reported an evolutionary mechanism that allows overlapping ORF proteins to evolve less restrictively at both the sequence and structural levels.157 For instance, studies analysing the CDSs of viral overlapping genes suggest that overlapping ORFs tend to encode structurally disordered proteins200 with codon-rich amino acids such as arginine, leucine and serine.201 Another overlapping ORF feature is the oscillating amino acid modification rate, which differs from that of single-CDS genes.202,203 Therefore, this mutation pattern can be used as an important parameter for overlapping smORF peptide detection.159
7.2.2. Examples of overlapping smORF peptides
Several overlapping smORFs have been discovered and previously annotated within mRNAs (brief review in Andrews and Rothnagel8) In 1996, the gp75 melanoma antigen transcript that contains an overlapping smORF encoding a 24 amino acid peptide was identified as a tumour rejection antigen that is recognized by T-cells.16 Since then, several tumour antigens encoded by overlapping smORFs have been discovered,204–210 indicating that overlapping smORFs are significant reservoirs of endogenous antigens.
Overlapping smORF antigens are commonly observed during viral infection or tumour cell growth.192,211 A ribosome-based mechanism evolved to provide cryptically translated peptides that can be strictly used as substrates for antigen processing has been suggested.212 Moreover, this mechanism could represent one of the most significant pathways for the generation of endogenous antigens associated with the major class I histocompatibility complex (MHC class I).213 Thus, this mechanism is an important topic of studies on immunological surveillance and new vaccine development.214,215
Importantly, peptides with immunogenic potential are not unique products derived from overlapping smORFs. For example, the PRNP gene encodes the mammalian cellular prion protein (PrP),196 a glycoprotein that is the causative agent of neurodegenerative diseases after deleterious structural folding.216 The physiological functions of PrP are not entirely known because the PrP transcript is widely expressed not only in nerve tissues but also in the heart, skeletal muscle, intestine, uterus, and testis (reviewed in Sarnataro et al.216). Interestingly, the diversity of the PrP transcript distribution may be associated with the highly conserved overlapping smORF that encodes the mammalian AltPrP peptide (73 amino acids). AltPrP is transported to mitochondria, and its stability is regulated by endoplasmic reticulum stress and proteasome inhibition.196 Some of the toxic or protective functions attributed to PrP may actually be triggered by AltPrP.196
The INK4a gene (also known as MTS1 or CDKN2) encodes a protein with tumour suppressor characteristics involved in cell cycle regulation called p16INKa.217 The INK4a transcript contains an overlapping smORF encoding a 132 amino acid peptide called p19ARF in humans. The ectopic expression of p19ARF in the fibroblast nucleus induces interphase G1 and G2 arrest, demonstrating that overlapping smORFs can also perform important roles in cell cycle control.218
7.3. smORFs as alternative CDSs in mRNAs: downstream smORFs
Downstream smORFs are ORFs located in 3′UTRs. The coding potential of downstream smORFs is underestimated and poorly explored in comparison to that of other alternative smORFs35; however, 3′UTRs contain important translational regulatory elements and subcellular localization signals, also contributing to eukaryotic transcript stability, tissue patterning processes, embryonic axis formation, mammalian spermatogenesis (reviewed in Wang et al.219) and cancer.170,220,221 In addition, 3′UTRs contain an SECIS (selenocysteine insertion sequence) element, a signal required for the insertion of the rare amino acid selenocysteine into UGA stop codons during the translation of canonical major ORFs.222,223
A recent study showed that downstream smORFs also represent a widespread potential translation regulatory mechanism among vertebrates because the translation of downstream smORFs itself is required for the increased translation of reference major ORFs, depending on the number of downstream smORFs in the mRNA. Importantly, the amino acid sequence and smORF peptide length do not influence this regulatory mechanism224; however, downstream smORF peptides have been scarcely reported in the literature.100,225 Although there is significant evidence of ribosomal coverage in 3′UTRs,226,227 these events are often associated with delays in ribosome decoupling after translation or stop codon read-through,155 where the ribosome does not recognize the refCDS stop codon and proceeds in the reading of the entire 3′UTR.228
7.3.1. Strategic insights into the investigation of downstream smORFs
The prediction of coding downstream smORFs is particularly challenging. Although the translation of downstream smORFs has been described,37,100,139,188 most of the previously reported examples exhibit a low translation efficiency in ribosome profiling analysis; however, some downstream smORFs are more highly translated,139 which suggests that rare coding downstream smORFs exist and await annotation.
Interestingly, an important phenomenon has been described wherein 3′UTRs are cleaved by an unknown post-transcriptional mechanism, but polyadenylation sites and downstream smORFs are retained in a new independent transcript.229 Importantly, 3′UTR transcripts usually exhibit different expression patterns than their parental mRNAs,230 as described for up to 50% of 3′UTR transcripts in rats.229 RNAs generated by 3′UTR cleavage have evolved non-coding activities,231 and traditional gene prediction approaches consider their coding capacity to be low, classifying them as potential ncRNAs229; however, the coding capacity of 3′UTR transcripts remains unclear because coding smORFs are usually dismissed by traditional gene prediction methods. Thus, studies designed to explore the coding potential of smORFs in 3′UTR transcripts could be promising, but the mapping of 3′UTR transcripts is difficult due to the low availability of 3′UTR read annotation, coverage and assembly data in public repositories, even for well-studied CDS transcripts.232
7.3.2. Examples of downstream smORF peptides
The first coding downstream smORF was identified in the H60 histocompatibility gene, encoding the eight amino acid antigen LYL8, which is presented by MHC class I to the immune system, thereby suppressing cytotoxic T-cell activation and inducing immunological self-tolerance against endogenous polypeptides.225,233 Interestingly, the observation of the insertion of stop codons between the refCDS and downstream smORF as well as the alternation of frames showed that LYL8 translation does not occur via stop codon read-through because LYL8 bioactivity remained unchanged.225,234 These data suggest that downstream smORF translation is possible via direct ribosome recognition, independent of the refCDS.
Other findings have shown that stop codon read-through is an important mechanism for downstream smORF translation. Interesting findings were obtained from studies involving aminoglycoside antibiotics, which increase translational frequency via premature termination codon (PTC) read-through and are indicated for use against disorders caused by defective proteins generated due to PTCs.235 Cells treated with the aminoglycoside gentamicin undergo an apparent autoimmune response involving the translation of downstream smORF antigens that are able to activate CD8+ T cells.235
The MRVI1 (murine retrovirus integration-site 1) gene encodes a protein with myeloid leukaemia suppressor activity.236 MRVI1 also encodes a downstream smORF peptide (∼95 amino acids) that interacts with the tumour suppressor BRCA1 (BReast CAncer type 1 susceptibility protein) in the HeLa cell nucleus.100 The MRVI1 downstream smORF peptide was possibly previously rejected as a bioactive product due to its out-of-frame position with respect to the refCDS.100 These data suggest that other functional downstream smORFs may have been dismissed for the same reason in large-scale analyses.
8. Conclusion and future prospects
The integration between omic sciences and bioinformatics has enabled the study of previously obscured topics in systems biology, such as junk DNA, wherein hundreds of smORFs have been discovered as new players in developmental biology, cancer, neuropathologies, transcription/translation control, tissue physiology, immunological surveillance, and responses to environmental stimuli, among other phenomena.
The emergence of smORF peptides as a significant and non-sporadic phenomenon occurred during mid-2010, when the NGS era allowed deep sequencing analysis. Since then, numerous smORFs have been discovered, especially during the last 5 years, when hundreds of lncRNAs were reannotated as smORF transcripts encoding important and essential functional peptides. Furthermore, housekeeping RNAs such as rRNAs, pri-miRNAs and circRNAs have been identified as coding smORF reservoirs.
In summary, the new smORF classification presented here will help to direct the further development of bioinformatics and functional studies. Genome annotation screenings have dismissed the coding potential of smORFs owing to the lack of knowledge about this new class of genes; thus, this review will help researchers uncover these important elements of molecular biology by offering many insights into the study of each smORF class. The functional characterization of even more smORF peptides in the future will provide evidence of the number of essential smORFs in different taxa, supporting comparative analysis. Future studies should also address the evolutionary trends of smORFs among different phyla, such as whether the classes described here differ among species. Studies comparing smORFs at the base of metazoan and non-bilaterian groups, such as sponges, cnidarians, placozoans and ctenophores, would be particularly interesting. Additionally, a comparison of the roles and conservation of smORFs during speciation and/or whole-genome duplication events might provide new insights into the origin and function of this interesting class of genes. Finally, the recent discussion about non-coding smORFs as precursors of coding gene birth32,33 is one of the new frontiers of evolutionary biology, which may lead to a whole new area of future research. Hence, understanding smORF diversity and its singularities is essential to discover evolutionary innovations in this hidden coding DNA world.
Authors’ contributions
D.G.-A., D.A.T. and R.N.-da-F. contributed equally to the writing of this manuscript.
Funding
D.G.-A. was a master’s student of Federal University of Rio de Janeiro with scholarship financed by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - CAPES. D.A.T. and R.N.-da-F. were supported by Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro - FAPERJ (D.A.T.: E-26/202.736/2019; R.N.-da-F.: E-26/211.169/2019, E-26/202.605/2019, E-210.264/2018) and Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq (grant numbers unavailable due to system lockdown)
Conflict of interest
None declared.
Contributor Information
Diego Guerra-Almeida, Integrated Laboratory of Morphofunctional Sciences, Institute of Biodiversity and Sustainability, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil.
Diogo Antonio Tschoeke, Health Systems Engineering Laboratory, Alberto Luiz Coimbra Institute of Graduate Studies and Engineering Research (COPPE), Federal University of Rio de Janeiro, Rio de Janeiro, Brazil.
Rodrigo Nunes-da-Fonseca, Integrated Laboratory of Morphofunctional Sciences, Institute of Biodiversity and Sustainability, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil; National Institute of Science and Technology in Molecular Entomology, Rio de Janeiro, Brazil.
References
- 1. Tattersall A., Grant M.J.. 2016, Big data - what is it and why it matters, Health Info. Libr. J., 33, 89–91. [DOI] [PubMed] [Google Scholar]
- 2. Mumtaz M.A.S., Couso J.P.. 2015, Ribosomal profiling adds new coding sequences to the proteome, Biochem. Soc. Trans., 43, 1271–6. [DOI] [PubMed] [Google Scholar]
- 3. Patraquim P., Mumtaz M.A.S., Pueyo J.I., Aspden J.L., Couso J.-P.. 2020, Developmental regulation of canonical and small ORF translation from mRNAs, Genome Biol., 21, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Z., Gerstein M., Snyder M.. 2009, RNA-Seq: a revolutionary tool for transcriptomics, Nat. Rev. Genet., 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ramamurthi K.S., Storz G.. 2014, The small protein floodgates are opening; now the functional analysis begins, BMC Biol., 12, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brylinski M. 2013, Exploring the “dark matter” of a mammalian proteome by protein structure and function modeling, Proteome Sci., 11, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sieber P., Platzer M., Schuster S.. 2018, The definition of open reading frame revisited, Trends Genet., 34, 167–70. [DOI] [PubMed] [Google Scholar]
- 8. Andrews S.J., Rothnagel J.A.. 2014, Emerging evidence for functional peptides encoded by short open reading frames, Nat. Rev. Genet., 15, 193–204. [DOI] [PubMed] [Google Scholar]
- 9. Chekulaeva M., Rajewsky N.. 2019, Roles of long noncoding RNAs and circular RNAs in translation, Cold Spring Harb. Perspect. Biol., 11, a032680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chu Q., Ma J., Saghatelian A.. 2015, Identification and characterization of sORF-encoded polypeptides, Crit. Rev. Biochem. Mol. Biol., 50, 134–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang X., Tschaplinski T.J., Hurst G.B., et al. 2011, Discovery and annotation of small proteins using genomics, proteomics, and computational approaches, Genome Res., 21, 634–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Storz G., Wolf Y.I., Ramamurthi K.S.. 2014, Small proteins can no longer be ignored, Annu. Rev. Biochem., 83, 753–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guo B., Zhai D., Cabezas E., et al. 2003, Humanin peptide suppresses apoptosis by interfering with Bax activation, Nature, 423, 456–61. [DOI] [PubMed] [Google Scholar]
- 14. Rohrig H., Schmidt J., Miklashevichs E., Schell J., John M.. 2002, Soybean ENOD40 encodes two peptides that bind to sucrose synthase, Proc. Natl. Acad. Sci. U S A., 99, 1915–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diba F., Watson C.S., Gametchu B.. 2001, 5’UTR sequences of the glucocorticoid receptor 1A transcript encode a peptide associated with translational regulation of the glucocorticoid receptor, J. Cell. Biochem., 81, 149–61. [PubMed] [Google Scholar]
- 16. Wang R.F., Parkhurst M.R., Kawakami Y., Robbins P.F., Rosenberg S.A.. 1996, Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen, J. Exp. Med., 183, 1131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sasaki K., Nakamura Y., Maki K., et al. 2004, Functional analysis of a dominant-negative ΔETS TEL/ETV6 isoform, Biochem. Biophys. Res. Commun., 317, 1128–37. [DOI] [PubMed] [Google Scholar]
- 18. Olexiouk V., Menschaert G.. 2016, Identification of small novel coding sequences, a proteogenomics endeavor, Proteogenomics Cham., 926, 49–64. [DOI] [PubMed] [Google Scholar]
- 19. Yeasmin F., Yada T., Akimitsu N.. 2018, Micropeptides encoded in transcripts previously identified as long noncoding RNAs: a new chapter in transcriptomics and proteomics, Front. Genet., 9, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Basrai M., Hieter P., Boeke J.D.. 1997, Small open reading frames: beautiful needles in the haystack, Genome Res., 7, 768–71. [DOI] [PubMed] [Google Scholar]
- 21. Prasse D., Thomsen J., De Santis R., Muntel J., Becher D., Schmitz R.A.. 2015, First description of small proteins encoded by spRNAs in Methanosarcina mazei strain Gö1, Biochimie, 117, 138–48. [DOI] [PubMed] [Google Scholar]
- 22. Albuquerque J.P., Tobias-Santos V., Rodrigues A.C., Mury F.B., da Fonseca R.N.. 2015, small ORFs: a new class of essential genes for development, Genet. Mol. Biol., 38, 278–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Finkel Y., Stern-Ginossar N., Schwartz M.. 2018, Viral short ORFs and their possible functions, Proteomics, 18, 1700255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Magny E.G., Pueyo J.I., Pearl F.M.G., et al. 2013, Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames, Science, 341, 1116–20. [DOI] [PubMed] [Google Scholar]
- 25. Matsumoto A., Pasut A., Matsumoto M., et al. 2017, MTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide, Nature, 541, 228–32. [DOI] [PubMed] [Google Scholar]
- 26. Merino-Valverde I., Greco E., Abad M.. 2020, The microproteome of cancer: from invisibility to relevance, Exp. Cell Res., 392, 111997. [DOI] [PubMed] [Google Scholar]
- 27. Rytömaa T. 2014, Identification of an exceptionally short open reading frame in the genome of man, encoding a decapeptide, which regulates granulopoiesis by negative feedback, Cell Prolif., 47, 287–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Slavoff S.A., Heo J., Budnik B.A., Hanakahi L.A., Saghatelian A.. 2014, A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining, J. Biol. Chem., 289, 10950–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu S., Wang J., He Y., Meng N., Yan G.-R.. 2018, Peptides/proteins encoded by non-coding RNA: a novel resource bank for drug targets and biomarkers, Front. Pharmacol., 9, 1295–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anderson D.M., Anderson K.M., Chang C.L., et al. 2015, A micropeptide encoded by a putative long noncoding RNA regulates muscle performance, Cell, 160, 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nelson B.R., Makarewich C.A., Anderson D.M., et al. 2016, A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle, Science, 351, 271–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ruiz-Orera J., Verdaguer-Grau P., Villanueva-Cañas J.L., Messeguer X., Albà M.M.. 2018, Translation of neutrally evolving peptides provides a basis for de novo gene evolution, Nat. Ecol. Evol., 2, 890–6. [DOI] [PubMed] [Google Scholar]
- 33. Guerra-Almeida D., Nunes-da-Fonseca R.. 2020, Small open reading frames: how important are they for molecular evolution? Front. Genet., 11, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Couso J., Patraquim P.. 2017, Classification and function of small open reading frames, Nat. Rev. Mol. Cell Biol., 18, 575–89. [DOI] [PubMed] [Google Scholar]
- 35. Chugunova A., Navalayeu T., Dontsova O., Sergiev P.. 2018, Mining for small translated ORFs, J. Proteome Res., 17, 1–11. [DOI] [PubMed] [Google Scholar]
- 36. Orr M.W., Mao Y., Storz G., Qian S.-B.. 2020, Alternative ORFs and small ORFs: shedding light on the dark proteome, Nucleic Acids Res., 48, 1029–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Slavoff S.A., Mitchell A.J., Schwaid A.G., et al. 2013, Peptidomic discovery of short open reading frame-encoded peptides in human cells, Nat. Chem. Biol., 9, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Samandi S., Roy A.V., Delcourt V., et al. 2017, Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins, Elife, 6, 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fesenko I., Kirov I., Kniazev A., et al. 2019, Distinct types of short open reading frames are translated in plant cells, Genome Res., 29, 1464–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Warren A.S., Archuleta J., Feng W., Setubal J.C.. 2010, Missing genes in the annotation of prokaryotic genomes, BMC Bioinformatics, 11, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rice P., Longden I., Bleasby A.. 2000, EMBOSS: the European Molecular Biology Open Software Suite, Trends Genet., 16, 276–7. [DOI] [PubMed] [Google Scholar]
- 42. Ladoukakis E., Pereira V., Magny E.G., Eyre-Walker A., Couso J.. 2011, Hundreds of putatively functional small open reading frames in Drosophila, Genome Biol., 12, R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hücker S.M., Ardern Z., Goldberg T., et al. 2017, Discovery of numerous novel small genes in the intergenic regions of the Escherichia coli O157:H7 Sakai genome, PLoS One, 12, e0184119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vanderpool C.K., Balasubramanian D., Lloyd C.R.. 2011, Dual-function RNA regulators in bacteria, Biochimie, 93, 1943–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen H., Xu Z., Liu D.. 2019, Small non‐coding RNA and colorectal cancer, J. Cell. Mol. Med., 23, 3050–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grosshans H., Filipowicz W.. 2008, The expanding world of small RNAs, Nature, 451, 414–6. [DOI] [PubMed] [Google Scholar]
- 47. Babski J., Maier L.-K., Heyer R., et al. 2014, Small regulatory RNAs in Archaea, RNA Biol., 11, 484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Friedman R.C., Farh K.K.-H., Burge C.B., Bartel D.P.. 2008, Most mammalian mRNAs are conserved targets of microRNAs, Genome Res., 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Waters L.S., Storz G.. 2009, Regulatory RNAs in bacteria, Cell, 136, 615–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Beisel C.L., Storz G.. 2011, Discriminating tastes, RNA Biol., 8, 766–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kantar M., Lucas S.J., Budak H.. 2011, miRNA expression patterns of Triticum dicoccoides in response to shock drought stress, Planta, 233, 471–84. [DOI] [PubMed] [Google Scholar]
- 52. Bronesky D., Wu Z., Marzi S., et al. 2016, Staphylococcus aureus RNAIII and its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression, Annu. Rev. Microbiol., 70, 299–316. [DOI] [PubMed] [Google Scholar]
- 53. Gimpel M., Brantl S.. 2016, Dual-function sRNA encoded peptide SR1P modulates moonlighting activity of B. subtilis GapA, RNA Biol., 13, 916–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mangold M., Siller M., Roppenser B., et al. 2004, Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule, Mol. Microbiol., 53, 1515–27. [DOI] [PubMed] [Google Scholar]
- 55. Friedman R.C., Kalkhof S., Doppelt-Azeroual O., et al. 2017, Common and phylogenetically widespread coding for peptides by bacterial small RNAs, BMC Genomics, 18, 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Morgado L., Johannes F.. 2019, Computational tools for plant small RNA detection and categorization, Brief. Bioinformatics, 20, 1181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. King A.M., Vanderpool C.K., Degnan P.H.. 2019, sRNA Target Prediction Organizing Tool (SPOT) integrates computational and experimental data to facilitate functional characterization of bacterial small RNAs, mSphere, 4, e00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Aspden J.L., Eyre-Walker Y.C., Phillips R.J., et al. 2014, Extensive translation of small open reading frames revealed by Poly-Ribo-Seq, Elife, 3, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li H., Xiao L., Zhang L., et al. 2018, FSPP: a tool for genome-wide prediction of smORF-encoded peptides and their functions, Front. Genet., 9, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bazzini A.A., Johnstone T.G., Christiano R., et al. 2014, Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation, Embo J., 33, 981–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mackowiak S.D., Zauber H., Bielow C., et al. 2015, Extensive identification and analysis of conserved small ORFs in animals, Genome Biol., 16, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jager D., Sharma C.M., Thomsen J., Ehlers C., Vogel J., Schmitz R.A.. 2009, Deep sequencing analysis of the Methanosarcina mazei Go1 transcriptome in response to nitrogen availability, Proc. Natl. Acad. Sci., 106, 21878–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Artimo P., Jonnalagedda M., Arnold K., et al. 2012, ExPASy: SIB bioinformatics resource portal, Nucleic Acids Res., 40, W597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. 1990, Basic local alignment search tool, J. Mol. Biol., 215, 403–10. [DOI] [PubMed] [Google Scholar]
- 65. Savard J., Marques-Souza H., Aranda M., Tautz D.. 2006, A segmentation gene in Tribolium produces a polycistronic mRNA that codes for multiple conserved peptides, Cell, 126, 559–69. [DOI] [PubMed] [Google Scholar]
- 66. Wadler C.S., Vanderpool C.K.. 2007, A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide, Proc. Natl. Acad. Sci. U S A., 104, 20454–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gimpel M., Heidrich N., Mäder U., Krügel H., Brantl S.. 2010, A dual-function sRNA from B. subtilis: SR1 acts as a peptide encoding mRNA on the gapA operon, Mol. Microbiol., 76, 990–1009. [DOI] [PubMed] [Google Scholar]
- 68. Williams R.E.O., Harper G.J.. 1947, Staphylococcal haemolysins on sheep-blood agar with evidence for a fourth hæmolysin, J. Pathol. Bacteriol., 59, 69–78. [DOI] [PubMed] [Google Scholar]
- 69. Verdon J., Girardin N., Lacombe C., Berjeaud J.-M., Héchard Y.. 2009, δ-hemolysin, an update on a membrane-interacting peptide, Peptides, 30, 817–23. [DOI] [PubMed] [Google Scholar]
- 70. Perry R.B.-T., Ulitsky I.. 2016, The functions of long noncoding RNAs in development and stem cells, Development, 143, 3882–94. [DOI] [PubMed] [Google Scholar]
- 71. Kageyama Y., Kondo T., Hashimoto Y.. 2011, Coding vs non-coding: translatability of short ORFs found in putative non-coding transcripts, Biochimie, 93, 1981–6. [DOI] [PubMed] [Google Scholar]
- 72. Cabili M.N., Trapnell C., Goff L., et al. 2011, Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses, Genes Dev., 25, 1915–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hezroni H., Koppstein D., Schwartz M.G., Avrutin A., Bartel D.P., Ulitsky I.. 2015, Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species, Cell Rep., 11, 1110–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Baboo S., Cook P.R.. 2014, “Dark matter” worlds of unstable RNA and protein, Nucleus, 5, 281–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Galindo M.I., Pueyo J.I., Fouix S., Bishop S.A., Couso J.P.. 2007, Peptides encoded by short ORFs control development and define a new eukaryotic gene family, PLoS Biol., 5, e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pauli A., Norris M.L., Valen E., et al. 2014, Toddler: an embryonic signal that promotes cell movement via apelin receptors. Science, 343, 1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Patop I.L., Wüst S., Kadener S.. 2019, Past, present, and future of circRNAs, Embo J., 38, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xu S., Zhou L., Ponnusamy M., et al. 2018, A comprehensive review of circRNA: from purification and identification to disease marker potential, PeerJ, 6, e5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yu C.-Y., Kuo H.-C.. 2019, The emerging roles and functions of circular RNAs and their generation, J. Biomed. Sci., 26, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li Q., Ahsan M.A., Chen H., Xue J., Chen M.. 2018, Discovering putative peptides encoded from noncoding RNAs in ribosome profiling data of Arabidopsis thaliana, ACS Synth. Biol., 7, 655–63. [DOI] [PubMed] [Google Scholar]
- 81. Iyer M.K., Niknafs Y.S., Malik R., et al. 2015, The landscape of long noncoding RNAs in the human transcriptome, Nat. Genet., 47, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Smith J.E., Alvarez-Dominguez J.R., Kline N., et al. 2014, Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae, Cell Rep., 7, 1858–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zanet J., Chanut-Delalande H., Plaza S., Payre F.. 2016, Small peptides as newcomers in the control of Drosophila development. Curr. Top. Dev. Biol., 199–219 [DOI] [PubMed] [Google Scholar]
- 84. Li L., Eichten S.R., Shimizu R., et al. 2014, Genome-wide discovery and characterization of maize long non-coding RNAs, Genome Biol., 15, R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pamudurti N.R., Bartok O., Jens M., et al. 2017, Translation of CircRNAs, Mol. Cell., 66, 9–21.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Legnini I., Di Timoteo G., Rossi F., et al. 2017, Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis, Mol. Cell., 66, 22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang M., Zhao K., Xu X., et al. 2018, A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma, Nat. Commun., 9, 4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tautz D., Domazet-Lošo T.. 2011, The evolutionary origin of orphan genes, Nat. Rev. Genet., 12, 692–702. [DOI] [PubMed] [Google Scholar]
- 89. Li J., Zhang X., Liu C.. 2020, The computational approaches of lncRNA identification based on coding potential: status quo and challenges, Comput. Struct. Biotechnol. J., 18, 3666–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kim K.H., Son J.M., Benayoun B.A., Lee C.. 2018, The mitochondrial-encoded peptide MOTS-c translocates to the nucleus to regulate nuclear gene expression in response to metabolic stress, Cell Metab., 28, 516–524.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lauressergues D., Couzigou J.-M., Clemente H.S., et al. 2015, Primary transcripts of microRNAs encode regulatory peptides, Nature, 520, 90–3. [DOI] [PubMed] [Google Scholar]
- 92. Prabakaran S., Hemberg M., Chauhan R., et al. 2014, Quantitative profiling of peptides from RNAs classified as noncoding, Nat. Commun., 5, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Fritsch C., Bernardo-Garcia F.J., Humberg T.-H., et al. 2019, Multilevel regulation of the glass locus during Drosophila eye development, PLOS Genet., 15, e1008269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Guo X., Chavez A., Tung A., et al. 2018, High-throughput creation and functional profiling of DNA sequence variant libraries using CRISPR–Cas9 in yeast, Nat. Biotechnol., 36, 540–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lee C., Zeng J., Drew B.G., et al. 2015, The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance, Cell Metab., 21, 443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lu H., Tang S., Xue C., et al. 2019, Mitochondrial-derived peptide MOTS-c increases adipose thermogenic activation to promote cold adaptation, Ijms, 20, 2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Li X., Yang L., Chen L.L.. 2018, The biogenesis, functions, and challenges of circular RNAs, Mol. Cell., 71, 428–42. [DOI] [PubMed] [Google Scholar]
- 98. Yang Y., Fan X., Mao M., et al. 2017, Extensive translation of circular RNAs driven by N6-methyladenosine, Cell Res., 27, 626–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zheng X., Chen L., Zhou Y., et al. 2019, A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling, Mol. Cancer, 18, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Vanderperre B., Lucier J.-F., Bissonnette C., et al. 2013, Direct detection of alternative open reading frames translation products in human significantly expands the proteome, PLoS One, 8, e70698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Dinger M.E., Pang K.C., Mercer T.R., Mattick J.S.. 2008, Differentiating protein-coding and noncoding RNA: challenges and ambiguities, PLoS Comput. Biol., 4, e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Fang J., Morsalin S., Rao V., Reddy E.S.. 2017, Decoding of non-coding DNA and non-coding RNA: pri-Micro RNA-encoded novel peptides regulate migration of cancer cells, J. Pharmaceut. Sci. Pharmacol., 3, 23–7. [Google Scholar]
- 103. Hanyu-Nakamura K., Sonobe-Nojima H., Tanigawa A., Lasko P., Nakamura A.. 2008, Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells, Nature, 451, 730–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bi P., Ramirez-Martinez A., Li H., et al. 2017, Control of muscle formation by the fusogenic micropeptide myomixer, Science, 356, 323–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. D'Lima N.G., Ma J., Winkler L., et al. 2017, A human microprotein that interacts with the mRNA decapping complex, Nat. Chem. Biol., 13, 174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kondo T., Hashimoto Y., Kato K., Inagaki S., Hayashi S., Kageyama Y.. 2007, Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA, Nat. Cell Biol., 9, 660–5. [DOI] [PubMed] [Google Scholar]
- 107. Yang Y., Gao X., Zhang M., et al. 2018, Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis, JNCI J. Natl. Cancer Inst., 110, 304–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhang M., Huang N., Yang X., et al. 2018, A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis, Oncogene, 37, 1805–14. [DOI] [PubMed] [Google Scholar]
- 109. Banfai B., Jia H., Khatun J., et al. 2012, Long noncoding RNAs are rarely translated in two human cell lines, Genome Res., 22, 1646–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Chew G.-L., Pauli A., Rinn J.L., Regev A., Schier A.F., Valen E.. 2013, Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs, Development, 140, 2828–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wilhelm M., Schlegl J., Hahne H., et al. 2014, Mass-spectrometry-based draft of the human proteome, Nature, 509, 582–7. [DOI] [PubMed] [Google Scholar]
- 112. Pueyo J.I., Couso J.P.. 2008, The 11-aminoacid long Tarsal-less peptides trigger a cell signal in Drosophila leg development, Dev. Biol., 324, 192–201. [DOI] [PubMed] [Google Scholar]
- 113. Pauli A., Valen E., Schier A.F.. 2015, Identifying (non-)coding RNAs and small peptides: challenges and opportunities, Bioessays, 37, 103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Chanut-Delalande H., Hashimoto Y., Pelissier-Monier A., et al. 2014, Pri peptides are mediators of ecdysone for the temporal control of development, Nat. Cell Biol., 16, 1035–44. [DOI] [PubMed] [Google Scholar]
- 115. Ray S., Rosenberg M.I., Chanut-Delalande H., et al. 2019, The mlpt/Ubr3/Svb module comprises an ancient developmental switch for embryonic patterning, Elife, 8, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Tobias-Santos V., Guerra-Almeida D., Mury F., et al. 2019, Multiple roles of the polycistronic gene tarsal-less/mille-pattes/polished-rice during embryogenesis of the kissing bug Rhodnius prolixus, Front. Ecol. Evol., 7, 1–16. [Google Scholar]
- 117. Pueyo J.I., Couso J.P.. 2011, Tarsal-less peptides control Notch signalling through the Shavenbaby transcription factor, Dev. Biol., 355, 183–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kondo T., Plaza S., Zanet J., et al. 2010, Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis, Science, 329, 336–9. [DOI] [PubMed] [Google Scholar]
- 119. Lynch M., Kewalramani A.. 2003, Messenger RNA surveillance and the evolutionary proliferation of introns, Mol. Biol. Evol., 20, 563–71. [DOI] [PubMed] [Google Scholar]
- 120. Pueyo J.I., Magny E.G., Sampson C.J., Amin U., Evans I.R., Bishop S.A., Couso J.P.. 2016, Hemotin, a regulator of phagocytosis encoded by a small ORF and conserved across metazoans, PLOS Biol., 14, e1002395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Billingsley M.L., Yun J., Reese B.E., Davidson C.E., Buck-Koehntop B.A., Veglia G.. 2006, Functional and structural properties of stannin: roles in cellular growth, selective toxicity, and mitochondrial responses to injury, J. Cell. Biochem., 98, 243–50. [DOI] [PubMed] [Google Scholar]
- 122. Zhang S., Reljić B., Liang C., et al. 2020, Mitochondrial peptide BRAWNIN is essential for vertebrate respiratory complex III assembly, Nat. Commun., 11, 1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kornblihtt A.R., Schor I.E., Alló M., Dujardin G., Petrillo E., Muñoz M.J.. 2013, Alternative splicing: a pivotal step between eukaryotic transcription and translation, Nat. Rev. Mol. Cell Biol., 14, 153–65. [DOI] [PubMed] [Google Scholar]
- 124. de Klerk E., 't Hoen P.A.C.. 2015, Alternative mRNA transcription, processing, and translation: insights from RNA sequencing, Trends Genet., 31, 128–39. [DOI] [PubMed] [Google Scholar]
- 125. Touriol C., Bornes S., Bonnal S., Audigier S., Prats H., Prats A.-C., Vagner S.. 2003, Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons, Biol. Cell., 95, 169–78. [DOI] [PubMed] [Google Scholar]
- 126. Matlin A.J., Clark F., Smith C.W.J.. 2005, Understanding alternative splicing: towards a cellular code, Nat. Rev. Mol. Cell Biol., 6, 386–98. [DOI] [PubMed] [Google Scholar]
- 127. Yu H., Tian C., Yu Y., Jiao Y.. 2016, Transcriptome survey of the contribution of alternative splicing to proteome diversity in Arabidopsis thaliana, Mol. Plant, 9, 749–52. [DOI] [PubMed] [Google Scholar]
- 128. Chaudhary S., Khokhar W., Jabre I., Reddy A.S.N., Byrne L.J., Wilson C.M., Syed N.H.. 2019, Alternative splicing and protein diversity: plants versus animals, Front. Plant Sci., 10, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wang E.T., Sandberg R., Luo S., et al. 2008, Alternative isoform regulation in human tissue transcriptomes, Nature, 456, 470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Stastna M., Van Eyk J.E.. 2012, Analysis of protein isoforms: can we do it better? Proteomics, 12, 2937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Barbosa-Morais N.L., Irimia M., Pan Q., et al. 2012, The evolutionary landscape of alternative splicing in vertebrate species, Science, 338, 1587–93. [DOI] [PubMed] [Google Scholar]
- 132. Yoshimura J., Ichikawa K., Shoura M.J., et al. 2019, Recompleting the Caenorhabditis elegans genome, Genome Res., 29, 1009–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Djebali S., Davis C.A., Merkel A., et al. 2012, Landscape of transcription in human cells, Nature, 489, 101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Blakeley P., Siepen J.A., Lawless C., Hubbard S.J.. 2010, Investigating protein isoforms via proteomics: a feasibility study, Proteomics, 10, 1127–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Hoff K.J., Stanke M.. 2019, Predicting genes in single genomes with AUGUSTUS, Curr. Protoc. Bioinforma, 65, e57. [DOI] [PubMed] [Google Scholar]
- 136. Baharlou Houreh M., Ghorbani Kalkhajeh P., Niazi A., Ebrahimi F., Ebrahimie E.. 2018, SpliceDetector: a software for detection of alternative splicing events in human and model organisms directly from transcript, Sci. Rep., 8, 5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Cass A.A., Xiao X.. 2019, mountainClimber identifies alternative transcription start and polyadenylation sites in RNA-Seq. Cell Syst., 9, 393–400.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Zhang Z., Carriero N., Zheng D., Karro J., Harrison P.M., Gerstein M.. 2006, PseudoPipe: an automated pseudogene identification pipeline, Bioinformatics, 22, 1437–9. [DOI] [PubMed] [Google Scholar]
- 139. Ji Z., Song R., Regev A., Struhl K.. 2015, Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins, Elife, 4, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Graeff M., Wenkel S.. 2012, Regulation of protein function by interfering protein species, Biomol. Concepts, 3, 71–8. [DOI] [PubMed] [Google Scholar]
- 141. Seo P.J., Hong S.Y., Kim S.G., Park C.M.. 2011, Competitive inhibition of transcription factors by small interfering peptides, Trends Plant Sci., 16, 541–9. [DOI] [PubMed] [Google Scholar]
- 142. Graeff M., Straub D., Eguen T., et al. 2016, MicroProtein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in Arabidopsis, PLOS Genet., 12, e1005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Staudt A., Wenkel S.. 2011, Regulation of protein function by ‘microProteins’, EMBO Rep., 12, 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Penfield S., Josse E.-M., Halliday K.J.. 2010, A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy, Plant Mol. Biol., 73, 89–95. [DOI] [PubMed] [Google Scholar]
- 145. Ingram V.M. 1957, Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin, Nature, 180, 326–8. [DOI] [PubMed] [Google Scholar]
- 146. Mouilleron H., Delcourt V., Roucou X.. 2016, Death of a dogma: eukaryotic mRNAs can code for more than one protein, Nucleic Acids Res., 44, 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Kiran A., Baranov P.V.. 2010, DARNED: a DAtabase of RNa EDiting in humans, Bioinformatics, 26, 1772–6. [DOI] [PubMed] [Google Scholar]
- 148. Kolakofsky D., Roux L., Garcin D., Ruigrok R.W.H.. 2005, Paramyxovirus mRNA editing, the ‘rule of six’ and error catastrophe: a hypothesis, J. Gen. Virol., 86, 1869–77. [DOI] [PubMed] [Google Scholar]
- 149. Ohno S. 1984, Birth of a unique enzyme from an alternative reading frame of the preexisted, internally repetitious coding sequence, Proc. Natl. Acad. Sci. U S A., 81, 2421–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Sanger F., Air G.M., Barrell B.G., et al. 1977, Nucleotide sequence of bacteriophage φX174 DNA, Nature, 265, 687–95. [DOI] [PubMed] [Google Scholar]
- 151. Keese P.K., Gibbs A.. 1992, Origins of genes: ‘big bang’ or continuous creation? Proc. Natl. Acad. Sci. U S A., 89, 9489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Klemke M., H., Kehlenbach R., Huttner W.B.. 2001, Two overlapping reading frames in a single exon encode interacting proteins–a novel way of gene usage, Embo J., 20, 3849–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Vattem K.M., Wek R.C.. 2004, Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells, Proc. Natl. Acad. Sci. U S A., 101, 11269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Kwun H.J., Toptan T., Ramos da Silva S., Atkins J.F., Moore P.S., Chang Y.. 2014, Human DNA tumor viruses generate alternative reading frame proteins through repeat sequence recoding, Proc. Natl. Acad. Sci. U S A., 111, E4342–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Arribere J.A., Cenik E.S., Jain N., et al. 2016, Translation readthrough mitigation, Nature, 534, 719–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Michel A.M., Choudhury K.R., Firth A.E., Ingolia N.T., Atkins J.F., Baranov P.V.. 2012, Observation of dually decoded regions of the human genome using ribosome profiling data, Genome Res., 22, 2219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Rancurel C., Khosravi M., Dunker A.K., Romero P.R., Karlin D.. 2009, Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation, J. Virol., 83, 10719–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Long M., Betrán E., Thornton K., Wang W.. 2003, The origin of new genes: glimpses from the young and old, Nat. Rev. Genet., 4, 865–75. [DOI] [PubMed] [Google Scholar]
- 159. Chung W.-Y., Wadhawan S., Szklarczyk R., Pond S.K., Nekrutenko A.. 2007, A first look at ARFome: dual-coding genes in mammalian genomes, PLoS Comput. Biol., 3, e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Tautz D. 2008, Polycistronic peptide coding genes in eukaryotes–how widespread are they? Brief. Funct. Genomics Proteomic., 8, 68–74. [DOI] [PubMed] [Google Scholar]
- 161. Kozak M. 1991, An analysis of vertebrate mRNA sequences: intimations of translational control, J. Cell Biol., 115, 887–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Pop C., Rouskin S., Ingolia N.T., et al. 2014, Causal signals between codon bias, scp mRNA structure, and the efficiency of translation and elongation, Mol. Syst. Biol., 10, 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Cardon T., Franck J., Coyaud E., et al. 2020, Alternative proteins are functional regulators in cell reprogramming by PKA activation, Nucleic Acids Res., 48, 7864–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Kochetov A.V. 2008, Alternative translation start sites and hidden coding potential of eukaryotic mRNAs, Bioessays, 30, 683–91. [DOI] [PubMed] [Google Scholar]
- 165. Hood H.M., Neafsey D.E., Galagan J., Sachs M.S.. 2009, Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi, Annu. Rev. Microbiol., 63, 385–409. [DOI] [PubMed] [Google Scholar]
- 166. Yamashita R., Suzuki Y., Nakai K., Sugano S.. 2003, Small open reading frames in 5′ untranslated regions of mRNAs, C R Biol., 326, 987–91. [DOI] [PubMed] [Google Scholar]
- 167. Kozak M. 1986, Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes, Cell, 44, 283–92. [DOI] [PubMed] [Google Scholar]
- 168. Rogozin I.B., Kochetov A.V., Kondrashov F.A., Koonin E.V., Milanesi L.. 2001, Presence of ATG triplets in 5’ untranslated regions of eukaryotic cDNAs correlates with a weak’ context of the start codon, Bioinformatics, 17, 890–900. [DOI] [PubMed] [Google Scholar]
- 169. Zhang H., Wang Y., Lu J.. 2019, Function and evolution of upstream ORFs in eukaryotes, Trends Biochem. Sci., 44, 782–94. [DOI] [PubMed] [Google Scholar]
- 170. Mehta A., Trotta C.R., Peltz S.W.. 2006, Derepression of the Her-2 uORF is mediated by a novel post-transcriptional control mechanism in cancer cells, Genes Dev., 20, 939–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Ebina I., Takemoto-Tsutsumi M., Watanabe S., et al. 2015, Identification of novel Arabidopsis thaliana upstream open reading frames that control expression of the main coding sequences in a peptide sequence-dependent manner, Nucleic Acids Res., 43, 1562–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Hsu P.Y., Benfey P.N.. 2018, Small but mighty: functional peptides encoded by small ORFs in plants, Proteomics, 18, 1700038. [DOI] [PubMed] [Google Scholar]
- 173. Parola A.L., Kobilka B.K.. 1994, The peptide product of a 5’ leader cistron in the beta 2 adrenergic receptor mRNA inhibits receptor synthesis, J. Biol. Chem., 269, 4497–505. [PubMed] [Google Scholar]
- 174. Jorgensen R.A., Dorantes-Acosta A.E.. 2012, Conserved peptide upstream open reading frames are associated with regulatory genes in angiosperms, Front. Plant Sci., 3, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Peccarelli M., Kebaara B.W.. 2014, Regulation of natural mRNAs by the nonsense-mediated mRNA decay pathway, Eukaryot. Cell, 13, 1126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Rodriguez C.M., Chun S.Y., Mills R.E., Todd P.K.. 2019, Translation of upstream open reading frames in a model of neuronal differentiation, BMC Genomics, 20, 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Akimoto C., Sakashita E., Kasashima K., et al. 2013, Translational repression of the McKusick–Kaufman syndrome transcript by unique upstream open reading frames encoding mitochondrial proteins with alternative polyadenylation sites, Biochim. Biophys. Acta, 1830, 2728–38. [DOI] [PubMed] [Google Scholar]
- 178. Nguyen H.L., Yang X., Omiecinski C.J.. 2013, Expression of a novel mRNA transcript for human microsomal epoxide hydrolase (EPHX1) is regulated by short open reading frames within its 5’-untranslated region, RNA, 19, 752–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Pendleton L.C., Goodwin B.L., Solomonson L.P., Eichler D.C.. 2005, Regulation of endothelial argininosuccinate synthase expression and NO production by an upstream open reading frame, J. Biol. Chem., 280, 24252–60. [DOI] [PubMed] [Google Scholar]
- 180. Yosten G.L.C., Liu J., Ji H., Sandberg K., Speth R., Samson W.K.. 2016, A 5′-upstream short open reading frame encoded peptide regulates angiotensin type 1a receptor production and signalling via the β-arrestin pathway, J. Physiol., 594, 1601–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Johnstone T.G., Bazzini A.A., Giraldez A.J.. 2016, Upstream ORFs are prevalent translational repressors in vertebrates, Embo J., 35, 706–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Calvo S.E., Pagliarini D.J., Mootha V.K.. 2009, Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans, Proc. Natl. Acad. Sci. U S A., 106, 7507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Iacono M., Mignone F., Pesole G.. 2005, uAUG and uORFs in human and rodent 5′untranslated mRNAs, Gene, 349, 97–105. [DOI] [PubMed] [Google Scholar]
- 184. Browning K.S., Bailey-Serres J.. 2015, Mechanism of cytoplasmic mRNA translation, Arab. B, 13, e0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Hayden C.A., Jorgensen R.A.. 2007, Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes, BMC Biol., 5, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Crowe M.L., Wang X.-Q., Rothnagel J.A.. 2006, Evidence for conservation and selection of upstream open reading frames suggests probable encoding of bioactive peptides, BMC Genomics, 7, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Ivanov I.P., Wei J., Caster S.Z., et al. 2017, Translation initiation from conserved non-AUG codons provides additional layers of regulation and coding capacity, MBio, 8, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Oyama M., Kozuka-Hata H., Suzuki Y., Semba K., Yamamoto T., Sugano S.. 2007, Diversity of translation start sites may define increased complexity of the human short ORFeome, Mol. Cell. Proteomics, 6, 1000–6. [DOI] [PubMed] [Google Scholar]
- 189. Law G.L., Raney A., Heusner C., Morris D.R.. 2001, Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase, J. Biol. Chem., 276, 38036–43. [DOI] [PubMed] [Google Scholar]
- 190. Combier J.P., de Billy F., Gamas P., Niebel A., Rivas S.. 2008, Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development, Genes Dev., 22, 1549–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Uenaka A., Ono T., Akisawa T., Wada H., Yasuda T., Nakayama E.. 1994, Identification of a unique antigen peptide pRL1 on BALB/c RL male 1 leukemia recognized by cytotoxic T lymphocytes and its relation to the Akt oncogene, J. Exp. Med., 180, 1599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Starck S.R., Shastri N.. 2011, Non-conventional sources of peptides presented by MHC class I, Cell. Mol. Life Sci., 68, 1471–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Ribrioux S., Brüngger A., Baumgarten B., Seuwen K., John M.R.. 2008, Bioinformatics prediction of overlapping frameshifted translation products in mammalian transcripts, BMC Genomics, 9, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Bab I., Smith E., Gavish H., et al. 1999, Biosynthesis of osteogenic growth peptide via alternative translational initiation at AUG 85 of histone H4 mRNA, J. Biol. Chem., 274, 14474–81. [DOI] [PubMed] [Google Scholar]
- 195. Torrance V., Lydall D.. 2018, Overlapping open reading frames strongly reduce human and yeast STN1 gene expression and affect telomere function, PLOS Genet., 14, e1007523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Vanderperre B., Staskevicius A.B., Tremblay G., et al. 2011, An overlapping reading frame in the PRNP gene encodes a novel polypeptide distinct from the prion protein, Faseb J., 25, 2373–86. [DOI] [PubMed] [Google Scholar]
- 197. Pavesi A., Vianelli A., Chirico N., et al. 2018, Overlapping genes and the proteins they encode differ significantly in their sequence composition from non-overlapping genes, PLoS One, 13, e0202513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Vanderperre B., Lucier J.-F., Roucou X.. 2012, HAltORF: a database of predicted out-of-frame alternative open reading frames in human, Database (Oxford), 2012, bas025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Xu H., Wang P., Fu Y., et al. 2010, Length of the ORF, position of the first AUG and the Kozak motif are important factors in potential dual-coding transcripts, Cell Res., 20, 445–57. [DOI] [PubMed] [Google Scholar]
- 200. Karlin D., Ferron F., Canard B., Longhi S.. 2003, Structural disorder and modular organization in Paramyxovirinae N and P, J. Gen. Virol., 84, 3239–52. [DOI] [PubMed] [Google Scholar]
- 201. Pavesi A., De Iaco B., Granero M.I., Porati A.. 1997, On the informational content of overlapping genes in prokaryotic and eukaryotic viruses, J. Mol. Evol., 44, 625–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Nekrutenko A., He J.. 2006, Functionality of unspliced XBP1 is required to explain evolution of overlapping reading frames, Trends Genet., 22, 645–8. [DOI] [PubMed] [Google Scholar]
- 203. Nekrutenko A., Wadhawan S., Goetting-Minesky P., Makova K.D.. 2005, Oscillating evolution of a mammalian locus with overlapping reading frames: an XLαs/ALEX relay, PLoS Genet., 1, e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Probst-Kepper M., Stroobant V., Kridel R., et al. 2001, An alternative open reading frame of the human macrophage colony-stimulating factor gene is independently translated and codes for an antigenic peptide of 14 amino acids recognized by tumor-infiltrating CD8 T lymphocytes, J. Exp. Med., 193, 1189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Rosenberg S.A., Tong-On P., Li Y., et al. 2002, Identification of BING-4 cancer antigen translated from an alternative open reading frame of a gene in the extended MHC class II region using lymphocytes from a patient with a durable complete regression following immunotherapy, J. Immunol., 168, 2402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Shichijo S., Nakao M., Imai Y., et al. 1998, A gene encoding antigenic peptides of human squamous cell carcinoma recognized by cytotoxic T lymphocytes, J. Exp. Med., 187, 277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Tykodi S.S., Fujii N., Vigneron N., et al. 2008, C19orf48 encodes a minor histocompatibility antigen recognized by CD8+ cytotoxic T cells from renal cell carcinoma patients, Clin. Cancer Res., 14, 5260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Graddis T.J., Diegel M.L., McMahan C.J., Tsavler L., Laus R., Vidovic D.. 2004, Tumor immunotherapy with alternative reading frame peptide antigens, Immunobiology, 209, 535–44. [DOI] [PubMed] [Google Scholar]
- 209. Wang R.F., Johnston S.L., Zeng G., Topalian S.L., Schwartzentruber D.J., Rosenberg S.A.. 1998, A breast and melanoma-shared tumor antigen: T cell responses to antigenic peptides translated from different open reading frames, J. Immunol., 161, 3598–606. [PubMed] [Google Scholar]
- 210. Ronsin C., Chung-Scott V., Poullion I., Aknouche N., Gaudin C., Triebel F.. 1999, A non-AUG-defined alternative open reading frame of the intestinal carboxyl esterase mRNA generates an epitope recognized by renal cell carcinoma-reactive tumor-infiltrating lymphocytes in situ, J. Immunol., 163, 483–90. [PubMed] [Google Scholar]
- 211. Shastri N., Cardinaud S., Schwab S.R., Serwold T., Kunisawa J.. 2005, All the peptides that fit: the beginning, the middle, and the end of the MHC class I antigen-processing pathway, Immunol. Rev., 207, 31–41. [DOI] [PubMed] [Google Scholar]
- 212. Yewdell J.W. 2007, Plumbing the sources of endogenous MHC class I peptide ligands, Curr. Opin. Immunol., 19, 79–86. [DOI] [PubMed] [Google Scholar]
- 213. Ho O., Green W.R.. 2006, Alternative translational products and cryptic T cell epitopes: expecting the unexpected, J. Immunol., 177, 8283–9. [DOI] [PubMed] [Google Scholar]
- 214. Koster J., Plasterk R.H.A.. 2019, A library of Neo Open Reading Frame peptides (NOPs) as a sustainable resource of common neoantigens in up to 50% of cancer patients, Sci. Rep., 9, 6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Starck S.R., Shastri N.. 2016, Nowhere to hide: unconventional translation yields cryptic peptides for immune surveillance, Immunol. Rev., 272, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Sarnataro D., Pepe A., Zurzolo C.. 2017, Cell biology of prion protein, Prog. Mol. Biol. Transl. Sci., 150, 57–82. [DOI] [PubMed] [Google Scholar]
- 217. Romagosa C., Simonetti S., López-Vicente L., Mazo A., Lleonart M.E., Castellvi J., Ramon y Cajal S.. 2011, p16Ink4a overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors, Oncogene, 30, 2087–97. [DOI] [PubMed] [Google Scholar]
- 218. Quelle D.E., Zindy F., Ashmun R.A., Sherr C.J.. 1995, Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest, Cell, 83, 993–1000. [DOI] [PubMed] [Google Scholar]
- 219. Kuersten S., Goodwin E.B.. 2003, The power of the 3′ UTR: translational control and development, Nat. Rev. Genet., 4, 626–37. [DOI] [PubMed] [Google Scholar]
- 220. Sandberg R., Neilson J.R., Sarma A., Sharp P.A., Burge C.B.. 2008, Proliferating cells express mRNAs with shortened 3’ untranslated regions and fewer MicroRNA target sites. Science, 320, 1643–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Jupe E.R., Liu X.T., Kiehlbauch J.L., McClung J.K., Dell’Orco R.T.. 1996, Prohibitin in breast cancer cell lines: loss of antiproliferative activity is linked to 3’ untranslated region mutations, Cell Growth Differ., 7, 871–8. [PubMed] [Google Scholar]
- 222. Low S.C., Berry M.J.. 1996, Knowing when not to stop: selenocysteine incorporation in eukaryotes, Trends Biochem. Sci., 21, 203–8. [PubMed] [Google Scholar]
- 223. Shen Q., Chu F.F., Newburger P.E.. 1993, Sequences in the 3’-untranslated region of the human cellular glutathione peroxidase gene are necessary and sufficient for selenocysteine incorporation at the UGA codon, J. Biol. Chem., 268, 11463–9. [PubMed] [Google Scholar]
- 224. Wu Q., Wright M., Gogol M.M., Bradford W.D., Zhang N., Bazzini A.A.. 2020, Translation of small downstream ORFs enhances translation of canonical main open reading frames, Embo J., 39, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Schwab S.R. 2003, Constitutive display of cryptic translation products by MHC class I molecules . Science, 301, 1367–71. [DOI] [PubMed] [Google Scholar]
- 226. Jungreis I., Lin M.F., Spokony R., et al. 2011, Evidence of abundant stop codon readthrough in Drosophila and other Metazoa, Genome Res., 21, 2096–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Juntawong P., Girke T., Bazin J., Bailey-Serres J.. 2014, Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis, Proc. Natl. Acad. Sci. U S A., 111, E203–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Miettinen T.P., Björklund M.. 2015, Modified ribosome profiling reveals high abundance of ribosome protected mRNA fragments derived from 3′ untranslated regions, Nucleic Acids Res., 43, 1019–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Mercer T.R., Wilhelm D., Dinger M.E., et al. 2011, Expression of distinct RNAs from 3′ untranslated regions, Nucleic Acids Res., 39, 2393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Kocabas A., Duarte T., Kumar S., Hynes M.A.. 2015, Widespread differential expression of coding region and 3′ UTR sequences in neurons and other tissues, Neuron, 88, 1149–56. [DOI] [PubMed] [Google Scholar]
- 231. Chao Y., Vogel J.. 2016, A 3′ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response, Mol. Cell, 61, 352–63. [DOI] [PubMed] [Google Scholar]
- 232. Harrison B.J., Park J.W., Gomes C., et al. 2019, Detection of differentially expressed cleavage site intervals within 3′ untranslated regions using CSI-UTR reveals regulated interaction motifs, Front. Genet., 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Malarkannan S., Shih P.P., Eden P.A., et al. 1998, The molecular and functional characterization of a dominant minor H antigen, H60, J. Immunol., 161, 3501–9. [PubMed] [Google Scholar]
- 234. Malarkannan S., Horng T., Shih P.P., Schwab S., Shastri N.. 1999, Presentation of out-of-frame peptide/MHC class I complexes by a novel translation initiation mechanism, Immunity, 10, 681–90. [DOI] [PubMed] [Google Scholar]
- 235. Goodenough E., Robinson T.M., Zook M.B., et al. 2014, Cryptic MHC class I-binding peptides are revealed by aminoglycoside-induced stop codon read-through into the 3’ UTR, Proc. Natl. Acad. Sci. U S A., 111, 5670–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Shaughnessy J.D., Largaespada D.A., Tian E., et al. 1999, Mrvil, a common MRV integration site in BXH2 myeloid leukemias, encodes a protein with homology to a lymphoid-restricted membrane protein Jaw1, Oncogene, 18, 2069–84. [DOI] [PubMed] [Google Scholar]