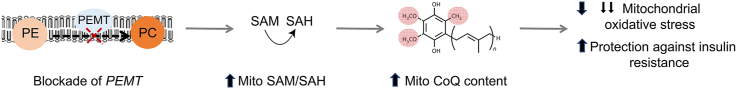
Abstract
Mitochondrial energy production and function rely on optimal concentrations of the essential redox-active lipid, coenzyme Q (CoQ). CoQ deficiency results in mitochondrial dysfunction associated with increased mitochondrial oxidative stress and a range of pathologies. What drives CoQ deficiency in many of these pathologies is unknown, just as there currently is no effective therapeutic strategy to overcome CoQ deficiency in humans. To date, large-scale studies aimed at systematically interrogating endogenous systems that control CoQ biosynthesis and their potential utility to treat disease have not been carried out. Therefore, we developed a quantitative high-throughput method to determine CoQ concentrations in yeast cells. Applying this method to the Yeast Deletion Collection as a genome-wide screen, 30 genes not known previously to regulate cellular concentrations of CoQ were discovered. In combination with untargeted lipidomics and metabolomics, phosphatidylethanolamine N-methyltransferase (PEMT) deficiency was confirmed as a positive regulator of CoQ synthesis, the first identified to date. Mechanistically, PEMT deficiency alters mitochondrial concentrations of one-carbon metabolites, characterized by an increase in the S-adenosylmethionine to S-adenosylhomocysteine (SAM-to-SAH) ratio that reflects mitochondrial methylation capacity, drives CoQ synthesis, and is associated with a decrease in mitochondrial oxidative stress. The newly described regulatory pathway appears evolutionary conserved, as ablation of PEMT using antisense oligonucleotides increases mitochondrial CoQ in mouse-derived adipocytes that translates to improved glucose utilization by these cells, and protection of mice from high-fat diet-induced insulin resistance. Our studies reveal a previously unrecognized relationship between two spatially distinct lipid pathways with potential implications for the treatment of CoQ deficiencies, mitochondrial oxidative stress/dysfunction, and associated diseases.
Keywords: Coenzyme Q, Mitochondria, PEMT, Insulin resistance, S-adenosylmethionine, S-adenosylhomocysteine, Reactive oxygen species
Graphical abstract
Highlights
-
•
Mitochondrial CoQ deficiency results in oxidative stress and a range of pathologies
-
•
The drivers of mitochondrial CoQ deficiency remain largely unknown
-
•
PEMT deficiency is the first identified positive regulator of mitochondrial CoQ
-
•
PEMT deficiency increases CoQ by increasing the mitochondrial SAM-to-SAH ratio
-
•
PEMT deficiency prevents insulin resistance by increasing mitochondrial CoQ
1. Introduction
Mitochondrial dysfunction is increasingly recognized as a central mediator of many common disorders, including cardiovascular and metabolic diseases via the key role mitochondria play in cellular energy homeostasis and oxidant production. One of the central determinants of mitochondrial function is ubiquinone (or coenzyme Q, CoQ), an essential, evolutionarily conserved, redox active lipid in aerobic organisms. CoQ is best known for its function in mitochondrial ATP production, antioxidant protection and cell survival. More recently, the role of mitochondrial CoQ has expanded to the regulation of ferroptosis [1,2] nitric oxide synthase [3] and the homeostasis of brown adipose tissue [4].
There are two forms of CoQ deficiency in humans, both resulting in mitochondrial dysfunction. Primary CoQ deficiency results from mutations in CoQ biosynthetic genes and presents as a genetically and clinically heterogenous disorder [5]. Secondary CoQ deficiency occurs without underlying genetic defects in CoQ biosynthesis, yet is strongly associated with aging, insulin resistance and cardiovascular diseases such as atherosclerosis, myopathy, and heart failure [[6], [7], [8], [9], [10], [11]]. Preclinical studies have established that increasing tissue CoQ can decrease mitochondrial oxidant production, improve mitochondrial function, and can ameliorate disease. Unfortunately however, translating this knowledge into the clinic has remained challenging, principally due to our inability to effectively restore CoQ concentrations in most human tissues as a result of the low bioavailability of orally supplemented CoQ [12]. A better understanding of the endogenous systems that regulate tissue CoQ content beyond the biosynthetic genes may help overcome this current limitation and identify new strategies to enhance endogenous CoQ biosynthesis.
Although CoQ was discovered more than 60 years ago, our understanding of how cells regulate their CoQ content has remained limited to individual factors required for optimal CoQ synthesis [[13], [14], [15]]. To date, a systemic approach aimed at identifying networks important for the regulation of cellular CoQ concentrations beyond the mevalonate pathway has not been carried out. A likely reason for this is the technically challenging nature of high-throughput CoQ extraction and analysis. To overcome this current limitation, we developed a large-scale screening method to analyze and quantify cellular CoQ using high-performance liquid chromatography coupled with electrochemical detection (HPLC-ECD). Using this platform, we used the Yeast Deletion Collection to conduct a genome-wide genetic screen and identified 30 new potential CoQ-regulating genes. Of these, we focused on phosphatidylethanolamine N-methyltransferase (PEMT) that plays a key role in phospholipid metabolism. We observed that cells deficient in PEMT contain strikingly higher concentrations of mitochondrial CoQ in multiple biological systems. Using genetic manipulations, lipidomics, and untargeted and targeted metabolomics, we discovered a previously unrecognized link between endoplasmic reticulum/mitochondrial associated membrane PEMT activity, and the mitochondrial methylation activity required for CoQ synthesis. Finally, we identified increased mitochondrial CoQ as the molecular basis for how PEMT deficiency protects mice against insulin resistance.
2. Methods
2.1. Genome-wide screen of mutants with altered total CoQ6 content
The homozygous diploid yeast knockout collection (BY4743; Euroscarf) was used to screen genes that affected total CoQ6 content. The collection is housed in a series of 96-well plates kept frozen at −80 °C. Briefly, cells were inoculated from −80 °C stocks with 2.4 μL of defrosted culture inoculated into 96-well plates containing 195 μL media. Cells were grown for 2 d at 30 °C with shaking. After 2 d, 2.4 μL of pre-culture was inoculated into 96-well plates containing 195 μL fresh media and cells were grown for 18 h at 30 °C with shaking. After 18 h, the OD600 was measured (Pherastar 96-well plate spectrophotometer) and the plate centrifuged (1500×g; 4 °C; 5 min) to pellet cells. 190 μL of supernatant was removed from each well and the cell pellet in each well resuspended in 50 μL of 155 mM ammonium acetate. The 96-well plate was frozen at −20 °C until CoQ6 extraction and analysis. On the day of analysis, plates were defrosted and 50 μL of cell suspension was transferred to a glass-coated 96 deep-well plate containing 50 μL of glass beads per well. As a control, a separate batch of WT cells grown to OD600 ∼1.0 and frozen in aliquots were used as quality control (QC) samples in each plate. Wells A1, E6 and H12 of each plate were used for the QC samples with 50 μL of QC sample put in each well on the day of analysis. For each plate to pass ‘QC’, the CoQ6 content of the QC wells had to be within 20% of each other. Cells were lysed and CoQ6 extracted and analyzed as outlined below.
2.2. Determination of CoQ6 and ergosterol content in yeast cells
The method used was adapted from Gay & Stocker 2004 [16]. 50 μL 0.155 mM ammonium acetate was added to yeast cell pellets and the suspension was transferred to a glass-coated 96 deep well plate containing 50 μL glass beads per well. Cells were lysed using a Thermomixer C (Eppendorf, 1400 rpm, 2 h, 4 °C). 200 μL of acidified methanol and 500 μL water-washed hexane was added to each sample, and the plate shaken (1000 rpm, 1 min, 4 °C). 300 μL of the hexane layer was removed and 500 μL hexane added, and the plate re-shaken (1000 rpm, 1 min, 4 °C). This was repeated five times with 800 μL hexane removed on the fifth repeat (a total of 2 mL hexane collected). The collected hexane was dried under nitrogen in a fume hood for 1 h at room temperature. The resulting dried lipids were re-dissolved in 150–180 μL ice-cold mobile phase (ethanol:methanol:isopropanol: ammonium acetate pH 4.4, 65:30:3:2, vol/vol/vol/vol) and transferred into HPLC vials. Samples were stored at 4 °C until analysis via HPLC coupled to UV and electrochemical detection (HPLC-UV/ECD) or liquid chromatography tandem mass spectrometry (LC-MS/MS). For HPLC-UV/EC analyses, 100 μL of samples were injected onto a Supelcosil LC-C18 column (5 μm, 250 mm × 4.6 mm) eluted at 1 mL/min and connected to UV and electrochemical (ESA CoulArray 5600A) detectors. Ergosterol was detected at 280 nm, while total CoQ6 (CoQ6 + CoQ6H2) was detected at −700, +700 and + 700 mV. For studies in which total CoQ6 (CoQ6 + CoQ6H2) was determined by LC-MS/MS, 2 μL of sample was injected onto an Agilent 1290 UHPLC system connected to an Agilent 6490 triple-quadrupole mass spectrometer. Analytes were separated on a Luna 5 μm C18 (2) 100 Å column (150 × 4.6 mm; Phenomenex, USA) by gradient elution using mobile phase A (2.5 mM ammonium formate in 95:5 methanol:isopropanol) and mobile phase B (2.5 mM ammonium formate in 100% isopropanol) at 0.4 mL/min. The gradient consisted of 50% mobile phase B from 0 to 15 min, 50–70% B from 15 to 17 min, 100% B from 17 to 19 min and back to 50% B from 19 to 24 min. Flow was then directed into the triple quadrupole mass spectrometer with parameters set as follows: gas temperature = 290 °C; gas flow = 14 L/min; nebuliser pressure = 25 psi; sheath gas heater = 400 °C; sheath gas flow = 11 L/min; capillary voltage = 3000 V. Detection of CoQ6, was by multiple reaction monitoring (MRM) in positive ion mode using the above general mass spectrometry parameters with fragmentor voltage at 380 V and cell accelerator voltage at 5 V. The fragment ion generated by collision-induced dissociation of the [M + H]+ ion was used for quantification. MRM settings for the target analytes were (parent ion → fragment ion); CoQ6 (m/z 591.3.1 → 197.1) with collision energy of 21 V and CoQ6H2 (m/z 593.3.1 → 197.1) with collision energy of 25 V. CoQ6, CoQ6H2 and ergosterol were quantified against authentic commercial standards obtained from Avanti Polar Lipids and Sigma Aldrich (USA) respectively.
2.3. Determination of S-adenosyl-l-methionine and S-adenosyl-l-homocysteine content in yeast cells and mitochondria
All work was carried out under an argon gas cloud. Yeast pellets were defrosted and 100 μL of 50% methanol containing 100 μM diethylenetriaminepentaacetic acid (DTPA) was added and cell pellets lysed at 40 kpsi (50 s on, 10 s off; 30 cycles at 20 °C) using a Barocycler 2320EXT (Pressure BioSciences Inc). 100 μL chloroform containing 100 μM DTPA was added to the lysate, the suspension mixed vigorously for 1 min, centrifuged (5000×g, 5 min, 4 °C), and 50 μL top aqueous layer removed and 100 μL 50% methanol containing 100 μM DTPA added to the sample. The sample was vortexed for 1 min, centrifuged (5000×g, 5 min, 4 °C), and 100 μL of the aqueous layer removed (total 150 μL sample collected). Samples were stored at −80 °C until analysis. 5 μL of sample was injected onto an Agilent 1290 UHPLC system connected to an Agilent 6490 triple-quadrupole mass spectrometer. Analytes were separated on a Polar RP 80 Å column 250 × 2.5 mm (Phenomenex, USA) by gradient elution using mobile phase A (0.1% formic acid, 5 μM medronic acid in water) and mobile phase B (0.1% formic acid, 5 μM medronic acid in 100% methanol) at 0.2 mL/min. The gradient consisted of 2% mobile phase B from 0 to 5 min, 2–90% B from 10 to 15 min, 2% B from 15 to 18 min. Flow was then directed into the triple quadrupole mass spectrometer with parameters set as follows: gas temperature = 220 °C; gas flow = 12 L/min; nebuliser pressure = 40 psi; sheath gas heater = 380 °C; sheath gas flow = 12 L/min; capillary voltage = 4000 V. Detection of SAM and SAH was by multiple reaction monitoring (MRM) in positive ion mode using the general mass spectrometry parameters listed above with fragmentor voltage at 380 V and cell accelerator voltage at 4 V. The fragment ion generated by collision-induced dissociation of the [M + H]+ ion was used for quantification. MRM settings for the target analytes were (parent ion → fragment ion); SAM (m/z 399.4 → 249.9) with collision energy of 17 and SAH (m/z 385.4 → 136.0) with collision energy of 25. SAM and SAH were quantified against commercial standards (Sigma-Aldrich).
For mitochondrial analyses of SAM and SAH, yeast mitochondria were isolated as described [17]. Briefly, cells were grown until; OD600 ∼1.0 and centrifuged (3000×g, 5 min, 4 °C) to pellet cells. Cells were resuspended in 100 mM Tris-H2SO4 buffer pH 9.4 containing 10 mM dithiothreitol (DTT) and incubated at 30 °C for 20 min. Cells were centrifuged at 3000×g for 5 min at 4 °C and resuspended in Zymolyase buffer (10 mM potassium phosphate pH 7.4, 1.2 M sorbitol). Cells were centrifuged (3000×g, 5 min, 4 °C), the supernatant removed, and pellets resuspended in Zymolyase buffer containing Zymolyase 100T and incubated at 30 °C shaking for 1 h. Following washing with Zymolyase buffer, cells were resuspended in homogenization buffer (10 mM Tris-Cl pH 7.4, 0.6 M sorbitol, 1 mM EDTA, 1 mM PMSF, 0.2% (w/v) bovine serum albumin (BSA)) and homogenized with 15 strokes of a Teflon-glass homogenizer. Following removal of cell debris by low-speed centrifugation (1500×g, 5 min, 4 °C), the supernatant was centrifuged at 4000×g for 5 min at 4 °C and the resulting supernatant centrifuged at 12,000×g for 15 min at 4 °C. The resulting mitochondrial pellet was gently resuspended in yeast mitochondrial buffer (1 mM MOPS-KOH pH 7.2, 250 mM sucrose). Protein concentration was determined by BCA assay as per manufacturer's protocol (ThermoFisher Scientific) and samples stored at −20 °C until analysis. On the day of analysis, 50 μg mitochondrial protein was resuspended in 100 μL of 50% methanol containing 100 μM DTPA and extracted as per yeast homogenates, and SAM and SAH concentrations determined by LC-MS/MS as outlined above.
2.4. McArdle-RH7777 hepatoma cell culture
Rat McArdle-RH7777 hepatoma cells stably transfected with a vector containing human PEMT [18] were grown in Dulbecco's Modified Eagle Medium (DMEM, Thermo Fisher Scientific, 11995) containing 10% fetal calf serum (FCS), 4.5 g/L glucose, 110 mg/L sodium pyruvate, 1X GlutaMAX™ (ThermoFisher Scientific) supplemented with penicillin and streptomycin in a humidified atmosphere with 5% CO2. Cells were incubated with 10 μM 3-deazaadenosine (DZA) or vehicle (water) for 24 h and cells were harvested and stored at −20 °C until mitochondrial CoQ, SAM and SAH were determined as outlined previously.
2.5. 3T3-L1 fibroblast culture and differentiation into adipocytes
Mycoplasma-free 3T3-L1 fibroblasts obtained from Howard Green (Harvard Medical School, Boston, MA) were maintained in Dulbecco's Modified Eagle Medium (DMEM) (Thermo Fisher Scientific, 11995) supplemented with 10% FCS (Thermo Fisher Scientific), 1% GlutaMAX™ (Thermo Fisher Scientific) in a humidified atmosphere with 5% CO2. Confluent 3T3-L1 cells were differentiated into adipocytes by the addition of DMEM supplemented with 10% FSC and 0.22 μM dexamethasone, 100 ng/mL biotin, 2 μg/mL insulin and 500 μM 3-isobutyl-1-methylxanthine (day zero). After 72 h, medium was replaced with DMEM/10% FCS/GlutaMAX™ containing 2 μg/mL insulin (day three post differentiation) and grown for a further 72 h (day six post differentiation). Cells were used between days 6 and 7 after the initiation of differentiation for all experiments.
2.6. Determination of CoQ content in differentiated 3T3-L1 adipocytes and McArdle RH7777 hepatoma cells
Briefly, cells were homogenized in mitochondrial isolation buffer (10 mM Tris-MOPS, pH 7.4, 1 mM EGTA, 200 mM sucrose containing protease inhibitors) for 30 s at 7000 rpm (Heidolph homogenizer) and samples kept at 4 °C subsequently. 100 μL of the homogenates was kept for total tissue CoQ9 and CoQ10 analyses, and the remaining homogenate was used to isolate mitochondria as previously described [19]. Briefly, homogenates were centrifuged at 700×g for 10 min and the supernatant isolated and centrifuged at 7000×g for 10 min to obtain a pellet containing the mitochondria. The pellet was re-suspended in mitochondrial isolation buffer and re-centrifuged at 7000×g for 10 min. The mitochondrial pellet was finally re-suspended in mitochondrial isolation buffer and protein concentration determined using the BCA assay. CoQ extraction was carried out as previously described [16]. Briefly, 50–100 μL of isolated mitochondria was incubated with 25 μL para-benzoquinone (2 mg/mL) for 30 min at room temperature, and 2 mL acidified methanol and 10 mL of water-washed hexane were added. The mixture was mixed vigorously for 1 min, centrifuged (1430×g, 5 min, 4 °C) and 9 mL of hexane layer collected and dried using a rotary evaporator. The resulting dried lipids were re-dissolved in 150–180 μL ice-cold mobile phase (ethanol:methanol:isopropanol: ammonium acetate pH 4.4, 65:30:3:2, vol/vol/vol/vol), transferred into HPLC vials and analyzed using HPLC connected to UV and electrochemical detectors as outlined above, or using liquid chromatography tandem mass spectrometry (LC-MS/MS). If LC-MS/MS was used, the dried lipids were resuspended in 150 μL ice-cold HPLC-grade ethanol and analyses carried out as per described previously [11].
2.7. Determination of S-adenosyl-l-methionine and S-adenosyl-l-homocysteine content in liver
Mitochondria from liver tissue was extracted as previously described for mitochondrial CoQ determination. Mitochondrial pellets were resuspended in 100 μL mitochondrial isolation buffer (10 mM Tris-MOPS, pH 7.4, 1 mM EGTA, 200 mM sucrose containing protease inhibitors) and stored at −20 °C until analysis. 50 μL of liver mitochondria were defrosted and 50 μL of 100% methanol containing 100 μM DTPA was added to mitochondria. 100 μL chloroform containing 100 μM DTPA was added to the sample and the suspension mixed vigorously for 1 min, centrifuged (5000×g, 5 min, 4 °C). 50 μL of the top aqueous layer was removed and 100 μL 50% methanol containing 100 μM DTPA added to the sample. The sample was vortexed for 1 min, centrifuged (5000×g, 5 min, 4 °C), and 100 μL of the aqueous layer removed (total 150 μL sample collected). All sample work up was done under a cloud of argon gas. Samples were stored at −80 °C until analysis. 5 μL of sample was injected onto an TSQ Altis™ Triple Quadrupole Mass Spectrometer (ThermoFisher Scientific). Analytes were separated on a Polar RP 80 Å column 250 × 2.5 mm (Phenomenex, USA) by gradient elution using mobile phase A (0.1% formic acid, 5 μM medronic acid in water) and mobile phase B (0.1% formic acid, 5 μM medronic acid in 100% methanol) at 0.2 mL/min. The gradient consisted of 2% mobile phase B from 0 to 5 min, 2–90% B from 10 to 15 min, 2% B from 15 to 18 min. The triple quadrupole mass spectrometer H-ESI source parameters were set as follows: sheath gas = 58.8 Arb, aux gas = 13.4 Arb, sweep gas = 2.9 Arb, Ion transfer tube temperature = 350 °C, static spray voltage with positive ion voltage set at 4936.36 V and negative ion voltage set at 3500 V. Detection of SAM and SAH, was by multiple reaction monitoring (MRM) in positive ion mode using the above general mass spectrometry parameters. The fragment ion generated by collision-induced dissociation of the [M + H]+ ion was used for quantification. MRM settings for the target analytes were (parent ion → fragment ion); SAM (m/z 399.088 → 136.125) with collision energy of 27.03 and RF lens 64 V, SAH (m/z 385.088 → 134.042) with collision energy of 19.49 and RF lens 58 V. SAM and SAH were quantified against commercial standards (Sigma-Aldrich). For samples analyzed on the TSQ Altis™ Triple Quadrupole Mass Spectrometer, data was analyzed using Thermo Fisher Scientific™ FreeStyle software.
2.8. Animal studies
All animal procedures were approved by the University of Alberta's Institutional Animal Care Committee (AUP00000175) in accordance with guidelines of the Canadian Council on Animal Care. Mice were exposed to a 12-h light/dark cycle with free access to food and drinking water. Male Pemt+/+ and Pemt–/– (C57BL/6J) mice were fed a standard chow diet (5001; LabDiet, St. Louis, MO, USA) or a semisynthetic high-fat diet (HFD, F3282; Bio-Serv, Flemington, NJ, USA) that contained 60 kcal% from lard. Mice were kept on the appropriate diet for 6–10 weeks and tissue collected and stored at −80 °C until analyses. As previously reported (20), for in vivo experiments using control (GFP) adeno-associated virus (AAV.GFP) and PEMT expressing adeno-associated virus (AAV.PEMT) [20], Pemt–/– mice were injected with 1 × 1010 genome copies per mouse of AAV.GFP or AAV.PEMT, and Pemt+/+ mice were injected with an equal dose of AAV.GFP. At 1-week post-AAV administration mice, were fed HFD for 10 weeks [20]. For in vivo antisense oligonucleotide (ASO) experiments, male C57BL/6J mice (purchased from Jackson Laboratory, Bar Harbor, ME, USA) were intra-peritoneally injected weekly (25 mg/kg b.w.) with either a scrambled control ASO (5′-GGCCAATACGCCGTCA-3′) or an ASO that inhibits PEMT (5′-CTTTATTAGTGTGTCG-3′), (5′-TTATTAGTGTGTCGGG-3′) or (5′-ACAACATGATTGGACC-3′) provided by Ionis Pharmaceuticals). One week after the first ASO injection, mice were fed the HFD for 10 weeks and tissue collected and stored at −80 °C until analyses. Glucose tolerance tests, insulin tolerance tests and PEMT activity assays in mice treated with ASO were carried out as previously described [20]. For glucose tolerance test data, area under the curve was computed using the trapezoid rule. For insulin tolerance test data, incremental area under the curve was computed. For this, each data point was baseline (0 time) corrected and Δ blood glucose was plotted over time. The area under the Δ blood glucose curve was then calculated using the trapezoid rule.
2.9. Determination of CoQ9 and CoQ10 content in mouse tissue and mitochondria
Tissues were homogenized in mitochondrial isolation buffer (10 mM Tris-MOPS, pH 7.4, 1 mM EGTA, 200 mM sucrose containing protease inhibitors) for 30 s at 7000 rpm (Heidolph homogenizer) and samples kept at 4 °C for all subsequent procedures. 100 μL of the homogenates was kept for total tissue CoQ9 and CoQ10 analyses, and the remaining homogenate was used to isolate mitochondria as previously described [19]. Briefly, homogenates were centrifuged at 700×g for 10 min, the supernatant isolated and centrifuged at 7000×g for 10 min to obtain a pellet containing mitochondria. The pellet was re-suspended in mitochondrial isolation buffer and re-centrifuged at 7000×g for 10 min. The mitochondrial pellet was finally re-suspended in mitochondrial isolation buffer and a BCA assay (Thermo Fischer Scientific) carried out to quantify protein concentration. 50–100 μL of homogenate or isolated mitochondria was placed in a 15 mL screw top tube, 25 μL para-benzoquinone (2 mg/ml) was added, incubated for 30 min at room temperature and then 2 mL ethanol: isopropanol (95:5) and 10 mL of water-washed hexane were added. The mixture was mixed vigorously for 1 min, centrifuged (1430×g, 5 min, 4 °C) and 9 mL of hexane was collected and dried using a rotary evaporator. The resulting dried lipids were re-dissolved in 150–180 μL ice-cold mobile phase (ethanol:methanol:isopropanol: ammonium acetate pH 4.4, 65:30:3:2, vol/vol/vol/vol) and transferred into HPLC vials. Cholesterol, CoQ9 and CoQ10 were determined by HPLC using a Supelcosil LC-C18 column (5 μm, 250 mm × 4.6 mm) eluted at 1 mL/min connected to UV and electrochemical (ESA CoulArray 5600A) detectors. Non-esterified cholesterol was detected at 214 nm, while CoQ9 and CoQ10 were detected at −700, +700 and + 700 mV and quantified against authentic commercial standards obtained from Sigma Aldrich (USA).
3. Results
3.1. Genome-wide screening identifies 30 novel regulators of CoQ content
To systematically identify the genetic networks controlling CoQ synthesis we conducted a quantitative genome-wide screen of all 5420 mutants contained in the Saccharomyces cerevisiae homozygous diploid Deletion Collection [21]. This approach represents an unbiased, global approach to interrogate the CoQ regulatory framework by identifying gene deletions that alter cellular CoQ concentration. To enable large-scale screening by quantitative HPLC coupled to electrochemical detection, we developed an optimized method for cell growth, lysis and CoQ extraction for small culture volumes (∼200 μL) in a 96-well plate format (Figs. S1A–C). Intra- and inter-day reproducibility of the method was 3 and 15%, respectively (Fig. S1D), and considered acceptable. The development of this screening method also increased the number of samples processed in one day by about five-fold, from 20 to 96. We then used this method to determine the cellular CoQ concentration in each knockout mutant, with results normalized to cell biomass. The screen was carried out in two phases (Fig. S2A). In both phases cells were grown in minimal media in the absence of any CoQ precursors such as para-aminobenzoic acid or 4-hydroxybenzoic acid to reduce confounding results caused by substrate availability. First, all mutants were screened (n = 1), identifying 140 mutants with CoQ content 2 standard deviations higher or lower than the plate population mean. Second, these mutants were re-screened (n = 4–6) against wild-type (WT) cells to confirm gene mutants with significantly altered CoQ content. The genetic screen revealed 30 mutants with significantly higher CoQ (‘high CoQ’) and seven with significantly lower CoQ (‘low CoQ’) than WT (Table 1). The ‘low CoQ’ mutants were comprised entirely of known genes of the biosynthetic pathway, as well as HFD1 which encodes a dehydrogenase involved in the production of the CoQ precursor, 4-hydroxybenzoic acid [22]. These results validate the approach chosen to identify mutants with altered cellular CoQ content. The 30 ‘high CoQ’ mutants identified were comprised entirely of genes not previously associated with cellular CoQ and represent potential novel CoQ regulators.
Table 1.
S. cerevisiae mutants identified with significantly decreased or increased CoQ6 content compared with WT.
| ‘Low CoQ’ mutants | |
|---|---|
| ∼Fold decrease in CoQ6vs WT | |
| coq2Δ | not detectable |
| coq3Δ | not detectable |
| coq4Δ | not detectable |
| coq6Δ | not detectable |
| coq8Δ | not detectable |
| coq9Δ | not detectable |
|
hfd1Δ |
2 |
|
‘High CoQ’ mutants | |
| ∼Fold increase in CoQ6 vs WT | |
| mss116Δ | 2 |
| sin3Δ | 2 |
| swa2Δ | 2 |
| trf5Δ | 2 |
| aim26Δ | 2 |
| rri1Δ | 2 |
| trf4Δ | 2 |
| vps54Δ | 2 |
| aim9Δ | 2 |
| drs2Δ | 2.5 |
| atp15Δ | 2.5 |
| pac10Δ | 2.5 |
| ctk3Δ | 2.5 |
| nup60Δ | 3 |
| arc1Δ | 3 |
| sac1Δ | 3 |
| ume6Δ | 3 |
| hof1Δ | 3 |
| bud31Δ | 3 |
| vps20Δ | 3 |
| mrpl4Δ | 4 |
| pho85Δ | 4 |
| rad51Δ | 5 |
| srs2Δ | 5 |
| cho2Δ | 5 |
| rrg7 Δ | 7 |
| cdc50Δ | 8 |
| ric1Δ | 11 |
| ort1 Δ | 11 |
| adk1 Δ | 12 |
We next asked whether the identified 30 ‘high CoQ’ regulators form part of a transcriptional response to altered CoQ by studying the transcriptional profiles of yeast mutants of the CoQ biosynthetic pathway (coq1Δ, coq3Δ, coq7Δ, coq8Δ and coq9Δ) grown in the absence and presence of exogenously supplemented CoQ. Gene expression changes in mutants compared to WT (≥2-fold change) were identified. (Supplementary Table 1). Notably, deficiency in any one of the COQ genes did not lead to the alteration of the expression of other COQ genes for the biosynthesis of CoQ. Of all mutants tested, only deletion of COQ7 resulted in downregulation of COQ1 gene expression, with no COQ transcripts differentially expressed in any other mutant. There was also no change in the expression of genes identified in our screen, genes previously known to contribute to CoQ synthesis, such as PTC7 and HFD1, or genes involved in isoprenoid or benzoquinone head group synthesis. Comparing gene expression profiles across all COQ mutants (Figs. S2B–C), only 5 genes were identified as differentially expressed in the absence of CoQ supplementation (ARN2, FIT2, FIT3, FDC1 and YDR514C) and 7 genes in CoQ supplemented conditions (ARN1, ARN2, FIT2, PHO84, SIT1, TIS11 and YDR514c).
Strikingly, the transcriptional changes observed in the various coq mutants were not able to be completely reversed by exogenous CoQ supplementation (Supplementary Table 1; Fig. S2C). Thus, exogenous CoQ supplementation may not be able to fully replace endogenous CoQ functions. In addition, CoQ supplementation did not lead to changes in any genes identified in our study, such as CHO2 or genes identified previously such as HFD1. From this it appears that exogenous CoQ supplementation does not, at least transcriptionally, affect endogenous CoQ regulatory pathways.
3.2. A deficiency in phosphatidylethanolamine (PE) methylation results in increased mitochondrial CoQ
Among the top hits was the cho2Δ mutant which lacks PEMT, encoded by the CHO2 gene. The methylation pathway is one of three distinct PC synthesis pathways, along with the Kennedy Pathway and lyso-PC to PC cycling (Fig. 1A). We focused on the cho2Δ mutant for the following reasons. First, PEMT activity is evolutionarily conserved in eukaryotes. Second, mice lacking PEMT are viable and have long been known to be protected from atherosclerosis, age-dependent cardiac dysfunction and insulin resistance [[23], [24], [25]], although the underlying reasons for this protective phenotype have remained obscure. Third, PEMT catalyzes the methylation of phosphatidylethanolamine (PE) to phosphatidylcholine (PC), and CoQ biosynthesis requires three methylation steps. Finally, a potential link between PEMT and CoQ has not been investigated to date.
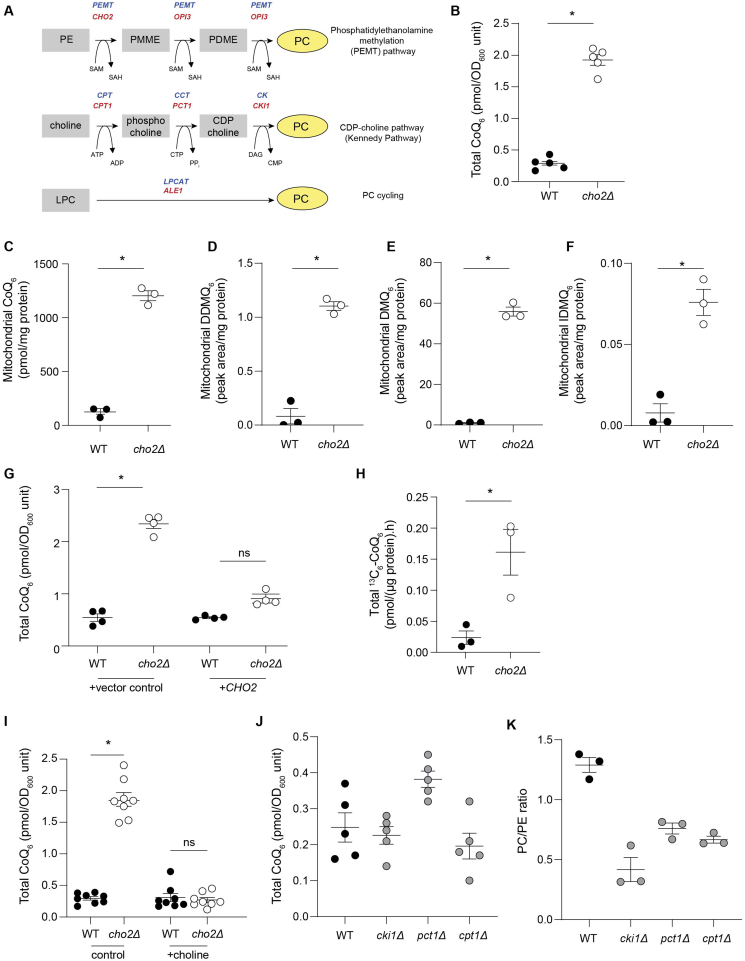
Fig. 1.
PEMT deficiency is a novel regulator of mitochondrial CoQ6content in S. cerevisiae. (A) Scheme of the three pathways that mediate phosphatidylcholine (PC) synthesis in yeast and mammals; red indicates genes in yeast, while blue indicates mammalian genes. (B) Total CoQ6 concentrations in cho2Δ mutants compared with WT (n = 5). (C) Mitochondrial CoQ6 concentrations in cho2Δ mutants compared with WT cells (n = 3). (D) Mitochondrial demethoxy-demethyl-Q6 (DDMQ6) concentrations in cho2Δ mutants compared with WT cells (n = 3). (E) Mitochondrial demethoxy-Q6 (DMQ6) concentrations in cho2Δ mutants compared with WT cells (n = 3). (F) Mitochondrial imino-demethoxy-Q6 (IDMQ6) concentrations in cho2Δ mutants compared with WT cells (n = 3). (G) Total CoQ6 concentrations in WT and cho2Δ mutants harboring either control vector or a single copy of CHO2 (n = 4). (H) CoQ6 biosynthetic rate in cho2Δ mutants and WT cells measured using the rate of 13C6-CoQ6 formed from supplemented 13C6-4-hydroxybenzoic acid (4HB) (n = 3). (I) Total CoQ6 concentrations in WT and cho2Δ mutants with or without 1 mM choline supplementation (n = 8). (J) Total CoQ6 concentrations in WT and mutants of the Kennedy Pathway (n = 5). (K) PC/PE ratio in WT and mutants of the Kennedy Pathway (n = 5). Data and error bars depict mean ± s.e.m. *depicts P ≤ 0.05 and ns indicates ‘not significant’ as determined by Mann-Whitney (B–F, H) or Kruskal-Wallis (G, I–K) test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The cho2Δ mutant accumulated five times more total CoQ than WT cells (Fig. 1B) and over ten times more mitochondrial CoQ than WT cells (Fig. 1C). Consistent with this, the cho2Δ mutant displayed significantly increased concentrations of the CoQ6 intermediates demethoxy-demethyl-Q6 (DDMQ6), demethoxy-Q6 (DMQ6) and imino-demethoxy-Q6 (IDMQ6) (Fig. 1D–F). The cho2Δ-CoQ relationship was confirmed in multiple yeast strain backgrounds (Figs. S3A and S3B). Mutants heterozygous for CHO2 did not have altered total or mitochondrial CoQ content (Figs. S3C and S3D). The observed increase in cellular CoQ in cho2Δ was reversed by re-expression of CHO2 (Fig. 1G), indicating that CoQ content is dependent on CHO2 expression. Isotope tracing studies using 13C6-4-hydroxybenzoic acid showed that cho2Δ cells have an increased rate of CoQ biosynthesis (Fig. 1H). These results establish that CHO2 deficiency is a positive regulator of mitochondrial CoQ content.
3.3. PEMT deficiency increases mitochondrial CoQ through a phosphatidylcholine-independent pathway
In yeast, conversion of PE to PC is carried out by two enzymes (Cho2 and Opi3) [26], whereas in mammals PEMT catalyzes all three methylation reactions [27]. Consistent with previous reports, cho2Δ mutants had decreased PC (∼2-fold) and increased PE concentrations (∼1.5-fold) compared to WT cells [28,29] reflecting the role of CHO2 in PC synthesis (Figs. S3E–G). To test whether altered cellular PC content was responsible for increased CoQ in the cho2Δ mutant, cho2Δ cells were supplemented with choline to bypass defects in the methylation pathway [28]. This significantly elevated PC concentrations, normalized PE contents (Figs. S3E–F) and restored WT CoQ concentrations (Fig. 1I), consistent with cellular PC or the PC-to-PE ratio regulating CoQ concentrations. Inconsistent with this interpretation, however, supplementation with monomethylethanolamine (MME) also restored WT CoQ content in cho2Δ (compare control in Fig. 1I with Fig. S3H) without restoring WT PC-to-PE ratios [28]. Moreover, the opi3Δ mutant, which lacks the ability to catalyze the last two methylation steps in PE formation from PC, had significantly lower PC and similar PC-to-PE ratios compared with cho2Δ cells (Fig. S3G and S3I-K), yet accumulated ∼ four-times less mitochondrial CoQ than cho2Δ cells (Fig. S3L). Mutants of the Kennedy pathway also did not have altered CoQ content despite having PC/PE ratios comparable to the cho2Δ mutant (Fig. 1J and K). Together, these results indicate that CHO2/PEMT modulates cellular CoQ independent of changes in PC and PE content, or changes to the PC-to-PE ratio.
3.4. Mitochondrial CoQ is selectively altered in response to PEMT deficiency
Mitochondrial CoQ synthesis requires three distinct building blocks (Fig. S4A): the ‘benzoquinone ring’ derived from a tyrosine/ring precursor; the polyisoprenoid ‘tail’ derived from the mevalonate pathway; and a methyl donor S-adenosylmethionine (SAM) required to produce the fully substituted benzoquinone ring in CoQ. These building blocks are transported from the cytosol into mitochondria where the synthesis of CoQ takes place. To determine if increased CoQ biosynthesis in cho2Δ cells could be explained by a simple increase in mitochondria that provide the ‘machinery’ for CoQ biosynthesis, mitochondrial mass was assessed by four independent assays: mitochondrial DNA, citrate synthase activity, porin content, and oxygen flux (Figs. S4B–E). All four parameters showed only a modest increase in mitochondrial mass in cho2Δ, well below the 10-fold increase in mitochondrial CoQ. We next asked whether mevalonate-derived lipids were altered. Untargeted lipidomics analyses revealed an increase in dolichols in cho2Δ (Fig. S4F) in parallel with an increase in expression of genes involved in the dolichol synthesis pathway (Fig. S4G). This suggested flux through the mevalonate pathway may be altered. However, concentrations of ergosterol (the yeast equivalent of mammalian cholesterol) were not altered in cho2Δ (Fig. S4H), indicating that it is unlikely an increase in mevalonate pathway flux or farnesyl pyrophosphate availability underpins the alterations in CoQ and dolichols in cho2Δ. Supporting this, there was no change in the expression of farnesyl diphosphate synthase (ERG20), or the genes of the mevalonate pathway in cho2Δ (Fig. S4I), indicating differential regulation of isoprenoid lipids by CHO2. Finally, the increased rate of CoQ biosynthesis was observed in the absence of changes in the transcript or protein levels of presently known members of the CoQ biosynthetic ‘machinery’ [30] (Figs. S5A–B), although cho2Δ displayed a more stabilized CoQ-synthome (Fig. S5C). The stabilization of the CoQ synthome is an intriguing observation and a direct interaction between Cho2p and the CoQ synthome may exist. However, we believe it is more likely that lack of CHO2 indirectly acts to stabilize the complex by virtue of increasing CoQ content which has been shown to stabilize some Coq polypeptides and potentially stabilize the CoQ synthome [31].
3.5. PEMT deficiency increases CoQ biosynthesis via increasing mitochondrial methylation capacity
To further understand how a deficiency in cho2 increases mitochondrial CoQ, we performed untargeted metabolomic analyses. This identified numerous metabolites significantly altered in cho2Δ compared with WT cells (Fig. 2A and B; Figs. S6A–B), with the greatest differences observed in S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH) (Fig. S6C). Whole-cell SAM was increased while SAH concentrations were decreased in cho2Δ, as reported previously [32]. Targeted LC-MS/MS analyses confirmed these results (Fig. 2C and D). Deficiencies in methyltransferases other than cho2 had no effect on cellular CoQ, SAM and SAH (Figs. S6D–F). Strikingly, mitochondrial SAM and SAH concentrations were both increased in cho2Δ (Fig. 2E and F), in sharp contrast to the situation in whole cells. As CoQ biosynthesis requires SAM-dependent methylation of the benzoquinone ring, WT cells were supplemented with the SAM precursor methionine. This caused a ∼20-fold increase in cellular and mitochondrial SAM (Figs. S7A–B) yet CoQ was unaffected (Fig. S7C). Methionine supplementation also increased cellular and mitochondrial SAM concentrations in cho2Δ mutants, and in this case, CoQ decreased significantly (Fig. S7C). Together, these results show that cellular or mitochondrial SAM concentrations alone do not determine CoQ concentrations.
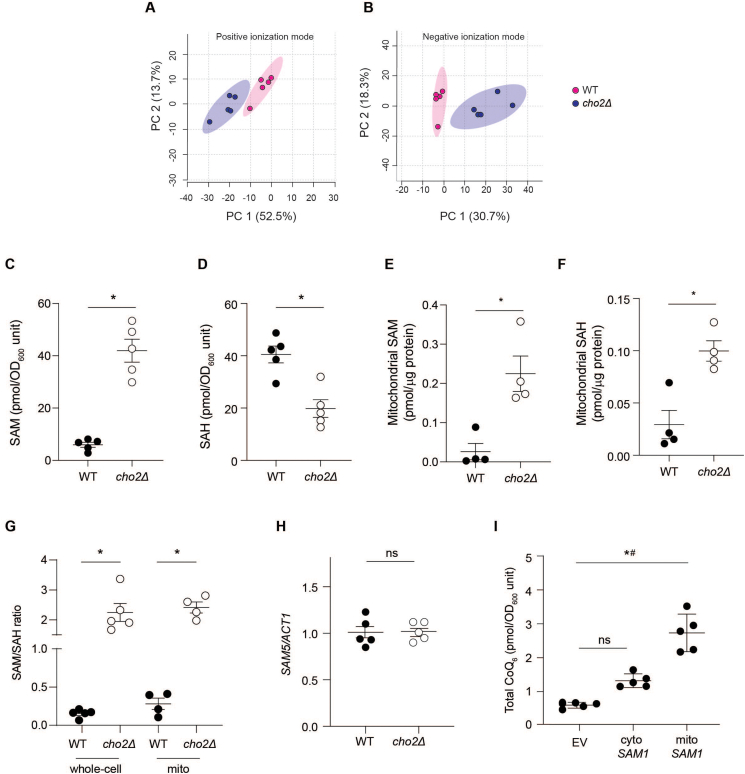
Fig. 2.
PEMT deficiency increases CoQ biosynthesis via altering mitochondrial S-adenosylmethionine and S-adenosylhomocysteine in S. cerevisiae. (A) Principal component analyses (PCA) plots of metabolites identified in WT and cho2Δ in both positive and negative ionization modes. (B) Whole cell S-adenosylmethionine (SAM) in WT and cho2Δ cells (n = 5). (C) Whole cell S-adenosylhomocysteine (SAH) in WT and cho2Δ cells (n = 5). (E) Mitochondrial SAM in WT and cho2Δ cells (n = 4). (F) Mitochondrial SAH in WT and cho2Δ cells (n = 4). (G) Whole-cell and mitochondrial SAM/SAH ratios in WT and cho2Δ cells (n = 4). (H) SAM5 gene expression in WT and cho2Δ mutants as measured by qPCR (n = 5). (I) The effect of overexpression of S-adenosylmethionine synthetase 1 (SAM1) either in the mitochondria (mito-SAM1) or cytosol (cyto-SAM1) compared to empty vector (EV) control on CoQ6 content in WT. # indicates significant difference between EV and mito-SAM1. Data and error bars depict mean ± s.e.m. *P ≤ 0.05 as determined by Mann-Whitney (A-H) or Kruskal-Wallis test (I).
SAH is a product inhibitor of methyltransferase activity, and the SAM-to-SAH ratio (rather than SAM concentrations alone) is considered to reflect methylation potential. Compared with WT cells, cho2Δ mutants had increased cellular and mitochondrial SAM-to-SAH ratios (Fig. 2G) in parallel with increased CoQ. In WT cells, methionine supplementation increased cellular and mitochondrial SAH (Figs. S7D–E) to an extent comparable to that of SAM (Figs. S7A–B), such that there was no change the SAM-to-SAH ratio (Fig. S7F). In cho2Δ cells, methionine supplementation had no effect on cellular SAH (Fig. S7D), whereas it significantly increased mitochondrial SAH concentrations (Fig. S7E), such that the mitochondrial SAM-to-SAH ratio decreased (Fig. S7G) in parallel with decreased CoQ. These results suggest that it is the mitochondrial SAM-to-SAH ratio i.e., the methylation capacity of mitochondria that regulates mitochondrial CoQ, although it remains unclear why methionine supplementation differentially affects cellular and mitochondrial SAM and SAH pools.
The methionine cycle, including SAM synthesis, takes place in the cytosol. Cytosolic SAM is then transported into mitochondria by SAM5 which itself is inhibited competitively by SAH. As SAM5 expression was not altered in cho2Δ mutants (Fig. 2H), we reasoned that their higher mitochondrial SAM-to-SAH ratio reflected a proportionally higher uptake of cytosolic SAM than SAH, thereby increasing the mitochondrial methylation capacity and, as a result, mitochondrial CoQ synthesis. To test this hypothesis, we overexpressed the SAM synthetase SAM1 in either the cytosol or mitochondria of WT cells. Strikingly, mitochondrial overexpression of SAM1 increased CoQ almost five-fold, whereas cytosolic overexpression of SAM1 did not change CoQ (Fig. 2I). These observations, together with the findings that i) increased mitochondrial SAM-to-SAH is a hallmark of cho2Δ and ii) decreasing this ratio decreases mitochondrial CoQ, indicate that the mitochondrial SAM-to-SAH ratio is a previously unrecognized key regulator of CoQ biosynthesis and mitochondrial CoQ content.
3.6. PEMT deficiency regulation of mitochondrial CoQ concentrations in evolutionarily conserved
The methylation pathway of PC synthesis is highly conserved in eukaryotes [33] and the impact of PEMT deficiency on phospholipids, triacylglycerols [29] and ceramides [34] (Figs. S8A–B) is mirrored in yeast and mammals. To test if the yeast phenotype was conserved across species, we investigated the effect of PEMT deficiency on mitochondrial CoQ in multiple rodent cell types in vitro and in vivo. Pharmacological inhibition of PEMT in PEMT-expressing McArdle 7777 rat hepatoma cells (PEMT-McA) with 3-deazaadenosine (DZA) [35] increased mitochondrial CoQ (Fig. 3A). In line with this, livers of Pemt–/– mice fed chow had a significantly increased total (Fig. S8C) and mitochondrial CoQ (Fig. 3B) compared with Pemt+/+ littermates. Plasma CoQ, and total and mitochondrial CoQ in skeletal muscle, kidney, brain and white adipose tissue were not changed (Fig. S8D-L), consistent with PEMT expression being limited to the liver in chow-fed animals [36]. Like the situation in yeast, PEMT deficiency did not alter hepatic expression of CoQ biosynthetic pathway genes (Fig. S8M). Consumption of a high fat diet (HFD) doubled total (Fig. S9A) and mitochondrial CoQ (Fig. 3C) in Pemt–/– mice again without changes in the expression of CoQ biosynthetic pathway genes (Fig. S9B). Normalization of hepatic PEMT activity in Pemt–/– mice using an adeno-associated viral (AAV) system [20] decreased mitochondrial CoQ in the liver to that of Pemt+/+ animals expressing control GFP plasmid (Fig. 3D). These results show that PEMT expression regulates mitochondrial CoQ content in hepatic cell lines and the liver of mice, without apparent changes in the canonical CoQ biosynthetic pathway.
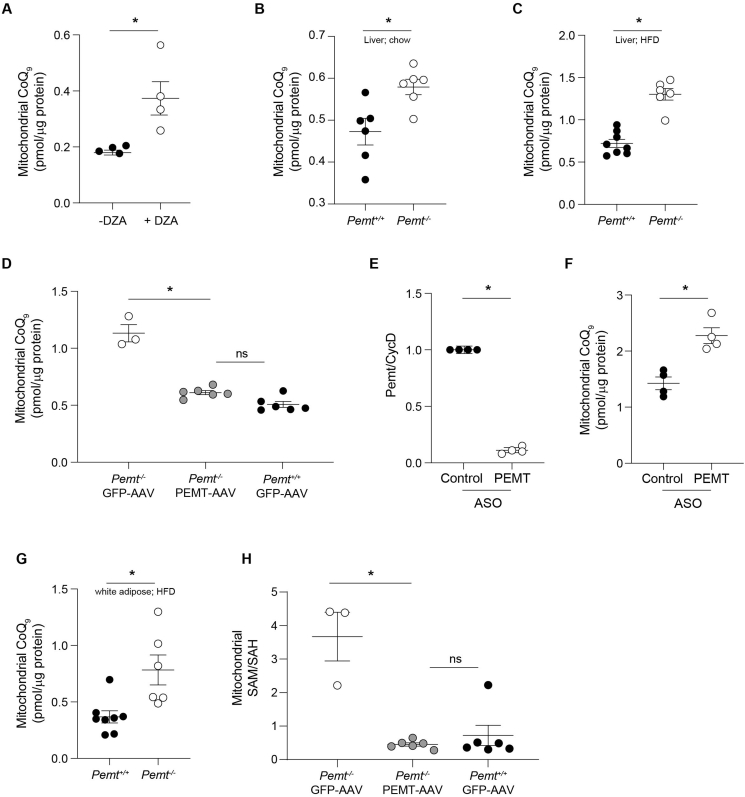
Fig. 3.
PEMT deficiency increases mitochondrial CoQ content in mammalian cells and tissues. (A) Effect of inhibiting PEMT activity in PEMT-expressing McArdle 7777 hepatoma cells by 3-deazaadenosine (DZA) on mitochondrial CoQ9 content (n = 4). (B) Hepatic mitochondrial CoQ9 content in Pemt+/+ and Pemt–/– mice fed chow (n = 6). (C) Hepatic mitochondrial CoQ9 content in Pemt+/+ and Pemt–/– mice fed HFD for 6 weeks (n = 6–8). (D) Mitochondrial CoQ9 content in mice treated with adeno-associated virus (AAV) expressing Pemt or GFP control (n = 3–6). (E) Effect of anti-sense oligonucleotide (ASO) mediated knockdown of PEMT in differentiated 3T3-L1 adipocytes on PEMT gene expression as determined by qPCR (n = 4). (F) Mitochondrial CoQ9 content in differentiated 3T3-L1 adipocytes treated with either control or anti-PEMT ASO (n = 4). (G) White adipose tissue mitochondrial CoQ9 content in Pemt+/+ and Pemt–/– mice fed HFD for 6 weeks (n = 6–8) (H) Mitochondrial SAM/SAH ratio in mice treated with adeno-associated virus (AAV) expressing Pemt or GFP control (n = 3–6). Data and error bars depict mean ± s.e.m. *P ≤ 0.05 and ns indicates ‘not significant’ as determined by Mann-Whitney (A-C, E-G) or Kruskal-Wallis test (D, H).
We next investigated a potential role of PEMT in the regulation of CoQ concentration in non-hepatic cells. PEMT expression was induced during adipocyte differentiation (Fig. S9C), consistent with previous studies [37]. Inhibition of Pemt expression in 3T3-L1 adipocytes using anti-sense oligonucleotides (ASO; Fig. 3E) significantly increased mitochondrial CoQ (Fig. 3F). Moreover, we observed a significant increase in total (Fig. S9D) and mitochondrial CoQ (Fig. 3G) in white adipose tissue of Pemt–/– mice fed a high fat diet, conditions where PEMT expression is induced in adipose tissue [37]. Together, these results show that PEMT expression can regulate mitochondrial CoQ content in hepatocytes and adipocytes in vitro and in vivo, as observed in yeast.
3.7. Mitochondrial SAM-to-SAH ratio is a central feature of PEMT deficiency
A previous commentary [38] suggested that hepatic PEMT was a major component of whole body SAM utilization. A separate investigation reported no difference in hepatic SAM and SAH concentrations in Pemt–-/– compared with Pemt+/+ mice [39] although mitochondrial SAM and SAH concentrations were not independently measured. We therefore examined whether PEMT expression regulated mitochondrial CoQ in liver through specific changes in the mitochondrial SAM-to-SAH ratio using the previously described AAV system. Compared with WT animals, livers from Pemt–/– mice expressing GFP (control) had significantly increased mitochondrial SAM-to-SAH ratio (Fig. 3H), paralleling their increase in mitochondrial CoQ (Fig. 3D). Expression of Pemt in Pemt–/– mice decreased the mitochondrial SAM-to-SAH ratio to levels observed in Pemt+/+ mice, again in parallel with the observed normalization of mitochondrial CoQ (Fig. 3D). Thus, PEMT expression affects the SAM-to-SAH ratio in mitochondria. Together the data indicate that lowering PEMT expression is sufficient to increase both the mitochondrial SAM-to-SAH ratio and mitochondrial CoQ content, and this relationship is maintained in a broad range of species.
3.8. PEMT deficiency protects against insulin resistance via modulating CoQ and decreasing mitochondrial oxidative stress
We next examined whether regulating mitochondrial CoQ via PEMT can be exploited to ameliorate disease associated with CoQ deficiency. To do this, we focused on insulin resistance for three reasons. First, Pemt–/– mice are protected from insulin resistance [25,40] via a currently unexplained mechanism. Second, decreased mitochondrial CoQ is an up-stream driver of insulin resistance in humans, mice, and cellular models [11,41,42], and CoQ supplementation reverses insulin resistance. Third, increasing mitochondrial CoQ decreases mitochondrial superoxide [11] another upstream driver of insulin resistance. We therefore first examined the effect of PEMT deficiency on mitochondrial superoxide. Compared with WT cells, cho2Δ mutants had decreased mitochondrial superoxide, as assessed by MitoSOX fluorescence (Fig. S9E). Similarly, cho2Δ cells were also protected from decreased viability induced by polyunsaturated fatty acids (Fig. S9F), consistent with superoxide being the primordial reactive oxygen species that can give rise to cell damage via oxidation of unsaturated fatty acids. These results indicate that PEMT deficiency decreases mitochondrial oxidative stress.
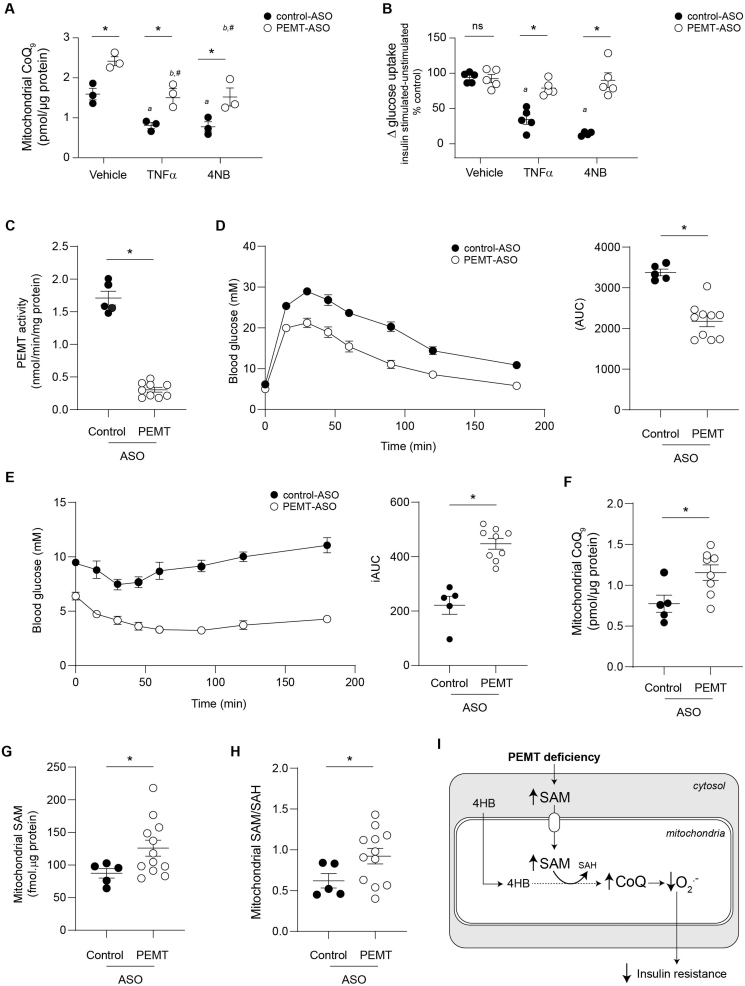
To more directly link mitochondrial CoQ and PEMT, we used antisense oligonucleotide (ASO) technology in differentiated 3T3-L1 adipocytes treated with tumour necrosis factor-α (TNFα) as a cellular model of insulin resistance [43]. Decreasing PEMT expression by ASO treatment increased mitochondrial CoQ concentrations above that seen in control ASO-treated cells (Fig. 4A) without a change in insulin-stimulated 2-deoxyglucose uptake (Fig. 4B). Exposure of control ASO-treated adipocytes to TNFα decreased mitochondrial CoQ (Fig. 4A) and insulin-stimulated 2-deoxyglucose uptake (Fig. 4B). Similarly, pharmacological blockade of CoQ synthesis with 4-nitrobenzoic acid (4NB, a competitive inhibitor of 4-hydroxybenzoate:polyprenyltransferase) [44], decreased mitochondrial CoQ (Fig. 4A) in control ASO-treated cells to a comparable extent to that seen with TNFα treatment. This was associated with a comparable decrease in insulin-stimulated glucose uptake (Fig. 4B), consistent with a causal link between mitochondrial CoQ and insulin sensitivity [11]. Replacing control with anti-Pemt ASO restored mitochondrial CoQ and insulin-stimulated 2-deoxyglucose uptake to control values in both TNFα- and 4NB-treated cells (Fig. 4A and B). The results suggest that enhanced CoQ biosynthesis is required for PEMT deficiency to improve glucose utilization in a cellular model of insulin resistance.
Fig. 4.
Protection against IR by PEMT deficiency depends on increased mitochondrial CoQ content. (A) Mitochondrial CoQ9 content in 3T3-L1 adipocytes treated with control or anti-Pemt ASO, and TNFα (chronic low dose inflammation inducer) or 4-nitrobenzoic acid (4NB, an inhibitor of CoQ biosynthesis) (n = 3). *Significant difference between control and PEMT-ASO in the same treatment group. aSignificant difference between TNFα and 4NB treatments compared with vehicle in control-ASO treated cells. bSignificant difference between TNFα and 4NB treatments compared with vehicle in PEMT-ASO treated cells. #No significant difference between cells treated with PEMT-ASO and TNFα or 4NB compared with vehicle-treated control-ASO cells. (B) Glucose uptake in 3T3-L1 adipocytes treated with control or anti-Pemt ASO, and TNFα (chronic low dose inflammation inducer) or 4-nitrobenzoic acid (4NB, an inhibitor of CoQ biosynthesis) as measured by uptake of 3H-2-deoxyglucose (n = 5). *ns denotes significant difference or lack thereof, respectively, between control and PEMT-ASO in the same treatment group. aSignificant difference between TNFα and TNFα+4NB treatments compared with vehicle in control-ASO treated cells. #No significant difference between cells treated with PEMT-ASO and TNFα or 4NB compared with vehicle-treated control-ASO cells. (C) PEMT activity in C57BL/6J mice fed HFD treated with either control or anti-PEMT ASO for 10 weeks (n = 5–9). (D) Glucose tolerance test (GTT) in C57BL/6J mice fed HFD treated with either control or anti-PEMT ASO for 10 weeks (n = 5–10). For area under the curve (AUC), each data point represents the area under the curve extrapolated per animal from blood glucose concentrations determined over 3 h post glucose challenge. AUC was determined as outlined in the Methods. (E) Insulin tolerance test (ITT) in C57BL/6J mice fed HFD treated with either control or anti-PEMT ASO for 10 weeks (n = 5–10). For incremental area under the curve (iAUC), blood glucose concentrations per animal were baseline (0 min) corrected, and iAUC calculated from Δ blood glucose concentrations determined over 3 h post insulin challenge. (F) Mitochondrial CoQ9 concentrations in C57BL/6J mice fed HFD treated with either control or anti-PEMT ASO for 10 weeks (n = 5–12). (G) Mitochondrial SAM concentrations in C57BL/6J mice fed HFD treated with either control or anti-PEMT ASO for 10 weeks (n = 5–12). (H), Mitochondrial SAM/SAH ratio in C57BL/6J mice fed HFD treated with either control or anti-PEMT ASO for 10 weeks (n = 5–12). (I) Schematic summary of results and proposed mechanism. PEMT deficiency increases cytosolic SAM resulting in increased mitochondrial SAM and SAM/SAH. Increased mitochondrial SAM increases CoQ biosynthesis by donating methyl groups to the benzoquinone head group of CoQ formed from 4-hydroxybenzoic acid (4HB). Increased mitochondrial CoQ results in decreased mitochondrial superoxide, an upstream driver of insulin resistance. Data and error bars depict mean ± s.e.m. *P ≤ 0.05 as determined by Mann-Whitney (C–H) or Kruskal-Wallis test (A, B).
Finally, we examined how improved insulin sensitivity in vivo via decrease in PEMT activity relates to the mitochondrial SAM-to-SAH ratio. Anti-Pemt ASO significantly decreased hepatic PEMT activity in mice fed HFD (Fig. 4C) and such mice had improved glucose clearance (Fig. 4D) and insulin sensitivity (Fig. 4E). This was evidenced by a decreased area under the curve (AUC) for the glucose tolerance test (GTT), and an increased incremental area under the curve (iAUC) for insulin tolerance test (ITT) (see Fig. S9G for baseline corrected ITT values). Improved glucose clearance and insulin sensitivity were associated with an increase in mitochondrial CoQ, SAM and SAM-to-SAH ratio (Fig. 4F–H), recapitulating the situation in global Pemt–/– mice. Together, these data provide a mechanism for how PEMT deficiency ameliorates insulin resistance, i.e., by increasing mitochondrial SAM-to-SAH ratio and CoQ synthesis, a known driver of insulin resistance (Fig. 4I).
4. Discussion
This study outlines the discovery of PEMT deficiency as the first identified positive regulator of mitochondrial CoQ content in organisms from budding yeast to mammals. PEMT regulates CoQ concentrations via modulation of the mitochondrial SAM-to-SAH ratio that determines the methylation reactions required for CoQ biosynthesis. Changes in CoQ are not dependent on altering cellular SAM concentrations or the SAM-to-SAH ratio per se, but rather a case of modulating the balance of mitochondrial metabolites, i.e., the ‘right ratio in the right place’. This interplay between SAM and PEMT is independent of the cellular PE-to-PC ratio, and it establishes a previously unrecognized functional relationship between PEMT, mitochondrial one-carbon metabolism and CoQ synthesis. Our discovery has potential medical implication because increasing mitochondrial CoQ via PEMT knockdown decreases mitochondrial oxidative stress and it maintains insulin sensitivity in vivo. At a mechanistic level, the biological benefit of PEMT knockdown is affected by increasing mitochondrial CoQ and a resulting decrease in superoxide, two known key drivers of insulin resistance. Together, this work enhances our understanding of how cells regulate mitochondrial CoQ synthesis and how this may be translated to a conceptually novel approach to treat insulin resistance.
Our key finding of PEMT as a conserved regulator of mitochondrial CoQ content is supported by work in budding yeast, mammalian cell culture models and in vivo mouse studies. Combining the classical genetic approach of gene knockout and re-expression with targeted LC-MS/MS analysis, a direct relationship between PEMT expression and mitochondrial CoQ content was established. Significantly, re-introduction of PEMT expression normalized mitochondrial CoQ concentrations in both budding yeast and mouse models. Acute PEMT disruption mediated by small molecules such as 3-deazaadenosine or antisense oligonucleotides was also sufficient to increase mitochondrial CoQ, recapitulating the results from the genetic approach. Collectively these data confirm that modulation of PEMT at the gene, mRNA or enzyme activity level increases mitochondrial CoQ in a variety of species.
Our studies establish that spatial compartmentalization of the one-carbon metabolites SAM and SAH in mitochondria is necessary for changes in CoQ concentrations and the underlying basis of the PEMT-CoQ relationship. Use of untargeted ‘global’ metabolomics and targeted sub-compartmental measurements led to the identification of increased SAM-to-SAH ratio in mitochondria across models of PEMT deficiency, paralleling the changes observed in mitochondrial CoQ. Changes (up or down) to the mitochondrial SAM-to-SAH ratio were sufficient to alter mitochondrial CoQ. Mitochondrial CoQ concentrations were unaffected by cytosolic expression of SAM synthetase (SAM1) whereas ectopic expression of SAM1 in mitochondria increased mitochondrial CoQ. These results demonstrate that it is mitochondrial rather than ‘cellular’ SAM and SAH per se, that are the key in modulating mitochondrial CoQ content, and more generally highlight the importance of measuring metabolites at subcellular resolution.
Several lines of evidence support the conclusion that increasing mitochondrial CoQ via attenuated PEMT and the enhanced mitochondrial SAM-to-SAH ratio is beneficial in the context of insulin resistance. We found increasing mitochondrial CoQ via ASO-mediated PEMT knockdown resulted in maintenance of glucose uptake in adipocytes in the presence of TNFα. Increasing mitochondrial CoQ via ASO-mediated PEMT knockdown also maintained glucose uptake in the presence of 4-nitrobenzoic acid. 4-Nitrobenzoic acid is an inhibitor of COQ2, but its inhibitory action on CoQ synthesis and hence CoQ content was ameliorated by the ASO-mediated PEMT knock down. These results suggest that improved glucose uptake in PEMT-deficient cells is causally related to increased (or preserved) CoQ content. Importantly, this causal relationship extends to the in vivo situation: modulation of PEMT activity by ASO in HFD-fed mice led to maintenance of insulin sensitivity with improved glucose and insulin responses, and this was associated with a parallel increase in mitochondrial SAM-to-SAH ratio and CoQ in the livers of these mice. As mitochondrial superoxide is a known driver of insulin resistance downstream of CoQ [11,42] and is decreased in PEMT deficiency, our results also uncover a previously unrecognized causal relationship between PEMT and mitochondrial oxidant(s).
In addition to PEMT, our initial screen in conjunction with the yeast knockout collection, identified 30 previously unrecognized regulators of CoQ content in yeast. The genes identified in our study add to the set of gene mutants identified previously [45] as altering total CoQ content. There is limited overlap between the groups of gene mutants identified in each study. This is likely explained by the different growth conditions used, with cellular CoQ content known to be affected by the abundance of benzoquinone head group precursors and cellular metabolic activity. This highlights the complexity of the metabolic factors that govern CoQ synthesis and the need to consider the interaction of metabolic and genetic regulators. Similarly in our study, mutants were selected for further analyses based on their relative CoQ content compared to other mutants grown on the same 96-well plate as meaningful intra-plate comparisons were not feasible. We cannot exclude the possibility that some mutants were missed in our screen, especially those that display a change in CoQ lower than the threshold of the screen.
While we developed the quantitative screening method for yeast cells, it could be applied to most biological samples and hence aid large-scale studies, e.g., clinical studies requiring CoQ measurement. Most of the yeast genes identified have a functional mammalian homolog, raising the possibility for these genes to be additional conserved regulators of cellular CoQ content in mammalian cells. Gene ontology (GO) analyses via FunSpec [46] identified 35 GO Biological Processes as enriched in the ‘high CoQ’ mutants, none of which have previously been associated with CoQ synthesis. The biological processes identified in the ‘high CoQ’ mutant groups are in various cellular compartments in addition to mitochondria, e.g., endoplasmic reticulum, the Golgi network, and the nucleus. This suggests that several extra-mitochondrial processes regulate cellular CoQ content, further emphasizing the complex interplay between spatially distinct pathways in the regulation of cellular CoQ concentrations, as we have demonstrated in the case of PEMT deficiency. The development of techniques such as MALDI-imaging to analyze CoQ distribution in vivo [47] could aid in the study of such issues, especially in more complex tissues.
Our study raises several interesting questions, e.g., how mitochondrial concentrations of SAM and SAH are regulated, and the potential of our findings for translation to treat CoQ deficiency and related pathologies. What is clear from our studies is that changes in cytosolic SAM concentrations alone are not sufficient to increase mitochondrial SAM or the mitochondrial SAM-to-SAH ratio, and that the mitochondrial SAM-to-SAH ratio does not always reflect the whole-cell situation. While PEMT deficiency increases mitochondrial SAM-to-SAH ratio to beneficially affect mitochondrial CoQ and cell function, ablation of PEMT as a therapeutic strategy poses significant challenges due to the impact of PEMT deficiency on hepatic lipid metabolism. Pemt–/– mice fed regular chow diet display no phospholipid defects [36], most likely due to the increased activity of the rate limiting step in the Kennedy Pathway. However, Pemt–/– mice fed a high-fat diet display decreased PC (∼25%) and a decreased PC/PE ratio [25]. These mice eventually develop severe triglyceride accumulation, endoplasmic reticulum stress and non-alcoholic steatohepatitis (NASH) most likely due to a loss of membrane integrity induced by the decreased PC/PE ratio [48]. Polymorphisms in the PEMT gene area associated with non-alcoholic fatty liver disease in humans [49]. Nevertheless, understanding how PEMT deficiency leads to increased mitochondrial SAM-to-SAH ratio may ultimately lead to novel strategies to increase CoQ concentrations in vivo.
The methionine cycle is a centre piece of the methylation pathway and occurs in the cytosol. SAM must be transported into mitochondria and the SAH generated must be transported back to the cytosol for either conversion back to SAM or for cysteine/glutathione synthesis. It is likely that other factors affect how much SAM is transported into the mitochondria in PEMT deficiency. One potential factor could be the sub-cellular localization of PEMT. PEMT is localized to ER-mitochondria associated membranes (MAMs) [50], an interface for crosstalk between the ER and mitochondria that is integral in the appropriate functioning of both organelles. Therefore, it seems plausible that PEMT deficiency increases SAM locally in ER-MAM microdomains and that this facilitates SAM accessibility to CoQ biosynthetic complex and mitochondrial CoQ synthesis. In support this, FKBP8 is a regulator of the formation of the ER-mitochondria interface [51] that interacts with COQ9 [52], a protein essential for the stabilization of the putative CoQ biosynthetic complex [53]. COQ polypeptides that participate in the CoQ synthome are also highly enriched at ER-MAM sites [54,55]. These enzymes include COQ3, a SAM-dependent methyltransferase responsible for both O-methylation steps in CoQ biosynthesis. We speculate that the increased SAM-SAH-ratio observed in mitochondria of PEMT-deficient cells is driven by the spatial co-enrichment of PEMT and CoQ biosynthetic enzymes in ER-MAM microdomains. If so, this may be an example of redox control at the ER-mitochondrial interface, a sub-cellular location also important in glucose metabolism and insulin signaling [56].
Undoubtedly further studies are necessary to fully understand the complex issues raised by the present study and to harness its potential to develop novel strategies to increase mitochondrial CoQ and overcome mitochondrial oxidative stress and dysfunction resulting from CoQ deficiency. Nevertheless, our work identifies a new network of genes that regulate CoQ, opening the door for further research in this area. In addition, the identification of the intersection of phospholipid metabolism, mitochondrial one-carbon metabolism, and CoQ synthesis in PEMT deficiency, and how these interact in the setting of insulin resistance represents an exciting advance in our understanding of one-carbon metabolism, lipid biology and mitochondrial function.
Author contributions
AA and RS conceived the work; AA, DJF, SGC, IWD, JWKH, DEV, CFC, RLJ and RS designed the experiments; AA, DJF, GJM, CS, DS, KJL, MCB, LFdR, AY, JWKH, ST, SMYK, and JNV acquired or analyzed data; AA, DJF, SGC, IWD, DEV, CFC, RLJ and RS interpreted data; AA and RS drafted the work or substantially revised the text.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
We thank Dr Gustav Dallner (Karolinska Institutet, Stockholm Sweden) for helpful advice. We also thank Ionis Pharmaceuticals (Carlsbad, CA, USA) for providing ASOs. This research was supported by grants from the Australian Research Council (DP150102408 to RS, CFC, IWD), the National Health and Medical Research Council of Australia (NHMRC1052616 to RS) and the Canadian Institutes of Health Research (MOP 5182 to DEV, RLJ, JnV; and MOP 33505 to RLJ). RS was supported by NH&MRC Senior Principal Research Fellowship (1111632), AY was supported by an Australian Post-graduate Award, and SGC was supported by a grant from the National Science Foundation (MCB-1714569). We thank New South Wales Government Office for Health and Medical Research for infrastructure support. We also thank David Fuentes and Atul Bhatnagar of Sydney Mass Spectrometry for assistance and advice with the mass spectrometry analysis carried out at Sydney Mass Spectrometry, a core research facility at the University of Sydney.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102127.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bersuker K. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mao C. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593:586–590. doi: 10.1038/s41586-021-03539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoogland H. A steady-state study on the formation of Compounds II and III of myeloperoxidase. Biochim. Biophys. Acta. 1988;955:337–345. doi: 10.1016/0167-4838(88)90213-0. [DOI] [PubMed] [Google Scholar]
- 4.Anderson C.M. Dependence of brown adipose tissue function on CD36-mediated coenzyme Q uptake. Cell Rep. 2015;10:505–515. doi: 10.1016/j.celrep.2014.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alcázar-Fabra M., Rodríguez-Sánchez F., Trevisson E., Brea-Calvo G. Primary coenzyme Q deficiencies: a literature review and online platform of clinical features to uncover genotype-phenotype correlations. Free Radic. Biol. Med. 2021;167:141–180. doi: 10.1016/j.freeradbiomed.2021.02.046. [DOI] [PubMed] [Google Scholar]
- 6.Folkers K., Vadhanavikit S., Mortensen S.A. Biochemical rationale and myocardial tissue data on the effective therapy of cardiomyopathy with coenzyme Q10. Proc. Natl. Acad. Sci. U.S.A. 1985;82:901–904. doi: 10.1073/pnas.82.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas S.R., Witting P.K., Stocker A role for reduced coenzyme Q in atherosclerosis? Biofactors. 1999;9:207–224. doi: 10.1002/biof.5520090216. [DOI] [PubMed] [Google Scholar]
- 8.Letters J.M. Time-dependent changes to lipids and antioxidants in plasma and aortas of apolipoprotein E knockout mice. J. Lipid Res. 1999;40:1104–1112. [PubMed] [Google Scholar]
- 9.Witting P.K., Pettersson K., Letters J., Stocker R. Anti-atherogenic effect of coenzyme Q10 in apolipoprotein E gene knockout mice. Free Radic. Biol. Med. 2000;29:295–305. doi: 10.1016/s0891-5849(00)00311-7. [DOI] [PubMed] [Google Scholar]
- 10.Molyneux S.L. Coenzyme Q10: an independent predictor of mortality in chronic heart failure. J. Am. Coll. Cardiol. 2008;52:1435–1441. doi: 10.1016/j.jacc.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 11.Fazakerley D.J. Mitochondrial CoQ deficiency is a common driver of mitochondrial oxidants and insulin resistance. Elife. 2018;7 doi: 10.7554/eLife.32111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayer A., Macdonald P., Stocker R. CoQ₁₀ function and role in heart failure and ischemic heart disease. Annu. Rev. Nutr. 2015;35:175–213. doi: 10.1146/annurev-nutr-071714-034258. [DOI] [PubMed] [Google Scholar]
- 13.Awad A.M. Chromatin-remodeling SWI/SNF complex regulates coenzyme Q(6) synthesis and a metabolic shift to respiration in yeast. J. Biol. Chem. 2017;292:14851–14866. doi: 10.1074/jbc.M117.798397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin-Montalvo A. The phosphatase Ptc7 induces coenzyme Q biosynthesis by activating the hydroxylase Coq7 in yeast. J. Biol. Chem. 2013;288:28126–28137. doi: 10.1074/jbc.M113.474494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veling M.T. Multi-omic mitoprotease profiling defines a role for Oct1p in coenzyme Q production. Mol. Cell. 2017;68:970–977 e911. doi: 10.1016/j.molcel.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gay C.A., Stocker R. Simultaneous determination of coenzyme Q10, cholesterol, and major cholesterylesters in human blood plasma. Methods Enzymol. 2004;378:162–169. doi: 10.1016/S0076-6879(04)78013-1. [DOI] [PubMed] [Google Scholar]
- 17.Meisinger C., Pfanner N., Truscott K.N. Isolation of yeast mitochondria. Methods Mol. Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- 18.Noga A.A., Zhao Y., Vance D.E. An unexpected requirement for phosphatidylethanolamine N-methyltransferase in the secretion of very low density lipoproteins. J. Biol. Chem. 2002;277:42358–42365. doi: 10.1074/jbc.M204542200. [DOI] [PubMed] [Google Scholar]
- 19.Daniel J.M., Friess S.D., Rajagopalan S., Wedt S., Zenobi R. Quantitative determination of noncovalent binding interactions using soft ionization mass spectrometry. Int. J. Mass Spectrom. 2002;216:1–27. [Google Scholar]
- 20.Wan S. Hepatic PEMT activity mediates liver health, weight gain, and insulin resistance. FASEB. J. 2019;33:10986–10995. doi: 10.1096/fj.201900679R. [DOI] [PubMed] [Google Scholar]
- 21.Winzeler E.A. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 22.Payet L.A. Mechanistic details of early steps in coenzyme Q biosynthesis pathway in yeast. Cell. Chem. Biol. 2016;23:1241–1250. doi: 10.1016/j.chembiol.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y. Lack of phosphatidylethanolamine N-methyltransferase alters plasma VLDL phospholipids and attenuates atherosclerosis in mice. Arterioscler. Thromb. Vasc. Biol. 2009;29:1349–1355. doi: 10.1161/ATVBAHA.109.188672. [DOI] [PubMed] [Google Scholar]
- 24.Cole L.K., Dolinsky V.W., Dyck J.R., Vance D.E. Impaired phosphatidylcholine biosynthesis reduces atherosclerosis and prevents lipotoxic cardiac dysfunction in ApoE-/- mice. Circ. Res. 2011;108:686–694. doi: 10.1161/CIRCRESAHA.110.238691. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs R.L. Impaired de novo choline synthesis explains why phosphatidylethanolamine N-methyltransferase-deficient mice are protected from diet-induced obesity. J. Biol. Chem. 2010;285:22403–22413. doi: 10.1074/jbc.M110.108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanipes M.I., Henry S.A. The phospholipid methyltransferases in yeast. Biochim. Biophys. Acta. 1997;1348:134–141. doi: 10.1016/s0005-2760(97)00121-5. [DOI] [PubMed] [Google Scholar]
- 27.Vance D.E. Phospholipid methylation in mammals: from biochemistry to physiological function. Biochim. Biophys. Acta. 2014;1838:1477–1487. doi: 10.1016/j.bbamem.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 28.Summers E.F., Letts V.A., McGraw P., Henry S.A. Saccharomyces cerevisiaecho2 mutants are deficient in phospholipid methylation and cross-pathway regulation of inositol synthesis. Genetics. 1988;120:909–922. doi: 10.1093/genetics/120.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thibault G. The membrane stress response buffers lethal effects of lipid disequilibrium by reprogramming the protein homeostasis network. Mol. Cell. 2012;48:16–27. doi: 10.1016/j.molcel.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Awad A.M. Coenzyme Q10 deficiencies: pathways in yeast and humans. Essays Biochem. 2018;62:361–376. doi: 10.1042/EBC20170106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He C.H., Xie L.X., Allan C.M., Tran U.C., Clarke C.F. Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim. Biophys. Acta. 2014;1841:630–644. doi: 10.1016/j.bbalip.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye C., Sutter B.M., Wang Y., Kuang Z., Tu B.P. A metabolic function for phospholipid and histone methylation. Mol. Cell. 2017;66:180–193.E8. doi: 10.1016/j.molcel.2017.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vance D.E., Walkey C.J., Agellon L.B. Why has phosphatidylethanolamine N-methyltransferase survived in evolution? Biochem. Soc. Trans. 1998;26:337–340. doi: 10.1042/bst0260337. [DOI] [PubMed] [Google Scholar]
- 34.Presa N. Vitamin E alleviates non-alcoholic fatty liver disease in phosphatidylethanolamine N-methyltransferase deficient mice. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2019;1865:14–25. doi: 10.1016/j.bbadis.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Pritchard P.H., Chiang P.K., Cantoni G.L., Vance D.E. Inhibition of phosphatidylethanolamine N-methylation by 3-deazaadenosine stimulates the synthesis of phosphatidylcholine via the CDP-choline pathway. J. Biol. Chem. 1982;257:6362–6367. [PubMed] [Google Scholar]
- 36.Walkey C.J., Donohue L.R., Bronson R., Agellon L.B., Vance D.E. Disruption of the murine gene encoding phosphatidylethanolamine N-methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12880–12885. doi: 10.1073/pnas.94.24.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hörl G. Sequential synthesis and methylation of phosphatidylethanolamine promote lipid droplet biosynthesis and stability in tissue culture and in vivo. J. Biol. Chem. 2011;286:17338–17350. doi: 10.1074/jbc.M111.234534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stead L.M., Brosnan J.T., Brosnan M.E., Vance D.E., Jacobs R.L. Is it time to reevaluate methyl balance in humans? Am. J. Clin. Nutr. 2006;83:5–10. doi: 10.1093/ajcn/83.1.5. [DOI] [PubMed] [Google Scholar]
- 39.Mudd S.H. Methyl balance and transmethylation fluxes in humans. Am. J. Clin. Nutr. 2007;85:19–25. doi: 10.1093/ajcn/85.1.19. [DOI] [PubMed] [Google Scholar]
- 40.Fu S. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoehn K.L. Insulin resistance is a cellular antioxidant defense mechanism. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17787–17792. doi: 10.1073/pnas.0902380106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fazakerley D.J. Mitochondrial oxidative stress causes insulin resistance without disrupting oxidative phosphorylation. J. Biol. Chem. 2018;293:7315–7328. doi: 10.1074/jbc.RA117.001254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoehn K.L. IRS1-independent defects define major nodes of insulin resistance. Cell Metab. 2008;7:421–433. doi: 10.1016/j.cmet.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forsman U., Sjöberg M., Turunen M., Sindelar P.J. 4-Nitrobenzoate inhibits coenzyme Q biosynthesis in mammalian cell cultures. Nat. Chem. Biol. 2010;6:515–517. doi: 10.1038/nchembio.372. [DOI] [PubMed] [Google Scholar]
- 45.Stefely J.A. Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat. Biotechnol. 2016;34:1191–1197. doi: 10.1038/nbt.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson M.D., Grigull J., Mohammad N., Hughes T.R. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinf. 2002;3:35. doi: 10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tatsuta Y. Imaging mass spectrometry analysis of ubiquinol localization in the mouse brain following short-term administration. Sci. Rep. 2017;7:12990. doi: 10.1038/s41598-017-13257-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Veen J.N. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta Biomembr. 2017;1859:1558–1572. doi: 10.1016/j.bbamem.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 49.Song J. Polymorphism of the PEMT gene and susceptibility to nonalcoholic fatty liver disease (NAFLD) FASEB J. 2005;19:1266–1271. doi: 10.1096/fj.04-3580com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vance J.E. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 1990;265:7248–7256. [PubMed] [Google Scholar]
- 51.Kwak C. Contact-ID, a tool for profiling organelle contact sites, reveals regulatory proteins of mitochondrial-associated membrane formation. Proc. Natl. Acad. Sci. U.S.A. 2020;117:12109–12120. doi: 10.1073/pnas.1916584117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Floyd B.J. Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol. Cell. 2016;63:621–632. doi: 10.1016/j.molcel.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lohman D.C. Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E4697–E4705. doi: 10.1073/pnas.1413128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eisenberg-Bord M. The endoplasmic reticulum-mitochondria encounter structure complex coordinates coenzyme Q biosynthesis. Contact. 2019;2 doi: 10.1177/2515256418825409. 2515256418825409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramanian K. Coenzyme Q biosynthetic proteins assemble in a substrate-dependent manner into domains at ER-mitochondria contacts. J. Cell Biol. 2019;218:1353–1369. doi: 10.1083/jcb.201808044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tubbs E. Disruption of mitochondria-associated endoplasmic reticulum membrane (MAM) integrity contributes to muscle insulin resistance in mice and humans. Diabetes. 2018;67:636–650. doi: 10.2337/db17-0316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.