Abstract
Background
Immune cells play a major role in the pathogenesis of COPD. Changes in the distribution and cellular functions of major immune cells, such as alveolar macrophages (AMs) and neutrophils are well known; however, their transcriptional reprogramming and contribution to the pathophysiology of COPD are still not fully understood.
Method
To determine changes in transcriptional reprogramming and lipid metabolism in the major immune cell type within bronchoalveolar lavage fluid, we analysed whole transcriptomes and lipidomes of sorted CD45+Lin−HLA-DR+CD66b−Autofluorescencehi AMs from controls and COPD patients.
Results
We observed global transcriptional reprogramming featuring a spectrum of activation states, including pro- and anti-inflammatory signatures. We further detected significant changes between COPD patients and controls in genes involved in lipid metabolism, such as fatty acid biosynthesis in GOLD2 patients. Based on these findings, assessment of a total of 202 lipid species in sorted AMs revealed changes of cholesteryl esters, monoacylglycerols and phospholipids in a disease grade-dependent manner.
Conclusions
Transcriptome and lipidome profiling of COPD AMs revealed GOLD grade-dependent changes, such as in cholesterol metabolism and interferon-α and γ responses.
Short abstract
AMs from COPD patients undergo GOLD grade-specific transcriptional reprogramming and acquire a complex activation profile. Among the observed changes are gene programmes involved in lipid metabolism that translate into alterations in the AM lipidome. https://bit.ly/3sYAqgd
Introduction
COPD is characterised by progressive airflow obstruction, inflammation in the airways and systemic comorbidities, such as cardiovascular diseases and diabetes [1]. It is a leading cause of morbidity and mortality worldwide [2], and induces a substantial and increasing economic and social burden [3, 4]. Current guidelines for COPD patient classification use the Global Initiative for Chronic Obstructive Lung Disease (GOLD) criteria on the basis of spirometry-estimated severity of airflow limitation and range from mild (GOLD grade 1) to very severe (GOLD grade 4) disease [5]. Cigarette smoking is the leading environmental risk factor for COPD, yet even for heavy smokers, fewer than 50% develop COPD during their lifetime, indicating that the disease initiates from a complex interaction between environment and genome. Some genetic factors, e.g. deficiency of α-1 antitrypsin [6] or single-nucleotide polymorphisms in MMP12 [7] have been associated with the risk to develop COPD.
Innate immune cells are key players in protecting the lung from airway infections and their impairment plays a major role in COPD pathogenesis and exacerbation episodes [1, 8, 9]. Although changes in cellular distribution and functions within the alveolar space have been described in COPD before [10], these findings are far from being complete. Furthermore, the advent of omics technologies even extending to metabolomics [11], as well as a better understanding of cell type classification requires revisiting these aspects of innate immune cell biology of the alveolar space, both in healthy individuals and COPD patients. Alveolar macrophages (AMs) are described to be the most abundant cells in humans [12]. Under physiological conditions, AMs are the major cell type on the luminal surface of the alveolar space and are major regulators of the initiation and resolution of inflammation [13, 14] and surfactant metabolism [15]. AMs catabolise pulmonary surfactants, which are composed of phospholipids and proteins and humans lacking functional AMs develop pulmonary proteinosis [16].
Recent reports have shown that in respiratory diseases, such as asthma, COPD and pneumonia a lipidomic remodelling of the bronchoalveolar lavage fluid (BALF) is observed, with changes in the levels of cholesterol, sphingomyelins, phospholipids and fatty acids being the most prominent [17–19]. In particular, the role of sphingolipids and cholesterol metabolism are believed to play a role in the pathogenesis of COPD [20, 21]. Nevertheless, changes of the lipid metabolism within AMs in COPD patients have not been addressed so far.
In this study, we hypothesised that transcriptional changes leading to altered lipid metabolism occur in a disease grade-dependent manner. Using multi-colour flow cytometry (MCFC), we defined AMs as the major cell type in COPD patients for which we provide clear evidence of global transcriptional reprogramming, which differed between GOLD grade 2 and GOLD grade 3/4 COPD patients. Among the major alterations in the AM transcriptome of COPD patients, we identified genes associated with lipid metabolism, which was linked to significant changes in several lipid species by lipidomics analysis.
Methods
Human specimens
Human studies were approved by the ethics committees of the University of Bonn and University Hospital Bonn (local ethics vote 076/16). All patients provided written informed consent according to the Declaration of Helsinki before specimens were collected. Patients with COPD were diagnosed and stratified according to the guidelines of GOLD [5]. Eligible patients were aged 18 years or older and were either current, past or nonsmokers (table E1). Age-matched individuals suffering from chronic idiopathic cough, demonstrating an exquisitely sensitive cough reflex without underlying pathology [22], served as control donors. Patients with other pulmonary diseases (termed “other” (figure 1a)) were diagnosed as asthma, asthma–COPD overlap, bronchiectasis, cancer, fibrosis, pneumonia and sarcoidosis (table E2), but were excluded from further evaluation within this study.
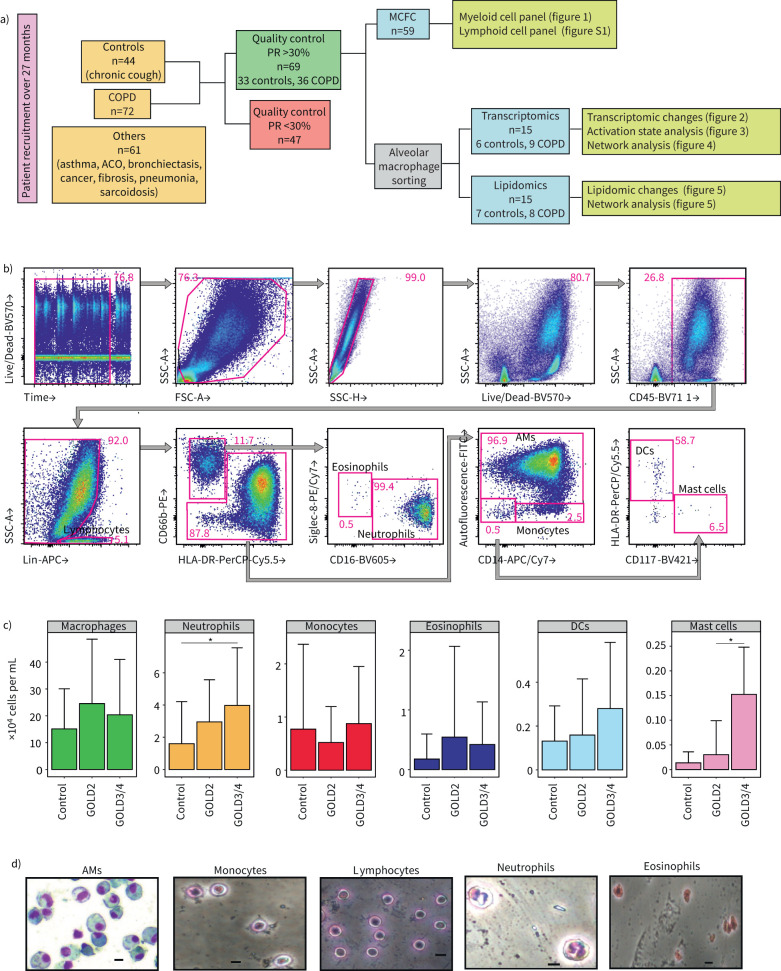
FIGURE 1.
Identification of myeloid immune populations in the bronchoalveolar lavage fluid (BALF) of COPD and control patients. Schema of the pipeline for collection and processing of human BALF samples. Of 177 BALF samples, 69 satisfied the quality criteria (>30% recovery rate and no blood and/or mucus contamination) and were included in the study for multiple colour flow cytometry, transcriptomics and/or lipidomics. a) 61 patients suffering from asthma, asthma–COPD overlap (ACO), bronchiectasis, cancer, fibrosis, pneumonia or sarcoidosis were excluded from the study. b) Representative multicolour flow cytometry (MCFC) analysis of the myeloid compartment of a control patient. c) Absolute numbers of myeloid immune cells in the BALF of control, Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage 2 and GOLD3/4 COPD patients calculated with traditional gating. Data are from 8–29 patients per group except for mast cells GOLD3/4 (n=2) and are represented as mean±sd. Data are represented as mean±sem and were analysed with an unpaired two-tailed t-test. d) Giemsa staining of cytospins for sorted BALF myeloid cell types. RR: recovery rate; AM: alveolar macrophage; DC: dendritic cell; . Scale bars=10 µm. *: p<0.05.
Bronchoscopy procedure
Bronchoscopy was performed as a part of the diagnostic workup by two bronchoscopists through oral access and with light conscious sedation and was performed in the middle lobe or, if not accessible, the lingular lobe. Warmed saline (six syringes of 20 mL each) was instilled into the airways to enable BALF recovery.
BALF processing
Human BALF was obtained from all patients included in the study (control, COPD, other) through bronchoscopy. BALF specimens were washed with PBS, suspended with 0.02% EDTA-2Na and washed again for final re-suspension with 2% fetal calf serum (FCS)/1 mM EDTA. CD45+Lin−HLA-DR+CD66−Autofluorescence+ AMs were sorted using a FACS Aria III cell sorter (BD Biosciences, USA).
Cell counting
Total cell counts were determined with (1:5) Trypan Blue exclusion (Sigma-Aldrich) under an optical microscope. BALF cells were diluted 1:10 in the Trypan Blue solution and counted in a Neubauer haemocytometer.
Flow cytometry/FACS
Single-cell suspensions were stained with Live/Dead yellow fluorescent dye (Thermo Fisher Scientific, USA) for 15 min at room temperature and were washed with PBS at 300×g for 5 min at 4°C. They were then re-suspended in 100 µL PBS and blocked with 5 µL human FcR blocking reagent (Miltenyi, Germany) for 15 min on ice and were subsequently stained with the listed anti-human antibodies (table E4) in buffer containing PBS, 2% FCS, 1 mM EDTA for 30 min on ice. Cells were spun at 300×g for 5 min at 4 °C and re-suspended in buffer containing PBS, 2% FCS, 1 mM EDTA for analysis. Data acquisition was performed on a FACS Aria III cell sorter. Analysis was performed with FlowJo v.10 software (Tree Star, USA).
Cytospin preparation
Cytospins were obtained by centrifuging 2×105 cells in 200 µL PBS on microscope slides at 20% power for 5 min. Excess buffer was carefully discarded and slides were air dried for 3 min followed by fixation with 100% methanol for 5 min at 4°C. The slides were subsequently washed with PBS and stained with 1:20 Giemsa solution (Sigma, USA) for 25 min at room temperature. A final rinsing step with H2O and air drying before mounting followed. Cell morphology was examined by microscopic evaluation of stained cells using an Axio Lab A1 microscope (Zeiss, Germany).
RNA extraction and library preparation
Total RNA was isolated from human AMs with the miRNeasy Micro kit (QIAGEN, Germany) according to the manufacturer's protocol. cDNA libraries were prepared from 5 ng total RNA with the SMART-seq2 protocol [23] and were tagmented with the Nextera XT kit (Illumina, USA). Library size selection was carried out with AMPure beads (Beckman-Coulter, USA). Libraries were sequenced for SR 75 cycles on a NextSeq500 system (Illumina) using High Output v2 chemistry. Base call files were converted to fastq format and demultiplexed using bcl2fastq v.2.20.
Data pre-processing and RNA-sequencing analysis
The 75 bp single-end reads were aligned to the human reference transcriptome hg38 from UCSC by kallisto v.0.44.0 using default parameters. Data were imported into DESeq2 (v.1.10.1; [24]) using the TXimport (v.1.2.0, [25]) package. DESeq2 was used for the calculation of normalised counts for each transcript using default parameters. All normalised transcripts with a maximum over all group means <10 were excluded resulting in 33 032 present transcripts. Unwanted or hidden sources of variation, such as batch, sex and smoking status were removed using the sva package [26]. The normalised rlog-transformed expression values were adjusted according to the five surrogate variables identified by sva using the function removeBatchEffect from the limma package [27]. Differentially expressed (DE) genes were defined by a p-value cut-off of 0.05 and an adjusted p-value (independent hypothesis weighting) <0.5.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) [28] was performed on all present genes of the dataset using the gene ontology set of biological processes. Information of gene ontology was obtained from the biological process gene set “c5.bp.v7.0.symbols.gmt”, downloaded from the Molecular Signatures Database (MSigDB). All present genes were used as background (universe).
Linear support vector regression
Linear support vector regression [29] was employed to characterise the relative contribution of 28 different activation signatures derived from Xue et al. [30] to the control and COPD patients (1000 permutations).
Filtering for transcription factors, epigenome, surfaceome and secretome
All present transcripts were filtered and sorted by their variance in the dataset. The 20 most variable genes of each category were selected and visualised in heat maps. Transcription factor (TF) lists were extracted from Fulton et al. [31], the epigenome gene list was derived from the literature, surface and secretome markers were extracted from the Human Protein Atlas [32, 33].
Construction of coexpressed network analysis: automated
Coexpressed network analysis (CoCena2) was performed to elucidate similarities and differences within the gene expression patterns of the three different patient groups. Pearson correlation was calculated on the 6000 most variable genes within the dataset using the R package Hmisc (v.4.3-0; [34]). Data were filtered for significant (p-value <0.05, Bonferroni correction p<0.05) and positive (r-value >0) correlation values. The group fold change (GFC) was calculated for each gene and each condition on the inverse logarithmic count data using the R package gtools (v.3.8.2; Unbiased clustering was performed using the R package igraph (v.1.2.4.1; [35]). The clustering algorithm “cluster_louvain” was selected as it achieves the highest modularity score. Clusters with fewer than 35 genes are not shown. Network generation was performed with the R package igraph. The network information was imported to and exported from Cytoscape using the R package RCy3 (v.2.6.2; [36]).
Biological function-related bioinformatic analysis of network modules
GSEA was performed on the patient group-related modules identified by CoCena2 using the R package ClusterProfiler (v.3.12.0; [37]). Information of hallmark genes was obtained from the hallmark gene set “h.all.v6.1.symbols.gmt”, downloaded from MSigDB. All genes present in the network were used as background (universe).
Lipidomics
Sorted AMs (5×104) pelleted and frozen at −80°C until analysis. Extraction mix (chloroform 1:5 methanol-containing internal standards) was spiked to the pellets before sonication. Samples were treated with chloroform and 1% acetic acid, the lower phase was transferred after centrifugation, and let evaporate in the vacuum concentrator (45°C for 10 min). After addition of spray buffer and sonication, samples were analysed separately with a Thermo Q Exactive Plus spectrometer equipped with the HESI II ion source for shotgun lipidomics.
Lipidomics analysis
LipidXplorer software was used for analysis using custom mfql files to identify sample lipids and internal standards. Absolute amounts were calculated using the internal standard intensities followed by normalisation on the sum of all measured lipid species per sample. %mol values were averaged for each patient group, log2-transformed and then used for fold change calculations. CoCena2 was used to find patient group-specific coregulated lipid species using Pearson correlation and clustering using the “cluster_louvain” algorithm.
Statistics
A two-tailed Welch's unpaired t-test was used to analyse data from two groups. Equality of population variance was assessed with the F-test statistic for two independent groups. A nonparametric Wilcoxon test was used to perform a pairwise comparison between patient groups for all enriched macrophage activation signatures. For more than two groups, normality and homoscedasticity were first assessed using the Shapiro–Wilk and Levene tests in R (v.3.6.1). A nonparametric Kruskal–Wallis test with Dunn's multiple correction post hoc was used in figure E3 because the data did not follow a normal distribution. Statistical significance was inferred when p<0.05.
For a full list of the methods, see the supplementary material.
Results
Macrophages are the most abundant cell type in the alveolar space in COPD
We first evaluated the cell type distribution in the BALF to ensure that AMs are the most abundant cell type in control and COPD samples, the latter of which an enrichment of neutrophils had been postulated [38]. We performed MCFC integrating markers to determine all major immune cell types expected within the alveolar space [39]. Individuals with chronic coughing served as controls (table E1). Only BALF samples of the highest quality following quality criteria established previously for the processing of BALF [40], such as recovery rate >30% and absence or minimal blood/mucus contamination were included in the study (figure 1a). Out of 177 screened BALF samples in the clinic, 72 were derived from COPD patients, 44 from control individuals, while patients suffering from asthma, ACO, bronchiectasis, cancer, fibrosis, pneumonia or sarcoidosis were excluded (table E2). 69 patients (36 COPD and 33 controls) finally qualified for further analysis (table E3).
We stained for markers of the myeloid cell compartment (figure 1b and table E5), whereas lymphocytes were labelled with antibodies against CD3, CD19 and CD56 (figure 1b). Our data show that AM numbers undergo a nonsignificant increase in COPD patients, whereas we could detect a statistically significant upregulation (p<0.05) in the neutrophil numbers of GOLD3/4 grade patients compared to controls (figure 1c). Eosinophil and monocyte numbers varied widely within groups and reached no significance, whereas mast cells were found to be significantly higher (p<0.05) in GOLD3/4 compared to GOLD2 COPD patients (figure 1c).
Lymphoid cells were of much lower frequency and using a second panel for lymphoid cell markers (figure S1a and table E5), we observed that only innate lymphoid cells (ILCs) were statistically significantly (p<0.05) changed in all COPD samples (figure S1b and c), with a similar trend when taking GOLD grades into account (figure S1b). Within ILCs, ILC1s were the only significantly increased (p<0.05) ILC population in COPD patients (figure S1d). Cell type classification was also confirmed on sorted cells by Giemsa staining of cytospins (figure 1d). Collectively, AMs are the most abundant cell type in BALF, which was then our focus for subsequent transcriptome and lipidome analyses.
Transcriptional changes in AMs within the alveolar space of COPD patients
To elucidate molecular changes of AMs as the most prevalent cells within the alveolar space, we sorted CD45+Lin−HLA-DR+CD66b−Autofluoreschencehi AMs from the BALF of nine COPD and six control patients and generated whole transcriptomes by RNA sequencing (figure 2a). Principal-component analysis (PCA) of all present genes (figure 2b) and hierarchical clustering of the 25% most variable genes within the dataset (figure S2a) revealed a clear disease grade-dependent structure within the data suggesting significant differences between COPD GOLD2, GOLD3/4 and control samples.
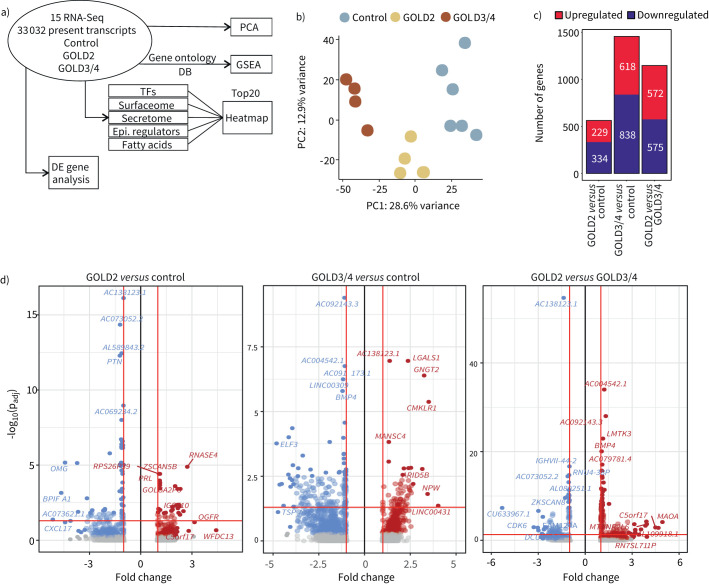
FIGURE 2.
Whole transcriptome analysis of alveolar macrophages (AMs) reveals lipidomic dysregulation in COPD patients. a) Schematic representation of the bioinformatics workflow and methods used for the transcriptome analysis of 15 RNA sequencing (RNA-Seq) samples from Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage 2, GOLD3/4 COPD and control patients. b) Principal component (PC) analysis of all 33 032 present transcripts. c) Distribution and amount of differentially expressed (DE) genes. d) Volcano plots of the comparisons GOLD2 versus control, GOLD3/4 versus control and GOLD2 versus GOLD3/4. Upregulated genes are marked in red, downregulated genes in blue. Fold change cut-off was set to 1.5, adjusted p-value cut-off was set to 0.05. TF: transcription factor; Epi.: epigenetic; GSEA: gene set enrichment analysis.
Next, we calculated DE genes between GOLD2 or GOLD3/4 COPD and control patients, respectively. There were 563 DE genes (229 up, 334 down, p<0.05, fold change (FC) >1.5) between GOLD2 and control, 1456 (618 up, 838 down, p<0.05, FC>1.5) between GOLD3/4 and control and 1147 (572 up, 575 down, p<0.05, FC>1.5) between GOLD2 and GOLD3/4 COPD patients (figure 2c), which we visualised in Volcano plots for all pairwise comparisons (figure 2d).
We first assessed overall transcriptional changes. We provide information for the top 20 altered TFs, epigenetic modulators, cell surface molecules (surfaceome) and soluble mediators as micro-environmental interactors (figure S2b–e). Upregulated TFs included ETS2 in COPD patients consistent with its function as a biomarker for loss of lung function [41] (figure S2b). Consistent with the reported dysregulation of histone deacetylase (HDAC) and sirtuin epigenetic signalling in AMs from COPD patients [42–45], the expression of histone-modifying enzymes HDAC9 and SETD1A was downregulated in GOLD3/4 COPD (Figure S2c). Regarding the surfaceome of the AMs, we measured decreased antigen presentation molecules (HLA-DQA2 and CD1B) [46, 47] and CCR2 expression in GOLD3/4 COPD patients (figure S2d). Notably, expression of the ligand chemokine to CCR2, CCL2, was elevated in the secretome-associated gene set of GOLD3/4 COPD patients, similar to other members of the CC family of chemokines, including CCL8 and CCL20 (figure S2d), highlighting the role of chemokines in COPD pathophysiology [48]. Finally, we detected the downregulation of MMP7 and MMP9 in GOLD3/4 COPD patients, with MMP12 being expressed significantly higher only in GOLD2 COPD patients (figure S2e), in agreement with the role of metalloproteinases in the progression of COPD [49–52].
Activation state analysis indicates changes in lipid metabolism
We have previously reported that macrophage activation follows a multi-dimensional model of cell activation [30]. However, for AMs in COPD, it has been proposed that AMs in COPD follow a dual polarisation model with deviation towards an anti-inflammatory phenotype [47, 49, 53, 54] with no hints on alterations in lipid metabolism. To analyse how the observed transcriptional changes in COPD AMs related to our multi-dimensional model of activation states, we performed linear support vector regression utilising our previous human macrophages activation spectrum model based on 29 conditions [30] (figure 3a). Our activation spectrum model could be grouped into nine major activation programs (c1–9) and were used here as the bait to deconvolute the AM transcriptomes from COPD and control patients. The COPD and control transcriptomes were enriched in a spectrum of activation signatures ranging from pro-inflammatory to anti-inflammatory, as well as those extracted from fatty acid-stimulated macrophages (c5 module signature), which show deviations in lipid metabolism. AMs from GOLD2 COPD patients exhibited significantly lower enrichment (p<0.05) of the c4 module signature (P3C/PGE2, P3C and PGE2 stimulation) compared to GOLD3/4 COPD and control patients and c8 module signatures (TNF-α/P3C, TNF-α and P3C stimulation) in comparison with GOLD3/4 COPD patients (figure 3a). In addition, COPD GOLD2 AMs showed a higher enrichment (p<0.05) of the c5 module signatures (lauric acid (LA), linoleic acid (LiA), oleic acid (OA), palmitic acid (PA) and stearic acid (SA) stimulation) compared to AMs from control patients (figure 3a).
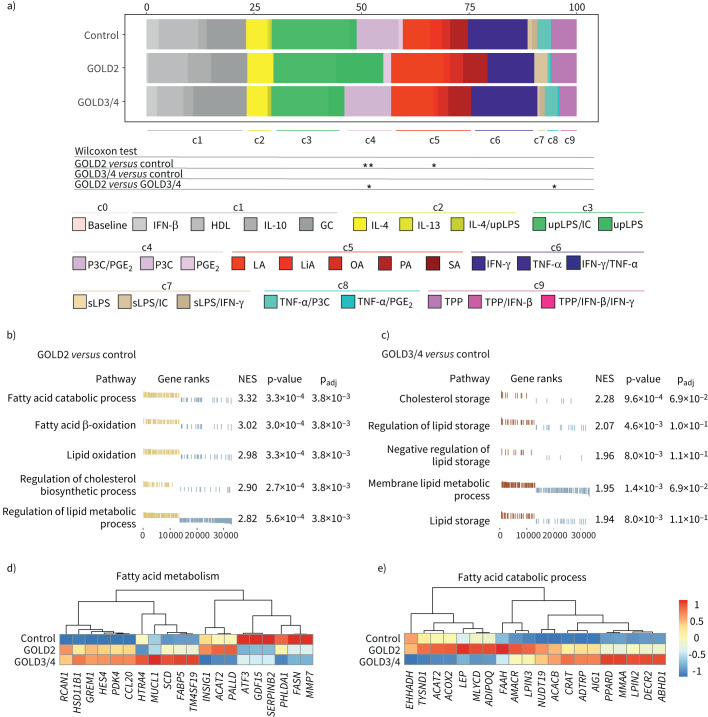
FIGURE 3.
Overlay of human macrophage activation states in alveolar macrophages (AMs) from COPD patients show lipid term enrichment. Overall, 29 macrophage activation signatures from [30] were used as input to identify the relative fraction of these activation signatures in COPD and control patients. Grouping of activation signatures in nine clusters was used as proposed in [30]. a) Data were statistically analysed with the nonparametric Wilcoxon test. Gene set enrichment analysis (GSEA) using the gene ontology (GO) database filtered for “fat” and “lipid” as reference signatures. b) Normalised enrichment scores (NES) and enrichment p-values for the five most enriched gene ontology (GO) terms for the comparison Global Initiative for Chronic Obstructive Lung Disease (GOLD)2 COPD versus control and c) GOLD3/4 COPD versus control patients. Heat map of the mean expression of the 20 most variable genes filtered by d) the GO term fatty acid metabolism and e) fatty acid catabolic process. IFN: interferon; HDL: high-density lipoprotein; IL: interleukin; GC: glucocorticoid; upLPS: ultra pure LPS; IC: poly(I:C); P3C: Pam3CSK4; PG: prostaglandin; LA: lauric acid; LiA: linoleic acid; OA: oleic acid; PA: palmitic acid; SA: stearic acid; TNF: tumour necrosis factor; sLPS: soluble LPS; TPP: TFNF/PGE2/P3CXXX;. *: p<0.05; **: p<0.01.
Based on these results indicating potential changes in lipid metabolism-associated-genes, we next performed GSEA for either GOLD2 or GOLD3/4 COPD patients compared to controls (figure 3b and c). Lipid-related gene sets, such as fatty acid catabolic process, fatty acid oxidation and regulation of cholesterol biosynthetic process were enriched in AMs from GOLD2 COPD patients (figure 3b). AMs from GOLD3/4 COPD patients presented with enrichment in cholesterol and lipid storage compared to controls (figure 3c).
To highlight the genes that are involved in the lipid-associated phenotype of COPD AMs, we visualised the 20 most variable genes for the gene ontology term “fatty acid metabolism” of the c5 module from figure 3a (figure 3d) and “fatty acid catabolic process” from figure 3b (figure 3e). Several genes that are involved in cholesterol homeostasis were dysregulated in COPD patients. For example, enzymes involved in fatty acid metabolism, such as ACAT2, FAAH, MLCYD and ACOX2 or enzymes that synthesise diacylglycerols (LPIN3) were expressed higher in GOLD2 COPD patients. On the contrary, the acyltransferase CRAT and the triglyceride synthesis enzyme LPIN2 were overexpressed in GOLD3/4 COPD patients. Fatty acid synthase FASN and the anti-inflammatory TF ATF3 [55, 56] were both downregulated in COPD patients irrespective of GOLD grade (figure 3d and e). These findings strongly support that AMs do not follow a simple polarisation model, but rather display a multi-activation phenotype with a shift towards lipid activation signatures.
Coexpression analysis reveals changes in cholesterol homeostasis in COPD
Coexpression network analysis is an alternative to statistical methods describing changes in transcriptome data, such as the calculation of DE genes based on a set fold change and p-value [57]. To further investigate active gene programs in AMs, we performed gene coexpression analysis using CoCena2 (figure 4a). The clustering of the genes within the constructed network led to the identification of groups of transcriptionally similarly regulated genes within the dataset. The mean GFC of these gene modules in GOLD2, GOLD3/4 COPD and control patients were visualised in a heat map (figure 4b). We detected 11 modules, three of which demonstrated GOLD grade-associated average expression; for instance, modules 7 and 8 had a higher GFC in GOLD2 COPD patients, modules 3 and 4 were GOLD3/4-related, while modules 9–11 had a higher GFC in AMs from control patients.
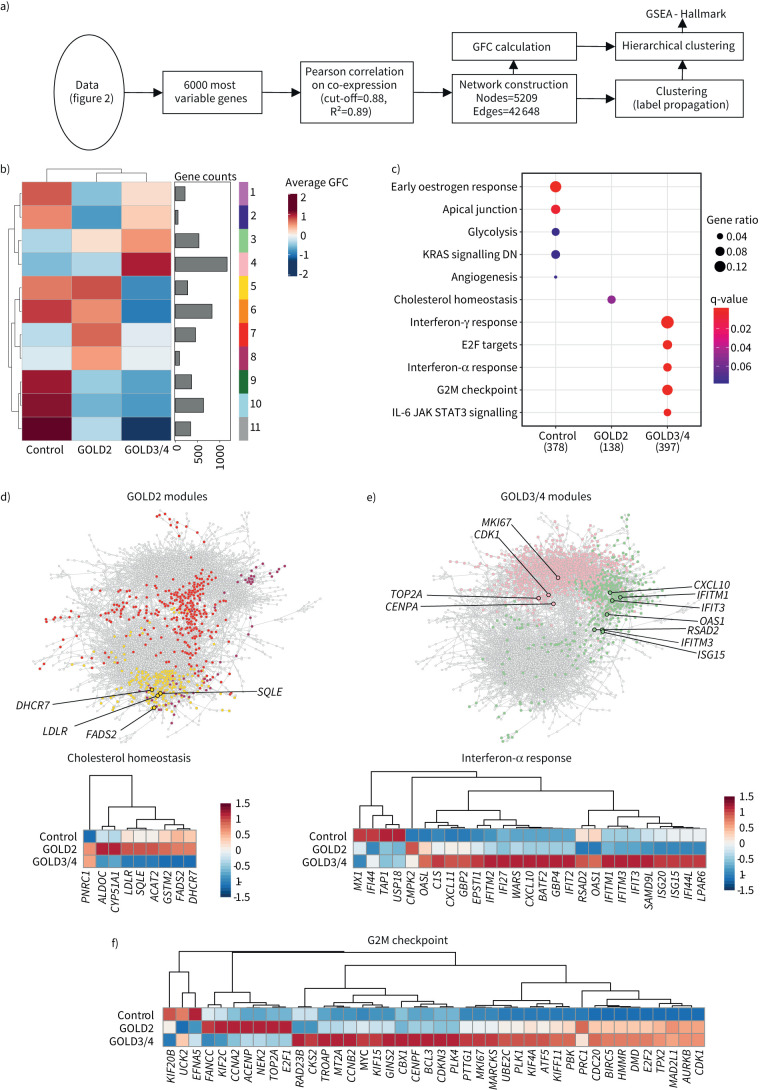
FIGURE 4.
Coexpression network analysis uncovers lipid metabolism-associated functions for alveolar macrophages (AMs) in COPD. a) Schematic representation of the bioinformatics workflow for coexpression network analysis of the 6000 most variable genes in the dataset by CoCena2. b) CoCena2 cluster-condition heat map. c) Significant enrichment (q-value <0.1) of hallmark genes within patient groups. Patient group modules consist of the respective cluster-condition heatmap modules. d) CoCena2 network with Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage 2-associated genes within the hallmark term “Cholesterol homeostasis” marked by black edging and coloured according to their cluster name. Each node represents one gene in the network and each edge represents a coexpression. The expression levels are presented in more detail in the following heat map. e) CoCena2 network with GOLD3/4-associated genes within the hallmark term “interferon-α response” marked by black edging and coloured according to their cluster name. Each node represents one gene in the network and each edge represents a coexpression. The expression levels are presented in more detail in the following heat map. f) Genes enriched within the hallmark term “G2M checkpoint” marked by black edging and coloured according to their cluster name. Each node represents one gene in the network and each edge represents a coexpression. The expression levels are presented in more detail in the following heat map. GFC: group fold change; GSEA: gene set enrichment analysis; DN: downregulated; IL: interleukin; JAK: Janus kinase; STAT: signal transducer and activation of transcription.
Next, GSEA was applied to concatenated modules per disease group (control: 1, 2, 6, 9, 10, 11, GOLD2: 5, 7, 8, GOLD3/4: 3, 4) to link the coexpressed genes to biological processes (figure 4c). While genes of control patients were enriched in apical junctions, genes from the modules enriched in AMs derived from GOLD2 patients suggested changes in cholesterol homeostasis in addition to the findings based on statistical analysis (figure 3). However, GSEA of modules enriched in AMs derived from GOLD3/4 patients were not revealing any enrichment of terms related to lipid metabolism, but rather terms associated with cell cycle and the secretion of pro-inflammatory cytokines.
The coexpression of genes associated with cholesterol homeostasis were not previously described to be altered in AMs of COPD patients. Therefore, we investigated the GOLD2-associated modules in more detail and identified genes involved in sterol biosynthesis (DHCR7, SQLE), cholesterol transportation to cells (LDLR) and fatty acid metabolism (FADS2) (figure 4d). In contrast, genes of the interferon response (CXCL10, IFIT3, IFITM1, IFITM3, OAS1, RSAD2, and ISG15) (figure 4e) and genes involved in the G2M checkpoint including the proliferation marker MKI67, cyclin-dependent kinases (CDK1) and DNA replication machinery (TOP2A, CENPA) (figure 4f) were highly expressed in GOLD3/4 AMs.
In summary, AMs derived from the BALF of COPD patients show numerous transcriptional changes that suggest broad reprogramming including genes involved in lipid metabolism with different functionalities that follow GOLD grade-specific patterns.
Lipidome analysis of AMs reveals GOLD-specific changes in COPD
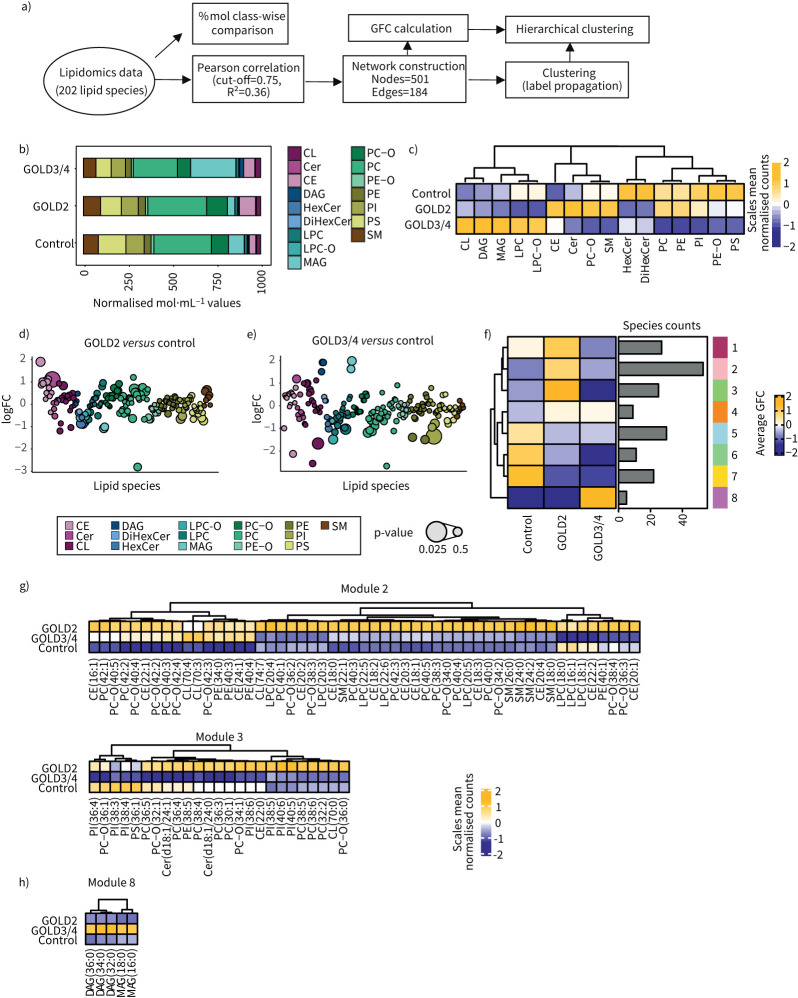
The AMs are the major cell type in the alveolar space being involved in lipid metabolism, e.g. of the surfactant [58]. Performing a transcriptome analysis of AMs derived from COPD and control patients using two different mathematical approaches identified reprogramming of lipid metabolism. To elucidate whether the changes we observed in these cells’ transcriptomes in COPD is associated with changes in their lipidome, CD45+Lin−HLA-DR+CD66b−Autofluoreschencehi AMs were sorted from the BALF of eight COPD and seven control patients and were used for mass spectrometry-based lipidomics (figure 5a). Seventeen different lipid classes including a total of 202 lipid species were quantified. Glycerophospholipids, especially the phosphatidylcholines (PCs), alongside phosphatidylethanolamines (PEs), phosphatidylinositols (PIs) and phosphatidylserines (PSs), the sphingolipids (ceramides (Cer) and sphingomyelins (SMs)) and the monoacylglycerols (MAGs) constituted the major lipid mass of AMs (figure 5b). Comparison of the AM lipidome of COPD with that of control patients indicated most prominent changes in the cholesterol ester (CE) and MAG classes (figure 5b) and quantification of the mean GFC revealed disease severity-specific changes in the analysed lipid classes (figure 5c). In accordance with the predicted change in cholesterol metabolism, AMs from GOLD2 COPD patients exhibited an upregulation of CE, but also downregulation of hexosylceramides (HexCer). In contrast, AMs from GOLD3/4 COPD patients showed an increase in MAG and diacylglycerols (DAGs) and a decrease in the PC class. GOLD grade-specific patterns were reflected as well on the single lipid species level, revealing the diverse changes within each lipid class (figure 5d and e), all in all prompting us to investigate this in greater detail.
FIGURE 5.
Quantitative lipidomics of alveolar macrophages (AMs) of COPD patients describe disease severity. a) Schematic representation of the analysis workflow for analysis of the 202 lipid species assessed by lipidomics of AMs from bronchoalveolar lavage fluid of 11 COPD and four control patients. b) Mean mol% per patient group for lipid class sums of lipid species normalised to total identified lipid content. c) Hierarchical clustering of average log2 fold change (FC) of mol% of lipid class sums. d) Average log2 FC of mol% for single lipid species per lipid class for Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage 2 versus control or e) GOLD3/4 versus control. Bubble size indicates p-value. f) CoCena2 cluster-condition heat map. g) Hierarchical clustering of the average lipid abundance for lipid species encompassed in the GOLD2-specific modules 2 and 3 and h) GOLD3/4-specific module 8. GFC: group fold change; CE: cholesteryl ester; Cer: ceramide; CL: cardiolipin; DAG: diacylglycerol; DiHexCer: dihexosylceramide; HexCer: hexosylceramide; LPC-O: ether-lysophosphatidylcholine; LPC: lysophosphatidylcholine; MAG: monoacylglycerol; PC-O: ether-phosphatidylcholine; PC: phosphatidylcholine; PE-O: ether-phosphatidylethanolamine; PE: phosphatidylethanolamine; PI: phosphatidylinositol.
For this purpose, we determined lipid-lipid level correlations using the CoCena2 pipeline. We identified nine different modules encompassing similarly regulated lipid species across the disease groups (figure 5f). Modules 1–3 showed enriched lipid species in GOLD2, whereas module 8 encompassed lipid species specifically enriched in GOLD3/4 COPD patients (figure 5f).
A closer look at the GOLD2-specific modules revealed enrichment for many unsaturated PC (e.g. PC(32:2, 36:3, 38:4, 38:5, 40:4) and CE (CE(16:1, 18:0, 18:1, 18:2, 18:3, 20:4)), as well as a few SM (e.g. SM(18:0, 22:1, 24:0,24:2)), lysophosphatidylcholines (LPCs) (e.g. LPC(20:4, 22:5, 22:6)) and Cer, such as Cer(d18:1/24:0) or (d18:1/24:1) (figure 5g). On the other hand, distinct lipid species were correlated in the module specific for GOLD3/4 AMs, which showed an accumulation of neutral lipids, such as saturated MAGs (MAG(16:0, 18:0)) and DAGs (DAG(32:0, 34:0, 36:0)) (figure 5h). Taken together, AMs from COPD patients display changes in their lipidome, which, in addition to their transcriptome, are GOLD grade-associated and could constitute a hallmark in the pathogenesis of COPD.
Discussion
Chronic inflammatory diseases such as COPD are associated with functional and transcriptional alterations of immune cells involved in the pathophysiological processes and disturbances of organ-related homeostatic functions, e.g. surfactant biology in the lung. Here we focused on the most abundant immune cell type in the alveolar space, namely AMs to determine transcriptional changes particularly in those genes associated with lipid metabolism. By defining the AM transcriptomes, we observed numerous GOLD grade-specific changes in COPD patients. Evident changes in gene expression related to lipid homeostasis were further evaluated by lipidome profiling of AMs, which demonstrated that prominent lipid changes correlate with disease severity.
We focused on AMs, the most prevalent immune cell in the alveolar space of COPD patients and investigated their transcriptomes. Previously, AMs were shown to convert to an anti-inflammatory phenotype in COPD smokers based on the simple paradigm of M1, M2 polarisation [47, 49, 54]. Additionally, a recent article found that some of those cells may carry a dual M1, M2 pattern in severe COPD [53]. However, we have previously demonstrated that macrophages follow a multi-dimensional model of activation [30]. Comparison of these various activation signatures with the transcriptome of AMs from COPD patients revealed that these cells can be better described by various activation signatures (including PGE2 (c4, c8) and lipid (c5) signatures). To further explore the overall transcriptional changes in AMs derived from COPD patients, we provide the data for further easy inspection at FASTGenomics.org. Here we focused on the changes related to lipid metabolism.
AMs maintain lung function by supporting the air–liquid interface on the alveolar surface, which, when compromised, results in augmented surface tension and pulmonary proteinosis [16]. It has been shown that lipid homeostasis in COPD is perturbed in both humans [21] and cigarette smoke-induced mouse models [59]. In addition, decreased BALF levels of cholesterol, SM and phospholipids correlate with loss in lung function in COPD patients [17]. Our analysis shows that AMs from GOLD2 COPD patients are enriched in PC and cholesterol esters, and express higher levels of lysosomal lipase LIPA, which suggests that in mild severity stages, AMs may contribute to the decrease in surfactant lipids in the alveolar space by increasing their surfactant catabolism rate. In contrast, in more severe stages, AMs are characterised by the enrichment in saturated MAGs and DAGs, which indicates a degradation defect is very likely due to the accumulation of glycerolipids.
Recently, lipidome analysis of perturbed macrophages showed that changes in lipid composition correlate with functional responses [60]. AMs exhibit a foam-like phenotype with augmented transforming growth factor-β secretion in response to silica dust and cholesterol uptake [61]. Other findings were changes in cholesterol biology. Here one could postulate that the balance between cholesterol synthesis and the liver X receptor (LXR) pathway acts as a rheostat that controls AM inflammatory responses. In GOLD2 patients, cholesterol synthesis was upregulated and LXR signalling was repressed. In contrast, in GOLD3/4 COPD patients, cholesterol synthesis was downregulated, which would be consistent with the upregulation of the LXR pathway and the induction of lipogenesis [62]. This switch could have implications in AM biology in COPD, as LXR activation confers antiviral protection [63] and inflammation resolution [64].
In addition, the observed heightened levels of CE and SM in AMs from GOLD2 COPD patients may be responsible for the dysregulation of AM functionalities, such as efferocytosis of apoptotic cells as shown before in rats exposed to cigarette smoke [65]. Dampened PI levels curtail viral infections in mice via Toll-like receptor pathway inhibition [66]. Our data provide the enriched interferon responses as additional mechanistic insight in AMs from GOLD3/4 patients.
As with every exploratory study, some of the limitations of our study need to be mentioned. First, the control cohort consisted of patients suffering from chronic idiopathic cough, which might be regarded as a potential confounder given the presence of lipid-laden macrophages in human BALF samples [67]. However, the assessment of these patients demonstrated that they suffered from an exquisitely sensitive cough reflex without any underlying lung pathology. On average, the control group was also younger than the COPD group and too small to thoroughly investigate differences between smokers and nonsmokers in COPD and control patients. To alleviate any confounding age, smoking status effects in our transcriptome analysis, we computationally removed technical and unknown variance before downstream analysis. We suggest that any effect of age or smoking on high-dimensional data, including lipidomics might be indeed interesting questions for future studies.
Taken together, although the numbers of AMs are not significantly changed in the alveolar space in COPD, we conclude that they undergo transcriptional reprogramming that leads to GOLD grade-dependent changes in lipid metabolism. Elucidation of the concomitant alterations in the transcriptome and lipidome of AMs aids the understanding of their role in COPD and provides druggable molecular pathways, such as cholesterol metabolism and interferon-α and γ responses.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00915-2020.SUPPLEMENT (589.4KB, pdf)
Figure S1 00915-2020.FIGURES1 (2.4MB, pdf)
Figure S2 00915-2020.FIGURES2 (2.9MB, pdf)
Table E1 00915-2020.TABLEE1 (129.7KB, pdf)
Table E2 00915-2020.TABLEE2 (106.7KB, pdf)
Table E4 00915-2020.TABLEE4 (71.7KB, pdf)
Table E5 00915-2020.TABLEE5 (51.9KB, pdf)
Table E6 00915-2020.TABLEE6 (606.5KB, pdf)
Table E7 00915-2020.TABLEE7 (113.7KB, pdf)
Acknowledgements
We thank Tamás Varga, German Center for Neurodegenerative Diseases, for helpful discussion.
Footnotes
This article has supplementary material available from openres.ersjournals.com.
Author contributions: W. Fujii, T.S. Kapellos and K. Baßler collected samples and performed experiments. K. Händler carried out RNA sequencing. C. Thiele carried out mass spectrometry for lipidomics. W. Fujii, T.S. Kapellos, T. Ulas, L. Holsten, R. Knoll, S. Warnat-Herresthal, M. Oestreich, J. Hasenauer and A.C. Aschenbrenner analysed data. C. Pizarro and D. Skowasch recruited patients. D. Skowasch and J.L. Schultze designed study. W. Fujii, T.S. Kapellos and J.L. Shultze wrote the manuscript with input from all authors. All authors were involved in discussing the results of the study.
Conflict of interest: All authors have no conflict of interest to disclose.
Support statement: Wataru Fujii was supported by a fellowship of the Alexander von Humboldt Foundation (JPN-1186019-HFST-P). This work was further supported by the German Research Foundation (Deutsche Forschungsgemeinschaft; SCHU 950,9-1, 639379, and under Germany's Excellence Strategy EXC2151, 390873048 and EXC2047) and Sparse2Big, a pilot project within the Helmholtz Association Information and Data Science Incubator. Jan Hasenauer was supported by the Hausdorff Center of Mathematics (EXC 2047) and Immunosensation2 (EXC 2151). Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Decramer M, Janssens W, Miravitlles M. Chronic obstructive pulmonary disease. Lancet 2012; 379: 1341–1351. doi: 10.1016/S0140-6736(11)60968-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012; 380: 2095–2128. doi: 10.1016/S0140-6736(12)61728-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vos T, Flaxman AD, Naghavi M, et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012; 380: 2163–2196. doi: 10.1016/S0140-6736(12)61729-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rennard SI, Vestbo J. COPD: the dangerous underestimate of 15%. Lancet 2006; 367: 1216–1219. doi: 10.1016/S0140-6736(06)68516-4 [DOI] [PubMed] [Google Scholar]
- 5.Global Initiative for Chronic Obstructive Lung Disease. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Pulmonary Disease. https://goldcopd.org/wp-content/uploads/2020/11/GOLD-REPORT-2021-v1.1-25Nov20_WMV.pdf Date last accessed: January 24, 2021. Date last updated: 2021.
- 6.Stoller JK, Aboussouan LS. α1-antitrypsin deficiency. Lancet 2005; 365: 2225–2236. doi: 10.1016/S0140-6736(05)66781-5 [DOI] [PubMed] [Google Scholar]
- 7.Hunninghake GM, Cho MH, Tesfaigzi Y, et al. MMP12, lung function, and COPD in high-risk populations. N Engl J Med 2009; 361: 2599–2608. doi: 10.1056/NEJMoa0904006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor AE, Finney-Hayward TK, Quint JK, et al. Defective macrophage phagocytosis of bacteria in COPD. Eur Respir J 2010; 35: 1039–1047. doi: 10.1183/09031936.00036709 [DOI] [PubMed] [Google Scholar]
- 9.Berenson CS, Wrona CT, Grove LJ, et al. Impaired alveolar macrophage response to Haemophilus antigens in chronic obstructive lung disease. Am J Respir Crit Care Med 2006; 174: 31–40. doi: 10.1164/rccm.200509-1461OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni L, Dong C. Roles of myeloid and lymphoid cells in the pathogenesis of chronic obstructive pulmonary disease. Front Immunol 2018; 9: 1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kan M, Shumyatcher M, Himes BE. Using omics approaches to understand pulmonary diseases. Respir Res 2017; 18: 149. doi: 10.1186/s12931-017-0631-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Y-RA, Hotten DF, Malakhau Y, et al. Flow cytometric analysis of myeloid cells in human blood, bronchoalveolar lavage, and lung tissues. Am J Respir Cell Mol Biol 2016; 54: 13–24. doi: 10.1165/rcmb.2015-0146OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwasaki A, Foxman EF, Molony RD. Early local immune defences in the respiratory tract. Nat Rev Immunol 2017; 17: 7–20. doi: 10.1038/nri.2016.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe S, Alexander M, Misharin AV, et al. The role of macrophages in the resolution of inflammation. J Clin Invest 2019; 129: 2619–2628. doi: 10.1172/JCI124615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agassandian M, Mallampalli RK. Surfactant phospholipid metabolism. Biochim Biophys Acta – Mol Cell Biol Lipids 2013; 1831: 612–625. doi: 10.1016/j.bbalip.2012.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki T, Trapnell BC. Pulmonary alveolar proteinosis syndrome. Clin Chest Med 2016; 37: 431–440. doi: 10.1016/j.ccm.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agudelo CW, Kumley BK, Area-Gomez E, et al. Decreased surfactant lipids correlate with lung function in chronic obstructive pulmonary disease (COPD). PLoS ONE 2020; 15: e0228279. doi: 10.1371/journal.pone.0228279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang YP, Lee WJ, Hong JY, et al. Novel approach for analysis of bronchoalveolar lavage fluid (BALF) using HPLC-QTOF-MS-based lipidomics: lipid levels in asthmatics and corticosteroid-treated asthmatic patients. J Proteome Res 2014; 13: 3919–3929. doi: 10.1021/pr5002059 [DOI] [PubMed] [Google Scholar]
- 19.Zheng Y, Ning P, Luo Q, et al. Inflammatory responses relate to distinct bronchoalveolar lavage lipidome in community-acquired pneumonia patients: a pilot study. Respir Res 2019; 20: 82. doi: 10.1186/s12931-019-1028-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kikuchi T, Sugiura H, Koarai A, et al. Increase of 27-hydroxycholesterol in the airways of patients with COPD: possible role of 27-hydroxycholesterol in tissue fibrosis. Chest 2012; 142: 329–337. doi: 10.1378/chest.11-2091 [DOI] [PubMed] [Google Scholar]
- 21.Telenga ED, Hoffmann RF, t'Kindt R, et al. Untargeted lipidomic analysis in chronic obstructive pulmonary disease. Uncovering sphingolipids. Am J Respir Crit Care Med 2014; 190: 155–164. doi: 10.1164/rccm.201312-2210OC [DOI] [PubMed] [Google Scholar]
- 22.Haque RA, Usmani OS, Barnes PJ. Chronic idiopathic cough: a discrete clinical entity? Chest 2005; 127: 1710–1713. doi: 10.1378/chest.127.5.1710 [DOI] [PubMed] [Google Scholar]
- 23.Picelli S, Björklund ÅK, Faridani OR, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 2013; 10: 1096–1098. doi: 10.1038/nmeth.2639 [DOI] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 2015; 4: 1521. doi: 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leek JT, Johnson WE, Parker HS, et al. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012; 28: 882–883. doi: 10.1093/bioinformatics/bts034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43: e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015; 12: 453–457. doi: 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue J, Schmidt SV, Sander J, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014; 40: 274–288. doi: 10.1016/j.immuni.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fulton DL, Sundararajan S, Badis G, et al. TFCat: the curated catalog of mouse and human transcription factors. Genome Biol 2009; 10: R29. doi: 10.1186/gb-2009-10-3-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Human Protein Atlas – surfaceome. www.proteinatlas.org/search/protein_class:Predicted+membrane+proteins+AND+NOT+protein_class:Predicted+secreted+proteins Date last accessed: January 23, 2020.
- 33.The Human Protein Atlas – secretome. www.proteinatlas.org/search/protein_class:Predicted+secreted+proteins+AND+NOT+protein_class:Predicted+membrane+proteins Date last accessed: January 23, 2020.
- 34.Harrell FE Jr. Hmisc: Harrell Miscellaneous. 2019. https://cran.r-project.org/package=Hmisc.
- 35.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal 2006; Complex Systems:1695 [Google Scholar]
- 36.Gustavsen JA, Pai S, Isserlin R, et al. RCy3: Network biology using Cytoscape from within R. F1000Res 2019; 8: 1774. doi: 10.12688/f1000research.20887.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R Package for comparing biological themes among gene clusters. Omi A J Integr Biol; 2012; 16: 284–287. doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H, Um S-J, Kim YS, et al. Association of the neutrophil-to-lymphocyte ratio with lung function and exacerbations in patients with chronic obstructive pulmonary disease. PLoS ONE 2016; 11: e0156511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tricas L, Echeverría A, Blanco MA, et al. Flow cytometry counting of bronchoalveolar lavage leukocytes with a new profile of monoclonal antibodies combination. Cytometry B Clin Cytom 2012; 82: 61–66. doi: 10.1002/cyto.b.20630 [DOI] [PubMed] [Google Scholar]
- 40.Meyer KC, Raghu G, Baughman RP, et al. An official American Thoracic Society clinical practice guideline: the clinical utility of bronchoalveolar lavage cellular analysis in interstitial lung disease. Am J Respir Crit Care Med 2012; 185: 1004–1014. doi: 10.1164/rccm.201202-0320ST [DOI] [PubMed] [Google Scholar]
- 41.Savarimuthu Francis SM, Larsen JE, Pavey SJ, et al. Genes and gene ontologies common to airflow obstruction and emphysema in the lungs of patients with COPD. PLoS ONE 2011; 6: e17442 doi: 10.1371/journal.pone.0017442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang S-R, Chida AS, Bauter MR, et al. Cigarette smoke induces proinflammatory cytokine release by activation of NF-κB and posttranslational modifications of histone deacetylase in macrophages. Am J Physiol Lung Cell Mol Physiol 2006; 291: L46–L57. doi: 10.1152/ajplung.00241.2005 [DOI] [PubMed] [Google Scholar]
- 43.Trocme C, Deffert C, Cachat J, et al. Macrophage-specific NOX2 contributes to the development of lung emphysema through modulation of SIRT1/MMP-9 pathways. J Pathol 2015; 235: 65–78. doi: 10.1002/path.4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mercado N, Thimmulappa R, Thomas CMR, et al. Decreased histone deacetylase 2 impairs Nrf2 activation by oxidative stress. Biochem Biophys Res Commun 2011; 406: 292–298. doi: 10.1016/j.bbrc.2011.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito K, Lim S, Caramori G, et al. Cigarette smoking reduces histone deacetylase 2 expression, enhances cytokine expression, and inhibits glucocorticoid actions in alveolar macrophages. FASEB J 2001; 15: 1110–1112. doi: 10.1096/fsb2fj000432fje [DOI] [PubMed] [Google Scholar]
- 46.Pons AR, Noguera A, Blanquer D, et al. Phenotypic characterisation of alveolar macrophages and peripheral blood monocytes in COPD. Eur Respir J 2005; 25: 647–652. doi: 10.1183/09031936.05.00062304 [DOI] [PubMed] [Google Scholar]
- 47.Hodge S, Matthews G, Mukaro V, et al. Cigarette smoke-induced changes to alveolar macrophage phenotype and function are improved by treatment with procysteine. Am J Respir Cell Mol Biol 2011; 44: 673–681. doi: 10.1165/rcmb.2009-0459OC [DOI] [PubMed] [Google Scholar]
- 48.Henrot P, Prevel R, Berger P, et al. Chemokines in COPD: from implication to therapeutic use. Int J Mol Sci 2019; 20 doi: 10.3390/ijms20112785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaykhiev R, Krause A, Salit J, et al. Smoking-dependent reprogramming of alveolar macrophage polarization: implication for pathogenesis of chronic obstructive pulmonary disease. J Immunol 2009; 183: 2867–2883. doi: 10.4049/jimmunol.0900473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woodruff PG, Koth LL, Yang YH, et al. A distinctive alveolar macrophage activation state induced by cigarette smoking. Am J Respir Crit Care Med 2005; 172: 1383–1392. doi: 10.1164/rccm.200505-686OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finlay GA, O'Driscoll LR, Russell KJ, et al. Matrix metalloproteinase expression and production by alveolar macrophages in emphysema. Am J Respir Crit Care Med 1997; 156: 240–247. doi: 10.1164/ajrccm.156.1.9612018 [DOI] [PubMed] [Google Scholar]
- 52.Wallace AM, Sandford AJ, English JC, et al. Matrix metalloproteinase expression by human alveolar macrophages in relation to emphysema. COPD J Chronic Obstr Pulm Dis 2008; 5: 13–23. doi: 10.1080/15412550701817789 [DOI] [PubMed] [Google Scholar]
- 53.Bazzan E, Turato G, Tinè M, et al. Dual polarization of human alveolar macrophages progressively increases with smoking and COPD severity. Respir Res 2017; 18: 40. doi: 10.1186/s12931-017-0522-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eapen MS, Hansbro PM, McAlinden K, et al. Abnormal M1/M2 macrophage phenotype profiles in the small airway wall and lumen in smokers and chronic obstructive pulmonary disease (COPD). Sci Rep 2017; 7. doi: 10.1038/s41598-017-13888-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Labzin LI, Schmidt S V, Masters SL, et al. ATF3 Is a key regulator of macrophage IFN responses. J Immunol 2015; 195: 4446–4455. doi: 10.4049/jimmunol.1500204 [DOI] [PubMed] [Google Scholar]
- 56.De Nardo D, Labzin LI, Kono H, et al. High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nat Immunol 2014; 15: 152–160. doi: 10.1038/ni.2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Dam S, Võsa U, van der Graaf A, et al. Gene co-expression analysis for functional classification and gene-disease predictions. Brief Bioinform 2018; 19: 575–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trapnell BC, Nakata K, Bonella F, et al. Pulmonary alveolar proteinosis. Nat Rev Dis Primers 2019; 5: 16. doi: 10.1038/s41572-019-0066-3 [DOI] [PubMed] [Google Scholar]
- 59.Morissette MC, Shen P, Thayaparan D, et al. Disruption of pulmonary lipid homeostasis drives cigarette smoke-induced lung inflammation in mice. Eur Respir J 2015; 46: 1451–1460. doi: 10.1183/09031936.00216914 [DOI] [PubMed] [Google Scholar]
- 60.Köberlin MS, Snijder B, Heinz LX, et al. A conserved circular network of coregulated lipids modulates innate immune responses. Cell 2015; 162: 170–183. doi: 10.1016/j.cell.2015.05.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hou X, Summer R, Chen Z, et al. Lipid uptake by alveolar macrophages drives fibrotic responses to silica dust. Sci Rep 2019; 9:399. doi: 10.1038/s41598-018-36875-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muse ED, Yu S, Edillor CR, et al. Cell-specific discrimination of desmosterol and desmosterol mimetics confers selective regulation of LXR and SREBP in macrophages. Proc Natl Acad Sci USA 2018; 115: E4680–E4689. doi: 10.1073/pnas.1714518115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeng J, Wu Y, Liao Q, et al. Liver X receptors agonists impede hepatitis C virus infection in an Idol-dependent manner. Antiviral Res 2012; 95: 245–256. doi: 10.1016/j.antiviral.2012.06.004 [DOI] [PubMed] [Google Scholar]
- 64.Körner A, Zhou E, Müller C, et al. Inhibition of Δ24-dehydrocholesterol reductase activates pro-resolving lipid mediator biosynthesis and inflammation resolution. Proc Natl Acad Sci USA 2019; 116: 20623–20634. doi: 10.1073/pnas.1911992116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petrusca DN, Gu Y, Adamowicz JJ, et al. Sphingolipid-mediated inhibition of apoptotic cell clearance by alveolar macrophages. J Biol Chem 2010; 285: 40322–40332. doi: 10.1074/jbc.M110.137604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Voelker DR, Numata M. Phospholipid regulation of innate immunity and respiratory viral infection. J Biol Chem 2019; 294: 4282–4289. doi: 10.1074/jbc.AW118.003229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palomo B, Belda J, Arias M, et al. Lipid laden macrophages in patients with chronic cough. Eur Respir J 2011; 38: Suppl. 55, p3864. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00915-2020.SUPPLEMENT (589.4KB, pdf)
Figure S1 00915-2020.FIGURES1 (2.4MB, pdf)
Figure S2 00915-2020.FIGURES2 (2.9MB, pdf)
Table E1 00915-2020.TABLEE1 (129.7KB, pdf)
Table E2 00915-2020.TABLEE2 (106.7KB, pdf)
Table E4 00915-2020.TABLEE4 (71.7KB, pdf)
Table E5 00915-2020.TABLEE5 (51.9KB, pdf)
Table E6 00915-2020.TABLEE6 (606.5KB, pdf)
Table E7 00915-2020.TABLEE7 (113.7KB, pdf)