Abstract
The development of Drug Delivery Systems (DDS) has led to increasingly efficient therapies for the treatment and detection of various diseases. DDS use a range of nanoscale delivery platforms produced from polymeric of inorganic materials, such as micelles, and metal and polymeric nanoparticles, but their variant chemical composition make alterations to their size, shape, or structures inherently complex. Genetically encoded protein nanocages are highly promising DDS candidates because of their modular composition, ease of recombinant production in a range of hosts, control over assembly and loading of cargo molecules and biodegradability. One example of naturally occurring nanocompartments are encapsulins, recently discovered bacterial organelles that have been shown to be reprogrammable as nanobioreactors and vaccine candidates. Here we report the design and application of a targeted DDS platform based on the Thermotoga maritima encapsulin reprogrammed to display an antibody mimic protein called Designed Ankyrin repeat protein (DARPin) on the outer surface and to encapsulate a cytotoxic payload. The DARPin9.29 chosen in this study specifically binds to human epidermal growth factor receptor 2 (HER2) on breast cancer cells, as demonstrated in an in vitro cell culture model. The encapsulin-based DDS is assembled in one step in vivo by co-expressing the encapsulin-DARPin9.29 fusion protein with an engineered flavin-binding protein mini-singlet oxygen generator (MiniSOG), from a single plasmid in Escherichia coli. Purified encapsulin-DARPin_miniSOG nanocompartments bind specifically to HER2 positive breast cancer cells and trigger apoptosis, indicating that the system is functional and specific. The DDS is modular and has the potential to form the basis of a multi-receptor targeted system by utilising the DARPin screening libraries, allowing use of new DARPins of known specificities, and through the proven flexibility of the encapsulin cargo loading mechanism, allowing selection of cargo proteins of choice.
Keywords: Encapsulin, Drug delivery system, Cytotoxic protein, DARPin
Abbreviations: HER2, Human Epidermal growth factor Receptor 2; SK-BR-3, Sloan-Kettering Breast cancer cell line/HER2-overexpressing human breast cancer cell line; MSCs, Mesenchymal Stem Cells; DARPin9.29, Designed Ankyrin Repeat Protein 9.29; mScarlet, a bright monomeric red fluorescent protein; iLOV, improved Light, Oxygen or Voltage-sensing flavoprotein; T. maritima, Thermotoga maritima; miniSOG, mini-Singlet Oxygen Generator; STII, StrepII-tag, an eight-residue peptide sequence (Trp-Ser-His-Pro-Gln-Phe-Glu-Lys) with intrinsic affinity toward streptavidin that can be fused to recombinant protein in various fashions; rTurboGFP, recombinant Turbo Green Fluorescent Protein; Annexin V-FITC, Annexin V-Fluorescein IsoThiocyanate Conjugate; His6, Hexahistidine; iGEM, international Genetically Engineered Machine; DDS, Drug Delivery System; EPR, Enhanced Permeability and Retention effect; VLPs, Virus-Like Particle; NPs, NanoParticles
1. Introduction
For decades, cytotoxic chemotherapy had been the predominant medical treatment for breast cancer. Chemotherapeutic drugs target rapidly dividing cells, a characteristic of most cancer cell types and certain normal tissues [1]. Although highly effective, cytotoxic cancer drugs, such as doxorubicin and paclitaxel, demonstrate significant detrimental off-target effects which limit the dosage of chemotherapeutic drugs [2,3]. The use of Drug Delivery Systems (DDS) can improve the clinical success of traditional chemotherapeutics by improving their pharmacological properties.
The advent of DDSs has had a pivotal impact on the field of biomedicine, and increasingly efficient therapies and diagnostic tools are now being developed for the treatment and detection of various diseases. Over the last decade, about 40,000 studies focusing on the development of potential targeting strategies and the interaction of nanoparticle-based DDSs with cells and tissues, were published [4]. The Nanomedicine approach to encapsulating cytotoxic therapeutic small molecules provides several benefits to pharmacological properties, most critically, the passive targeting to the tumour site via the associated leaky vasculature, called the Enhanced Permeability and Retention (EPR) effect [5]. Other nanoparticle (NPs)- associated benefits include longer circulation times, slow clearance, greater formulation flexibility [6], tumour penetration and facilitated cellular uptake [7]. All of these factors raise the therapeutic index of the administered chemotherapy drugs [8].
An immense range of nanoscale delivery platforms have been investigated as efficient drug delivery vehicles for diagnostic or therapeutic purposes, including liposomes, micelles, metal and polymeric nanoparticles, and protein cages [[9], [10], [11], [12]]. However, these DDSs are often synthetically developed using polymeric or inorganic materials, and their highly variant chemical compositions make any alterations to their size, shape or structures inherently complex. Further, successful biotherapeutics must meet three major requirements: high end-product quality, economic viability, and accessibility to the public. Therefore, manufacturing platforms which allow robust and cost-effective production must be developed. Additional key challenges include: high production costs, toxicity, immunogenicity, inability to release drug cargo on demand, and low drug carrying capacity.
Protein nanoparticles (PNPs) are promising candidates to address these challenges. They have been extensively studied as delivery systems for chemical or biological drugs such as anticancer drugs and therapeutic proteins. PNPs have several advantages over polymeric and inorganic materials including biocompatibility of size, biodegradability, defined fate, morphological uniformity, atomistic detail, self-assembly and scalability. Moreover, mild conditions are used in the preparation of PNPs, bypassing the need for toxic chemicals or organic solvents. PNPs can be classed into coalescing proteins forming nanoparticles, native self-assembling and de novo designed particles. Coalescing PNPs can be generated by chemical and physical methods using proteins, such as the silk protein fibroin, human serum albumin, gelatin and others [13]. Native self-assembling PNPs are natural structures (ferritins, small heat shock proteins, vaults, encapsulins and lumazine synthase) that perform biological roles in living cells [[14], [15], [16], [17]]; and virus-like particles (VLP) of which prominent examples are cowpea chlorotic mottle virus (CCMV), bacteriophage MS2, hepatitis B virus (HBV), bacteriophage P22 and many others [18]. De novo designed PNPs such as those developed by the Baker [19,20], Yeates [21] and King [22] groups are also self-assembling nanocages but they are developed by computational programming and simulations.
Large number of studies are available on VLP-based PNP for therapeutic applications such as targeted cancer therapeutics, these are comprehensively summarised elsewhere [23]. Examples of VLPs that have been utilised to deliver synthetic chemotherapy drugs include the bacteriophage VLP MS2 [24], bacteriophage P22 VLP [25], multiple plant VLPs [26,27] and mammalian VLPs [28,29]. VLPs have also been designed to encapsulate therapeutic protein cargo such as metalloproteins to convert untargeted prodrugs to their active forms at the site of interest [30]. Yet, the encapsulation of protein cargos in traditional VLPs is a multi-step process normally requiring disassembly and reassembly and electrostatic interactions between the cargo molecule and the capsid or specific DNA stem loops conjugations. This can involve expensive and non-scalable chemistries and processes.
The proposed DDS in this work is based on the encapsulin. Encapsulins are highly promising candidates for use in multifunctional DDS due to their well-defined structures and biodegradability. Encapsulins are 20–45 nm self-assembling microbial nano-compartments formed from 60, 180 or 240 copies of a single capsid monomer [31,32]. In prokaryotes, encapsulins function to mitigate oxidative stress through packaging enzymatic cargo, iron mineralising ferritin-like proteins or peroxidase [31]. Encapsulin systems are widespread in nature with operons observed in approximately 1% of prokaryotic genomic sequences, most still uncharacterised [33].
Encapsulins have been employed in a broad range of biotechnological applications by functionalising the single protomer and exploiting the characterised cargo loading system [34,35]. The crystal structures of a number of encapsulins have been resolved to an atomic resolution [[36], [37], [38]], giving researchers greater control when bio-engineering these particles. Key applications include the use of encapsulins as imaging agent [39,40], chimeric vaccines [41], immunotherapeutic [42], functional nanoarchitectures [43], as well as the demonstration of functionalisation by chemical conjugation and protein-protein interactions [17,44,45]. Recently, Diaz et al. (2021) reported the re-engineering of encapsulins as light-responsive nanoreactor for photodynamic therapy, showing loading of a cytotoxic agent which has been the inspiration for the cytotoxic model protein used in this work [46].
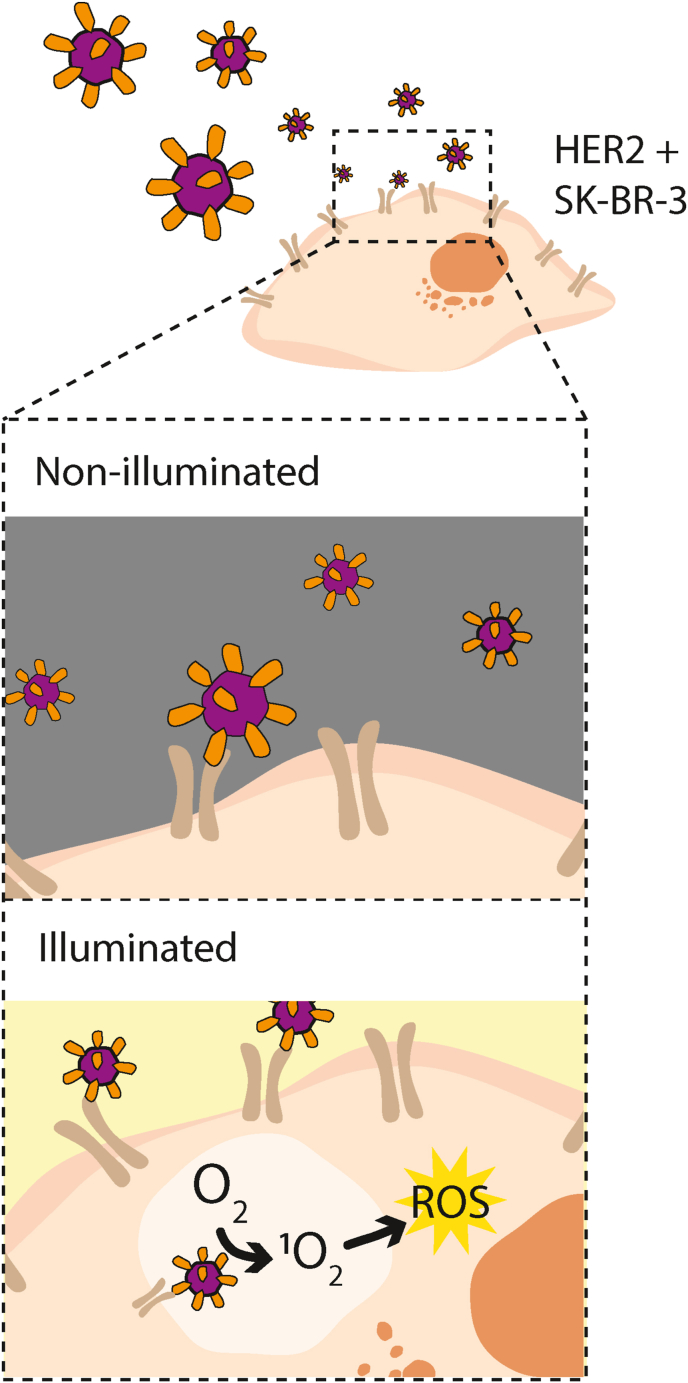
In this proof or concept study, using International Genetically Engineered Machine (iGEM) principles, we demonstrate the redesign and characterisation of the naturally existing encapsulin from Thermotoga maritima as a functional targeted drug delivery system specific to breast cancer cells (Fig. 1), as a step towards the development of a modular platform for targeted delivery of therapies.
Fig. 1.
Schematic drawing showing the concept of the genetically encoded targeted drug delivery system this study aimed to develop. The genetically engineered antibody mimetic protein DARPin9.29 (orange) is fused to the capsid protein of the T. maritima encapsulin (purple) and loaded with the cytotoxic protein miniSOG (not shown). This drug delivery system binds specifically to breast cancer cells on the HER2 receptor (brown) and upon uptake and illumination releases reactive oxygen species (ROS, yellow) which trigger apoptosis of the targeted cell.
2. Materials and methods
2.1. Construction of plasmids
Plasmids used in this study were created as shown in Table A.1. The DNA for the T. maritima encapsulin was ordered from Twist. DNA for all other constructs were ordered as gBlocks from IDT. All parts were condon-optimised for expression in Escherichia coli. Parts were cloned into pSB1C-FB via the BsaI sites. The miniSOG fused with the targeting peptide of T. maritima ferritin-like protein (GGSENTGGDLGIRKL) was sub-cloned into plasmids containing encapsulin genes, including a separate T7 expression cassette, using standard BioBrick assembly [47].
2.2. Expression and purification of recombinant proteins
Plasmids were transformed into competent E. coli BL21Star(DE3) (Thermo Fisher Scientific). Cells were grown in 50 ml (400 ml for repeat experiments) of Luria-Bertani (LB) broth (containing 34 mg/L chloramphenicol) at 37 °C, shaking at 225 rpm. Protein expression was induced for 16 h with 400 μΜ isopropyl β-d-1-thiogalactopyranoside (IPTG) (Thermo Fisher Scientific) when the OD600 reached 0.6. The cells were cooled to 4 °C and harvested by centrifugation at 5000×g for 10 min. The pellet was resuspended in 1 ml (25 ml for 400 ml culture) of buffer W (0.1 M Tris-Cl, 0.15 M NaCl, 1 mM EDTA, pH 8.0) and the cells were lysed using sonication (5 cycles for 30 s pulse followed by 30 s off at 50% the amplitude; 400 ml culture sample was sonicated for 15 cycles at 10 s on 10 s off). The cell debris was removed via centrifugation at 18000×g for 10 min. StrepII (STII)-tagged proteins were then purified using either 1 ml (50 ml culture) or 5 ml (400 ml culture) Strep-Tactin®XT Superflow columns® (IBA Lifesciences GmbH, Germany) and eluted in BXT buffer (0.1 M Tris-Cl, 0.15 M NaCl, 50 mM Biotin, pH 8.0). A typical encapsulin purification from 400 ml culture yielded approximately 1 mg of protein after pooling all fractions from the 5 ml Strep-Tactin column (0.2 mg/ml). Darpin fusion to encapsulins did not impact the concentration of the eluted samples. It should be noted that the encapsulin yield was significantly lower than the yield of mScarlet-DARPin-STII, DARPin-mScarlet-STII and mScarlet alone, which yielded 2.5–4.7 mg from a 1 ml Strep-Tactin column. miniSOG-STII yielded 0.6–1.1 mg protein when purified on a 1 ml Strep-Tactin column. Lastly, purified proteins were concentrated through Amicon Ultra 0.5 ml centrifugal filters with a 10 KDa cut-off to a final concentration of 3 μM.
Hexahistidine (His6)-tagged mScarlet was similarly expressed and purified via Immobilized Metal Affinity Chromatography (IMAC) using Chelating Fast Flow Sepharose resin (GE Healthcare) in a gravity flow column (PD10). Wash steps followed a stepwise imidazole gradient from 10 to 100 mM with final elution in 250 mM imidazole. Elution was visually confirmed, and the eluted sample buffer exchanged using a GE PD10 desalting column into 50 mM Tris-Cl, 150 mM NaCl buffer, pH 7.5.
To provide evidence for miniSOG loading, the Step-tag purified and concentrated TmEnc-DARPin-STII_miniSOG sample was further purified via size exclusion chromatography (SEC), using a HiPrep 16/60 Sephacryl S-500 HR column (Cyitva, USA) on an Äkta Explorer (GE Healthcare). The injection volume was 1 ml, the flow rate 0.5 ml/min in 100 mM Tris-Cl, 150 mM NaCl, pH 8.0 buffer.
2.3. Cell culture of SK-BR-3 and mesenchymal stem cells
The SK-BR-3 HER2 overexpressing cancer cell line was obtained from ATCC, and mesenchymal stem cells (MSCs) were isolated from patient's fat in the Department of Biochemical Engineering (UCL, London). The cell lines were cultured in Dulbecco's Modified Eagle Medium DMEM (Gibco) supplemented with 10% fetal bovine serum and incubated in a humidified atmosphere containing 5% CO2 at 37 °C. The cells were grown in a monolayer up to 70–80% confluence. They were detached using trypsin and split every 3 days at a ratio of 1: 4. The cells were passaged in the same way. When seeding cells for experiments, 10 μL of cell culture were mixed with 10 μL of trypan blue and counted using a hemacytometer to check the cell viability and density.
2.4. Binding and internalisation studies with DARPin9.29
SK-BR-3 cells were plated in 6-well plates and incubated at 5% CO2 at 37 °C until a cell density of 100 × 106 cells/mL was reached. To observe binding, the cells were washed with Phosphate-Buffered Saline (PBS) once and incubated with purified mScarlet-DARPin-STII or DARPin-mScarlet-STII at a final concentration of 3 μM for 60 min at 5% CO2 and 37 °C. The cells were then washed three times with PBS, stained with 1 ml nuclear stain 4′,6-diamidino-2-phenylindole (DAPI) with a dilution of 1:10,000 and observed using an EVOS fluorescence (FL) inverted microscope. The same process was also repeated with non-target MSC (HER2 negative) to demonstrate specific binding of DARPin9.29 to HER2. The negative controls, His-mScarlet, recombinant Turbo green fluorescent protein (rTurboGFP) and T. maritima encapsulin displaying improved light, oxygen, or voltage-sensing (iLOV) fluorescent protein were incubated with SK-BR-3 following the same experimental protocol. To determine mScarlet-DARPin9.29 binding under hypoxic conditions, the cells were incubated at 5% CO2 and 37 °C but 2% O2 while the rest of the protocol was followed as before.
For quantitative determination of the cell population that bound DARPin9.29 or control samples (His-mScarlet, rTurboGFP, T. maritima_iLOV), the SK-BR-3 and MSCs cells were washed once with PBS after 60-min incubation and detached with 500 μL EDTA to prevent disturbing interaction of DARPin9.29-HER2 and then centrifuged at 1500 rpm at 4 °C for 5 min. The cells were resuspended in PBS and flow cytometry analysis was performed on a BD Accuri C6 cytometer (Becton Dickinson, USA).
2.5. Binding and cytotoxicity of TmEnc-DARPin_miniSOG
To determine binding of the DDS, SK-BR-3 and MSCs (negative control) cells from T-flasks were seeded into 96-well plates in duplicates. Cells were incubated at 37 °C and 20% oxygen and 5% CO2 for one day to allow formation of a confluent monolayer. Cells were washed once with PBS before purified TmEnc-DARPin-STII_miniSOG and control samples (TmEnc-STII, TmEnc-STII_miniSOG, miniSOG-STII). were added at a final concentrations of 3 μM. The plates were then incubated at the above conditions for 30 min to allow binding of the DARPin9.29 fused to the encapsulin, after which half of the cells were illuminated using a white flashlight of 40 lumens/cm2 (for the repeat experiment this was a done with 1W Samsung LH351B LED with luminous flux of 177 lm at 350 mA), to allow activation of the photosensitizer miniSOG for 60 min. At the end of the 90 min the cells were subjected to flow cytometry analysis. To observe binding of TmEnc-DARPin-STII_miniSOG, cells were imaged using the green gate-GFP channel of EVOS FL microscope to detect miniSOG's green fluorescence. As control, a set of SK-BR-3 and MSCs was not incubated with sample.
2.6. Annexin V-FITC assay for assessment of cytotoxicity of TmEnc-DARPin-STII_miniSOG
To detect percentage loss in viability and apoptosis the SK-BR-3 and MSCs cells were collected after incubation with the various samples (section 2.5), treated using an Annexin V-fluorescein isothiocyanate conjugate (FITC) apoptosis detection kit (Abcam, cat. no. ab4085) and analysed via flow cytometry. The samples were prepared according to the manufacturer's protocol. Cells were washed with 500 μL of PBS, detached using 100 μL of EDTA and centrifuged at 1500 rpm for 4 min. The cell pellets were suspended in 500 μL of 1x Binding buffer from the kit and then 5 μL of Annexin-V and Propidium iodide (PI) (50 mg/ml) were added and incubated for 5 min at room temperature in the dark. The samples were analysed using flow cytometry. Annexin V is a Ca2+-dependent phospholipid-binding protein that has a high affinity for phosphatidylserine, which is translocated from the cytoplasmic side of the cell membrane to the extracellular side of the cell membrane upon apoptosis. The cell membrane is impermeable to PI, and hence PI is excluded from living cells. Cells that stain negative for Annexin V-FITC and negative for PI are considered living cells. Cells that stain positive for Annexin V-FITC and negative for PI are early apoptotic, or if the other way around they are necrotic. If both are positive, cells are in late stage of apoptosis. For Annexin V-FITC-PI apoptosis testing, detection parameters were as follows: 20 mV laser power and appropriate detector channel position for Annexin-V-FITC (Ex = 488 nm; Em = 530 nm) and PI (585/40 bandpass filter).
2.7. Dynamic light scattering
To validate assembly, the hydrodynamic diameter of purified encapsulins was determined by dynamic light scatter (DLS) using the Malvern Zetasizer Nano ZS. All measurements were performed at 0.2 mg/ml in 0.1 M Tris-Cl, 0.15 M NaCl, 50 mM D-biotin, pH 8.0 at 25 °C and averaged over three measurements. Volume particle size distribution results were automatically plotted using Malvern Zetasizer Software version 7.13.
2.8. SDS and native polyacrylamide gel electrophoresis (PAGE)
For SDS-PAGE, purified proteins were mixed 1:1 with 2x Laemmli buffer and incubated at 95 °C for 10–20 min. The samples were loaded on precast Novex™ 12% Tris-Glycine mini gels (Thermo Fisher Scientific) and run at 90 V for 15 min to stack the proteins then 160 V for 50 min or until the running front reached the bottom of the gel. Native PAGE of encapsulin construct (TmEnc-STII and TmEnc-DARPin-STII) were run on handcast discontinuous gels with a 3% acrylamide stacking (0.5 M Tris-Cl, pH 6.8) and running gel (1.5 M Tris-Cl, pH 8.8) with 10% acrylamide running gel footing. Prior to loading, samples were mixed 1:1 in loading buffer (62.5 mM Tris-HCl, pH 6.8, 40% glycerol, 0.01% bromophenol blue) and then ran with ice packs at 100 V, 15 mA for 160 min. Gels were incubated with InstantBlue™ (Sigma Aldrich) and visualised with a Trans Illuminator (GE Healthcare).
2.9. Western blot
SDS-PAGE fractionated gel samples were transferred to a PVDF membrane using a Trans-Blot Turbo Transfer System (Bio-Rad) according to the manufacturer's protocol. Membranes were then incubated overnight at 4 °C with 20 ml of PBS blocking buffer (4 mM KH2PO4, pH 7.4, 16 mM Na2HPO4, 115 mM NaCl). The blocking buffer was discarded, and the membranes were washed three times with 20 ml of PBS-Tween 20 buffer (PBS buffer with 0.1% v/v Tween 20). Strep-Tactin horseradish peroxidase (HRP) conjugate (IBA Lifesciences GmbH, Germany) diluted 1:100 in enzyme buffer (PBS with 0.2% BSA and 0.1% Tween 20) was added to the membrane and incubated for an hour at room temperature. The membrane was then washed twice using PBS-Tween20 buffer, and twice with PBS. The membrane was incubated for 5 min with 10 ml of peroxide/luminol enhancer solution and imaged using a chemiluminescent imager (GE Healthcare - Imager 600) according to the manufacturer's protocol.
2.10. Transmission electron microscope (TEM) imaging
For sample preparation, 5 μL of purified protein sample in BXT buffer was applied onto a carbon/formvar-coated copper grid (300 mesh, Generon, Slough, UK) and allowed to dry for 2 min. The grid sample face was then washed to remove excess sodium ions by touching it to a droplet of distilled water for 5 s, gently drained, and then negatively stained with 2% uranyl acetate in distilled water for 30 s and allowed to dry. When dry, samples were viewed on a JEM1010 transmission electron microscope (Welwyn Garden City, UK), with a Gatan Orius camera. Images were taken at a magnification of 150,000x. Figures show representative areas without further image processing.
3. Results
3.1. Fusing DARPin9.29 to a fluorescent protein and binding to SK-BR-3 breast cancer cells
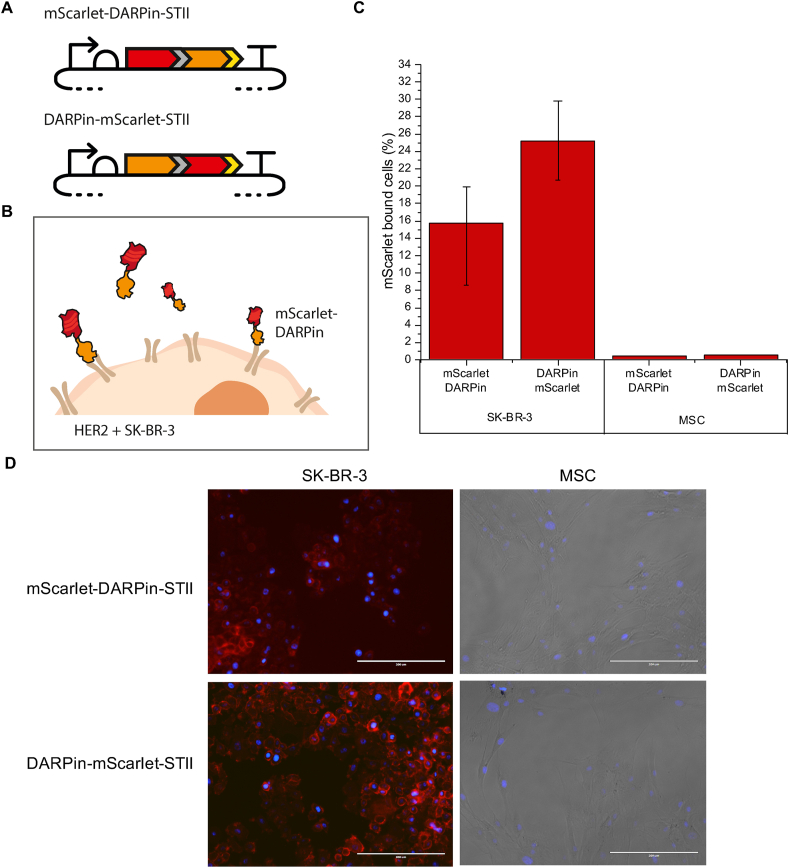
In this work encapsulins were coupled with the designed ankyrin repeat protein DARPin9.29 which was selected for specific binding to the human epidermal growth factor receptor 2 (HER2) overexpressed by the human breast cancer cell line SK-BR-3 [48]. Prior to display on an encapsulin, DARPin9.29 was fused to the C terminus of the fluorescent protein mScarlet (mScarlet-DARPin-STII), in order to demonstrate specificity to the laboratory SK-BR-3 cells and to show that binding is not inhibited by fusion of DARPin9.29 to another protein. The reverse orientation fusion protein, DARPin-mScarlet-STII (fusion of DARPin9.29 to the N terminus of mScarlet), was included as a positive control as it had previously been shown that a similar fusion protein can bind to the HER2 receptor [49].
Following expression and purification (Figure A.1), 3 μM of each of the two fusion proteins were incubated for 1 h at 20% oxygen and 37 °C with SK-BR-3 cells expressing HER2 and MSCs, which do not express the HER2 receptor. Both fusion proteins were capable of binding to SK-BR-3 cells, which indicates that DARPin9.29 tolerates fusion to another protein without abolishing binding to the receptor. Interestingly, the DARPin9.29 followed by mScarlet fusion (DARPin-mScarlet-STII) resulted in higher binding efficiency compared to the mScarlet-DARPin-STII orientation (Fig. 2C and D). The lower binding efficiency of the mScarlet-DARPin-STII is likely due to restraints caused by the orientation of the fusion and interference with the DAPRin9.29 repeat motif binding to the receptor. Different linkers and linker lengths could be screened to test this hypothesis and improve binding. Nevertheless the mScarlet-DARPin-STII fusion orientation was viable which indicates that fusion of DARPin9.29 to the C terminus of the T. maritima encapsulin shell protein should not disrupt interactions with the HER2 receptor.
Fig. 2.
Binding of DARPin9.29 fusion proteins to SK-BR-3. (A) mScarlet-DARPin-STII and DARPin-mScarlet-STII plasmid designs, DARPin in orange, mScarlet in red, (GSG)2 in grey, STII in yellow. (B) Schematic representation of DARPin binding to HER2 positive SK-BR-3. (C) Flow cytometry analysis of cells with mScarlet signal for SK-BR-3 and MSC at 37 °C and 20% O2 after 1 h. Error bars showing the range of values from two technical repeats. (D) Confocal microscopy images of SK-BR-3 and MSC cells incubated with DARPin-mScarlet-STII and mScarlet-DARPin-STII. Red = DARPins represented by the red fluorescence of mScarlet; blue = cell nuclei are stained with DAPI (4′,6-diamidino-2-phenylindole). Images were taken at 20× magnification using an Evos Fluorescence Microscope. Scale bars = 200 μm.
To ascertain that binding was specific to DARPin9.29, the experiments were repeated with mScarlet only as a control and two other control samples, rTurboGFP and T. maritima encapsulins fused with iLOV. None of the control samples bound to either SK-BR-3 or MSC cells confirming the selective targeting capabilities of the DARPin9.29 fusion proteins (Figures A.2 and A.3). A repeat of the fusion protein incubations was carried out after completion of the iGEM project (Figure A.2). Although a lower proportion of cells was found to bind DARPin9.29, a similar trend as before was observed (Figure A.2 and Fig. 2C); the fusion proteins binding to SK-BR-3 but not to MSC, and DARPin-mScarlet-STII displaying better binding ability than mScarlet-DARPin-STII. The variability in the repeat experiment may be attributed to biological variation in primary cell cultures, especially handling of the cells.
Finally, binding of the mScarlet-DAPRPin9.29 fusion proteins to HER2 was also examined at 2% O2 and 37 °C to mimic the hypoxic conditions of the tumour microenvironment. The data shows that binding was still possible at hypoxic conditions (Figure A.4). This warrants further investigation into the behaviour of the drug delivery system in low oxygen tension as it represents the common situation in a solid tumour microenvironment.
3.2. Design and construction of a targeted drug delivery system (DDS) based on the T. maritima encapsulin
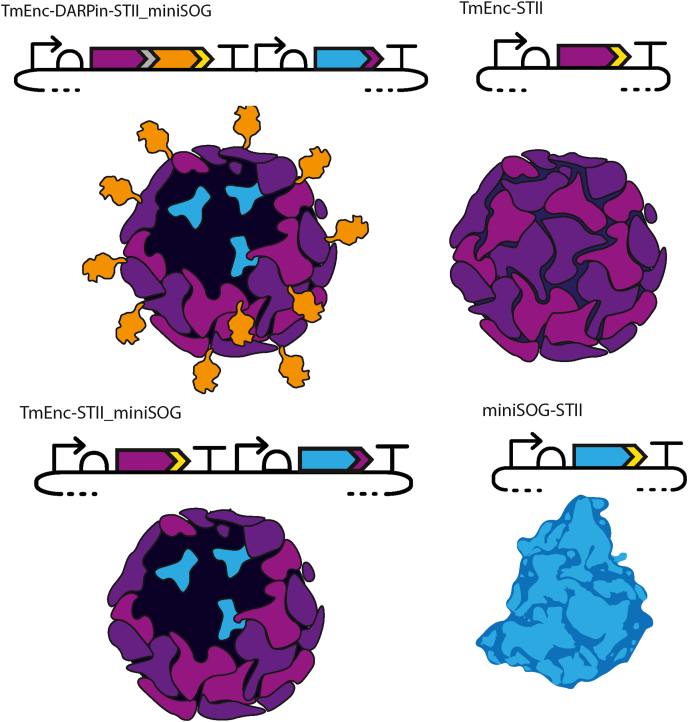
The targeted DDS was designed to be expressed from a single plasmid in E. coli and to self-assemble in vivo from only two components - the capsid displaying DARPin9.29 and a cytotoxic protein of interest. The DDS can then be isolated via single-step purification using an affinity tag. Plasmids expressing the DDS and control constructs used in this work were constructed using BioBrick conventions. First, DARPin9.29 was cloned in frame with the T. maritima encapsulin gene to produce the TmEnc-DARPin-STII fusion protein with the aim to assemble an icosahedral capsid, with T number = 1, from 60 protomers displaying 60 copies of the DARPin9.29 molecule on its surface (Fig. 3). Simple structural modelling (Figure A.5) showed that a standard flexible linker of 8 amino acids should allow sufficient space for the DARPin9.29 on the surface of the encapsulin and limit structural clashes. The selection of the linker was predominantly based on the assumption that a shorter linker than this may lead to crowding and restrict room for rotation for optimal receptor binding.
Fig. 3.
Design and assembly of the targeted drug delivery system and control samples. Plasmid designs and schematic representation of the protein assembly products. TmEnc-DARPin-STII_miniSOG = encapsulin displaying DARPin loaded with miniSOG; TmEnc-STII = encapsulin only; TmEnc-STII-miniSOG = encapsulin loaded with miniSOG, and miniSOG-STII = miniSOG only. Plasmid component symbols comply with Synthetic Biology Open Language (SBOL) convention. TmEnc (purple) = T. maritima encapsulin gene with His6 insertion between amino acid 42 and 43; DARPin (orange) = DARPin9.29 gene; STII (yellow) = Strep-tag; miniSOG (blue) = mini Singlet Oxygen Generator; small purple arrow at the 3′end of miniSOG denotes targeted peptide derived from T. maritima ferritin-like cargo protein for recruitment of miniSOG into the capsid; grey = 8 amino acid linker.
Next, a cytotoxic model protein called miniSOG (mini Singlet Oxygen Generator), was encapsulated by fusing the minimal targeting peptide region from the T. maritima ferritin-like cargo protein onto the C terminus of miniSOG, separated by a short flexible linker [50]. MiniSOG is a biological photosensitizer that when activated by blue light, generates reactive oxygen species (ROS), primarily singlet oxygen (1O2) [51]. It has recently been used to generate a light-responsive encapsulin nanoreactor for photodynamic therapy [46]. The cytotoxic ROS generated by miniSOG can readily diffuse through the pores of the encapsulin shell, making it an ideal therapeutic protein candidate for encapsulation as the release of cargo proteins from an encapsulin's shell either requires disassembly under extreme conditions or advanced capsid engineering, and in vivo endosomal escape and cargo release remain a major barrier for DDSs [4].
The focus of our work is to demonstrate the potential of a biomarker-specific encapsulin-based system to target specific cells/tissues. DARPins are highly attractive protein targeting moieties, readily expressible and amenable to screening technologies, with large combinatorial flexibility. To build new variants of our targeted DDS, the HER2-specific DARPin9.29 can potentially be exchanged for any other DARPin from the extensive DARPin library [52]. Likewise, flexibility of the encapsulin loading system to encapsulate heterologous protein means that the miniSOG model protein can be replaced by another protein of choice.
In addition to TmEnc-DARPin-STII_miniSOG, we constructed encapsulins fused with a Strep-tag and loaded with miniSOG (TmEnc-STII_miniSOG), and miniSOG fused with a Strep-tag (miniSOG-STII) as control samples for a non-targeted empty capsid, non-targeted loaded capsid and free cytotoxic protein control samples, respectively (Fig. 3).
3.3. Selection of encapsulin with His6 insertion
In the process of selecting a T. maritima encapsulin candidate for the DDS, we compared the wild type T. maritima encapsulin to an encapsulin that contains a His6 insertion with a linker (GGGGGGHHHHHHGGGGG) between residues 42 and 43 of the wild type encapsulin. The His6 linker has been shown to convey thermostability, an attractive property that could allow encapsulins to withstand harsh processing conditions during downstream processing and extend their storage stability [53]. To this end, plasmids (kindly gifted by the EPFL 2018 iGEM team) encoding for encapsulins without (BBa_K2686001) and with the internal His6 insert (BBa_K2686002) were expressed in E. coli BL21Star(DE3).
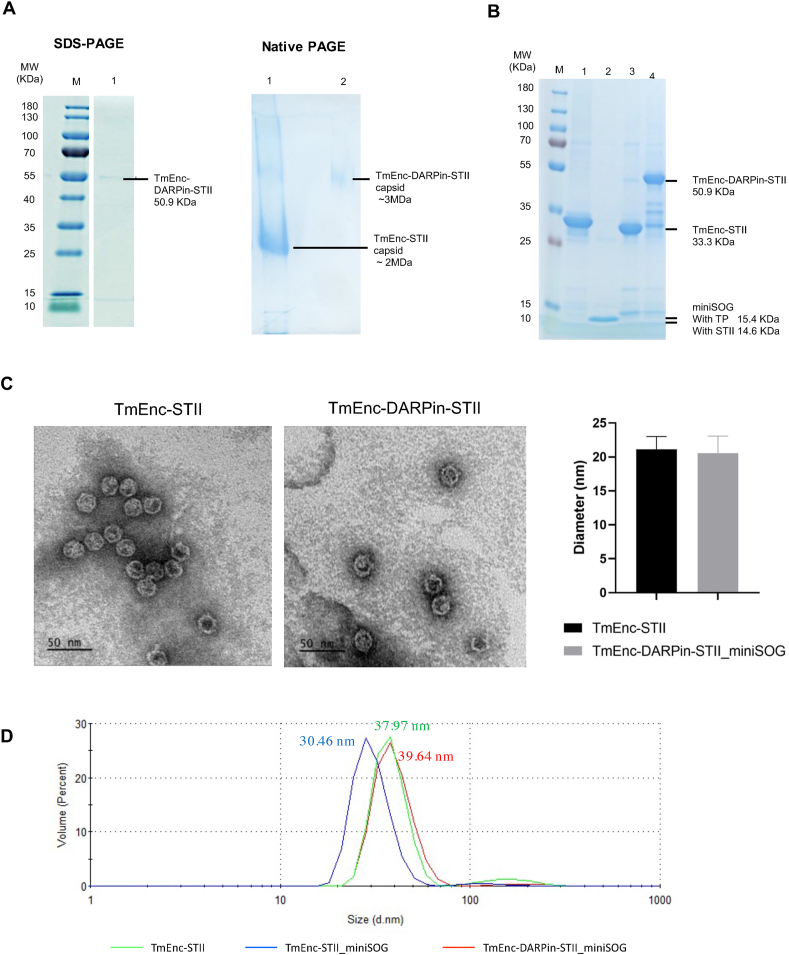
In our hands the expression levels of the constructs and yields were low. To still benefit from increased stability and to circumvent heat-purification, the two BioBrick parts were modified by inserting a Strep-tag at the C terminus, resulting in T. maritima encapsulins with Strep-tag on the outer surface (BBa_K3111102) and T. maritima encapsulins with His6 insert with Strep-tag (BBa_K3111103). This modification allowed successful expression and purification of the proteins from the soluble fraction of the cell lysate. Whilst the wild type T. maritima encapsulin was only partially soluble at the post-induction temperature of 37 °C, its solubility was improved when lowering the induction temperature to 18 °C (Figure A.6A and B). The T. maritima encapsulin with His6 insert produced a considerably higher soluble to insoluble protein ratio than the wild type encapsulin at induction temperature of 37 °C (Figure A.6C). Therefore, the variant with the His6 insert (and Strep-tag) was selected for building the drug delivery system. Production and assembly of Strep-tag-purified encapsulins with His6 insert was demonstrated via TEM where particles of 21.14 ± 1.87 nm in diameter were observed (Fig. 4C).
Fig. 4.
Biochemical/biophysical analysis of T. maritima encapsulin variants. (A) Left: SDS-PAGE, lane M = molecular weight marker (kDa), lane 1 = TmEnc-STII-DARPin-STII. Right: non-reducing PAGE, lane 1 = TmEnc-STII, lane 2 = TmEnc-STII-DARPin-STII. (B) SDS-PAGE loaded with 3.75 μg protein per well: lane M = molecular weight marker (kDa), lane 1 = TmEnc-STII, lane 2 = miniSOG-STII, lane 3 = TmEnc-STII_miniSOG, lane 4 = TmEnc-DARPin-STII_miniSOG. (C) TEM of TmEnc-STII on the left and TmEnc-DARPin-STII on right, histograph shows average diameter and SD of 21.14 ± 1.87 nm (n = 106) for TmEnc-STII and 20.58 ± 2.50 nm (n = 106) for TmEnc-DARPin-STII. (D) Dynamic light scatter graph showing size distribution by volume, red line = TmEnc-DARPin-STII_miniSOG (39.64 nm), green line = TmEnc-STII (37.97 nm), blue line = TmEnc-STII_miniSOG (30.46 nm). Note, the hydrodynamic diameter of the capsid is expected to be larger than the diameter of dried samples measured by TEM.
3.4. Production and assembly of targeted DDS
Next, encapsulins with His6 insert fused with DARPin9.29 were successfully expressed and purified. Correct assembly was verified using SDS-PAGE, non-reducing PAGE gel (Fig. 4A right) and TEM (Fig. 4C). On SDS-PAGE the TmEnc_DARPin-STII fusion protein migrated at approximately the expected molecular weight of 50.9 kDa. As expected, the encapsulins fused with DARPin9.29 migrated slower through the non-reducing PAGE gel than the encapsulins without DARPin9.29, indicating an increase in molecular weight consistent with the presence of the DARPin9.29. Purified particles measured 20.58 ± 2.50 nm in diameter from negative stain TEM images, similar to encapsulins without DARPin9.29 fusion (Fig. 4C), indicating that the overall size has not significantly changed due to fusion on the surface. This was slightly unexpected but maybe be due to the flexibility of the DARPin9.29 fusion protein.
The final sample, miniSOG loaded into these TmEnc-DARPin-STII encapsulins, was also successfully expressed and purified. Assembly was confirmed by the presence of two bands with expected sizes for TmEnc-DARPin-STII (50.9 kDa) and miniSOG (15.4 kDa) on SDS-PAGE (Fig. 4B, lane 4). Co-purification of the miniSOG with the capsid protein provides evidence for encapsulation because miniSOG does not contain a Strep-tag. The two bands also co-eluted from the size exclusion column (SEC) (Figure A.7). The DLS showed particles of similar hydrodynamic diameter (Fig. 4D, red line) to unmodified capsids (TmEnc-STII, Fig. 4D, green line) indicating correct particle formation.
In addition, the control samples, miniSOG alone (miniSOG-STII) and encapsulins loaded with miniSOG but without DARPin9.29 (TmEnc-STII_miniSOG) were also purified and run out alongside the DDS on the SDS-PAGE (Fig. 4B, lanes 2 and 3). The DLS showed assembly of the TmEnc-STII_miniSOG particle with a slightly smaller hydrodynamic diameter than that of the unloaded encapsulin (TmEnc-STII, green line) and the full DDS (TmEnc-DARPin-STII_miniSOG, blue line). The reason for this size difference is unknown.
3.5. The DDS (TmEnc-DARPin-STII_miniSOG) is targeting SK-BR-3 cells and triggers apoptosis
To demonstrate the delivery of the cytotoxic cargo specifically to HER2 receptor expressing cells, SK-BR-3 cells were incubated with the DDS (TmEnc-DARPin-STII_miniSOG) for 60 min at 37 °C and 20% oxygen without illumination while in a parallel sample white light was applied for 60 min in order to activate the encapsulated miniSOG. At the end of the experiment, the cells were visualised by confocal microscopy to observe uptake of the encapsulins. Following that, cell samples were stained using the Annexin V-PI staining kit to determine potential cell death and percentage loss in viability was measured using flow cytometry. To examine the specificity of the cytotoxic effect, MSCs were incubated alongside as negative control.
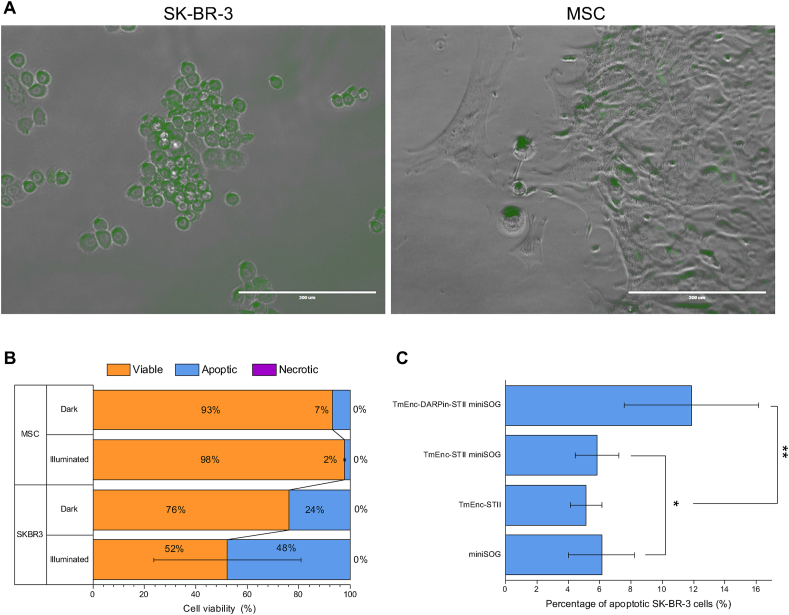
After incubation, green fluorescence from miniSOG was localised within SK-BR-3 cells, some fluorescence signal was also detected in MSCs (Fig. 5A). We hypothesize that non-specific passive uptake into the MSCs has taken place in the absence of the HER2 receptor. It cannot be ruled out that fluorescence is located on the surface of the cells rather than inside the cells. Regardless, the higher fluorescence signal observed in SK-BR-3 cells demonstrates substantial binding and indicates internalisation of the drug delivery system, enhanced by HER2 overexpression and HER2 mediated uptake (Fig. 5A).
Fig. 5.
Specific binding and apoptosis of SK-BR-3 by the DDS (TmEnc-DARPin-STII_miniSOG). (A) Confocal Microscopy image of SK-BR-3 and MSCs after 60-min incubation with DDS showing increased fluorescence intensity correlation to SK-BR-3 cells; Scalebar: 200 μm. It should be noted that SK-BR-3 and MSCs have different morphologies, MSCs are elongated with fibroblastic morphology while the SK-BR-3 have hexagonal shapes and grow in colonies. (B) Flow cytometry analysis showing cell viability percentages from AnnexinV-PI staining after 1 h incubation with the DDS with and without light. Error bars indicate SD across two biological repeats. (C) Percentage apoptotic SK-BR-3 from AnnexinV-PI staining after 1 h incubation in light with control samples (TmEnc-STII_miniSOG, TmEnc-STII and miniSOG-STII). Error bars show SD across triplicate experiments across two biological repeats. T test carried out between * and ** samples returned a P value of 0.031 < 0.05.
The confocal microscopy observations aligned well with flow cytometry analysis that showed a considerable increase of apoptotic cells (48% of cells) in SK-BR-3 incubations, particularly after illumination, leading to reduction in cell viability (Fig. 5B) as was expected if the DDS was functional. A shift in SK-BR-3 cell population incubated in the dark towards apoptosis (24%) was also observed. It was not expected that miniSOG becomes activated in the dark. It can be speculated that light exposure during sample processing has triggered activation and resulted in this loss of cell viability. It is also possible that internalized bacterial proteins in general caused apoptosis. Only a small percentage of apoptotic cells (2% light, 7% dark) was detected in the control MSCs. As the DDS is not expected to bind to those cells, the loss of viability in MSC through apoptosis could be attributed to the greater sensitivity of such stem cells to environmental condition fluctuation, in this instance, strong illumination or the handling of the cells required for imaging and staining. Variation in cell viability was observed in repeat experiments which were carried out after completion of the iGEM project with different passage numbers of SK-BR-3 and a different donor for the MSCs. As before, post-incubation with DDS apoptosis was triggered in SK-BR-3 cells, however apoptosis and necrosis were also observed in MSCs in the light and in the dark, respectively (Figure A.8). Investigations into these variations was out of the scope of this iGEM project and requires careful addressing in future.
Finally, to determine that apoptosis is specifically caused by encapsulins being targeted to the HER2 receptor for uptake into the cells, the DDS incubation experiment was repeated, and the SK-BR-3 cell line was incubated with 3 μM purified sample of encapsulins only (TmEnc-STII), encapsulins loaded with miniSOG (TmEnc-STII_miniSOG) and purified miniSOG (miniSOG-STII). All three control samples showed a similar percentage of apoptotic cells (4–6%), however the percentage of apoptotic cells was significantly higher (12%) after incubation with the targeted DDS (TmEnc-DARPin-STII_miniSOG) (Fig. 5C). This supports the hypothesis that the DDS is capable of specific binding to the HER2 receptor followed by internalisation and release of the cytotoxic payload. It is conceivable that unbound encapsulins (TmEnc-STII), miniSOG (miniSOG-STII) and combined TmEnc-STII_miniSOG sample may still exert a cytotoxic effect on the cells, leading some cells into apoptosis.
4. Discussion
Encapsulins have previously been demonstrated to be viable DDS, where they have been shown to decrease the viability of various cancer targets compared to free drugs. For example, genetic insertion of a short hepatocellular carcinoma (HCC) targeting peptide into the T. maritima encapsulin shell resulted in selective targeting to HCC cells. Subsequent thiol-maleimide conjugation of the synthetic aldoxorubicin drug to the outside surface created a functional targeted, pH-mediated cytotoxic DDS [54]. Recently, Diaz et al. (2021) demonstrated the dynamics of photodynamic therapy using miniSOG loaded encapsulins, which has inspired the use of this cytotoxic protein in our work [46].
Here we describe a breast cancer-targeting DDS system that is entirely genetically encoded and does not require chemical modification. We have fused a genetically engineered antibody mimetic protein (DARPin9.29) to the capsid protein of the T. maritima encapsulin and loaded the cytotoxic protein miniSOG into the lumen of the encapsulin (TmEnc-DARPin-STII_miniSOG).
Using an in vitro cell culture model we first confirmed that DARPin9.29 exhibits specificity for the HER2 receptor of the SK-BR-3 breast cancer cell line when fused to another protein. We observed that binding efficiency was lowered when fusing DARPin9.29 to the C terminus of the fluorescent protein as opposed to the other orientation of the fusion. Nevertheless, the mScarlet-DARPin-STII fusion was still viable (1%–16% of cells bound mScarlet-DARPin-STII) and binding, even to a small number of cells, is likely to reduce the side effects caused to other cells/healthy organs of the human body and may significantly reduce drug concentration needed.
After assembly of the full DDS, we observed effective uptake via the HER2 receptor and activity of the miniSOG. This was evidenced by a significant increase in apoptosis in breast cancer cells treated with the DDS compared to cells treated with non-targeted encapsulins encapsulating miniSOG, free miniSOG and encapsulins without modifications.
Diaz et al. (2021) recently showed passive uptake of otherwise unmodified encapsulins loaded with miniSOG and subsequent ROS generation in human lung adenocarcinoma cells [46]. Incubation for 8 h with miniSOG-loaded encapsulin, followed by a 10 min light pulse, caused a large loss in cell viability (34%) associated with a 2.3-fold increase in internal ROS. We incubated for a significantly shorter time, to maintain cell viability and avoid significant passive uptake of the DDS and non-targeted encapsulins containing miniSOG. Greater effect of our DDS might be expected when allowing for longer incubation times and could be investigated further.
Our results and other group's data also suggested that effective delivery of miniSOG as a phototherapeutic relies on encapsulation or targeting [55,56]. We observed that free miniSOG is not taken up or not at a rate sufficient to stimulate cell death comparable to our DDS. Similarly, encapsulins on their own did not significantly affect cell viability. The same has been observed by Diaz et al. (2021), no significant cell death was caused by T. maritima encapsulins over a PBS control when exposed to light.
Another targeted deliver approach showed that a direct genetic fusion of DARPin9.29 to miniSOG, specifically targeted HER2 and caused phototoxicity [55]. The DARPin miniSOG fusion protein was taken up quickly (5–7 min to localise in the endosome) but affected SK-BR-3 cell viability via necrosis rather than apoptosis. This indicates a different cell death pathway in the same cell line (SK-BR-3). Packaging of miniSOG likely changes the protein uptake rate and the reactive oxygen species release rate and this may affect cell death mechanisms.
When we compared SK-BR-3 and MSCs (control cells) in the cell killing assay we observed higher percentages of apoptotic cells in the SK-BR-3 compared to MSCs, with the highest rate of apoptosis when cells were illuminated, as was expected. However, direct comparison of cell viability has been challenging and a more stable control cell line (other than the in-house MSCs) should be used in future before investigating the functionality and efficacy of the system in vivo.
5. Conclusion
While we have demonstrated the cytotoxic activity of miniSOG when delivered to HER2 breast cancer cells, the key finding of this paper is the successful ‘one-pot’ production of a targeted DDS from a single plasmid and one-step purification of the entire DDS. Self-assembling nanoparticles such as virus like particles (VLPs) and in this study encapsulins can be highly sensitive to direct genetic fusions to capsid proteins. We have shown direct fusion of the T. maritima encapsulin monomer with an 18.4 kDa protein (DARPin-STII), half of the encapsulin monomeric mass, and successful in vivo assembly of the encapsulin-DARPin fusion protein into particles. This is to the best of our knowledge the largest external encapsulin fusion to date and demonstrates high assembly robustness and stability of the T. maritima encapsulin. With small modifications, such as tag-less purification, such a system may have potential for large-scale manufacturing in a robust and cost-effective process.
Lastly, DARPins represent a library of antibody-like specific interactions and could theoretically be combined with encapsulins of different sizes, packed with cargo of choice. The approach described here could form the basis of a modular and multimodal targeted drug delivery platform with high affinity for tumour cells, reducing off-target effects and enhancing safety, with prospects for the development of personalised and targeted therapeutics.
CRediT authorship contribution statement
Alexander Van de Steen: Data curation, Formal analysis, Writing – review & editing, Visualization. Rana Khalife: Data curation, Formal analysis, Writing – review & editing, Visualization. Noelle Colant: Writing – review & editing, Supervision. Hasan Mustafa Khan: Data curation, Formal analysis. Matas Deveikis: Data curation, Formal analysis, Writing – review & editing. Saverio Charalambous: Data curation, Formal analysis, Writing – review & editing. Clare M. Robinson: Data curation, Formal analysis, Writing – review & editing, Visualization. Rupali Dabas: Writing – review & editing. Sofia Esteban Serna: Data curation, Formal analysis, Writing – review & editing. Diana A. Catana: Data curation, Formal analysis, Writing – review & editing. Konstantin Pildish: structural modelling. Vladimir Kalinovskiy: Laboratory safety, Data curation. Kenth Gustafsson: Conceptualization, Project administration, Supervision. Stefanie Frank: Conceptualization, Project administration, Supervision, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors of this paper have no actual or potential conflict of interest including any financial, personal or other relationships with other people or organizations within three years of beginning the submitted work that could inappropriately influence, or be perceived to influence, our work.
Acknowledgements
We thank Mark Turmaine for help with TEM sample preparation and imaging; Ludmila Ruban for mammalian cell culture training; Samir Aoudjane for technical support in the laboratory; Chris Barnes, Darren Nesbeth and Andrew Care (University of Technology Sydney) for project advise; Giovanni Maddalena, Martina Sebastian, Maria Parau, Chileab Redwood-Sawyerr and Moeez Ahmed Khan for general wet lab supervision.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2021.09.001.
Funding acquisition
We thank the EPSRC for funding Stefanie Frank (EP/R013756/1) through the Future Vaccine Manufacturing Research Hub (Vax-Hub); the EPSRC DTP for funding Alexander Van De Steen (EP/R513143/1); Wellcome, BBSRC and EPSRC for financial support for iGEM summer bursaries and the Faculty of Engineering and Department of Biochemical Engineering at UCL for general iGEM project funding. We also thank iGEM sponsors: SnapGene, Twist Bioscience, Integrated DNA Technologies (IDT), New England Biolabs (NEB).
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Gerber D.E. Targeted therapies: a new generation of cancer treatments. Am Fam Phys. Feb. 2008:311–319. [PubMed] [Google Scholar]
- 2.Thorn C.F., Oshiro C., Marsh S., Hernandez-Boussard T., McLeod H., Klein T.E. Doxorubicin pathways: pharmacodynamics and adverse effects. Pharmacogenetics Genom. 2011;21(7):440–446. doi: 10.1097/FPC.0b013e32833ffb56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desai N., Trieu V., Hwang L., Wu R., Soon-Shiong P., Gradishar W. Improved effectiveness of nanoparticle albumin-bound (nab) paclitaxel versus polysorbate-based docetaxel in multiple xenografts as a function of HER2 and SPARC status. Anti Canc Drugs. 2008;19(9):899–909. doi: 10.1097/CAD.0b013e32830f9046. [DOI] [PubMed] [Google Scholar]
- 4.Rosenblum D., Joshi N., Tao W., Karp J.M., Peer D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat Commun. 2018;9:1410. doi: 10.1038/s41467-018-03705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco E., Shen H., Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol. 2015;33(9):941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kakkar A., Traverso G., Farokhzad O., Weissleder R., Langer R. Evolution of macromolecular complexity in drug delivery systems. Nat. Rev. Chem. 2017;1:1–18. doi: 10.1038/s41570-017-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golombek S.K., May J.N., Theek B., Appold L., Drude N., Kiessling F. Tumor targeting via EPR: strategies to enhance patient responses. Adv Drug Deliv Rev. 2018;130:17–38. doi: 10.1016/j.addr.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Shaughnessy J. Pegylated liposomal doxorubicin in the treatment of breast cancer. Clin Breast Canc. 2003;4(5):318–328. doi: 10.3816/cbc.2003.n.037. [DOI] [PubMed] [Google Scholar]
- 9.Allen T., Cullis P. Liposomal drug delivery systems: from concept to clinical applications. Adv Drug Deliv Rev. 2013;65(1):36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 10.Gong J., Chen M., Zheng Y., Wang S., Wang Y. Polymeric micelles drug delivery system in oncology. J Contr Release. 2012;159(3):312–323. doi: 10.1016/j.jconrel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Wang A., Langer R. Nanoparticle delivery of cancer drugs. Annu Rev Med. 2012;63:185–198. doi: 10.1146/annurev-med-040210-162544. [DOI] [PubMed] [Google Scholar]
- 12.Ma Y., Nolte R., Cornelissen J. Virus-based nanocarriers for drug delivery. Adv Drug Deliv Rev. 2012;64(9):811–825. doi: 10.1016/j.addr.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Hong S., Choi D.W., Kim H.N., Park C.G., Lee W., Park H.H. Protein-based nanoparticles as drug delivery systems. Pharmaceutics. 2020;12(7):1–28. doi: 10.3390/pharmaceutics12070604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi S., Kwon I., Hwang K., Kim I., Ahn H. Small heat shock protein as a multifunctional scaffold: integrated tumor targeting and caspase imaging within a single cage. Biomacromolecules. 2011;12(8):3099–3106. doi: 10.1021/bm200743g. [DOI] [PubMed] [Google Scholar]
- 15.Min J., Kim S., Lee J., Kang S. Lumazine synthase protein cage nanoparticles as modular delivery platforms for targeted drug delivery. RSC Adv. Oct. 2014;4(89):48596–48600. doi: 10.1039/c4ra10187a. [DOI] [Google Scholar]
- 16.Han J.A., Kang Y.J., Shin C., Ra J.S., Shin H.H., Hong S.Y. Ferritin protein cage nanoparticles as versatile antigen delivery nanoplatforms for dendritic cell (DC)-based vaccine development. Nanomed Nanotechnol Biol Med. 2014;10(3):561–569. doi: 10.1016/j.nano.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Moon H., Lee J., Min J., Kang S. Developing genetically engineered encapsulin protein cage nanoparticles as a targeted delivery nanoplatform. Biomacromolecules. Oct. 2014;15(10):3794–3801. doi: 10.1021/bm501066m. [DOI] [PubMed] [Google Scholar]
- 18.Lee E.J., Lee N.K., Kim I.S. Bioengineered protein-based nanocage for drug delivery. Adv Drug Deliv Rev. Nov. 2016;106:157–171. doi: 10.1016/J.ADDR.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 19.King N.P., Bale J.B., Sheffler W., McNamara D.E., Gonen S., Gonen T. Accurate design of co-assembling multi-component protein nanomaterials. Nature. 2014 doi: 10.2210/pdb4nwn/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang P.S., Boyken S.E., Baker D. The coming of age of de novo protein design. Nature. Sep. 2016;537(7620):320–327. doi: 10.1038/NATURE19946. [DOI] [PubMed] [Google Scholar]
- 21.Cannon K.A., Park R.U., Boyken S.E., Nattermann U., Yi S., Baker D. Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering. Protein Sci. Apr. 2020;29(4):919–929. doi: 10.1002/PRO.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khmelinskaia A., Wargacki A., King N.P. Structure-based design of novel polyhedral protein nanomaterials. Curr Opin Microbiol. Jun. 2021;61:51–57. doi: 10.1016/J.MIB.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung Y.H., Cai H., Steinmetz N.F. Viral nanoparticles for drug delivery, imaging, immunotherapy, and theranostic applications. Adv Drug Deliv Rev. 2020;156:214–235. doi: 10.1016/j.addr.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashley C.E., Carnes E.C., Phillips G.K., Durfee P.N., Buley M.D., Lino C.A. Cell-specific delivery of diverse cargos by bacteriophage MS2 virus-like particles. ACS Nano. Jul. 2011;5(7):5729–5745. doi: 10.1021/NN201397Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim H., Choi H., Bae Y., Kang S. Development of target-tunable P22 VLP-based delivery nanoplatforms using bacterial superglue. Biotechnol Bioeng. Nov. 2019;116(11):2843–2851. doi: 10.1002/BIT.27129. [DOI] [PubMed] [Google Scholar]
- 26.Le D.H.T., Lee K.L., Shukla S., Commandeur U., Steinmetz N.F. Potato virus X, a filamentous plant viral nanoparticle for doxorubicin delivery in cancer therapy. Nanoscale. Feb. 2017;9(6):2348–2357. doi: 10.1039/C6NR09099K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao J., Guenther R.H., Sit T.L., Opperman C.H., Lommel S.A., Willoughby J.A. Loading and release mechanism of red clover necrotic mosaic virus derived plant viral nanoparticles for drug delivery of doxorubicin. Small. Dec. 2014;10(24):5126–5136. doi: 10.1002/SMLL.201400558. [DOI] [PubMed] [Google Scholar]
- 28.Shan W., Zhang D., Wu Y., Lv X., Hu B., Zhou X. Modularized peptides modified HBc virus-like particles for encapsulation and tumor-targeted delivery of doxorubicin. Nanomed Nanotechnol Biol Med. Apr. 2018;14(3):725–734. doi: 10.1016/J.NANO.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Yan D. Foot-and-mouth disease virus-like particles as integrin-based drug delivery system achieve targeting anti-tumor efficacy. Nanomed Nanotechnol Biol Med. Apr. 2017;13(3):1061–1070. doi: 10.1016/J.NANO.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Sánchez-Sánchez L. Chemotherapy pro-drug activation by biocatalytic virus-like nanoparticles containing cytochrome P450. Enzym Microb Technol. Jun. 2014;60:24–31. doi: 10.1016/J.ENZMICTEC.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Nichols R.J., Cassidy-Amstutz C., Chaijarasphong T., Savage D.F. vol. 52. Taylor and Francis Ltd; 03-Sep-2017. pp. 583–594. (“Encapsulins: molecular biology of the shell,” Critical Reviews in Biochemistry and molecular biology). 5. [DOI] [PubMed] [Google Scholar]
- 32.Giessen T.W. Large protein organelles form a new iron sequestration system with high storage capacity. Elife. 2019;8(Jul) doi: 10.7554/eLife.46070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giessen T.W., Silver P.A. Widespread distribution of encapsulin nanocompartments reveals functional diversity. Nat Microbiol. Jun. 2017;2(6):17029. doi: 10.1038/nmicrobiol.2017.29. [DOI] [PubMed] [Google Scholar]
- 34.Jones J.A., Giessen T.W. vol. 118. John Wiley and Sons Inc; 01-Jan-2021. pp. 491–505. (“Advances in encapsulin nanocompartment biology and engineering,” Biotechnology and Bioengineering). 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodríguez J.M., Allende-Ballestero C., Cornelissen J.J.L.M., Castón J.R. Nanotechnological applications based on bacterial encapsulins. Nanomaterials. Jun. 2021;11(6):1467. doi: 10.3390/nano11061467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutter M. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol. 2008;15(9):939–947. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- 37.Lafrance B., Cassidy-Amstutz C., Nichols R.J., Oltrogge L.M., Nogales E., Savage D.F. The encapsulin from Thermatoga maritima is a flavoprotein with a symmetry matched ferritin-like cargo protein. bioRxiv. Apr. 2021:2021. doi: 10.1101/2021.04.26.441214. 04.26.441214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochoa J.M., Bair K., Holton T., Bobik T.A., Yeates T.O. MCPdb: the bacterial microcompartment database. PloS One. Mar. 2021;16(3 March) doi: 10.1371/journal.pone.0248269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sigmund F. Iron-sequestering nanocompartments as multiplexed electron microscopy gene reporters. ACS Nano. 2019 doi: 10.1021/acsnano.9b03140. [DOI] [PubMed] [Google Scholar]
- 40.Putri R.M., Fredy J.W., Cornelissen J.J.L.M., Koay M.S.T., Katsonis N. Labelling bacterial nanocages with photo-switchable fluorophores. ChemPhysChem. Jun. 2016;17(12):1815–1818. doi: 10.1002/cphc.201600013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lagoutte P. Simultaneous surface display and cargo loading of encapsulin nanocompartments and their use for rational vaccine design. Vaccine. Jun. 2018;36(25):3622–3628. doi: 10.1016/j.vaccine.2018.05.034. [DOI] [PubMed] [Google Scholar]
- 42.Choi B. Effective delivery of antigen-encapsulin nanoparticle fusions to dendritic cells leads to antigen-specific cytotoxic T cell activation and tumor rejection. ACS Nano. Aug. 2016;10(8):7339–7350. doi: 10.1021/acsnano.5b08084. [DOI] [PubMed] [Google Scholar]
- 43.Giessen T.W., Silver P.A. Converting a natural protein compartment into a nanofactory for the size-constrained synthesis of antimicrobial silver nanoparticles. ACS Synth Biol. Dec. 2016;5(12):1497–1504. doi: 10.1021/acssynbio.6b00117. [DOI] [PubMed] [Google Scholar]
- 44.Choi H., Eom S., Kim H., Bae Y., Jung H.S., Kang S. Load and display: engineering encapsulin as a modular nanoplatform for protein-cargo encapsulation and protein-ligand decoration using split intein and SpyTag/SpyCatcher. Biomacromolecules. Jun. 2021 doi: 10.1021/acs.biomac.1c00481. [DOI] [PubMed] [Google Scholar]
- 45.Bae Y., Kim G.J., Kim H., Park S.G., Jung H.S., Kang S. Engineering tunable dual functional protein cage nanoparticles using bacterial superglue. Biomacromolecules. Jul. 2018;19(7):2896–2904. doi: 10.1021/acs.biomac.8b00457. [DOI] [PubMed] [Google Scholar]
- 46.Diaz D., Vidal X., Sunna A., Care A. Bioengineering a light-responsive encapsulin nanoreactor: a potential tool for in vitro photodynamic therapy. ACS Appl Mater Interfaces. Feb. 2021;13(7):7977–7986. doi: 10.1021/acsami.0c21141. [DOI] [PubMed] [Google Scholar]
- 47.Knight T. MIT Synthetic Biology Working Group; 2003. “Idempotent vector design for standard assembly of biobricks,” MIT artificial intelligence laboratory. [Google Scholar]
- 48.Zahnd C., Pecorari F., Straumann N., Wyler E., Plückthun A. Selection and characterization of Her2 binding-designed ankyrin repeat proteins. J Biol Chem. Nov. 2006;281(46):35167–35175. doi: 10.1074/jbc.M602547200. [DOI] [PubMed] [Google Scholar]
- 49.Mironova K.E., Chernykh O.N., Ryabova A., Stremovskiy O.A., Proshkina G.M., Deyev S.M. Highly specific hybrid protein DARPin-mCherry for fluorescent visualization of cells overexpressing tumor marker HER2/neu. Biokhimiya. 2014;79(12):1698–1704. doi: 10.1134/S0006297914120141. [DOI] [PubMed] [Google Scholar]
- 50.Cassidy-Amstutz C. Identification of a minimal peptide tag for in vivo and in vitro loading of encapsulin. Biochemistry. Jun. 2016;55(24):3461–3468. doi: 10.1021/acs.biochem.6b00294. [DOI] [PubMed] [Google Scholar]
- 51.Torra J. Tailing miniSOG: structural bases of the complex photophysics of a flavin-binding singlet oxygen photosensitizing protein. Sci Rep. Dec. 2019;9(1):1–10. doi: 10.1038/s41598-019-38955-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Binz H.K. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol. May 2004;22(5):575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 53.Moon H., Lee J., Min J., Kang S. 2014. Developing genetically engineered encapsulin protein cage nanoparticles as a targeted delivery nanoplatform. [DOI] [PubMed] [Google Scholar]
- 54.Moon H., Lee J., Min J., Kang S. Developing genetically engineered encapsulin protein cage nanoparticles as a targeted delivery nanoplatform. Biomacromolecules. Oct. 2014;15(10):3794–3801. doi: 10.1021/bm501066m. [DOI] [PubMed] [Google Scholar]
- 55.Proshkina G.M., Shilova O.N., Ryabova A.V., Stremovskiy O.A., Deyev S.M. A new anticancer toxin based on HER2/neu-specific DARPin and photoactive flavoprotein miniSOG. Biochimie. 2015;118:116–122. doi: 10.1016/j.biochi.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 56.Shilova O.N., Proshkina G.M., Lebedenko E.N., Deyev S.M. Internalization and recycling of the HER2 receptor on human breast adenocarcinoma cells treated with targeted phototoxic protein DARPinminiSOG. Acta Naturae. 2015;7(3):126–132. doi: 10.32607/20758251-2015-7-3-126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web references
- Annexin V-FITC Apoptosis Staining/Detection Kit (ab14085) https://www.abcam.com/annexin-v-fitc-apoptosis-staining--detection-kit-ab14085.htmlhttps://www.abcam.com/annexin-v-fitc-apoptosis-staining--detection-kit-ab14085.html Abcam (no date). Available at: (Accessed: 20 January 2020)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.