Abstract
Sister chromatid exchange (SCE) frequency is a commonly used index of chromosomal stability in response to environmental or genetic mutagens. However, the mechanism generating cytologically detectable SCEs and, therefore, their prognostic value for chromosomal stability in mitotic cells remain unclear. We examined the role of the highly conserved homologous recombination (HR) pathway in SCE by measuring SCE levels in HR-defective vertebrate cells. Spontaneous and mitomycin C-induced SCE levels were significantly reduced for chicken DT40 B cells lacking the key HR genes RAD51 and RAD54 but not for nonhomologous DNA end-joining (NHEJ)-defective KU70−/− cells. As measured by targeted integration efficiency, reconstitution of HR activity by expression of a human RAD51 transgene restored SCE levels to normal, confirming that HR is the mechanism responsible for SCE. Our findings show that HR uses the nascent sister chromatid to repair potentially lethal DNA lesions accompanying replication, which might explain the lethality or tumorigenic potential associated with defects in HR or HR-associated proteins.
Symmetrical exchanges between newly replicated chromatids and their sisters can be visualized cytologically in vertebrate cells if the DNA of one chromatid is labelled with 5-bromodeoxyuridine (BUdR) during synthesis. Sister chromatid exchanges (SCEs) can be induced by various genotoxic treatments (10), suggesting that SCEs reflect a DNA repair process. Cytological assessment of SCE levels in peripheral blood lymphocytes is used as an index of the mutagenic potential of environmental factors. More importantly, ∼10 SCEs occur spontaneously in normally cycling human cells (5, 8), suggesting a link between SCE and DNA replication. Elevated spontaneous SCE levels are observed in cells from Bloom syndrome patients (9), in mouse cells that lack poly(ADP-ribose) polymerase (29) or KU70 (15), and in hamster cells with defects in XRCC1 (28), but the causal relationships between these enzymes and SCE are not clear.
While the phenomenon of SCE has long been established (27) and many observations about the induction of SCEs have been made, their molecular basis remains obscure. SCE is intimately associated with DNA replication, and eukaryotic cells exposed to DNA-damaging agents in G2 show elevated SCE levels only after completing a subsequent replication cycle (32). Homologous recombination (HR) was suggested as one of the mechanisms responsible (13, 14). While HR occurs between sister chromatids in yeast as a means to replicate around UV-induced lesions (12), it has not been considered constitutively active during metazoan mitosis, perhaps because of the predominance of the nonhomologous DNA end-joining (NHEJ) pathway (30). In addition, the lack of recombinational repair mutants precluded direct testing of HR’s involvement in SCE, so other models evolved. It was proposed that SCEs result from strand switching at stalled replication forks (20). Another model involved topoisomerase II action at coincident breaks at replication forks on both sister chromatids and subsequent rejoining (4, 11, 19).
The two double-strand break (DSB) repair pathways of HR and NHEJ are highly conserved between yeast and vertebrate cells. HR uses a homologous chromosome or a sister chromatid as a template to effect precise repair of a DNA lesion, while the NHEJ pathway carries out repair with lower fidelity and no requirement for homology. To test the idea that HR between sister chromatids is the primary mechanism for SCE, we used reverse genetics in the hyperrecombinogenic chicken B-cell line DT40 (3) to genetically ablate the HR enzymes Rad51 (23, 24) and Rad54 (1, 7) and then measured SCE levels. We found that spontaneous and mitomycin C (MMC)-induced SCE levels were significantly reduced in HR-deficient RAD51−/− or RAD54−/− DT40 cells but not in NHEJ-defective KU70−/− cells. These findings suggest that HR is one of the principal mechanisms responsible for SCE in vertebrate cells.
MATERIALS AND METHODS
Cells and cell culture.
The generation of RAD54−/−, RAD51−/−/tet-hRAD51+, and KU70−/− mutant DT40 cells has been described (1, 24, 25). RAD51−/− clone 104 was obtained by the same method as 110 cells (24). Cells were cultured in RPMI 1640 medium supplemented with 10−5 M β-mercaptoethanol, 10% fetal calf serum, and 1% chicken serum (Sigma, St. Louis, Mo.) at 39.5°C.
Gene targeting assay.
The targeting construct for the ovalbumin locus was prepared by insertion of an 8-kb PmaCI-PshAI genomic fragment into pSP72 followed by the insertion of a puromycin resistance cassette into the vector’s unique HindIII site (3). The targeting construct was linearized by PvuI and electroporated (550 V, 25 μF) into 5 × 106 cells of each clone. Southern analysis following selection was performed as described previously (3).
Western blot analysis.
Western blot analysis of Rad51 and Ku was performed as described previously (24). Briefly, 106 cells were lysed in 20 ml of sodium dodecyl sulfate lysis buffer. Following sonication and boiling, aliquots were subjected to sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis. After transfer to a nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany), proteins were detected by polyclonal rabbit anti-human Rad51 polyclonal serum or rabbit anti-chicken Ku70 polyclonal serum (25) and horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin (Santa Cruz Biotechnology, Santa Cruz, Calif.) with a Super Signal CL substrate (Pierce, Rockford, Ill.).
Measurement of SCE levels.
For BUdR labelling, cells were cultured in the presence of 10 μM BUdR for 18 to 21 h (two cell cycle periods) and pulsed with 0.1 μg of Colcemid per ml for the last 2 h (3 h in the case of RAD51−/− cells). MMC (50 ng/ml) was added 8 h before harvest. The MMC sensitivities of wild-type and RAD54−/− DT40 clones, as measured by colony formation in methylcellulose plates (0.2% survival relative to untreated cells), were comparable. Harvested cells were treated with 75 mM KCl for 15 to 30 min and subsequently fixed with methanol:acetic acid (3:1) for at least 30 min. Cells were fixed onto wet (50% ethanol) glass slides and dried on a 40 to 42°C plate. Dried slides were incubated with 10 μg of Hoechst 33258 per ml in phosphate buffer (pH 6.8) for 20 min, followed by rinsing with MacIlvaine solution (164 mM Na2HPO4, 16 mM citric acid [pH 7.0]). Slides were irradiated with black light (λ = 352 nm) for 60 min and incubated in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) solution at 62°C for 1 h before staining with 3% Giemsa solution (pH 6.8) and subsequent microscopy.
RESULTS AND DISCUSSION
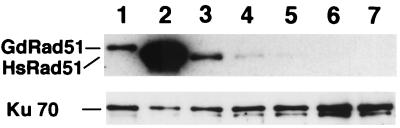
To measure the frequency of SCE, RAD54−/− and KU70−/− DT40 cells were labelled with BUdR for two cell cycle periods (18 h). Since RAD51 is an essential gene, we used RAD51−/− cells expressing a tet-repressible human RAD51 (hRAD51) transgene (24) and labelled cells with BUdR simultaneously with repression of the transgene, so that the rapid cell death following Rad51 depletion would not interfere with SCE analysis; the time-dependent repression of this transgene is confirmed in Fig. 1. The rate of spontaneous SCE in wild-type DT40 cells is 3.0 ± 1.7 exchanges per metaphase (Fig. 2 and 3A), while chicken and human cells display ∼3 and ∼10 exchanges per mitosis on average, respectively (5, 8, 10, 18, 31). As the human genome is three times larger than the chicken genome, the numbers of SCEs per unit length of genomic DNA are comparable among DT40 cells, chicken embryonic B lymphocytes, and human cells.
FIG. 1.
Western blot analysis of Rad51 expression in DT40 cells. Each lane contains 10 μg of protein visualized with anti-hRad51 and anti-Ku70 antisera as described previously (24). Lanes: 1, parental DT40; 2, RAD51−/− cells expressing high levels of hRad51 (clone 104: hRad51hi); 3, RAD51−/− cells expressing low levels of hRad51 (clone 110: hRad51lo); 4 to 7, clone 110 cells at 6, 12, 18, 21 h, respectively, after the repression of the tet-controlled hRAD51 transgene.
FIG. 2.
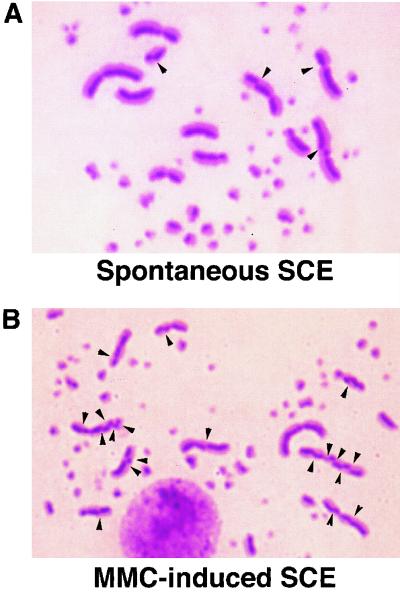
SCE in wild-type DT40 cells. Arrowheads indicate the sites of SCE.
FIG. 3.
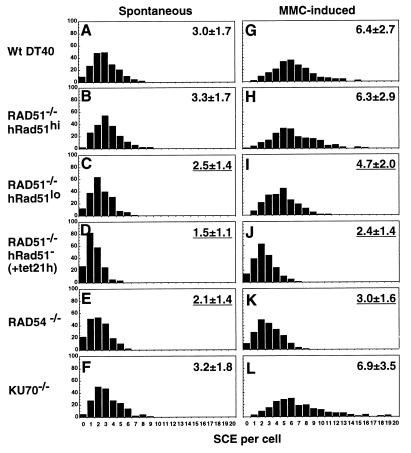
Reduced levels of SCE in cells deficient in HR. Cells were labelled with BUdR during two cell cycle periods with or without MMC treatment (50 ng/ml) for the last 8 h. Spontaneous and MMC-induced SCEs in the macrochromosomes of 200 metaphase cells were counted. Histograms show the frequency of cells with the indicated numbers of SCEs per cell. The mean number of SCEs per cell ± the standard deviation is shown in the upper right corner of each histogram; underlined values differ significantly (P < 0.002) from wild-type (wt) control SCE levels; statistical significance was calculated by the Mann-Whitney nonparametric U test.
Compared with wild-type cells (Fig. 3A; 3.0 SCEs/cell), the level of spontaneous SCE was significantly reduced in RAD54−/− cells (Fig. 3E; 2.1 SCEs/cell, P < 0.0001), which have a low level of HR as measured by targeted integration frequency (Table 1) (1). Similarly, the RAD51−/− clone 110, which expresses a low level of hRAD51 transgene (24), showed significantly reduced SCE frequency (Fig. 3C; 2.5 SCEs/cell, P = 0.0013) as well as a reduction in targeted integration (Table 1) compared with wild-type cells. The inhibition of the human RAD51 transgene with tetracycline further reduced the level of SCE in 110 cells (Fig. 3D; 1.5 SCEs/cell, P < 0.0001). The reduced level of SCE found with RAD51 deficiency is likely an underestimate, as this method detects SCE between chromatids labelled during DNA synthesis, when diminishing amounts of Rad51 are still present. As it does for spontaneous SCE, RAD51 or RAD54 deficiency causes a statistically significant reduction in MMC-induced SCE (Fig. 3I to K). The residual HR and SCE activity in RAD51−/− and RAD54−/− cells may be attributable to other Rad51 and Rad54 homologues, such as XRCC2, XRCC3, Rad51B, and Rad54B. Strikingly, overexpression of hRAD51 in the RAD51−/− clone 104 restored both targeted integration and spontaneous and induced SCE frequencies to wild-type levels (Fig. 3B and H; Table 1). To assess whether NHEJ contributes to SCE, we counted SCEs in KU70−/− DT40 cells (25) (Fig. 3F and L) and found a slight increase in SCE frequency, without statistical significance, despite a previous report of elevated SCE in Ku70-deficient mouse fibroblasts (15). We suggest that the elevated HR rate in DT40 cells (3) may mask any minor effects of Ku70 deficiency on SCE. In summary, these observations reveal that HR between sister chromatids is principally responsible for SCE in higher eukaryotic cells.
TABLE 1.
Transfection and targeted integration frequencies in wild-type and HR-deficient DT40 cells
| Cell type | No. of clones
|
Transfection efficiency | |
|---|---|---|---|
| Targeteda | Analyzed | ||
| Wild-type DT40 | 32 | 43 | 6.0 × 10−5 |
| RAD51−/− hRad51hi | 30b | 44 | 6.0 × 10−5 |
| RAD51−/− hRad51lo | 6c | 43 | 6.8 × 10−5 |
| RAD54−/− | 0 | 44 | 1.2 × 10−4 |
Ovalbumin targeting construct was transfected into cells of the genotypes indicated, and targeted integration events following selection were determined by Southern blot analysis.
Not significantly different from wild-type levels (χ2 = 0.413, P = 0.5204).
Significantly different from wild-type levels (χ2 = 31.873, P < 0.0001).
The involvement of HR in SCE is a little surprising, because the presence of spontaneous SCEs in vertebrate cells indicates the presence of active HR during mitosis. However, the presence of Rad51 foci in S-phase nuclei (26), as well as spontaneous chromosomal breaks in HR-deficient RAD51−/− (24) and RAD54−/− (25) cells, has suggested the essential involvement of HR in the maintenance of chromosomal integrity in vertebrate cells. Spontaneous DSBs that necessitate repair by the Rec proteins and occur during replication in Escherichia coli (17, 22) and the recently described formation and resolution of Holliday junctions in Saccharomyces cerevisiae mitosis (33) demonstrate that the role of HR in ensuring complete replication of the genome has been conserved throughout evolution.
DNA replication across a nick is likely to produce a DSB in one of the sister chromatids, while replication stalled at a damaged base may produce a single-strand gap between the damaged base and a new Okazaki fragment initiating downstream. However, the nature and origin of the DNA lesions generated during normal DNA replication and eventually repaired by HR, resulting in SCEs, remain to be determined. Chemical modifications of the genomic DNA, such as hydrolysis, oxidation, and nonenzymatic methylation, occur at significant rates in vivo (16). Although such covalently modified bases are usually repaired before DNA replication, it is possible that unrepaired lesions and nicks are encountered by replication forks and result in single-strand gaps and DSBs in one of the sister chromatids. Such strand discontinuities can be repaired postreplicationally by HR with the sister chromatid, as is the case for recombinational repair in yeast cells (12). Indeed, the modification of genomic DNA by 3-methyladenine specifically induces S-phase arrest, SCEs, and chromosome breaks (6). DSBs may also result from the breakage of arrested replication forks, following the forks’ being impeded by DNA secondary structures and DNA-bound proteins (2).
The genetic analysis of recombination between sister chromatids in yeast, which is useful in detecting nonmutagenic carcinogens (21), can detect unequal SCE and gene conversion but not equal exchange of sister chromatids. The analysis of SCE in vertebrate cells provides a striking insight not only into the cellular response to potential carcinogens but also into the frequency of spontaneous lesions necessitating HR-mediated repair during DNA replication and the accuracy with which the HR machinery repairs them.
ACKNOWLEDGMENTS
We thank M. Hashishin, Y. Sato, O. Koga, and M. Hirao for their excellent technical assistance and Y. Ejima (Kyoto University) and T. Horiuchi (Okazaki National Institutes) for helpful discussions. We are indebted to J. Haber (Brandeis University) and W. F. Morgan (UCSF) for their critical readings of the manuscript.
C.M. is the recipient of a JSPS Postdoctoral Fellowship. The Bayer-Chair Department of Molecular Immunology and Allergology is supported by Bayer Yakuhin, Kyoto, Japan. This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, by CREST of JST (Japan Science and Technology), and by a grant from the Mochida Memorial Foundation for Medical and Pharmaceutical Research.
REFERENCES
- 1.Bezzubova O Y, Silbergleit A, Yamaguchi-Iwai Y, Takeda S, Buerstedde J M. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell. 1997;89:185–193. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- 2.Bierne H, Michel B. When replication forks stop. Mol Microbiol. 1994;13:17–23. doi: 10.1111/j.1365-2958.1994.tb00398.x. [DOI] [PubMed] [Google Scholar]
- 3.Buerstedde J M, Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- 4.Cleaver J E. Correlations between sister chromatid exchange frequencies and replicon sizes. A model for the mechanism of SCE production. Exp Cell Res. 1981;136:27–30. doi: 10.1016/0014-4827(81)90034-3. [DOI] [PubMed] [Google Scholar]
- 5.Crossen P E, Drets M E, Arrighi F E, Johnston D A. Analysis of the frequency and distribution of sister chromatid exchanges in cultured human lymphocytes. Hum Genet. 1977;35:345–352. doi: 10.1007/BF00446625. [DOI] [PubMed] [Google Scholar]
- 6.Engelward B P, Allan J M, Dreslin A J, Kelly J D, Wu M M, Gold B, Samson L D. A chemical and genetic approach together define the biological consequences of 3-methyladenine lesions in the mammalian genome. J Biol Chem. 1998;273:5412–5418. doi: 10.1074/jbc.273.9.5412. [DOI] [PubMed] [Google Scholar]
- 7.Essers J, Hendriks R W, Swagemakers S M A, Troelstra C, de Wit J, Bootsma D, Hoeijmakers J H J, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 8.Galloway S M, Evans H J. Sister chromatid exchange in human chromosomes from normal individuals and patients with ataxia telangiectasia. Cytogenet Cell Genet. 1975;15:17–29. doi: 10.1159/000130495. [DOI] [PubMed] [Google Scholar]
- 9.German J, Ellis N A. Bloom syndrome. In: Vogelstein B, Kinzler K, editors. The genetic basis of human cancer. New York, N.Y: McGraw-Hill; 1998. pp. 301–315. [Google Scholar]
- 10.Hagmar L, Bonassi S, Stromberg U, Brogger A, Knudsen L E, Norppa H, Reuterwall C. Chromosomal aberrations in lymphocytes predict human cancer: a report from the European Study Group on Cytogenetic Biomarkers and Health (ESCH) Cancer Res. 1998;58:4117–4121. [PubMed] [Google Scholar]
- 11.Ishii Y, Bender M A. Effects of inhibitors of DNA synthesis on spontaneous and ultraviolet light-induced sister-chromatid exchanges in Chinese hamster cells. Mutat Res. 1980;79:19–32. doi: 10.1016/0165-1218(80)90144-5. [DOI] [PubMed] [Google Scholar]
- 12.Kadyk L C, Hartwell L H. Replication-dependent sister chromatid recombination in rad1 mutants of Saccharomyces cerevisiae. Genetics. 1993;133:469–487. doi: 10.1093/genetics/133.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato H. Possible role of DNA synthesis in formation of sister chromatid exchanges. Nature. 1974;252:739–741. doi: 10.1038/252739a0. [DOI] [PubMed] [Google Scholar]
- 14.Kuzminov A. Recombinational repair in eukaryotes. In: Kuzminov A, editor. Recombinational repair of DNA damage. New York, N.Y: Springer-Verlag; 1996. pp. 185–203. [Google Scholar]
- 15.Li G C, Ouyang H, Li X, Nagasawa H, Little J B, Chen D J, Ling C C, Fuks Z, Cordon-Cardo C. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol Cell. 1998;2:1–8. doi: 10.1016/s1097-2765(00)80108-2. [DOI] [PubMed] [Google Scholar]
- 16.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 17.Michel B, Ehrlich S D, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Natarajan A T, van Zeeland A A, Verdegaal-Immerzeel E A, Filon A R. Studies on the influence of photoreactivation on the frequencies of UV-induced chromosomal aberrations, sister-chromatid exchanges and pyrimidine dimers in chicken embryonic fibroblasts. Mutat Res. 1980;69:307–317. doi: 10.1016/0027-5107(80)90095-0. [DOI] [PubMed] [Google Scholar]
- 19.Painter R B. A replication model for sister-chromatid exchange. Mutat Res. 1980;70:337–341. doi: 10.1016/0027-5107(80)90023-8. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki M S. Chromosome aberration formation and sister chromatid exchange in relation to DNA repair in human cells. In: Generoso W M, Shelby M D, De Serres F J, editors. DNA repair and mutagenesis in eukaryotes. New York, N.Y: Plenum Press; 1980. pp. 285–313. [DOI] [PubMed] [Google Scholar]
- 21.Schiestl R H. Nonmutagenic carcinogens induce intrachromosomal recombination in yeast. Nature. 1989;337:285–288. doi: 10.1038/337285a0. [DOI] [PubMed] [Google Scholar]
- 22.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 23.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 24.Sonoda E, Sasaki M S, Buerstedde J-M, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. Rad51 deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takata M, Sasaki M S, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tashiro S, Kotomura N, Shinohara A, Tanaka K, Ueda K, Kamada N. S phase specific formation of the human Rad51 protein nuclear foci in lymphocytes. Oncogene. 1996;12:2165–2170. [PubMed] [Google Scholar]
- 27.Taylor J H. Sister chromatid exchanges in tritium-labeled chromosomes. Genetics. 1958;43:515–529. doi: 10.1093/genetics/43.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson L H, Brookman K W, Jones N J, Allen S A, Carrano A V. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol Cell Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z Q, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner E F. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weaver D T. What to do at an end: DNA double-strand-break repair. Trends Genet. 1995;11:388–392. doi: 10.1016/s0168-9525(00)89121-0. [DOI] [PubMed] [Google Scholar]
- 31.Wilmer J L, Colvin O M, Bloom S E. Cytogenetic mechanisms in the selective toxicity of cyclophosphamide analogs and metabolites towards avian embryonic B lymphocytes in vivo. Mutat Res. 1992;268:115–130. doi: 10.1016/0027-5107(92)90089-k. [DOI] [PubMed] [Google Scholar]
- 32.Wolff S, Bodycote J, Painter R B. Sister chromatid exchanges induced in Chinese hamster cells by UV irradiation of different stages of the cell cycle: the necessity for cells to pass through S. Mutat Res. 1974;25:73–81. doi: 10.1016/0027-5107(74)90220-6. [DOI] [PubMed] [Google Scholar]
- 33.Zou H, Rothstein R. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]