Highlights
-
•
Some gynecologic tumors harbor a POLE pathogenic variant, raising prognostic and therapeutic issues.
-
•
Tumors harboring a POLE pathogenic variant exhibit multiple BRCA1/2 variants, reflecting the high tumor mutational burden.
-
•
Tumor BRCA testing could be a way to detect tumors harboring a highly mutagenic POLE pathogenic variant.
Keywords: Oncogenetics, POLE, Tumor BRCA testing
Abstract
Objective
Tumors harboring a POLE pathogenic variant, associated with high tumor mutational burden, are good candidates for immunotherapy. However, POLE pathogenic variants are not currently screened in routine clinical practice. Can these tumors be identified by means of an already available test?
Methods
We describe seven tumors harboring a POLE pathogenic variant, among eight patients with tumors harboring multiple BRCA1/2 variants (from 4 to 20). All patients were managed at Institut Curie, Paris. Five patients were selected because of unexpected tumor BRCA testing results with multiple variants and another three patients were selected because of a POLE pathogenic variant detected by large tumor testing. We looked for other tumor variants by Next-Generation Sequencing in tumors harboring multiple BRCA1/2 variants, and for multiple BRCA1/2 variants in tumors harboring a POLE pathogenic variant.
Results
Four of the five tumors selected because of multiple BRCA1/2 variants exhibited a POLE pathogenic variant, and all three tumors selected for POLE pathogenic variants exhibited multiple BRCA1/2 variants.
Conclusions
Tumor BRCA testing could be a way to detect tumors harboring a highly mutagenic POLE pathogenic variant.
1. Introduction
Tumor genetic testing, allowing personalized treatment, is a rapidly growing field. Tumor BRCA testing is already performed routinely, especially in high-grade ovarian cancers, as identification of a pathogenic variant (PV) of BRCA1/2 genes may guide treatment towards PARP inhibitors. (Konstantinopoulos et al., 2020) Tumors usually harbor no more than one BRCA1/2 variant.
POLE and POLD1 genes both code for DNA polymerases involved in DNA proofreading and repair. Some PV in the exonuclease domain have been described as drivers in various types of ultramutated tumors. (Campbell et al., 2017, Kandoth et al., 2013, Network, 2012, Briggs and Tomlinson, 2013 Jun) These tumors exhibit a specific molecular signature: tumors with POLE PV are enriched in [TCT → A] and [TCG → T] and tumors with POLD1 PV are enriched in [TCT → A] and [TCA → A]. Their high tumor mutational burden (TMB) makes them good candidates for immunotherapy. (Alexandrov et al., 2013, Mehnert et al., 2016) They may also be associated with a better prognosis. (Van Gool et al., 2018, Domingo et al., 2016, Hoang et al., 2015) Identification of these tumors therefore has prognostic and therapeutic implications. Families with germline POLE PV, responsible for predisposition to various types of cancers, including colorectal and gynecologic tumors, have also been described. (Briggs and Tomlinson, 2013 Jun, Hansen et al., 2015) However, most tumor POLE PV are detected in the context of research studies, as they are not currently screened in routine clinical practice, but what if these tumors could be identified by means of an already available test: tumor BRCA testing? For the first time, we describe seven tumors with multiple BRCA1/2 variants associated with a POLE PV.
2. Materials and methods
Patient selection:
Patients were identified in two different ways.
First, we selected patients with more than 4 BRCA1/2 gene variants from a group composed of 1300 patients who underwent routine tumor BRCA testing at Institut Curie between December 2017 and July 2020 (mainly for ovarian cancers). We subsequentially looked for other variants in their tumors using a large Next-Generation Sequencing (NGS) panel.
Second, we selected patients with a POLE or POLD1 PV from a group composed of 1009 patients with different types of cancer who underwent large NGS tumor analysis between July 2019 and July 2020 and reviewed their BRCA1/2 results.
All patients included in this study were managed at Institut Curie and provided written consent for research including genetic analysis. This project was approved by the local IRB.
Tumor analysis:
Tumor DNA was extracted from formalin-fixed and paraffin-embedded (FFPE) tumor tissue.
Tumor BRCA testing was performed by targeted NGS encompassing the full coding sequences of BRCA1/2 genes. All variants with depth ≥ 300X and variant allelic frequency (VAF) ≥ 5% were retained, regardless of the ACMG class, as we wanted to detect passenger variants reflecting TMB and mutagenesis (excluding polymorphisms which can be germline variants).
Large tumor analysis was performed with an in-house NGS panel, which allows molecular analysis of tumors for microsatellite instability, TMB and mutational signatures. (Alexandrov, 2020) All variants with depth ≥ 200X, VAF ≥ 5% and population frequency less than 0.5% in gnomAD were validated.
Methods are described in detail in the supplementary material (s1).
3. Results
In the first group (1300 patients with routine tumor BRCA testing), we identified 5 tumors with more than 4 different BRCA1/2 variants. Use of the large NGS panel detected a POLE PV in 4 of these 5 tumors. No POLD1 PV was detected.
In the second group (1009 patients with large NGS panel analysis), we identified 3 tumors with a POLE PV, all of which exhibited at least 4 BRCA1/2 variants. No POLD1 PV was detected.
We therefore report a total of 7 tumors (6 gynecologic and 1 colorectal) with co-occurrence of a POLE PV and multiple BRCA1/2 variants. Patient characteristics are shown in Table 1 and described in detail in supplementary material (s2).
Table 1.
Main patient and tumor characteristics at diagnosis.
| Patient | Sex | Cancer site (age) | Cancer type | Cancer grade | Cancer stage | MMR IHC | Unstable markersa | Other significant medical historyb(age) | Family history of cancer (age) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | F | Ovary (53) | Endometrioid | 2 | FIGO IA | No loss | NA | No | Maternal grandfather: lung cancer (55), maternal uncle: prostate cancer (60), maternal niece: borderline ovarian cancer (47), maternal niece: renal cancer (35) |
| 2 | F | Ovary (43) | Endometrioid | 3 | FIGO IA | No loss | 0/5 | No | Paternal grandfather: colorectal cancer, paternal uncle: lymphoma, paternal uncle: gastric cancer |
| 3 | F | Endometrium (52) | Mixed, predominantly endometrioid | NA | FIGO IA | NA | NA | G0P0, hypertension, type 2 diabetes, overweight | Sister: lung cancer |
| 4 | F | Endometrium (65) | Endometrioid | 2 | FIGO IIIC1 | No loss | 0/5 | G0P0, hypertension, smoking, invasive ductal breast carcinoma (65), small-cell lung carcinoma (71) | No |
| 5 | F | Endometrium (51) | Endometrioid | 3 | FIGO IIIC2 | Loss of MSH6 | 2/5 | No | Maternal uncle: pancreas cancer (68), paternal uncle: chronic lymphocytic leukemia (61) + vesical cancer (63) |
| 6 | F | Endometrium + ovary (54) | Mixed, predominantly endometrioid | 3 | FIGO IA + FIGOA IA | No loss | 0/5 | G0P0 | Father: colorectal cancer (67), paternal uncle: prostate cancer (74), maternal grandmother: endometrial cancer (74), maternal aunt: endometrial cancer (80) |
| 7 | M | Rectum (31) | Adenocarcinoma | 1 | T4N0M0 | No loss | 0/5 | No | No |
F: female, M: male, MMR IHC: Mismatch repair immunohistochemistry, NA: not available
BRCA testing was performed first in patients 1, 2, 4 and 6 and large NGS tumor analysis was performed first in patients 3,5 and 7.
by Pentaplex analysis
only other cancers and risk factors for the cancer studied are reported here
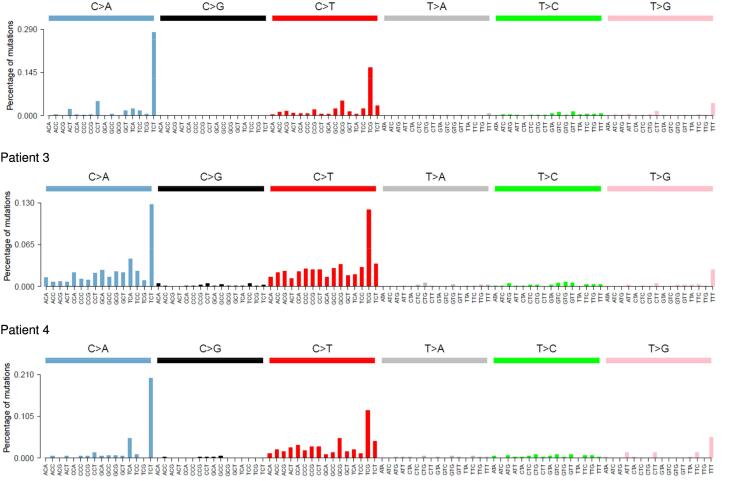
Molecular results are shown in Table 2. The 7 tumors harbored multiple BRCA1/2 variants (from 4 to 20) with a mean of 8.4 variants per tumor, mostly corresponding to variants of uncertain significance (78%). All tumors harbored a POLE PV with a VAF ranging from 25% to 39%. All tumors but two (unavailable signature) had an Alexandrov mutational signature 10, corresponding to the POLE signature (Fig. 1) and all had a high TMB (from 92 to 841 variants/Mb) with a mean of 312 variants/Mb per tumor (greater than 15 variants/Mb cut-off).
Table 2.
Molecular results of BRCA1/2 and POLE testing.
| Patient |
POLE pathogenic variant (NM_006231.2) |
TMB (variants/Mb) |
BRCA1 variants (NM_007294.3) |
BRCA2 variants (NM_000059.3) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variant | VAF (%) | Variant | VAF (%) | Classa | Variant | VAF (%) | Classa | |||||
| 1 | c.890C>T | p.(Ser297Phe) | 25 | 841 | c.5338C>A | p.(Leu1780Met) | 6.8 | 3 | c.1475C>T | p.(Ser492Phe) | 13.7 | 3 |
| c.4485-31G>A | p.? | 36.3 | 3 | c.2046C>A | p.(=) | 33.8 | 3 | |||||
| c.3890C>A | p.(Ser1297Tyr) | 19.4 | 3 | c.2246G>T | p.(Ser749Ile) | 7.6 | 3 | |||||
| c.2432A>C | p.(Lys811Thr) | 40.7 | 3 | |||||||||
| c.3364G>T | p.(Gly1122*) | 15.3 | 5 | |||||||||
| c.3614C>A | p.(Ser1205Tyr) | 5.1 | 3 | |||||||||
| c.5362T>A | p.(Ser1788Thr) | 34.9 | 3 | |||||||||
| c.6430G>T | p.(Glu2144*) | 16.2 | 5 | |||||||||
| c.7987G>T | p.(Glu2663*) | 19.0 | 5 | |||||||||
| c.8360G>A | p.(Arg2787His) | 33.6 | 2 | |||||||||
| c.9132T>G | p.(Ile3044Met) | 18.6 | 3 | |||||||||
| c.9239C>T | p.(Ser3080Phe) | 6.3 | 3 | |||||||||
| 2 | c.857C>G | p.(Pro286Arg) | 36 | 566 | c.5468-25C>T | p.? | 11.8 | 3 | c.632-16A>C | p.? | 39.6 | 3 |
| c.5280C>A | p.(=) | 6.0 | 3 | c.2458G>T | p.(Asp820Tyr) | 12.8 | 2 | |||||
| c.5238C>A | p.(His1746Gln) | 15.3 | 3 | c.3337G>T | p.(Glu1113*) | 16.7 | 5 | |||||
| c.2961G>T | p.(Lys987Asn) | 12.7 | 3 | c.3912T>G | p.(=) | 18.5 | 3 | |||||
| c.2162T>G | p.(Phe721Cys) | 15.3 | 3 | c.4219G>T | p.(Glu1407*) | 16.9 | 5 | |||||
| c.2152C>A | p.(Leu718Ile) | 15.8 | 3 | c.4939A>G | p.(Thr1647Ala) | 35.4 | 3 | |||||
| c.1788C>T | p.(=) | 15.1 | 3 | c.5071A>C | p.(Lys1691Gln) | 12.7 | 3 | |||||
| c.5228G>A | p.(Ser1743Asn) | 16.9 | 3 | |||||||||
| c.5634C>T | p.(=) | 13.5 | 3 | |||||||||
| c.5801A>C | p.(Gln1934Pro) | 17.4 | 3 | |||||||||
| c.6952C>T | p.(Arg2318*) | 18.2 | 5 | |||||||||
| c.8009C>T | p.(Ser2670Leu) | 17.4 | 5 | |||||||||
| c.9625C>A | p.(Pro3209Thr) | 18.7 | 3 | |||||||||
| 3 | c.857C>G | p.(Pro286Arg) | 27 | 170 | c.3798C>T | p.(=) | 33.1 | 2 | ||||
| c.2286A>G | p.(=) | 29.5 | 3 | |||||||||
| c.1934C>T | p.(Ser645Phe) | 7.2 | 3 | |||||||||
| c.842G>T | p.(Ser281Ile) | 6.0 | 3 | |||||||||
| 4 | c.857C>G | p.(Pro286Arg) | 39 | 106 | c.1342C>A | p.(His448Asn) | 24.1 | 3 | c.4790C>A | p.(Ser1597Tyr) | 8.4 | 3 |
| c.5719T>G | p.(Ser1907Ala) | 8.1 | 3 | |||||||||
| c.8524C>T | p.(Arg2842Cys) | 32.8 | 3 | |||||||||
| 5 | c.1231G>T | p.(Val411Leu) | 34 | 283 | c.4358-2782C>T | p.? | 32.3 | 2 | c.2841G>A | p.(Leu947=) | 32.2 | 3 |
| c.426-82G>T | p.? | 24.9 | 2 | |||||||||
| c.5602G>T | p.(Asp1868Tyr) | 36.8 | 3 | |||||||||
| c.2191G>T | p.(Glu731Ter) | 33.7 | 3 | |||||||||
| c.5982A>C | p.(Gln1994His) | 31.8 | 3 | |||||||||
| 6 | c.857C>G | p.(Pro286Arg) | 30 | 92 | c.2451G>T | p.(Lys817Asn) | 25.6 | 3 | ||||
| c.3662C>T | p.(Ser1221Phe) | 24.2 | 3 | |||||||||
| c.5550A>C | p.(Lys1850Asn) | 12.0 | 3 | |||||||||
| c.7850G>T | p.(Arg2617Ile) | 27.5 | 3 | |||||||||
| 7 | c.857C>G | p.(Pro286Arg) | 26 | 128 | c.2586G>T | p.(Lys862Asn) | 29.2 | 3 | c.7008-63C>T | p.? | 5.1 | 2 |
| c.1762A>C | p.(Ser588Arg) | 12.9 | 3 | c.4177G>A | p.(Ala1393Thr) | 6.8 | 3 | |||||
| c.2419G>A | p.(Val807Ile) | 6.7 | 3 | |||||||||
| c.6755C>A | p.(Ser2252Tyr) | 5.8 | 3 | |||||||||
VAF: variant allelic frequency
variants are classified according to ACMG guidelines: class 1 = benign, class 2 = likely benign, class 3 = uncertain significance, class 4 = likely pathogenic, class 5 = pathogenic
Fig. 1.
Tumor signatures. All five signatures exhibit a peak in [TCT → A] and [TCG → T], corresponding to the POLE signature. Signatures could not be computed for 2 patients, i.e. patients 1 and 7. Patient 2 had ovarian cancer, patients 3, 4 and 5 had endometrial cancers, patient 6 had ovarian and endometrial cancer.
Three different POLE variants were identified in these 7 tumors: c.890C > T, p.(Ser297Phe), c.857C > G, p.(Pro286Arg) and c.1231G > T, p.(Val411Leu). These variants have already been classified as drivers in ultramutated tumors. The c.857C > G, p.(Pro286Arg) variant was found in 5 different tumors, which is not surprising as it is a known hotspot variant. (Campbell et al., 2017)
All gynecologic tumors were of the endometrioid subtype (or mixed with a predominant endometrioid subtype). Pathology review of all cases failed to identify any characteristic pathologic features for these POLE-associated tumors.
Five of the 7 patients also underwent germline analysis, which did not reveal any POLE or BRCA1/2 variants. Moreover, the breast tumor of patient 4, who developed multiple cancers, was also analyzed by the large NGS panel and no POLE PVs, BRCA1/2 variants or high TMB were detected.
MSH2 and MSH6 PVs were also found in the tumor of patient 5 together with microsatellite instability, although germline analysis of these genes was normal. The tumor mutational signature is typical of a tumor driven by the POLE variant p.(Val411Leu) (i.e. signature 5; Fig. 1), but the contribution of a mutational signature associated with loss of mismatch repair cannot be excluded, as described by Haradhvala et al. (Haradhvala et al., 2018)
4. Discussion
We report 7 patients with ovarian, endometrial or colorectal cancer exhibiting a tumor POLE PV associated with multiple tumor BRCA1/2 variants. Each tumor also harbored a high TMB, MSS status (except for one tumor) and the molecular signature associated with POLE exonuclease deficiency was detected in all of the 5 tumors for which this signature could be computed, confirming the role of POLE in the oncogenesis of these tumors.
Five out of 7 tumors presented the c.857C > G, p.(Pro286Arg) POLE PV. This recurrent variant is found in 122 samples in the Cosmic database, including 87 endometrial cancers.
As we did not observe any characteristic pathologic features associated with these tumors, pathologic examination does not appear to be an informative tool to distinguish these tumors. However, it could be useful to investigate a deep learning-based approach on a greater number of tumors. (Courtiol et al., 2019) It should be noted that all of the gynecologic tumors described here were of the endometrioid subtype (including the ovarian tumors).
A new classification of endometrial cancers based on immunohistochemistry and variant analysis identified four groups of patients including the group of endometrial carcinomas with POLE PVs, characterized by their excellent prognosis (Alexa et al., 2021). However, they often have an aggressive presentation and patients usually receive intensive therapy based on stage and pathology. (Kandoth et al., 2013, Van Gool et al., 2018) It is therefore essential to identify these tumors in order to consider de-escalation of adjuvant therapy to avoid overtreatment. Tumor BRCA testing could be integrated into the new classification strategy for endometrial cancer.
POLE alterations seem to be a rare event in endometrioid ovarian cancers, but they are also suspected to be associated with a better prognosis, although this association has not been formally demonstrated. (Hoang et al., 2015) Tumor BRCA testing is already performed routinely and recommended for all ovarian cancers, as PARP inhibitor treatment may be indicated when a BRCA1/2 PV is detected. (Konstantinopoulos et al., 2020) Tumors usually harbor no more than one BRCA1/2 variant. We suggest that 4 or more BRCA1/2 tumor variants should indicate the need for further investigations, especially screening for an associated POLE PV, which could allow identification of tumors that may have a better prognosis, as well as potential good candidates for immunotherapy. These analyses could be the basis for a new classification for endometrioid ovarian cancers, mimicking endometrial cancers. To the best of our knowledge, immunotherapy has never been used for ovarian cancer harboring a POLE PV, but could constitute a new treatment option for these patients. Moreover, a combination of immunotherapy and PARP inhibitors could be proposed in patients with a tumor harboring BRCA1/2 PV associated with POLE PV.
Like endometrial carcinomas, colorectal cancers with POLE PVs appear to constitute an immunogenic subset of colorectal cancers characterized by a good prognosis. It may also be clinically relevant to identify these tumors. (Domingo et al., 2016)
Families with germline POLE PVs, responsible for a predisposition to various cancers, including colorectal and gynecologic tumors, have been described. (Briggs and Tomlinson, 2013 Jun, Hansen et al., 2015) Patients with tumors harboring a POLE PV should therefore have access to germline genetic testing.
In conclusion, tumor BRCA testing appears to be an easy, affordable and already available way to detect ultramutated tumors harboring a POLE PV who would be eligible for immunotherapy, as the number of BRCA1/2 variants reflects the TMB. We believe that this approach should be considered in other tumor types, especially in endometrioid endometrial carcinomas.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gore.2021.100855.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Alexa M., Hasenburg A., Battista M.J. The TCGA Molecular Classification of Endometrial Cancer and Its Possible Impact on Adjuvant Treatment Decisions. Cancers (Basel). 2021 Mar 23;13(6):1478. doi: 10.3390/cancers13061478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov, LB., Nik-Zainal, S., Wedge, DC., Aparicio, SA., Behjati, S., Biankin, AV., Bignell, GR., Bolli, N., Borg, A., Børresen-Dale, AL., Boyault, S., Burkhardt, B., Butler, AP., Caldas, C., Davies, HR., Desmedt, C., Eils, R., Eyfjörd, JE., Foekens, JA., Greaves, M., Hosoda, F., Hutter, B., Ilicic, T., Imbeaud, S., Imielinski, M., Jäger, N., Jones, DT., Jones, D., Knappskog, S., Kool, M., Lakhani, SR., López-Otín, C., Martin, S., Munshi, NC., Nakamura, H., Northcott, PA., Pajic, M., Papaemmanuil, E., Paradiso, A., Pearson, JV., Puente, XS., Raine, K., Ramakrishna, M., Richardson, AL., Richter, J., Rosenstiel, P., Schlesner, M., Schumacher, TN., Span, PN., Teague, JW., Totoki, Y., Tutt, AN., Valdés-Mas, R., van, Buuren, MM., van 't Veer, L., Vincent-Salomon, A., Waddell, N., Yates, L.R. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain, Zucman-Rossi, J., Futreal, P.A., McDermott, U., Lichter, P., Meyerson, M., Grimmond, S.M., Siebert, R., Campo, E., Shibata, T., Pfister, S.M., Campbell, P.J., Stratton, M.R. 2013. Signatures of mutational processes in human cancer. Nature. 2013 Aug 22;500(7463):415-21. doi: 10.1038/nature12477. Epub 2013 Aug 14. Erratum in: Nature. 2013 Oct 10;502(7470):258. Imielinsk, Marcin [corrected to Imielinski, Marcin]. PMID: 23945592; PMCID: PMC3776390. [DOI] [PMC free article] [PubMed]
- Alexandrov, LB., Kim, J., Haradhvala, NJ., Huang, MN., Tian, Ng, AW., Wu, Y., Boot, A., Covington, KR., Gordenin, DA., Bergstrom, EN., Islam, SMA., Lopez-Bigas, N., Klimczak, LJ., McPherson, JR., Morganella, S., Sabarinathan, R., Wheeler, DA., Mustonen, V., PCAWG, Mutational Signatures Working Group., Getz, G., Rozen, SG., Stratton, M.R. 2020. PCAWG Consortium. The repertoire of mutational signatures in human cancer. Nature. 2020 Feb;578(7793):94-101. doi: 10.1038/s41586-020-1943-3. Epub 2020 Feb 5. PMID: 32025018; PMCID: PMC7054213. [DOI] [PMC free article] [PubMed]
- Briggs S., Tomlinson I. Germline and somatic polymerase ε and δ mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol. 2013 Jun;230(2):148–153. doi: 10.1002/path.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell B.B., Light N., Fabrizio D., Zatzman M., Fuligni F., de Borja R., Davidson S., Edwards M., Elvin J.A., Hodel K.P., Zahurancik W.J., Suo Z., Lipman T., Wimmer K., Kratz C.P., Bowers D.C., Laetsch T.W., Dunn G.P., Johanns T.M., Grimmer M.R., Smirnov I.V., Larouche V., Samuel D., Bronsema A., Osborn M., Stearns D., Raman P., Cole K.A., Storm P.B., Yalon M., Opocher E., Mason G., Thomas G.A., Sabel M., George B., Ziegler D.S., Lindhorst S., Issai V.M., Constantini S., Toledano H., Elhasid R., Farah R., Dvir R., Dirks P., Huang A., Galati M.A., Chung J., Ramaswamy V., Irwin M.S., Aronson M., Durno C., Taylor M.D., Rechavi G., Maris J.M., Bouffet E., Hawkins C., Costello J.F., Meyn M.S., Pursell Z.F., Malkin D., Tabori U., Shlien A. Comprehensive Analysis of Hypermutation in Human Cancer. Cell. 2017;171(5):1042–1056.e10. doi: 10.1016/j.cell.2017.09.048. Epub 2017 Oct 19. PMID: 29056344; PMCID: PMC5849393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtiol P., Maussion C., Moarii M., Pronier E., Pilcer S., Sefta M., Manceron P., Toldo S., Zaslavskiy M., Le Stang N., Girard N., Elemento O., Nicholson A.G., Blay J.Y., Galateau-Sallé F., Wainrib G., Clozel T. Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat Med. 2019 Oct;25(10):1519–1525. doi: 10.1038/s41591-019-0583-3. Epub 2019 Oct 7 PMID: 31591589. [DOI] [PubMed] [Google Scholar]
- Domingo E, Freeman-Mills L, Rayner E, Glaire M, Briggs S, Vermeulen L, Fessler E, Medema JP, Boot A, Morreau H, van Wezel T, Liefers GJ, Lothe RA, Danielsen SA, Sveen A, Nesbakken A, Zlobec I, Lugli A, Koelzer VH, Berger MD, Castellví-Bel S, Muñoz J; Epicolon consortium, de Bruyn M, Nijman HW, Novelli M, Lawson K, Oukrif D, Frangou E, Dutton P, Tejpar S, Delorenzi M, Kerr R, Kerr D, Tomlinson I, Church DN. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol. 2016 Nov;1(3):207-216. doi: 10.1016/S2468-1253(16)30014-0. Epub 2016 Jul 20. PMID: 28404093. [DOI] [PubMed]
- Hansen M.F., Johansen J., Bjørnevoll I., Sylvander A.E., Steinsbekk K.S., Sætrom P., Sandvik A.K., Drabløs F., Sjursen W. A novel POLE mutation associated with cancers of colon, pancreas, ovaries and small intestine. Fam Cancer. 2015 Sep;14(3):437–448. doi: 10.1007/s10689-015-9803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haradhvala N.J., Kim J., Maruvka Y.E., Polak P., Rosebrock D., Livitz D., Hess J.M., Leshchiner I., Kamburov A., Mouw K.W., Lawrence M.S., Getz G. Distinct mutational signatures characterize concurrent loss of polymerase proofreading and mismatch repair. Nat Commun. 2018 May 1;9(1):1746. doi: 10.1038/s41467-018-04002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang L.N., McConechy M.K., Köbel M., Anglesio M., Senz J., Maassen M., Kommoss S., Meng B., Postovit L., Kelemen L.E., Staebler A., Brucker S., Krämer B., McAlpine J.N., Gilks C.B., Huntsman D.G., Lee C.H. Polymerase Epsilon Exonuclease Domain Mutations in Ovarian Endometrioid Carcinoma. Int J Gynecol Cancer. 2015 Sep;25(7):1187–1193. doi: 10.1097/IGC.0000000000000492. PMID: 26166557. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, Kandoth, C., Schultz, N., Cherniack, A.D., Akbani, R., Liu, Y., Shen, H., Robertson, A.G., Pashtan, I., Shen, R., Benz, C.C., Yau, C., Laird, P.W., Ding, L., Zhang, W., Mills, G.B., Kucherlapati, R., Mardis, E.R., Levine, D.A. Integrated genomic characterization of endometrial carcinoma. Nature. 2013 May 2;497(7447):67-7doi: 10.1038/nature1211Erratum in: Nature. 2013 Aug 8;500(7461):242. PMID: 23636398; PMCID: PMC3704730. [DOI] [PMC free article] [PubMed]
- Konstantinopoulos P.A., Norquist B., Lacchetti C., Armstrong D., Grisham R.N., Goodfellow P.J., Kohn E.C., Levine D.A., Liu J.F., Lu K.H., Sparacio D., Annunziata C.M. Germline and Somatic Tumor Testing in Epithelial Ovarian Cancer: ASCO Guideline. J Clin Oncol. 2020;38(11):1222–1245. doi: 10.1200/JCO.19.02960. Epub 2020 Jan 27. PMID: 31986064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnert J.M., Panda A., Zhong H., Hirshfield K., Damare S., Lane K., Sokol L., Stein M.N., Rodriguez-Rodriquez L., Kaufman H.L., Ali S., Ross J.S., Pavlick D.C., Bhanot G., White E.P., DiPaola R.S., Lovell A., Cheng J., Ganesan S. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest. 2016;126(6):2334–2340. doi: 10.1172/JCI84940. Epub 2016 May 9. PMID: 27159395; PMCID: PMC4887167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network C.G.A. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012 Jul 18;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gool, I.C., Ubachs, J.E.H., Stelloo, E., de Kroon, C.D., Goeman, J.J., Smit, V.T.H.B.M., Creutzberg, C.L., Bosse, T. Blinded histopathological characterisation of POLE exonuclease domain-mutant endometrial cancers: sheep in wolf's clothing. Histopathology. 2018 Jan;72(2):248-25doi: 10.1111/his.1333Epub 2017 Oct 10. PMID: 28795426. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.