Highlights
-
•
siRNA-SP1-loaded AuNPs (AuNPs-si-SP1) inhibits SP1 expression in lung cancer cells.
-
•
GZMB expression is elevated by silencing SP1 in lung cancer cells.
-
•
AuNPs-si-SP1 combined with radiotherapy inhibits lung cancer growth effectively.
-
•
AuNPs-si-SP1 enhances radiosensitivity of lung cancer via SP1/GZMB axis.
-
•
AuNPs-si-SP1 provides a novel target for radiotherapy of lung cancer.
Keywords: Lung cancer, AuNPs, siRNA-SP1, SP1, GZMB
Abstract
Radioresistance is a major challenge that largely limits the efficacy of radiotherapy in lung cancer. Gold nanoparticles (AuNPs) are emerging as novel radiosensitizers for cancer patients. Therefore, this study was designed to explore the radiosensitizing effect and mechanism of AuNPs loaded with small interfering RNA (siRNA)-SP1 (AuNPs-si-SP1) on lung cancer. AuNPs-si-SP1 was prepared by the noncovalent binding between AuNPs and siRNA-SP1. The adsorption capacity of AuNPs to siRNA-SP1 was analyzed by gel electrophoresis. The cell uptake of AuNPs-si-SP1 was observed under a laser confocal microscopy. Silencing efficacy of AuNPs-si-SP1 was validated by RT-qPCR and Western blot analysis. Cell viability was determined by CCK-8 assay, radiosensitization by plate colony formation assay, cell apoptosis and cell cycle by flow cytometry, and DNA double strand breaks by immunofluorescence in the presence or absence of AuNPs-si-SP1 or GZMB. The downstream mechanism of SP1 was predicted by bioinformatics analysis, followed by verification by Western blot analysis. Subcutaneous tumorigenesis in nude mice was established to verify the radiosensitization of AuNPs-si-SP1 and GZMB in vivo. AuNPs-si-SP1 effectively absorbed SP1 siRNA and was highly internalized by A549 cells to reduce SP1 protein expression. AuNPs-si-SP1 or GZMB overexpression promoted cells to G2/M phase, DNA double strand breaks, and enhanced radiosensitivity. SP1 could repress GZMB expression in lung cancer cells. In vivo experiments manifested that AuNPs-si-SP1 could inhibit the growth of solid tumor in nude mice to achieve radiosensitization by inhibiting SP1 to upregulate GZMB. AuNPs-si-SP1 might increase the radiosensitivity of lung cancer by inhibiting SP1 to upregulate GZMB.
Introduction
It was estimated by GLOBOCAN that there were 2.09 million new cases (11.6% of total cancer cases) and 1.76 million deaths (18.4% of total cancer deaths) of lung cancer in 2018 [1]. The high mortality rate in lung cancer patients correlates to a common diagnosis at late stage that precludes curative treatment, with a poor 5-year survival rate at 15% of all stages combined [2]. Approximately 80%−90% lung cancer develops in smokers, so smoking is the major risk factor for lung cancer [3]. Radiotherapy is a principle therapy for lung cancer, but radioresistance is a formidable challenge that largely limits the efficacy of radiotherapy [4]. Therefore, it is strongly needed to decipher the molecular mechanisms behind radioresistance and elaborate novel therapeutic targets for individualized radiotherapy for lung cancer.
Gold nanoparticles (AuNPs) consist of an inorganic core containing gold which is typically surrounded by an organic monolayer [5]. AuNPs are emerging as novel radiosensitizers for cancer patients because of their synthetic versatility, unique chemical, electronic, and optical properties, and high X-ray absorption [6]. AuNPs have been documented to exert anti-oncogenic activity in lung cancer [7]. More importantly, AuNPs are widely recognized to be a comparatively reliable small interfering RNA (siRNA) delivery strategy due to their biocompatibility and capacity to protect siRNA against degradation and versatility offered by their tunable shape, size and optical properties [8]. As a member of the 26 strong Sp/KLF family of transcription factors, Specificity Protein 1 (SP1) is a paradigm of a ubiquitously expressed transcription factor, which assumes a pivotal role in manipulation of the expression of genes correlating to various cell processes in mammalian cells [9]. Furthermore, a prior study detected that SP1 was overexpressed in liquid-based cells from bronchial brushings in patients with lung cancer [10]. Notably, it was predicted by JASPAR database in our research that there was a binding site between SP1 and granzyme B (GZMB). Interestingly, a prior work manifested that GZMB overexpression could increase the susceptibility of non-small cell lung cancer (NSCLC) cells to natural killer (NK) cell-mediated killing [11]. In this context, we hypothesized that AuNPs-loaded siRNA-SP1 (AuNPs-si-SP1) might participate in alleviation of lung cancer. Thus, this research was designed to figure out whether AuNPs could load siRNA-SP1 and whether AuNPs-si-SP1 could exert radiosensitive effect in lung cancer in regulation of GZMB.
Materials and methods
Ethics statement
This study was performed with approval of the Ethics Committee of Harbin Medical University Tumor Hospital by conforming to the Declaration of Helsinki. All participants or their guardians provided signed informed consent prior to enrollment. Animal experiments were ratified by Animal Ethics Committee of Harbin Medical University Tumor Hospital in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. Adequate measures were taken to minimize suffering of the included animals.
Sample collection
Lung cancer tissues and adjacent normal tissues (5 cm away from the cancer area) were collected from 40 patients who were confirmed as lung cancer by histopathology in Harbin Medical University Tumor Hospital. The patients did not receive any treatment (including radiotherapy and chemotherapy) before operation. All the resected specimens were taken from the non-necrotic bleeding area in the center of the cancer tissue and the normal mucosa 5 cm distal to the cancer tissues. The specimens were stored at −80 °C.
Sodium citrate (Na3C6H5O7•2H2O) and Hoechst 33,342 were purchased from Sigma-Aldrich Chemical Company (St Louis, MO, USA). Tetrachlorogenic acid (HAuCl4•3H2O) was obtained from Aurat (Moscow, Russia). Fetal bovine serum (FBS), Dulbecco's Modified Eagle Medium (DMEM) medium, ethylene diamine tetraacetic acid (EDTA, trypsin), and penicillin and streptomycin were attained from HyClone Company (Logan, UT, USA). Cell counting kit (CCK)−8 was harvested from Dojindo Molecular Technologies (Kumamoto, Japan). The 4′,6-Diamidino-2-Phenylindole (DAPI) was from Yeasen Company (Shanghai, China). Annexin V-fluoresceinisothiocyanat (FITC)/propidium iodide (PI) apoptosis detection kit was ordered from Shanghai BestBio Biotechnology Co., Ltd. (Shanghai, China). Cell cycle detection kit (PI and RNaseA) was acquired from MultiSciences (Lianke) Biotechnology Corporate Limited (Zhejiang, China). siRNA-SP1, Cy5-labeled siRNA-SP1 (Cy5-siRNA-SP1), and siRNA-GZMB were purchased from Thermo Fisher Scientific Inc. (Rockford, IL, USA). Anti-SP1 antibody (ab227383) was bought from Abcam Inc. (Cambridge, UK). Anti-GZMB (4275) antibody was got from Cell Signaling Technologies (CST) (Beverly, MA, USA). Anti-β-actin antibody (1:10,000, BS6007M) was from Bioworld Technology (St. Louis Park, MN, USA). Anti-γH2AX antibody (05–636, 1:1000) was purchased from Merck KGaA (Darmstadt, Germany). CF-568 goat-anti-mouse immunoglobulin G (IgG) (H + L) secondary antibodies (1: 500, Cat20100, Lot16C11108) was purchased from Biotium (Hayward, CA, USA).
Synthesis and characterization of AuNPs-si-SP1
The (COONa) (CH2COOONa)2 × 3H2O solution (5 mL, 38.8 mM) was added to the stirring HAuCl4 × 3H2O (45 mL, 1 mm) boiling solution, stirred well for 20 min, and cooled down to room temperature. After storage for 24 h, the solution was filtered through a filter with a pore size of 45 μm. Based on that, the AuNP suspension was stored as a concentration of 3.5 × 10−9 M at 4 °C (ε260 = 8.78 × 108 М−1 сm−1). The synthesized AuNP suspension (135 μL) was added into siRNA solution (4 μL, 25 μM). The reaction mixture was stirred at 1000 rpm and 25 °C for 24 h and centrifuged at 16,000 g and 25 °C for 30 min. Then, the unbound siRNA was washed with sodium citrate. The washing method was used in the whole experiment. The soliquid was diluted to 5 μL with sodium citrate.
The absorbance curves of AuNPs and AuNPs-si-SP1 were measured respectively by a UV spectrophotometer (DU800, Beckman Coulter Inc., Chaska, MN, USA). Transmission Electron Microscope (TEM) (HT7700, HITACHI, Japan) and Malvern Zetasizer Nano ZS (Malvern Instruments, Worcestershire, UK) were used to evaluate the morphology, particle size and potential of AuNPs-si-SP1. The adsorption capacity of AuNPs with different mass ratios (0.2, 0.5, 1, 2, 3) to siRNA-SP1 was researched by gel electrophoresis. In this experiment, the remaining siRNA in the supernatant of each group was collected by centrifugation. The adsorption capacity was evaluated by electrophoresis with pure siRNA as negative control.
Cell culture
Human lung adenocarcinoma cell line A549 was purchased from Shanghai Tongpai Biotechnology Co., Ltd. (Shanghai, China) as a moderately radioresistant cell line. A549 cells were cultured in DMEM containing 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin, and placed in a 5% CO2 incubator at 37 °C. According to the experimental needs, the cells were seeded in a laser confocal dish, a 96 well plate or a 6 well plate separately.
Cell absorption in vitro
The 5′ end of the sense strand of SP1 siRNA was labeled with Cy5 dye (excitation/emission at 649/670 nm) (Cy5-siRNA-SP1) to observe cell uptake. A549 cells were cultured in a laser confocal dish (20 mm) with a cell density of 5 × 104 cells per well for 24 h. The cells were treated with siRNA-SP1 and AuNPs-si-SP1 (equivalent to 100 nm siRNA) respectively for 12 h, and fixed with 4% paraformaldehyde (200 μL) at 4 °C for 10 min. Cells were stained with DAPI (10 μg/mL in PBS) for 10 min. Then, cells were washed with PBS to remove the free dye. The cell uptake was observed by a laser confocal microscopy (CLSM, Leica TCSNT1, Leica Microsystems, Heidelberg, Germany) with 633 nm laser.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells by TRIzol (Invitrogen Inc., Carlsbad, CA, USA). According to the instructions, the total RNA was reverse transcribed into cDNA by using TaqMan MicroRNA Assays Reverse Transcription primer (4427975, Applied Biosystems, Carlsbad, CA, USA)/PrimeScript RT reagent Kit (RR047A, Takara Holdings Inc., Kyoto, Japan). SP1 and GZMB primers were designed and synthesized by Takara Holdings Inc. (Table S1). TaqMan Multiplex Real-Time Solution (4461882, Thermo Fisher Scientific Inc.) and ABI7500 quantitative PCR instrument (7500, ABI, Foster City, CA, USA) were adopted for real-time PCR detection. Relative quantitative method (2−△△CT method) was employed to calculate the relative transcription level of target gene with β-actin as an internal reference. In addition, the follow-up experiments also evaluated the expression of genes in lung cancer tissues and adjacent normal tissues according to the above procedures.
Western blot analysis
The cultured cells were lysed with Radio-Immunoprecipitation assay cell lysis buffer containing phenylmethylsulfonyl fluoride on ice for 10 min. After centrifugation at 14,000 rpm and 4 °C, the supernatant was taken out. The protein concentration was determined by bicinchoninic acid method (Pierce, Rockford, IL, USA). Then 4% concentrated gel and 10% separated gel were prepared, and protein was transferred to the membrane after gel electropheresis. The membrane was sealed with 5% skimmed milk powder at room temperature for 1 hour, and then probed overnight with anti-SP1 antibody (ab227383, 1:10,000, Abcam Inc.) at 4 °C. After that, the membrane was reprobed with horseradish peroxidase-labeled rabbit secondary antibody (Invitrogen Inc., G-21,234, 1:2000) for 1 hour at room temperature. The membrane was put into electrogenerated chemiluminescence (ECL) reaction solution for coloring at room temperature. The results were observed by a ChemiDoc XRS system (Bio-Rad, Richmond, Cal., USA). The image J analysis software was applied to quantify the protein band intensity with β-actin (1:10,000, BS6007M) as an internal reference. Besides, the expressions of SP1 and GZMB in different tumor tissues were gauged according to the above methods.
CCK-8 assay for radiosensitizing effect of AuNPs-si-SP1 on cells
The killing ability of AuNPs-si-SP1 to A549 cells was studied by CCK-8. Cell viability was measured after A549 cells were treated with different concentrations of AuNPs-si-SP1 (0 nM, 5 nM, 10 nM, and 20 nM) for 24, 48 and 72 h, respectively. Then the radiosensitizing effect of AuNPs-si-SP1 on A549 cells was detected. A549 cells were cultured in a 96 well culture plate (1 × 104 cells/well) for 24 h, and then AuNPs-si-SP1 with different concentration gradients (0 nM, 5 nM, 10 nM, and 20 nM) was added. After incubation for 24 h, X-ray irradiation was performed for 20 min at a dose of 6 Gy (Varian 23EX linear accelerator, 6MV X-ray, 100 cm SSD, plus 2 cm equivalent). Fresh medium was replaced and incubated for 24 h, and then 10 μL CCK-8 was supplemented to the culture plate. After incubation for 4 h, the absorbance at 450 nm was measured by a microplate reader (BioTek synergy H4). Based on the above steps, the cell viability of subsequent AuNPs binding with different siRNA was surveyed. By setting the optical density (OD) value of untreated cells as blank and that of PBS as control, the cell viability was calculated using the following formula: cell viability (%) = [OD (sample) - OD (blank)]/[OD (control) - OD (blank)].
Cell colony-forming experiment for radiosensitizing effect of AuNPs-si-SP1 on cells
The experiment included four groups: radiotherapy group and groups of AuNPs-si-SP1 with different concentrations (5 nM, 10 nM, and 20 nM) in combination with radiotherapy. A549 cells in logarithmic growth phase were irradiated at doses of 0 Gy, 2 Gy, 4 Gy and 6 Gy respectively. The cells were seeded in 6-well plates according to the dose gradient. After overnight incubation, serum alone and serum containing AuNPs-si-SP1 (5 nM, 10 nM, and 20 nM) were added. After incubation for 24 h, cells were irradiated according to different dose gradients. After treatment, the cells were placed in the incubator to culture for 10–14 days with culture medium changed regularly. When the clone cells visible to the naked eye appeared in the 6-well plate, the culture was stopped. After discarding the culture medium, the cells were fixed with pure methanol for 15 min. After staining with Gimsa staining solution for 20 min, the staining solution was washed off and cells were dried. The images were taken by a camera, and the number of clones with 250 cells was observed and counted under the microscope. According to that, the cell activity of subsequent AuNPs combined with different siRNA was estimated. The survival fraction was calculated and the cell survival curve was drawn according to the following formula: plating efficiency (PE) = (the number of clones in the control group/the number of inoculated cells) × 100%, and survival fraction (SF) = the number of clones in the experimental group/(the number of inoculated cells × PE).
The cell survival curve was fitted by single hit multi target model and the effect of radiosensitization was expressed as sensitizer enhancement ratio (SER), which was defined as the ratio of the mean inactive dose (MID) of the radiotherapy group to that of the combined group, that is, the area under curve (AUC).
Flow cytometry for cell apoptosis
The cell apoptosis was determined by an Annexin V-FITC/PI apoptosis detection kit. A549 cells were seeded in 6-well plates with 2 × 105 cells/well. The experiment included four groups: control group (untreated group), AuNPs-si-SP1 group (20 nM), radiotherapy group, and combination group (AuNPs-si-SP1 + radiotherapy). After 24 h of transfection, cells in group 3 and group 4 were irradiated at doses of 6 Gy. After treatment, the cells were cultured for 24 h. Subsequently, the cells were centrifuged at 2000 rpm for 5 min. According to the kit instructions, the cells were resuspended in 200 μL binding buffer, added with 5 μL Annexin V-FITC in the dark, mixed completely, and incubated at 4 °C in dark for 15 min. Then 10 μL PI was added, mixed, and incubated at 4 °C for 5 min in dark. Finally, apoptosis was detected by a flow cytometer (BD FACScalibur).
Flow cytometry for cell cycle distribution
In order to study whether AuNPs-si-SP1 can promote the transformation of cells to G2/M phase, A549 cells in logarithmic growth phase were seeded in 6-well plates at a density of 2 × 105 cells/well. The cells were cultured overnight, and the old culture medium was discarded after the cells adhered to the wall. Then culture medium encompassing AuNPs-si-SP1 (20 nM) with the best sensitization effect and PBS were added respectively. After 24 h of culture, the cells were collected. The cells were dispersed after adding 1 mL PBS. The cell suspension was slowly dropped into 3 mL pure ethanol, mixed with high-speed vortex, and kept at 4 °C for 24 h. Before the test, cells were centrifuged, with the ethanol supernatant discarded. Cells were added with 5 mL PBS (room temperature), and allowed to stand for 15 min, and then rehydrated. Then cells were harvested by centrifugation, added with 1 mL DNA staining solution, whirled for 5 - 10 s, and kept in dark for 30 min at room temperature. The cell suspension was filtered using a 200-mesh filter before operation, and each sample was analyzed for 10,000 cells. Cell cycle was analyzed by cell cycle fitting software. The Modfit software was adopted to analyze cell cycle, followed by drawing of cell cycle distribution histogram.
Immunofluorescence staining
A549 cells were seeded on a cover glass. After treatment of the control group, the AuNPs-si-SP1 group (20 nM), the radiotherapy group, and the combination group (AuNPs-si-SP1 + radiotherapy) group, the cells were fixed with 4% paraformaldehyde at 4 °C for 30 min, and then permeabilized with 0.2% Triton X-100 for 10 min. The cells were probed with anti-γH2AX antibody (05–636, Merck KGaA, Darmstadt, Germany; 1:1000) at 4 °C. Then the cells were incubated with CF-568 Goat-anti-mouse IgG (H + L) secondary antibody (1:5000, Biotium, Hayward, CA, USA; Cat20100, Lot16C11108) at room temperature and stained with Hoechst 33,342 (1 μg/mL; Sigma-Aldrich Chemical Company). Finally, the immunofluorescence images were obtained with a laser confocal microscope (IX81; Olympus, Tokyo, Japan; 40 ×).
Bioinformatics analysis
The lung cancer-related genes were retrieved through GeneCards database (https://www.genecards.org/), Comparative Toxicogenomics Database (CTD) (http://ctdbase.org/detail.go?acc=C452899&type=chem), and Phenolyzer database (http://phenolyzer.wglab.org/). Totally 59 lung cancer candidate targets from the intersection of the results of these databases were obtained by the jvenn tool (http://jvenn.toulouse.inra.fr/app/example.html). The candidate targets underwent enrichment analysis by Metascape (https://metascape.org/gp/index.html#/main/step1) to get the main transcription factors among the candidate targets.
Lung cancer radiotherapy-related gene expression dataset GSE73095 was obtained from Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), including 3 lung cancer samples and 3 lung cancer samples after radiotherapy. The differentially expressed genes (DEGs) with |logFC| > 1 and p < 0.05 were selected through R language "limma" package with lung cancer samples as controls. The R scripts used are shown in the Supplementary Material. The interaction of DEGs was analyzed using STRING database (https://string-db.org/), and then the gene interaction network was constructed by cytoscape v3.7.1. GEPIA2 database (http://gepia2.cancer-pku.cn/#analysis) was utilized to analyze the relationship between GZMB gene in lung adenocarcinoma of The Cancer Genome Atlas (TCGA) and survival of patients. The binding sites of transcription factor were gained from JASPAR database (http://jaspar.genereg.net/).
Subcutaneous tumorigenesis in nude mice
Specific pathogen-free (SPF) grade female BALB/c nude mice (aged 5 weeks, weighing 18–20 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) for tumorigenesis experiment. The animals were individually reared in SPF facilities during the experiment.
Subcutaneous tumor model: PBS containing A549 cells (1 × 106 cells/200 μL) was injected into the left side of female BALB/c mice. The mice were arranged into four groups with 5 mice in each group, including control group (PBS group), AuNPs-si-SP1 group (60 mg/kg), radiotherapy group, combination group (AuNPs-si-SP1 + radiotherapy), and AuNPs group. When the tumor volume of subcutaneous tumor model reached about 100 mm3, mice were injected with 100 μL PBS or AuNPs-si-SP1 via tail vein. After 24 h, the tumor site was irradiated with 6 Gy X-ray for 20 min in radiotherapy groups. Subsequently, tumor size and mice weight were monitored daily. The tumor volume was calculated as follows: (tumor length) × (tumor width)2/2 (unit: mm3). On the 20th day after irradiation, the mice were euthanized. The tumor tissues of mice were photographed and weighed.
Hematoxylin-eosin (H&E) staining
The tumor tissue was fixed in 10% formalin buffer, embedded in paraffin, and sectioned into 5 μm. After H&E staining, the morphological damage was observed under a microscope.
TdT-mediated dUTP-biotin nick end-labeling (TUNEL) staining for apoptosis
The TUNEL immunofluorescence kit (Millipore Corp., Billerica, MA, USA) was used to assess cell apoptosis according to the manufacturer's protocols. Subsequent to nuclei staining with DAPI, the Olympus fluorescence microscope (Olympus) was employed to image the tumor sections.
Statistical analysis
SPSS 21.0 (IBM Corp. Armonk, NY, USA) was adopted for statistical analysis. The measurement data were summarized by mean ± standard deviation, with p < 0.05 considered to be statistically significant difference. Firstly, the tests of normal distribution and homogeneity of variance was carried out, which confirmed that data conformed to normal distribution and homogeneity of variance. Unpaired t-test was implemented for comparison between the two groups. One-way analysis of variance (ANOVA) was applied for comparison among multiple groups, followed by Tukey's post-hoc test. The repeated measurement ANOVA was utilized to compare the data at different time points, followed by Bonferroni post-hoc test. Kaplan-Meier method was applied to calculate the survival rate.
Results
Characterization of AuNPs-si-SP1 was observed
SP1 was overexpressed in a variety of tumors such as lung cancer [10], which was an unfavorable prognostic factor for survival. SP1 siRNA was prepared to inhibit the expression of SP1 to study whether it could affect the radiosensitivity of lung cancer, and further explore the corresponding mechanism. However, pure siRNA could not be effectively transfected into cells to achieve gene silencing effect. AuNPs possess the advantages of small size, good biocompatibility, and low cytotoxicity, which can effectively improve the accumulation of drugs in tumor sites. Therefore, AuNPs were chosen to combine with SP1 siRNA through non covalent interaction to synthesize AuNPs-si-SP1 for subsequent experiments. Under TEM images, AuNPs-si-SP1 was spherical with a diameter of 12.78 ± 0.29 nm (Fig. 1A). The AuNPs-si-SP1 was highly dispersed, arranged on the support film in the form of single particles or short chains, and separated by a clear visible space of 2.11 ± 0.23 nm. Importantly, even if some chains formed inconsistent "patterns", there was always a certain space between the particles, which clearly indicated the existence of siRNA layer. In addition, the charge of the main part (≈ 90% of the average zeta potential) of AuNPs-si-SP1 was −39.01 ± 2.46 mV (The main peaks are shown in Fig. 1B). Besides, there were particles with charge value of −105.27 ± 9.61 mV, which accounted for 9–11% of the average zeta potential. The average potential value depended on the charge of the total siRNA molecules bound by AuNPs, suggesting that a small number of AuNPs bound to a large number of siRNA molecules. The UV–Vis absorption spectrum (Fig. 1C) showed that there was a characteristic absorption peak of AuNPs at 513 nm, while the maximum absorption peak of AuNPs-si-SP1 was red shifted at 524 nm, which might be due to the shift of surface plasmon resonance after the adsorption of siRNA.
Fig. 1.
Characterization of AuNPs-si-SP1. A, TEM images of AuNPs-si-SP1. The illustration was an enlarged view of the sample. The scale bar was equivalent to 100 nm. B, Zeta potential distribution of AuNPs-si-SP1. Red and green arrows indicated the major and minor peaks, respectively. C, The absorption peaks of AuNPs and AuNPs-si-SP1 characterized by the UV spectrophotometer. The maximum absorption peak of AuNPs (513 nm, solid arrow) and AuNP-si-SP1 (524 nm, dashed arrow). D, The loading capacity of AuNPs with different mass ratios (0.2, 0.5, 1, 2, 3) to siRNA-SP1 studied by agarose gel electrophoresis.
Next, the best mass ratio of AuNPs to siRNA-SP1 was optimized by gel electrophoresis. As manifested in Fig. 1D, pure siRNA as control had obvious fluorescence. On the contrary, the fluorescence gradually decreased with the increase of mass ratio, indicating that AuNPs absorbed more siRNA to reduce the remaining siRNA in the supernatant after centrifugation. When the mass ratio exceeded 1, there was almost no fluorescence in siRNA bands, which meant that there was a strong binding affinity for siRNA. In addition, the loading rate of siRNA was about 90% by calculating the fluorescence of Cy5-siRNA. The results of the serum stability test and in vivo rapid clearance of si-SP1 and AuNP-si-SP1 from kidney are shown in Figure S1.
AuNPs-si-SP1 could be internalized by A549 cells and be adopted for gene silencing
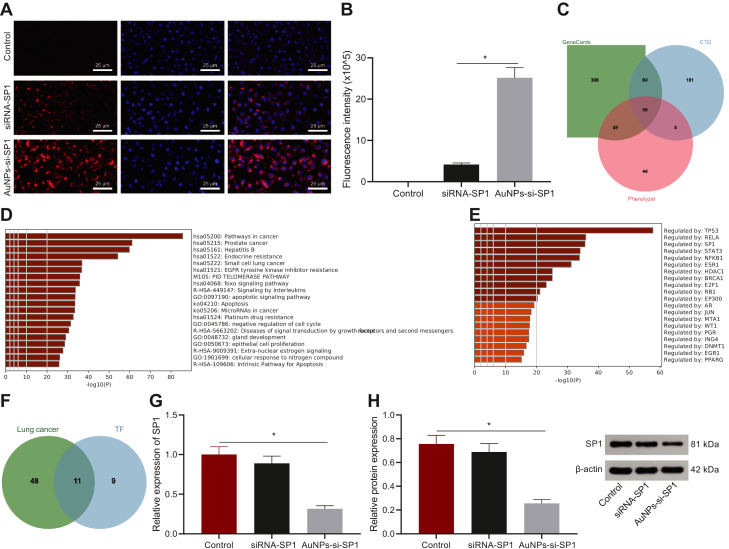
In order to study the cell uptake ability of siRNA, A549 cells were incubated with pure siRNA-SP1 and AuNP-si-SP1 for 12 h to investigate the uptake of si-SP1. The 5′ end of the sense strand of siRNA-SP1 was labeled with Cy5 dye (excitation/emission at 649/670 nm) (Cy5-siRNA-SP1) for visualization. The results showed that the amount of AuNP-si-SP1 entering eA549 cells was 5.5 times more than that of free siRNA (Fig. 2A-B). These results suggested that AuNPs could promote the uptake of si-SP1 by cells.
Fig. 2.
Cell uptake and gene silencing efficacy of AuNPs-si-SP1. A, Uptake of free siRNA and AuNPs-si-SP1 by A549 cells. The siRNA was labeled with Cy5 dye (Cy5-siRNA-SP1) and incubated with the cells for 12 h. The cells were observed by the confocal microscope at 633 nm laser excitation. The nuclei were stained with DAPI (blue). Scale bar = 20 mm. B, The fluorescence intensity of siRNA in A549 cells analyzed by ImageJ software. * p < 0.05. C, The Venn map of lung cancer-related genes by screening through GeneCards database, CTD database and Phenolyzer database. D, The pathway result map of enrichment analysis of lung cancer-related genes screened by Metascape. E, Transcription factors from Metascape enrichment analysis of lung cancer-related genes. F, The Venn map of the intersection of transcription factors obtained by enrichment analysis and lung cancer-related genes. G, Expression of SP1 mRNA in A549 cells after transfection analyzed by RT-qPCR. * p < 0.05 compared with the control group. H, The expression of SP1 protein including [SP1 (90 kDa) and β-actin (42 kDa)] in A549 cells detected by Western blot analysis. * p < 0.05 compared with the control group. The data results were measurement data, which were expressed by mean ± standard deviation. One-way ANOVA or repeated measurement ANOVA was applied for comparison among multiple groups, followed by Tukey's post-hoc test. The experiment was repeated three times.
At the same time, bioinformatics analysis was carried out to further research the relationship between SP1 and lung cancer and the mechanism of downregulated SP1 in radiosensitivity. The lung cancer-related genes were obtained by GeneCards database (the top 500 related genes), CTD database (genes with inference Score > 130), and Phenolyzer database (genes with Score > 0.3). The jvenn tool was employed to get 59 lung cancer candidate targets from the intersection of the results of these databases (Fig. 2C). Based on the result of Metascape, the target genes were mainly involved in the signal pathways such as Prostate cancer, Hepatitis B, Endocrine resistance, Small cell lung cancer, and EGFR tyrosine kinase inhibitor resistance (Fig. 2D). The target genes were mainly manipulated by transcription factors such as TP53, RELA, SP1, STAT3, NFKB1, and ESR1 (Fig. 2E). The top 20 transcription factors screened were intersected with candidate target genes of lung cancer to gain 11 transcription factors (Fig. 2F). Radiotherapy-related gene expression dataset GSE73095 for lung cancer was gained by GEO database. The differential analysis of GSE73095 was carried out, among which SP1 was downregulated after radiotherapy (Table S2).
The knockout effect of AuNPs-si-SP1 complex was evaluated by measuring SP1 gene silencing in A549 cells. RT-qPCR was performed to verify the inhibitory effects of AuNPs-si-SP1 at different concentrations (5 nM, 10 nM, and 20 nM) on the expression of SP1 in A549 cells, of which 20 nM had the strongest inhibitory effect on SP1 expression (Figure S2), which was selected for the subsequent experiments. Compared with the untreated controls, AuNPs-si-SP1 (equivalent to 20 nM siRNA) significantly inhibited the expression of SP1 mRNA and downregulated SP1 mRNA by 75% (Fig. 2G). Free siRNA showed a slight inhibition, and the expression of SP1 mRNA decreased by 17%. The expression of SP1 protein in A549 cells treated with AuNPs-si-SP1 was also significantly lower than that in untreated control A549 cells and free siRNA-treated A549 cells (Fig. 2H). These results suggested that gene silencing mediated by AuNPs-si-SP1 could effectively inhibit SP1 expression.
AuNPs-si-SP1 enhanced the radiosensitization of A549 cells
Based on the excellent gene silencing effect of AuNPs-si-SP1, the radiosensitivity of A549 cells after inhibiting SP1 was further investigated. The optimal transfection time was optimized by CCK-8 method. A549 cells were treated with different concentrations of AuNPs-si-SP1 for 24 h, 48 h and 72 h, respectively. Then cell viability was measured. It was displayed in Fig. 3A that different concentrations of AuNPs-si-SP1 slightly reduced the survival rate of tumor cells, because the down-regulation of SP1 mediated by AuNPs-si-SP1 inhibited the proliferation of tumor cells. In addition, when AuNPs-si-SP1 was transfected into A549 cells for 48 h and 72 h, there was no significant difference in cell viability, so the co-transfection time was 48 h in the next experiment.
Fig. 3.
AuNPs-si-SP1 augments radiosensitization of A549 cells. A, CCK-8 results of A549 cells treated with different concentrations of AuNPs-si-SP1 for 24, 48 and 72 h. B, Colony formation assay to detect the number of clones of A549 cells after different treatments. C, Colony formation curve in colony formation experiment. D, The cell viability of A549 cells treated with AuNPs-si-SP1 (20 nM), 6 Gy X-ray and different concentrations of AuNPs-si-SP1 (5, 10 and 20 nm) combined with X-ray evaluated by CCK-8 method. E, Apoptosis of A549 cells in different treatment groups. ♯ p < 0.05 compared with the AuNPs-si-SP1 + RT group. The data results were measurement data, which were expressed by mean ± standard deviation. The one-way ANOVA or repeated measurement ANOVA was used for comparison among multiple groups, followed by Tukey's post-hoc test. The repeated measurement ANOVA was adopted to compare the data for each group at different time points, followed by Bonferroni post-hoc test. The experiment was repeated three times.
The radiosensitization ability of AuNPs-si-SP1 was determined by clonal survival assay, and the survival fraction was calculated. The cell survival curve was drawn to explore the optimal X-ray radiation dose. Fig. 3B shows the degree of cell formation in each group. Compared with the radiotherapy group, the AuNPs-si-SP1 + radiotherapy group had the strongest inhibitory effect on cell formation, which was positively correlated with the concentration of AuNPs-si-SP1. The cell survival curve is shown in Fig. 3C. Radiotherapy obviously inhibited A549 cells at 6 Gy compared with 0 Gy, 2 Gy, and 4 Gy, and A549 survival rate decreased with the increase of radiation dose and concentration (Fig. 3C, black line). However, when the cells were treated with AuNPs-si-SP1 at different concentrations combined with radiotherapy, the survival rate of A549 cells decreased potently, and the survival curve of A549 cells moved down as a whole in contrast to the radiotherapy group. Among them, 20 nM AuNPs-si-SP1 + radiotherapy showed the best ability to inhibit cell survival, so 6 Gy radiation dose was selected for subsequent experiments. Moreover, the SER of AuNPs-si-SP1 at 10 nM and 20 nM were 2.09 and 2.13 respectively, which further improved the sensitivity efficiency compared with 5 nM. Then, the experiment was carried out with 20 nM AuNPs-si-SP1. It can be seen that AuNPs-si-SP1 had obvious radiosensitization effect on A549 cells.
CCK-8 method was employed to evaluate the viability of X-ray irradiated-cells after the cells were treated with different concentrations of AuNPs-si-SP1 (Fig. 3D). The cell growth was inhibited by treating with 10 nM AuNPs-si-SP1, and the survival rate was about 85.58% ± 2.06%. After irradiation alone, the cell survival rate abated to 82.05 ± 1.45%. Nevertheless, when the cells were treated with AuNPs-si-SP1 + irradiation, the cell survival was decreased notably as compared with the AuNPs-si-SP1 group. The inhibition rate ascended with the increase of AuNPs-si-SP1 concentration. The killing rate of 5 nM, 10 nM, and 20 nM AuNPs-si-SP1 combined with irradiation increased respectively by 2.02, 2.44 and 3.08 times in comparison with radiotherapy alone, which suggested that AuNPs-si-SP1 could promote the killing effect of radiotherapy in a dose dependent manner.
Furthermore, Annexin V-FITC and PI double staining were employed to detect apoptosis by flow cytometry. As shown in Fig. 3E, the number of apoptotic cells treated with AuNPs-si-SP1 or radiotherapy alone was higher than that of control cells. Importantly, the percentage of apoptotic cells after treatment with AuNPs-si-SP1 combined with radiotherapy increased by 13.33% compared with radiotherapy alone, suggesting that AuNPs-si-SP1 (20 nM) could effectively promoted the radiosensitivity of lung cancer cells.
AuNPs-si-SP1 enhanced radiosensitization of A549 cells by upregulating GZMB
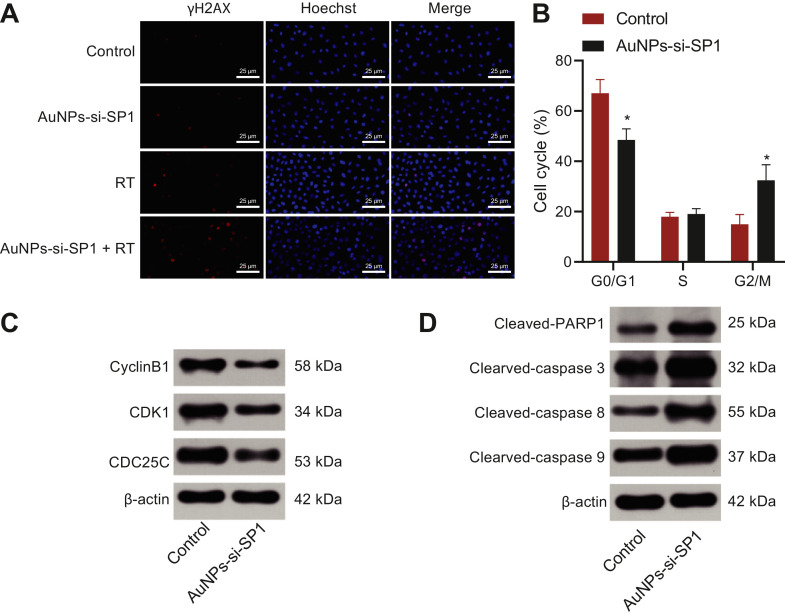
Based on the excellent radiosensitization effect of AuNPs-si-SP1, it was necessary and beneficial to explore its mechanism. Fig. 4A indicated that compared with control cells, A549 cells treated with AuNPs-si-SP1 or radiotherapy alone showed only a small amount of red fluorescence in the nucleus. However, when cells were treated with AuNPs-si-SP1 combined with radiotherapy, more red fluorescence was observed, most of which was overlapped with nuclear fluorescence. It clearly showed that DNA double strand breaks (DSBs) induced by radiotherapy were aggravated by the presence of AuNPs-si-SP1, and it also proved the radiosensitization of AuNPs-si-SP1 in lung cancer cells.
Fig. 4.
AuNPs-si-SP1 increases DNA double strand damage and the transformation of cells from S phase to G2/M phase of A549 cells. A, The representative images of A549 cells in different treatment groups stained with γΗ2ΑΧ. The inserted images were enlarged images. Hoechst 33,342 was used as a nuclear marker (scale bar = 25 nm). B, Cell cycle distribution of control A549 cells. C, The expression of G2/M block-related proteins (CDK1, CDC25C and CyclinB1) in A549 cells determined with Western blot analysis. D, The expression of apoptosis-related proteins (Cleaved-PARP1, Cleaved-Caspase3, Caspase-8 and Cleaved-Caspase-9) in A549 cells determined with Western blot analysis. * p < 0.05. The data results were measurement data, which were expressed by mean ± standard deviation. The two groups were analyzed by independent sample t-test, and the experiment was repeated 3 times.
Next, the effect of AuNPs-si-SP1 on cell cycle was studied. A549 cells were arranged into two groups including the PBS group (the control group) and the serum-free medium encompassing 20 nM AuNPs-si-SP1 group. Then cell cycle was surveyed by PI staining. Cell cycle distributions of the control group and the AuNPs-si-SP1 group are shown in Fig. 4B respectively. The number of cells in G0/G1 phase in the AuNPs-si-SP1 group was slightly lower than that in the control group, but no statistically significant difference was found in S phase between two groups. The sensitive phase of cells to irradiation is G2/M phase. Meanwhile, the G2/M ratio of A549 cells increased evidently after incubation with AuNPs-si-SP1 for 24 h. Western blot analysis was performed to determine the expression of G2/M block-related proteins (CDK1, CDC25C, and CyclinB1) and apoptosis-related proteins (Cleaved-PARP1, Cleaved-Caspase3, Caspase-8, and Cleaved-Caspase-9) in the control group and AuNPs-si-SP1 group (Fig. 4C, D), which revealed that compared with the control group, the protein expression of CDK1, CDC25C and CyclinB1 was significantly reduced, while Cleaved-PARP1, Cleaved-Caspase3, Caspase-8 and Cleaved-Caspase-9 levels were increased in the AuNPs-si-SP1 group. Collectively, AuNPs-si-SP1 could promote the transformation of cells from S phase to G2/M phase to enhance the radiosensitivity and apoptosis of A549 cells.
Bioinformatics analysis was performed to explore the mechanism of downregulated SP1 to increase radiosensitivity. The gene expression dataset GSE73095 for lung cancer before and after radiotherapy was obtained from GEO database. The DEGs were screened, which gained 71 DEGs (Fig. 5A). The interaction of these 71 genes was analyzed to construct the gene interaction network (Fig. 5B). The degree value of each gene in the network was calculated (Fig. 5C). The results found that the degree value of 7 genes was greater than 5. Among these 7 genes, the differential expression of GZMB in GSE73095 dataset was the most notable (Fig. 5D, Table S3), illustrating that GZMB was related to radiotherapy. The survival analysis of GZMB in lung adenocarcinoma samples collected by TCGA displayed that the overall survival rate of patients with high expression of GZMB was strikingly higher than that of patients with low expression of GZMB (Fig. 5E), which demonstrated that GZMB could effectively improve the prognosis and survival of patients. The binding sites of transcription factor SP1 and gene GZMB were predicted by JASPAR database (Fig. 5F). According to the above result, SP1 could regulate GZMB to influence the prognosis, survival and radiosensitivity of lung cancer patients.
Fig. 5.
Bioinformatics analysis demonstrates that SP1 may orchestrate GZMB in lung cancer. A, Heat map of DEGs in lung cancer radiotherapy gene expression dataset GSE73095. The abscissa represented the sample number, the ordinate represented the gene name, the tree on the left represented the gene expression level clustering, and the histogram on the upper right indicated the color scale. B, Network diagram of DEGs interaction analysis. Each circle in the graph represented a gene, and the lines between circles indicated the interaction between genes. If there were more interacting genes in a gene, the degree value of the gene was higher and the color was darker. C, Statistical analysis of degree values of core genes in DEG interaction network. The abscissa represented the degree value, and the ordinate represented the gene name. D, Box diagram of differential expression of 7 DEGs in GSE73095 screened by gene interaction analysis. Green indicated the lung cancer sample, and red indicated the sample after radiotherapy. E, Survival analysis of GZMB gene in lung adenocarcinoma of TCGA. The abscissa and ordinate represented survival time and survival rate respectively. Red represented samples with high expression of GZMB and blue represented samples with low expression of GZMB. F, The binding sites of transcription factor SP1 and gene GZMB predicted by JASPAR database. Kaplan-Meier method was used to count the survival rate of patients with lung cancer.
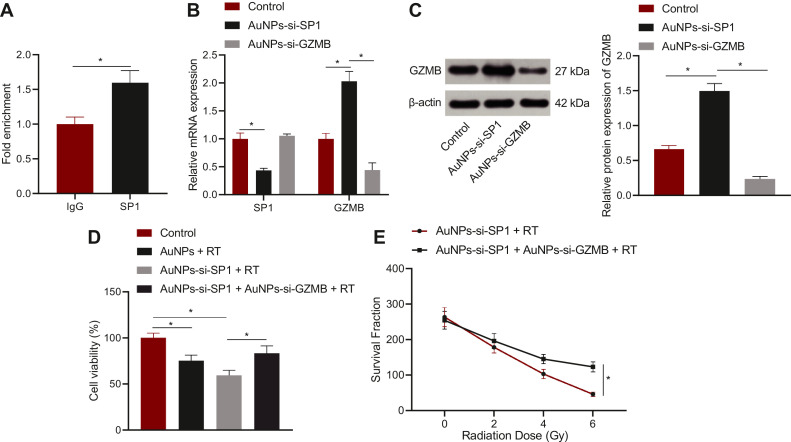
The DNA binding ability of SP1 to gene GZMB detected by ChIP assay is depicted in Fig. 6A. The amount of GZMB enriched in the SP1 group was notably more than that in the IgG group. The mRNA and protein expression and the killing effect of radiotherapy were analyzed through RT-qPCR, Western blot analysis, and CCK-8 assay and plate cloning experiment to verify the effect of SP1 on radiosensitivity by mediating GZMB. At this time, AuNPs combined with different siRNA were selected for the experiment. A549 cells were grouped into the control group, the AuNPs-si-SP1 group, and the AuNPs-si-SP1 + AuNPs-si-GZMB group. Lower mRNA expression of SP1 (Fig. 6B) and higher GZMB expression (Fig. 6B, C) in the AuNPs-si-SP1 group were observed in contrast to the control group, suggesting that silencing SP1 could upregulate GZMB in lung cancer cells. In comparison with the AuNPs-si-SP1 group, the mRNA expression of SP1 in the AuNPs-si-SP1 + AuNPs-si-GZMB group had no markedly change, but the expression of GZMB was remarkably decreased. A549 cells in the control group, the AuNPs-si-SP1 group, and the AuNPs-si-SP1 + AuNPs-si-GZMB group were irradiated at single dose of 6 Gy X-ray. As reflected by CCK-8 assay, compared with the AuNPs-si-SP1 + radiotherapy group, the cell survival rate in the AuNPs-si-SP1 + AuNPs-si-GZMB + radiotherapy group was strikingly augmented (Fig. 6D), indicating that simultaneous downregulation of SP1 and GZMB could reduce the radiosensitivity of lung cancer cells. Subsequently, A549 cells in the AuNPs-si-SP1 group and the AuNPs-si-SP1 + AuNPs-si-GZMB group were irradiated with different doses of X-ray. The result of plate cloning experiment exhibited that with the increase of radiation dose, the proliferation of cells in AuNPs-si-SP1 + AuNPs-si-GZMB group decreased more slowly (Fig. 6E). After simultaneous silencing SP1 and GZMB, the sensitivity of A549 cells to X-ray irradiation dropped, indicating that down-regulation of GZMB reversed the role of silencing of SP1. Collectively, SP1 silencing could upregulate GZMB to strengthen the radiosensitivity of lung cancer cells.
Fig. 6.
AuNPs-si-SP1 increases radiosensitivity of lung cancer cells by upregulating GZMB. A, ChIP assay to detect the binding ability of SP1 to GZMB promoter region. * p < 0.05 compared with IgG. B, The mRNA expression of SP1 and GZMB in A549 cells determined by RT-qPCR. C, The protein expression of GZMB in A549 cells checked by Western blot analysis. D, The survival rate of A549 cells measured by CCK-8 assay. E, The proliferation of A549 cells assessed by plate cloning assay. The data results were measurement data, which were expressed by mean ± standard deviation. Unpaired t-test was implemented for comparison between the two groups. The one-way ANOVA or repeated measurement ANOVA was applied for comparison among multiple groups, followed by Tukey's post-hoc test. The repeated measurement ANOVA was utilized to compare the data for each group at different time points, followed by Bonferroni post-hoc test. The experiment was repeated 3 times.
AuNPs-si-SP1 enhanced the radiosensitivity of A549 subcutaneous tumor
In view of the excellent radiosensitization effect of AuNPs-si-SP1 in vitro and its mechanism, the tumor growth retardation experiment was carried out in subcutaneous tumor bearing mice. As for the Fig. 7A, AuNPs-si-SP1 slightly delayed tumor growth compared with PBS or AuNPs group. However, all radiation groups showed different degrees of inhibition on tumor growth. Specifically, compared with radiotherapy alone, the tumorigenicity of radiotherapy-treated nude mice injected with AuNPs-si-SP1 was significantly reduced, showing the optimal anti-tumor effect. It indicated that AuNPs-si-SP1 markedly increased radiosensitization of mice with lung cancer during radiotherapy.
Fig. 7.
AuNPs-si-SP1 augments the radiosensitivity of lung cancer in vivo by upregulating GZMB. Mice were treated with PBS, 6 Gy radiotherapy alone, AuNPs-si-SP1 alone and 6 Gy radiation after intravenous injection of AuNPs-si-SP1 for 4 h separately. A, Tumor volume. B, Body weight of mice. C, Tumor weight of mice on the 20th day after various treatments. D, On the 20th day after treatment, HE staining and TUNEL test were performed on the tumor site of mice. E, Western blot to detect the expression of SP1 and GZMB protein in tumor tissues after treatment. n = 5 mice/group. The data results were measurement data, which were expressed by mean ± standard deviation. The one-way ANOVA or repeated measurement ANOVA was applied for comparison among multiple groups, followed by Tukey's post-hoc test. The repeated measurement ANOVA was utilized to compare the data for each group at different time points, followed by Bonferroni post-hoc test.
As described in Fig. 7C, AuNPs-si-SP1 or radiotherapy alone could inhibit tumor formation respectively. When AuNPs-si-SP1 combined with radiotherapy, the weight of tumor was the lightest, which also authenticated the radiosensitization effect of AuNPs-si-SP1. H&E staining (Fig. 7D) documented that the tumor cells in the PBS or AuNPs group had obvious cellular atypia because cells were closely arranged with large nucleus and few cytoplasm. In other groups, tumor cells were relatively reduced and replaced by fibrous tissue, especially in AuNPs-si-SP1 + radiotherapy group. TUNEL results (Fig. 7D) also depicted that the number of red fluorescence was the largest in AuNPs-si-SP1 + radiotherapy group, which caused severe tumor cell apoptosis, further certificating its ideal radiosensitivity effect. The weight changes of mice treated with different methods were observed, and no significant weight changes were observed during the experiment (Fig. 7B). H&E staining indicated that there was no obvious histopathological abnormality and pathological changes in normal tissues of mice after injection of AuNPs-si-SP1Q for 24 h (Figure S3). Meanwhile, the results of blood analysis and biochemical tests stated that there was no obvious physiological toxicity in mice treated with AuNPs-siRNA for 24 h (Figure S4). Conclusively, there was no side effect using AuNPs-si-SP1.
The protein expression of SP1 and GZMB in solid tumors was investigated to research the mechanism of AuNPs-si-SP1 increasing radiosensitivity through GZMB. It could be seen from Fig. 7E that after treatment with AuNPs-si-SP1 or radiotherapy alone, the expression of SP1 protein in solid tumors was strikingly decreased, while the expression of GZMB protein was rose. In particular, AuNPs-si-SP1 + radiotherapy group had decrease of the expression of SP1 and upregulation of GZMB. Taken together, AuNPs-si-SP1 could increase the radiosensitivity through the SP1/GZMB axis, and the system could be used as a radiosensitizer with excellent radiotherapy effect and low side effects.
Discussion
Radioresistance usually occurs and causes failure of radiotherapy for patients with lung cancer, so it is crucial to elucidate the mechanism of radioresistance in lung cancer [12]. AuNPs have been widely employed in biomedical fields including biosensing, diagnosis, and drug delivery thanks to their unique optical, electronic, sensing, and biochemical characteristics [13]. Moreover, AuNPs are able to a serve as a radiosensitizer in cancer radiation therapy [14]. In this context, our research was implemented to investigate the role of AuNPs in radiosensitivity for lung cancer via siRNA-SP1 delivery. Consequently, we observed that AuNPs-si-SP1 decreased SP1 expression to upregulate GZMB, thus facilitating radiosensitivity of lung cancer in vivo and in vitro.
Initially, our data demonstrated that AuNPs could load siRNA-SP1, which were internalized by A549 cells, and that compared with free siRNA-SP1, AuNPs-si-SP1 (equivalent to 100 nm siRNA) better suppressed SP1 mRNA and protein expression. AuNPs act as multifunctional therapeutics for cancer therapy through targeted delivery systems (anticancer agents, nucleic acids, biological proteins, and vaccines) [15]. Importantly, AuNPs possess the potential of targeted delivery of therapeutic siRNAs because they are capable of protecting siRNA against degradation [8]. For instance, a prior research exhibited that AuNPs loaded with EpCAM siRNA were obviously internalized in the retinoblastoma cells to downregulate EpCAM expression [16]. Another work manifested that siRNA designed against an oncogene c-Myc could be delivered by AuNPs into liver cancer cells to function as a candidate for anti-liver cancer therapy [17]. Also, the research conducted by Labala et al. elucidated that STAT3 siRNA-loaded AuNPs better reduced STAT3 protein expression and elevated cell apoptosis in B16F10 murine melanoma cells in contrast to also STAT3 siRNA treatment [18]. These data supported the delivery capacity of AuNPs for siRNAs into recipient cells.
Moreover, we also observed in our study that AuNPs-si-SP1 augmented radiosensitivity of lung cancer cells, accompanied by decreased cell viability and survival but increased cell cycle arrest, cell apoptosis and SER. It was widely recognized that AuNPs could be applied for radio-enhancement [19]. Also, it has been identified that AuNPs enhanced a multimodal synergistic cancer therapy strategy by co-delivering thermo-chemo-radio therapy [20]. Besides, radio-sensitization efficacy of AuNPs has been clarified in inhalational nanomedicine of lung cancer treatment [21]. Furthermore, Pandey et al. uncovered that AuNPs enhanced radio-sensitization of lewis lung cancer cells by diminishing cell survival, which was coincident with our result [22]. Notably, SP1 expression was high in lung cancer tissues, and that SP1 silencing repressed lung cancer cell proliferation, and accelerated cell cycle arrest and apoptosis [23]. A prior work ascertained depressed NSCLC cell proliferation and metastasis and blocked cell cycle on G1 phase after SP1 silencing [24]. In the present study, we adopted 6 Gy radiation. Consistently, 6 Gy radiation could achieve the killing effect at a dose of 6 Gy [25, 26].
As a transcription factor, SP1 bind to promoter, DNA tracts, and compete for binding of genes to repress gene expression [27]. The final finding in our research was that SP1 acted as transcription factor to suppress GZMB expression, thus repressing radiosensitivity of lung cancer cells by elevating cell viability and survival but diminishing cell cycle arrest, cell apoptosis and SER. SP1 elevation occurred in liquid-based cells from bronchial brushings in patients with lung cancer [10]. It was detected in a prior work that GZMB was upregulated in B16 melanoma cells after treatment with anti-PD-1 [28]. Moreover, it was indicated by another research that GZMB upregulation caused the susceptibility of NSCLC cells to NK cell-orchestrated killing [11]. Importantly, GZMB downregulation was discovered to be associated with progression-free survival of NSCLC patients [29]. In line with our finding, a previous research unraveled that GZMB knockdown could slow the Jurkat cell apoptosis during hand, foot, and mouth disease [30]. Also, GZMB activated p53 to interact with Bcl-2 to enhance cytotoxic lymphocyte-mediated apoptosis [31]. More importantly, Carnevalli et al. found that chemotherapeutic agent 5-fluorouracil (5-FU) treatment accelerated GZMB production in hematopoietic stem cells (HSCs), and that GZMB-deficient mice were more resistant to serial 5-FU treatments by increasing proliferation [18].
Conclusions
This study, for the time, developed a novel nanocomposite combining SP1 siRNA with AuNPs and investigated the radiosensitizing potential of AuNPs-si-SP1 for lung cancer (Fig. 8). AuNPs-si-SP1 caused upregulation of GZMB to slow cell viability and survival but accelerating cell cycle arrest, cell apoptosis and SER. Both in vitro and in vivo experiments unveiled that AuNPs-si-SP1 could facilitate radiosensitivity of lung cancer and have a therapeutic potential for lung cancer.
Fig. 8.
Radiosensitizing effect of AuNPs-si-SP1 on lung 721 cancer by up-regulating GZMB.
Ethics approval and consent to participate
This study was performed with approval of the Ethics Committee of Harbin Medical University Tumor Hospital by conforming to the Declaration of Helsinki. All participants or their guardians provided signed informed consent prior to enrollment. Animal experiments were ratified by Animal Ethics Committee of Harbin Medical University Tumor Hospital in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. Adequate measures were taken to minimize suffering of the included animals.
Consent for publication
Not applicable.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Availability of Data and Materials
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Funding
This work was supported by Haiyan Youth Foundation of Harbin Medical University Tumor Hospital (No. JJQN2019–08).
Authors’ Contributions
Ming Zhuang conceived and designed research. Shan Jiang performed experiments and interpreted results of experiments. Anxin Gu analyzed data and prepared figures. Ming Zhuang, Shan Jiang and Xuesong Chen drafted paper. Mingyan E edited and revised manuscript. All authors read and approved final version of manuscript.
Acknowledgements
We would like to give our sincere appreciation to the reviewers for their helpful comments on this article.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101210.
Contributor Information
Xuesong Chen, Email: chenxuesong2021@126.com.
Mingyan E, Email: emingyan2021@126.com.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Duruisseaux M., Esteller M. Lung cancer epigenetics: from knowledge to applications. Semin. Cancer Biol. 2018;51:116–128. doi: 10.1016/j.semcancer.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Nasim F., Sabath B.F., Eapen G.A., Cancer Lung. Med. Clin. North Am. 2019;103(3):463–473. doi: 10.1016/j.mcna.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Long L., Zhang X., Bai J., Li Y., Wang X., Zhou Y. Tissue-specific and exosomal miRNAs in lung cancer radiotherapy: from regulatory mechanisms to clinical implications. Cancer Manag. Res. 2019;11:4413–4424. doi: 10.2147/CMAR.S198966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darweesh R.S., Ayoub N.M., Nazzal S. Gold nanoparticles and angiogenesis: molecular mechanisms and biomedical applications. Int. J. Nanomed. 2019;14:7643–7663. doi: 10.2147/IJN.S223941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Her S., Jaffray D.A., Allen C. Gold nanoparticles for applications in cancer radiotherapy: mechanisms and recent advancements. Adv. Drug. Deliv. Rev. 2017;109:84–101. doi: 10.1016/j.addr.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., Xu J., Shi L., Yang H. Recent advances in the antilung cancer activity of biosynthesized gold nanoparticles. J. Cell. Physiol. 2020;235(12):8951–8957. doi: 10.1002/jcp.29789. [DOI] [PubMed] [Google Scholar]
- 8.Artiga A., Serrano-Sevilla I., De Matteis L., Mitchell S.G., de la Fuente J.M. Current status and future perspectives of gold nanoparticle vectors for siRNA delivery. J. Mater. Chem. B. 2019;7(6):876–896. doi: 10.1039/c8tb02484g. [DOI] [PubMed] [Google Scholar]
- 9.O'Connor L., Gilmour J., Bonifer C. The role of the ubiquitously expressed transcription factor Sp1 in tissue-specific transcriptional regulation and in disease. Yale J. Biol. Med. 2016;89(4):513–525. [PMC free article] [PubMed] [Google Scholar]
- 10.Cha N., Lv M., Zhao Y.J., Yang D., Wang E.H., Wu G.P. Diagnostic utility of VEGF mRNA and SP1 mRNA expression in bronchial cells of patients with lung cancer. Respirology. 2014;19(4):544–548. doi: 10.1111/resp.12272. [DOI] [PubMed] [Google Scholar]
- 11.Yao C., Ni Z., Gong C., Zhu X., Wang L., Xu Z. Rocaglamide enhances NK cell-mediated killing of non-small cell lung cancer cells by inhibiting autophagy. Autophagy. 2018;14(10):1831–1844. doi: 10.1080/15548627.2018.1489946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y.M., Liao X.Y., Chen X.W., Li D.Z., Sun J.G., Liao R.X. Regulation of miRNAs affects radiobiological response of lung cancer stem cells. Biomed. Res. Int. 2015;2015 doi: 10.1155/2015/851841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W., Cao Z., Liu R., Liu L., Li H., Li X. AuNPs as an important inorganic nanoparticle applied in drug carrier systems. Artif. Cells Nanomed. Biotechnol. 2019;47(1):4222–4233. doi: 10.1080/21691401.2019.1687501. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X.D., Wu D., Shen X., Chen J., Sun Y.M., Liu P.X. Size-dependent radiosensitization of PEG-coated gold nanoparticles for cancer radiation therapy. Biomaterials. 2012;33(27):6408–6419. doi: 10.1016/j.biomaterials.2012.05.047. [DOI] [PubMed] [Google Scholar]
- 15.Mioc A., Mioc M., Ghiulai R., Voicu M., Racoviceanu R., Trandafirescu C. Gold Nanoparticles as Targeted Delivery Systems and Theranostic Agents in Cancer Therapy. Curr. Med. Chem. 2019;26(35):6493–6513. doi: 10.2174/0929867326666190506123721. [DOI] [PubMed] [Google Scholar]
- 16.Mitra M., Kandalam M., Rangasamy J., Shankar B., Maheswari U.K., Swaminathan S. Novel epithelial cell adhesion molecule antibody conjugated polyethyleneimine-capped gold nanoparticles for enhanced and targeted small interfering RNA delivery to retinoblastoma cells. Mol. Vis. 2013;19:1029–1038. [PMC free article] [PubMed] [Google Scholar]
- 17.Shaat H., Mostafa A., Moustafa M., Gamal-Eldeen A., Emam A., El-Hussieny E. Modified gold nanoparticles for intracellular delivery of anti-liver cancer siRNA. Int. J. Pharm. 2016;504(1–2):125–133. doi: 10.1016/j.ijpharm.2016.03.051. [DOI] [PubMed] [Google Scholar]
- 18.Labala S., Jose A., Venuganti V.V. Transcutaneous iontophoretic delivery of STAT3 siRNA using layer-by-layer chitosan coated gold nanoparticles to treat melanoma. Colloids Surf. B Biointerfaces. 2016;146:188–197. doi: 10.1016/j.colsurfb.2016.05.076. [DOI] [PubMed] [Google Scholar]
- 19.Kim H., Sung W., Ye S.J. Microdosimetric-kinetic model for radio-enhancement of gold nanoparticles: comparison with LEM. Radiat. Res. 2021;195(3):293–300. doi: 10.1667/RADE-20-00223.1. [DOI] [PubMed] [Google Scholar]
- 20.Alamzadeh Z., Beik J., Mirrahimi M., Shakeri-Zadeh A., Ebrahimi F., Komeili A. Gold nanoparticles promote a multimodal synergistic cancer therapy strategy by co-delivery of thermo-chemo-radio therapy. Eur. J. Pharm. Sci. 2020;145 doi: 10.1016/j.ejps.2020.105235. [DOI] [PubMed] [Google Scholar]
- 21.Gadoue S.M., Toomeh D. Radio-sensitization efficacy of gold nanoparticles in inhalational nanomedicine and the adverse effect of nano-detachment due to coating inactivation. Phys. Med. 2019;60:7–13. doi: 10.1016/j.ejmp.2019.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Pandey A., Vighetto V., Di Marzio N., Ferraro F., Hirsch M., Ferrante N. Gold Nanoparticles Radio-Sensitize and Reduce Cell Survival in Lewis Lung Carcinoma. Nanomaterials (Basel) 2020;10(9) doi: 10.3390/nano10091717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang R., Xu J., Xu J., Zhu W., Qiu T., Li J. MiR-326/Sp1/KLF3: a novel regulatory axis in lung cancer progression. Cell Prolif. 2019;52(2):e12551. doi: 10.1111/cpr.12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Fu Y., Xia X., Zhang X., Xiao K., Zhuang X. Knockdown of SP1/Syncytin1 axis inhibits the proliferation and metastasis through the AKT and ERK1/2 signaling pathways in non-small cell lung cancer. Cancer Med. 2019;8(12):5750–5759. doi: 10.1002/cam4.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh S., Narang H., Sarma A., Kaur H., Krishna M. Activation of DNA damage response signaling in lung adenocarcinoma A549 cells following oxygen beam irradiation. Mutat. Res. 2011;723(2):190–198. doi: 10.1016/j.mrgentox.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X., Yu J., Li C., Sun X., Meng X. In-vivo comparison of (18)F-FLT uptake, CT number, tumor volume in evaluation of repopulation during radiotherapy for lung cancer. Sci. Rep. 2017;7:46000. doi: 10.1038/srep46000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vizcaino C., Mansilla S., Portugal J. Sp1 transcription factor: a long-standing target in cancer chemotherapy. Pharmacol. Ther. 2015;152:111–124. doi: 10.1016/j.pharmthera.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 28.D'Alterio C., Buoncervello M., Ierano C., Napolitano M., Portella L., Rea G. Targeting CXCR4 potentiates anti-PD-1 efficacy modifying the tumor microenvironment and inhibiting neoplastic PD-1. J. Exp. Clin. Cancer Res. 2019;38(1):432. doi: 10.1186/s13046-019-1420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurkmans D.P., Basak E.A., Schepers N., Oomen-De Hoop E., Van der Leest C.H., El Bouazzaoui S. Granzyme B is correlated with clinical outcome after PD-1 blockade in patients with stage IV non-small-cell lung cancer. J. Immunother. Cancer. 2020;8(1) doi: 10.1136/jitc-2020-000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M., Chen Y., Cheng X., Cai Z., Qiu S. GATA1/SP1 and miR-874 mediate enterovirus-71-induced apoptosis in a granzyme-B-dependent manner in Jurkat cells. Arch. Virol. 2020;165(11):2531–2540. doi: 10.1007/s00705-020-04783-4. [DOI] [PubMed] [Google Scholar]
- 31.Ben Safta T., Ziani L., Favre L., Lamendour L., Gros G., Mami-Chouaib F. Granzyme B-activated p53 interacts with Bcl-2 to promote cytotoxic lymphocyte-mediated apoptosis. J. Immunol. 2015;194(1):418–428. doi: 10.4049/jimmunol.1401978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.