Abstract
Disabled gene products are important for nervous system development in drosophila and mammals. In mice, the Dab1 protein is thought to function downstream of the extracellular protein Reln during neuronal positioning. The structures of Dab proteins suggest that they mediate protein-protein or protein-membrane docking functions. Here we show that the amino-terminal phosphotyrosine-binding (PTB) domain of Dab1 binds to the transmembrane glycoproteins of the amyloid precursor protein (APP) and low-density lipoprotein receptor families and the cytoplasmic signaling protein Ship. Dab1 associates with the APP cytoplasmic domain in transfected cells and is coexpressed with APP in hippocampal neurons. Screening of a set of altered peptide sequences showed that the sequence GYXNPXY present in APP family members is an optimal binding sequence, with approximately 0.5 μM affinity. Unlike other PTB domains, the Dab1 PTB does not bind to tyrosine-phosphorylated peptide ligands. The PTB domain also binds specifically to phospholipid bilayers containing phosphatidylinositol 4P (PtdIns4P) or PtdIns4,5P2 in a manner that does not interfere with protein binding. We propose that the PTB domain permits Dab1 to bind specifically to transmembrane proteins containing an NPXY internalization signal.
The disabled (dab) family of genes is conserved among nematodes, insects, and vertebrates. The drosophila dab gene is implicated in tyrosine kinase signaling pathways that regulate development of the embryonic central nervous system and the adult eye (15, 16, 31), suggesting that it is part of a system that interprets positional information during nervous system development. In the mouse, the disabled 1 (dab1) gene is required for correct positioning of neurons within layered structures of the brain (23, 48, 55). Mice that lack functional dab1 have defects in the organization of the cerebral cortex, hippocampus, and cerebellum. Recently obtained evidence suggests that the Dab1 protein is tyrosine phosphorylated in response to positional signals and likely functions in a tyrosine kinase signaling pathway (24). The functions of the mouse disabled 2 and nematode dab genes are unknown, although expression of disabled 2 is decreased in many tumors (37).
Comparison of the members of the Dab protein family shows the highest homology over the N-terminal 200 residues, which contains a predicted phosphotyrosine-binding (PTB) domain or phosphotyrosine-interacting domain (4). The mouse dab1 and dab2 genes are alternatively spliced, but all of the alternative proteins identified contain the PTB domain at their N termini (22, 56). The more C-terminal regions of Dab proteins are quite divergent, with only short regions of high similarity between the mouse Dab1 p80 and Dab2 p96 proteins and very little similarity to invertebrate proteins.
The high conservation of the PTB domain among members of the Dab family suggests a conserved function in relaying tyrosine kinase signals. PTB domains were first identified through their ability to bind to specific tyrosine-phosphorylated proteins and peptides (1, 18, 26). The PTB-containing proteins Shc and IRS-1 bind to tyrosine-phosphorylated growth factor receptors and then become tyrosine phosphorylated themselves, creating binding sites for other cell proteins (39). The Shc and IRS-1 PTB domains bind to a common phosphorylated sequence, Asn.Pro.X.pTyr (NPXpY), but differ in their preference for more N-terminal residues (18, 25, 38, 51). On the other hand, the brain proteins FE65 and X11 have PTB domains that bind to either nonphosphorylated or phosphorylated Asn.Pro.X.Tyr (NPXY) sequences (2, 12). Thus, not all PTB domains require tyrosine phosphorylation of their recognition sequences for binding.
Besides binding to proteins, some PTB domains may bind phospholipids. The three-dimensional structures of PTB domains resemble those of pleckstrin homology (PH) domains (10, 59, 60), which bind phosphoinositides with high specificity (11, 20, 32). The Shc PTB domain was also shown to bind phosphoinositides (60). A mutant Shc unable to bind phosphoinositides is unable to signal (42), suggesting that phosphoinositide binding is important for signaling by Shc.
We previously tested whether the Dab1 PTB domain would bind to phosphotyrosine-containing proteins from mouse brain extracts. Although unidentified phosphotyrosine-containing proteins were detected, we had no evidence that the binding was direct (22). We have now screened for other proteins that bind to the Dab1 PTB domain. We found that the Dab1 PTB domain binds with high affinity to a GYXNPXY sequence present in amyloid precursor protein and binds to FXNPXY sequences present in low-density lipoprotein (LDL) receptor family proteins and Ship, an inositol polyphosphate 5′ phosphatase. In each of these cases, binding is strongly inhibited if the tyrosine residues in the ligands are phosphorylated. The PTB domain also binds to phospholipid bilayers containing phosphoinositides. Phosphoinositide binding is stereospecific and can occur simultaneously with peptide binding. Thus, a membrane-anchored protein containing ΦXNPXY (where Φ is Y or F) could bind Dab1 with high affinity.
MATERIALS AND METHODS
Plasmids.
To construct the pBTM116-Dab1 PTB and the pBTM116-Dab1 PTB(Fms KIN) plasmids, the parental vectors pBTM116 (21, 53) and pBTM116(Fms KIN) (34), respectively, were digested with the restriction enzyme SalI and ligated with the XhoI-restricted PCR products of dab1 cDNA using the oligonucleotides Dab-BTM5 (GCGCTCGAGGGATGTCAACTGAGACAGA) and Dab-BTM3 (GCGCTCGAGCTAGCTTATCCTTTTGTGCCTT). This construct encodes a fusion protein containing an N-terminal LexA DNA binding domain followed by residues 1 through 178 of Dab1.
The construction of a glutathione S-transferase (GST)-Dab1 PTB domain and a GST-Dab1 PTB (R56E) domain has been described previously (22), and they encode fusions between GST and residues 29 to 197 of wild-type Dab1 or a mutant Dab1 with glutamic acid in place of arginine at residue 56, respectively. The GSTag-Dab1 PTB domain contains a protein kinase A (PKA) phosphorylation site between the GST and Dab1 PTB protein elements and was constructed similarly by using the GSTag parental vector (44).
The F158V mutation was introduced into the open reading frame of Dab1 p80 by inverse PCR with mutagenic oligonucleotides and Pfu polymerase (Stratagene). Following the PCR and ligation, template DNA was destroyed with DpnI. The mutation was confirmed by sequencing.
The plasmid pCS3+MT-APPC was a gift from Shlomo Handeli (Fred Hutchinson Cancer Research Center). The amyloid precursor protein (APP) partial cDNA was generated by reverse transcription from HeLa cell RNA, followed by PCR with the oligonucleotides AGATCTCGACAGTGATCGTCATCACC and AGATCTAGACTATTCTATAAATGGACACCG, and was cloned into the BglII site in the pCS3+MT vector (gift from Anne Vojtek). The plasmid expresses the last 59 residues of human APP including the entire cytoplasmic domain and 12 amino acids from the transmembrane domain fused C terminally to six reiterated copies of the Myc tag epitope.
Yeast two-hybrid screen.
A detailed protocol for the modified yeast two-hybrid screen is provided elsewhere (52). Briefly, the L40 strain of yeast, which has LexA operator sequences driving both HIS3 and lacZ reporter genes (21, 53), was transformed with pBTM116-Dab1(Fms KIN) and library DNAs. The activity of the Fms kinase in the yeast strains was confirmed by Western blotting of cell lysates made in radioimmunoprecipitation assay buffer (0.15 M NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 10 mM sodium phosphate [pH 7.0], 2 mM EDTA, 14 mM 2-mercaptoethanol, 50 mM NaF, 2 mM Na3VO4, 1 mM phenylarsine oxide, 20 μg of aprotinin/ml, 10 μg of pepstatin/ml, 10 μg of leupeptin/ml) and probing with antiphosphotyrosine antibody 4G10.
The libraries used were (i) neonatal brain cDNAs expressed as fusion proteins with the GAL4 activation domain from the pGAD-GH vector (Clontech) and (ii) EML cell cDNAs expressed as fusion proteins with the VP16 activation domain from the pVP16 vector (34) (gift from S. Tsai). Transformants (3 × 106 from library i and 3 × 105 from library ii) were plated on minimal medium lacking tryptophan and leucine to select for the LexA- and activation domain-encoding plasmids, respectively, and lacking histidine but containing 5 mM 3-amino-1,2,4-triazole to assay for transactivation of the HIS3 reporter. Colonies were picked after 3 days of growth. Yeasts were tested for β-galactosidase activity by filter lift assays (22), and colonies that produced stronger signals than the control yeast containing the pBTM116-Dab1(Fms KIN) and pVP16 vectors alone after 2 h were identified for further analysis. Library isolates (48 from library i and 24 from library ii) were retested for transactivation in yeast expressing either the LexA-lamin control fusion or the LexA-Dab1 PTB domain fusion in the absence of the Fms kinase. Those isolates that expressed less β-galactosidase activity than the vector-alone controls with LexA-lamin were not analyzed further. The remainder were isolated as plasmid DNAs and sequenced by using a BioSequencer (Applied Biosystems) with the pGAD-GH5′ oligonucleotide (CTATTCGATGATGAAGATACC; Clontech) or the M13 Forward oligonucleotide (Stratagene). Database searches were done with Blast, which is available at the National Center for Biotechnology Information website (37a).
Binding assays.
P19 embryonal carcinoma cells were grown as described elsewhere (45) and were treated with 10−6 M all-trans-retinoic acid and then grown on bacteriological-grade plastic petri dishes for indicated times. Cells (5 × 106) were washed with phosphate-buffered saline (PBS), lysed in TX-IPB (0.1 M NaCl, 1% Triton X-100, 10 mM HEPES [pH 7.4], 2 mM EDTA, 0.1% 2-mercaptoethanol, 20 μg of aprotinin per ml, 50 mM NaF, 0.2 mM phenylmethylsulfonyl fluoride, 2 mM Na3VO4, 1 mM phenylarsine oxide), and incubated at 0°C for 10 min. Lysates were clarified by centrifugation at 20,000 × g for 30 min, and equal aliquots were mixed with 1 μg of the indicated GST fusion protein bound to glutathione-agarose beads for 90 min at 4°C. Complexes were washed four times with TX-IPB and eluted by incubation with gel loading buffer (2% SDS, 20% glycerol, 0.1 M Tris-HCl [pH 6.8], 2.8 M 2-mercaptoethanol, 2.5 mM EDTA, 0.01% bromophenol blue) at 100°C for 5 min prior to analysis by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting with the antibodies indicated.
293T cells (5 × 106) grown on 100-mm-diameter tissue culture dishes in Dulbecco’s modified Eagle medium (Gibco BRL) containing 10% fetal bovine serum were transfected with 15 μg of each of the indicated DNAs by the calcium phosphate procedure (47). After 48 h, cells were lysed in TX-IPB and lysates were prepared as indicated above. Immunoprecipitation with the anti-Dab1 antibody B3 or preimmune antibodies that were cross-linked to protein A-Sepharose (Pharmacia) prior to assay has been described previously (22). Immunoprecipitates were analyzed by SDS-PAGE and Western blotting as described above.
Filter binding assays.
Arrays of peptides were synthesized on cellulose membranes as described previously (13, 14) with an ABIMED ASP 222 automated SPOT-robot. Filters were blocked with 10% fetal bovine serum in TBST (100 mM Tris Cl [pH 7.5], 150 mM NaCl, 1% Tween 20) for 1 h at 25°C, incubated with the 32P-labelled GSTag-Dab1 PTB domain (0.1 μg/ml, 2.5 × 106 cpm/ml) for 18 h at 4°C, and then washed several times with TSBT prior to autoradiography or quantitation using a PhosphorImager (Molecular Dynamics). To prepare the 32P-labelled GSTag-Dab1 PTB domain, 10 μg of the fusion protein immobilized on glutathione agarose was incubated with PKA (New England Biolabs) and [γ-32P]ATP at 10 μCi/μl for 30 min at 30°C in the buffer provided by the manufacturer. Unincorporated radioactivity was removed, and the labelled fusion protein was eluted with 20 mM reduced glutathione in PBS.
Fluorescence polarization.
Peptide binding to the GST-PTB domain fusion protein was measured by using fluorescence polarization (also known as fluorescence anisotropy [33]). An APP 17-mer peptide (acetyl-QNGYENPTYAFFEQGK-amide) and its phosphorylated analog (phosphate on the second tyrosine, residue 9) were synthesized with an AMS 222 Multiple Peptide Synthesizer using TenetaGel S resin (Rapp Polymere) and purified by high-pressure liquid chromatography. Peptide concentrations were determined by spectrophotometry and confirmed by amino acid analysis. Fifty nanomoles of peptide was reacted with 100 nmol of fluorescein-C6-succinimidyl ester (FXS; PanVera Corporation, Madison, Wis.) in 50 μl of 10% dimethyl sulfoxide–0.1 M potassium phosphate (pH 8.2) for 1 h at 37°C. Reactions were stopped with 5 μmol of Tris HCl (pH 8.0) and analyzed by thin-layer chromatography (Silica gel 60; methanol-acetic acid-water [4:1:1]). A fluorescent product at an Rf of 0.3 was eluted in 10 mM Tris HCl (pH 8.0)–1 mM EDTA. Based on fluorescence intensity, the fluorescent peptide (probe) concentration in binding reaction mixtures was estimated to be 20 nM.
Binding measurements were performed in a Beacon 2000 Variable Temperature Fluorescence Polarization System (PanVera Corporation). Essentially, a constant concentration of fluorescent probe peptide was incubated with various concentrations of the GST-PTB domain or GST in 100 μl of 0.5% Triton X-100–20 mM glutathione–200 mM Tris HCl (pH 8.0) at 4°C and fluorescence polarization was measured. Steady-state probe-to-protein binding was calculated from the linear relationship between polarization and the proportion of the probe bound. Actual polarization values ranged from 92.4 units (free probe) to 254.1 mP (100-fold excess of GST-PTB domain). Binding reached 95% of maximum after 5 min at 4°C. A Hill plot was linear with a slope of 1.0, indicating noncooperative binding. Competition experiments used a constant mixture of GST-PTB domain (1.1 μM), probe (20 nM), and buffer to which various concentrations of nonfluoresceinated synthetic peptide or its phosphorylated analog were added. To dephosphorylate the competitor phosphopeptide, we added potato acid phosphatase (0.05 U; Sigma, St. Louis, Mo.) to selected tubes and incubated them for 60 min at room temperature before cooling them to 4°C and measuring the polarization. To determine the effect of added phospholipids on GST-PTB domain binding to the probe, we added the probe to a standard unilamellar vesicle (LUV)–GST-PTB domain binding reaction mixture (see below) before reading the fluorescence polarization.
Phospholipid binding.
Phospholipid binding was assayed by mixing soluble GST-PTB domain or control proteins with large LUVs containing defined phospholipids (7, 43, 60). Following binding, the LUVs were isolated by centrifugation and the proportion of GST-PTB domain associated with the LUVs was determined by SDS-PAGE and Western blotting. If desired, the GSTag-PTB domain was first labelled with [γ-32P]ATP and the proportion associated with the LUVs was determined by SDS-PAGE and quantitation of radioactive bands on a PhosphorImager (Molecular Dynamics).
Lipids were from Sigma or Avanti (Ottawa, Ontario, Canada), except for PtdIns3,4,5P3, which was from C. S. Chen (54). Chloroform or methanol solutions containing 1 mg each of phosphatidylserine (PtdSer), phosphatidylcholine (PtdCho), and phosphatidylethanolamine (PtdEth) and 0 or 150 μg of phosphatidylinositol (PtdIns), PtdIns4P, PtdIns4,5P2, or PtdIns3,4,5P3 were dried under N2. One milliliter of 180 mM sucrose–20 mM KCl–3 mM EGTA–5 mM HEPES (pH 7.4) was then added, and the lipids were suspended by five cycles of freeze-thawing. The suspension was then passed 10 times through two 0.1-μm-pore-size polycarbonate membranes under N2 pressure of 400 lb/in2 to form LUVs. LUVs were stored at 4°C for up to a week before use.
In a standard binding experiment, 10 μl of LUVs was mixed with 10 μl of the GST-PTB domain or control proteins and 80 μl of buffer (100 mM KCl, 25 mM HEPES [pH 7.4], 3 mM EGTA, 1 mg of gelatin/ml) for 10 min at room temperature before centrifugation at 4°C at top speed in an Airfuge (Beckman Instruments) for 30 min. The supernatant and pellet were adjusted to equal volumes and analyzed by SDS-PAGE. Competing inositol polyphosphates (Sigma) or peptides were included as needed. To test whether the GST moiety participated in binding, 7 μg of GST-PTB domain fusion protein containing a thrombin cleavage site was incubated with 0.16 μg of thrombin at 30°C for 50 min before use in a standard binding experiment. GST and PTB domain fragments were detected by using anti-mDab antibody B3, which was raised to a GST-PTB domain fusion protein. To determine whether the GST-PTB domain could bind simultaneously to LUVs and to peptide, fluorescent probe peptide (20 nM) was added to a standard LUV–GST-PTB domain binding reaction mixture before centrifugation and the amounts of the fluorescent probe in the supernatant and pellet were quantified by fluorimetry.
Cell culture and immunocytochemistry.
Primary hippocampal neurons were prepared from an embryonic day 16 (E16) mouse essentially as described for E18 rats (17). Briefly, hippocampi were dissected, incubated with 0.25% trypsin, and dissociated by using a fire-polished Pasteur pipet. Cells were plated onto poly-l-lysine-coated coverslips in plating medium (minimal essential medium [MEM], 10% horse serum, 0.6% glucose) and allowed to adhere for 3 h; the coverslips were then transferred to dishes of glia in maintenance medium (MEM with the modified N2 supplement described in reference 17). After 24 h, cells were fixed in cold 4% paraformaldehyde (PFA)–PBS, blocked with 10% bovine serum albumin–PBS, and permeabilized with 0.2% Triton X-100–PBS. Antibody incubations were done in 1% bovine serum albumin–PBS. The primary antibodies were polyclonal anti-Dab1 and monoclonal anti-APP-A4 antibodies (Boehringer Mannheim). Oregon-green phalloidin (Molecular Probes) was used to stain filamentous actin (F-actin). The secondary antibodies were Cy-5–goat anti-rabbit and Texas red-donkey anti-mouse antibodies (Jackson Laboratory). Coverslips were mounted with DABCO (1,4-diazobicyclo octane) in polyvinyl alcohol and imaged by using a Deltavision deconvolution imaging microscope.
Glial cultures were prepared from postnatal day 0 mouse cortices. Briefly, cortices were dissected, cut into small pieces, and incubated with 0.25% trypsin and 1% DNase I in Hanks balanced salt solution. Tissues were then dissociated by trituration, and cells were plated onto tissue culture dishes in plating medium (see above). Cells were passaged by trypsin dissociation and maintained for up to 3 months. For hippocampal cultures, plating medium was replaced with maintenance medium (see above) 1 to 3 days before neuronal cultures were prepared.
RESULTS
Yeast two-hybrid screen for PTB ligands.
To identify tyrosine-phosphorylated ligands for the Dab1 PTB domain, we made use of a modified yeast two-hybrid system with a yeast strain that also expressed a protein tyrosine kinase (27, 34). The Dab1 PTB domain was expressed as a fusion protein with the LexA DNA binding domain, and cDNA libraries were expressed as fusion proteins with a transcriptional activator domain. Dab1 expression has been observed in brain and hematopoietic cells (22), so cDNA libraries derived from both sources were used. From the brain library, 48 clones were sequenced and found to represent six proteins, including APP (represented 21 times), the LDL receptor-related protein (LRP)–alpha-2 macroglobulin receptor (represented 10 times), and four novel sequences. From the hematopoietic library, we analyzed 24 clones which fell into two classes, which included the inositol polyphosphate-5′-phosphatase, Ship (represented 18 times), and one novel sequence. These clones were retested for interaction with the LexA-Dab1 PTB domain fusion protein in the presence or absence of the kinase to determine if phosphorylation is required for the interaction. Surprisingly, in all instances, lacZ expression was the same or slightly greater in the absence of kinase.
APP, LRP, and Ship share very little sequence homology, except for an NPXY sequence. Sequence motifs of this sort have been identified as ligands of a number of other PTB domains (2, 25), and we therefore investigated these potential ligands further.
Dab1 PTB domain binding to the cytoplasmic tail of APP in vitro and in cells.
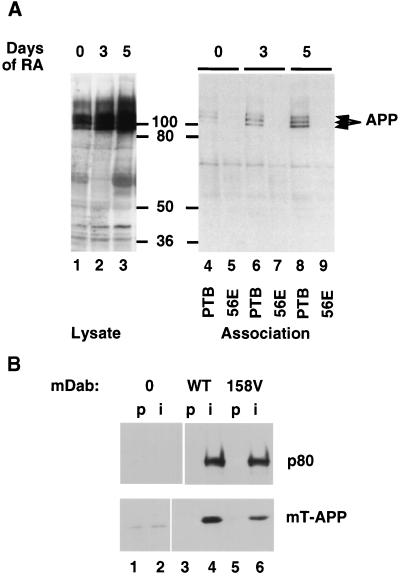
To test whether Dab1 would bind to the cytoplasmic tail of APP, we made use of P19 cells (45), which express Dab1 p80 (22) and differentiate into neuronal cells after retinoic acid treatment. Extracts of control or differentiated P19 cells were incubated with a GST fusion protein containing the Dab1 PTB domain, and bound proteins were detected by SDS-PAGE and Western blotting with APP antibodies. APP bound to the wild-type, but not to a mutant, PTB domain (Fig. 1A). To test whether association also occurs in vivo, we transfected 293T cells with expression vectors for p80 and for an epitope-tagged form of the cytoplasmic domain of APP (mT-APP). p80 was immunoprecipitated with an anti-Dab1 antibody, and the associated mT-APP was detected by SDS-PAGE and Western blotting with antibodies to the mT epitope tag (Fig. 1B). mT-APP was only immunoprecipitated from cells expressing Dab1 p80. These experiments suggest that the PTB domain of full-length Dab1 p80 can bind to the cytoplasmic domain of APP in cells.
FIG. 1.
Association between Dab1 and APP. (A) Retinoic acid treatment of P19 EC cells induces their differentiation into postmitotic neurons and glia and induces increased expression of APP as detected by Western blotting of total cell lysates (lanes 1 through 3; days of retinoic acid treatment are indicated above). Cell lysates were incubated with a GST fusion protein containing the wild-type Dab1 PTB domain (PTB; lanes 4, 6, and 8) or a fusion protein containing the 56E mutant PTB domain (lanes 5, 7, and 9). Bound APP was detected by Western blotting. The values between the gels are molecular sizes in kilodaltons. (B) Vector control (lanes 1 and 2) and wild-type (WT; lanes 3 and 4) and 158V mutant (lanes 5 and 6) Dab1 p80 expressed in 293T cells together with the Myc-tagged APP cytoplasmic domain (mT-APP). Immunoprecipitates were prepared by using preimmune serum (p; lanes 1, 3, and 5) or an anti-Dab1 (B3) antibody (i; lanes 2, 4, and 6) and analyzed by SDS-PAGE and Western blotting with antibodies to Dab1 (upper) or the Myc tag (lower).
Optimal peptide sequence for binding to the Dab1 PTB domain.
We tested the ability of the Dab1 PTB domain to interact directly with synthetic peptides based on the sequences identified in the two-hybrid screen. A GST-Dab1 PTB domain fusion protein was purified and labelled by phosphorylation with PKA and radioactive ATP. The purified, radioactive fusion protein was then incubated with a sheet of cellulose paper on which different 15-residue peptides had been synthesized directly (14). Each sheet contained up to 100 different peptide sequences. After incubation, the sheets were washed and exposed to film and bound radioactivity was quantified (Fig. 2A).
FIG. 2.
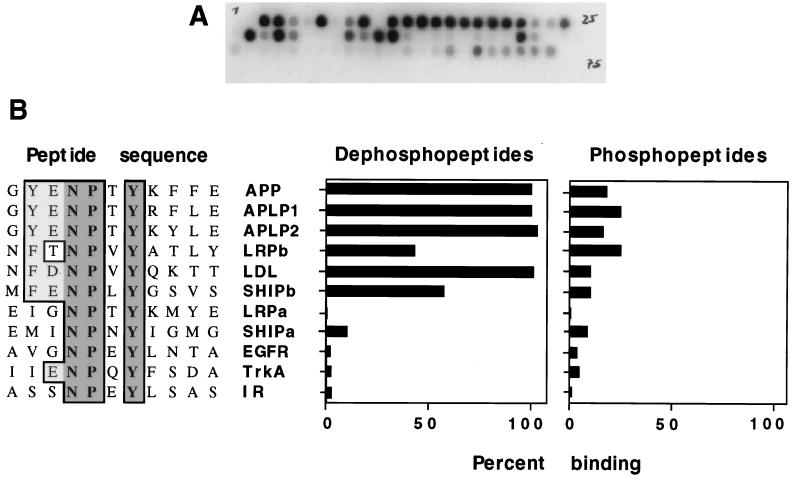
Direct binding between the Dab1 PTB domain and synthetic peptides. Various peptides, 15 to 17 residues in length, were synthesized on cellulose filters in a grided array. Filters were incubated with a 32P-labelled GSTag-PTB domain fusion protein, and the amount of the bound PTB domain was determined by autoradiography (A) or PhosphorImager (B). (A) Typical filter. (B) Comparison between interacting and noninteracting peptide sequences (left) showing invariant residues (dark shade) and conservative changes (light shade). Percent binding of the PTB domain to the indicated peptides, either unphosphorylated (left histogram) or phosphorylated (right histogram) at tyrosine, indicated by dark shading, is expressed relative to binding to the APP dephosphopeptide (set at 100%). Peptide sequences are designated by protein abbreviations. IR, insulin receptor; EGFR, epidermal growth factor receptor; LDL, low-density lipoprotein receptor.
We first tested the ability of the Dab1 PTB domain to bind to peptides containing NPXY motifs from APP, the APP relatives APP-like protein 1 (APLP1) and APLP2, LRP (N and C terminal [LRPa and LRPb, respectively]), the LDL receptor, Ship (N and C terminal [SHIPa and SHIPb, respectively]), and known ligands for the PTB domains of Shc and IRS-1, namely, the epidermal growth factor receptor, the nerve growth factor receptor (TrkA), and the insulin receptor (Fig. 2B). Peptides with phosphotyrosine in place of the tyrosine of the NPXY sequence (i.e., NPXpY peptides) were also tested (Fig. 2B).
The Dab1 PTB domain bound to peptides containing the NPXY regions of APP, APLP1, and APLP2. In all three proteins, the sequence GYENPTYXXEXXXX is conserved. Substitution of Phe for the upstream Tyr and Asp or Met for the Gly did not prevent binding in the context of the LRPb, LDL, and SHIPb dephosphopeptides. Phosphorylation of the tyrosine of the NPXY motif in these peptides inhibited binding. Known Shc PTB domain binding sites from the epidermal growth factor receptor and TrkA and the IRS-1 binding site on the insulin receptor failed to interact with the Dab1 PTB domain, whether phosphorylated or not (Fig. 2B). The peptides that bind to the Dab1 PTB domain share a tyrosine or phenylalanine at the position five residues N terminal to the tyrosine of the NPXY sequence (i.e., the −5 position), which is not conserved in the unbound peptides, suggesting that this residue is important for binding. Phosphorylation of the peptides on NPXY tyrosine inhibited binding in every case.
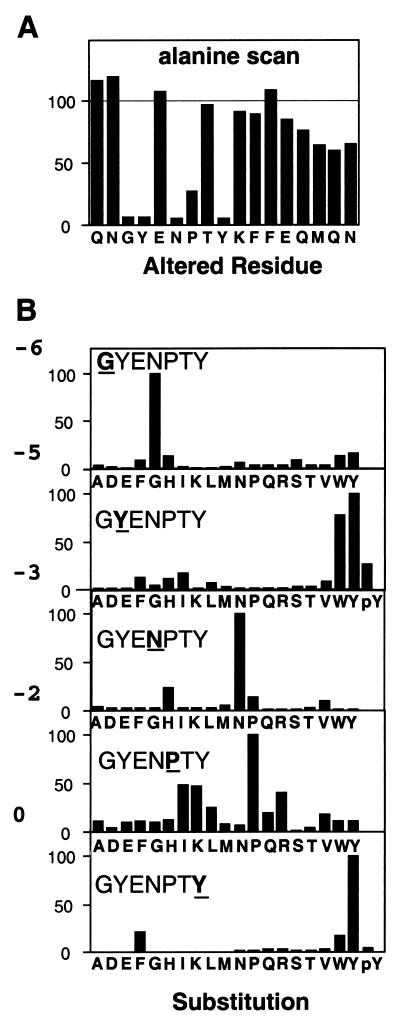
To determine which residues in the APP sequence might be involved in the interaction, we synthesized an array of peptides based on the APP sequence with an alanine substitution at each position in turn. The ability of each peptide to bind to the PTB domain was compared to that of the wild type (Fig. 3A). Most of the residues could be replaced with alanine with little or no effect on the amount of the Dab1 PTB domain that bound. However, alanine substitutions at positions −6, −5, −3, and −0 relative to the tyrosine inhibited binding by more than 90%. This suggests that the side chains of these residues are involved either in the interaction with the PTB domain or in the formation of a secondary structure that is required for the PTB domain interaction. As for other PTB domains, only the side chains of residues on the amino-terminal side of the NPXY motif appear to influence the strength of the interaction.
FIG. 3.
Relative binding of the PTB domain to 17-residue peptides based on the APP sequence. (A) Substitution of alanine for each amino acid in turn. The original amino acid is indicated below the histogram. (B) Retesting of each alanine substitution that resulted in reduced binding with 19 amino acids. The positions of the substitutions are indicated in each window by underlining of the wild-type residue. Substituted amino acids are listed below. Phosphotyrosine (pY) was substituted at positions −5 and 0.
In order to determine what features of the side chains in the APP peptide were recognized by the PTB domain, each of the five critical residues identified in Fig. 3A was replaced with all 19 amino acids except cysteine. In all cases, the Dab1 PTB domain bound better to the wild-type APP sequence than to any altered sequence (Fig. 3B). Most substitutions inhibited PTB domain binding by greater than 80%. However, the proline normally present at −2 could be replaced with isoleucine, lysine, or arginine with only 60% loss of binding, suggesting some tolerance for substitutions at this position. Surprisingly, replacement of Tyr-5 with phenylalanine resulted in a substantial reduction of binding, although tryptophan was tolerated. Phosphotyrosine and 16 other residues were not tolerated in place of Tyr-5. These results show that the wild-type APP sequence is optimal for binding to the Dab1 PTB domain but does not exclude the possibility that a distinct sequence containing multiple substitutions relative to the APP sequence would bind equally or better. For example, the significant binding of the Ship, LRP, and LDL receptor peptides implies that replacement of Tyr-5 with phenylalanine, and of Gly-6 with asparagine or methionine, is permitted, provided other changes are also made (Fig. 2B).
Binding to the APP sequence is high affinity and inhibited by phosphorylation.
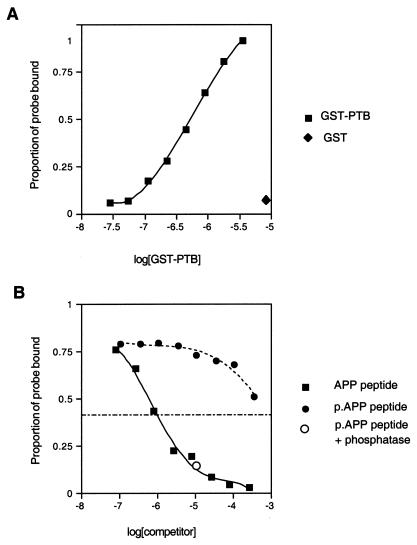
We used fluorescence polarization to determine the affinity of interaction between the Dab1 PTB domain and the APP synthetic peptide (see Materials and Methods). Binding was hyperbolic and noncooperative (Fig. 4A, squares). The concentration of the Dab1 PTB domain required for half-maximal binding, i.e., the dissociation constant, was 0.55 μM. GST alone did not bind (Fig. 4A, diamond). Binding was also evaluated by competition assay (Fig. 4B). Reaction mixtures contained 1.1 μM PTB domain, tracer fluorescent peptide, and different concentrations of nonfluorescent peptide (filled squares) or phosphopeptide (filled circles). Addition of either 2 μM peptide or 500 μM phosphopeptide reduced binding of the fluorescent tracer to the PTB domain by approximately 50%. This suggests that phosphorylation of Tyr-0 reduces the affinity of the APP peptide approximately 250-fold. Dephosphorylation of the phosphopeptide with a phosphatase turned it into a competitor as good as the synthetic peptide (Fig. 4B, open circle). This experiment shows that binding of the APP peptide is high affinity and strongly inhibited by phosphorylation.
FIG. 4.
Binding of the GST-PTB domain to the APP C-terminal peptide in solution. (A) Direct binding assay. Increasing amounts of the GST-PTB domain were added to a solution containing a fixed amount of fluorescently labelled APP peptide (probe). The proportion of probe bound to the PTB domain was determined from the fluorescence polarization (squares). The hyperbolic binding curve was fitted with a Kd of 0.55 μM. GST protein alone did not bind (diamond). (B) Nonfluorescent peptide or phosphopeptide competitors added to a mixture of the GST-PTB domain (1.1 μM) and a fixed amount of the fluorescent APP peptide. Addition of increasing amounts of the unphosphorylated APP peptide resulted in a decrease in the fraction of fluorescent APP bound, with half-maximal inhibition at 2 μM (closed squares). The Tyr-0-phosphorylated APP (p.APP) peptide competed less well, with half-maximal inhibition at 500 μM (closed circles). Phosphatase treatment of the phosphopeptide restored its ability to compete with the fluorescent probe for binding (open circle).
The PTB domain also binds to phosphoinositides.
The PTB domain three-dimensional fold resembles a PH domain, and the Shc PTB domain has been shown to bind to phospholipids (60). To determine if Dab1 might bind to phospholipids, we assayed for association between the GST-Dab1 PTB domain fusion protein and large LUVs with different lipid compositions. Following an incubation with the soluble PTB domain, LUVs and bound protein were pelleted by centrifugation. The fusion proteins were detected by SDS-PAGE and Western blotting with antibodies to GST.
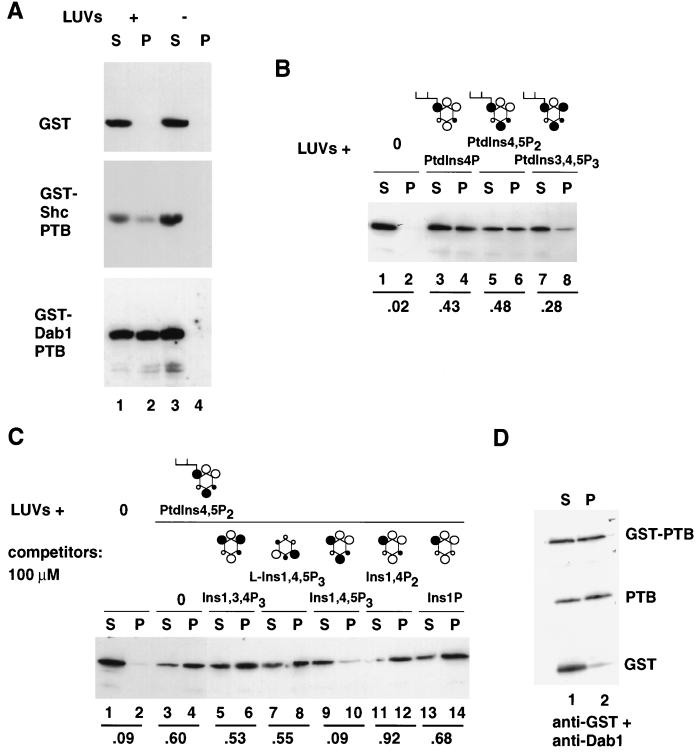
We first compared GST fusion proteins containing the Shc or Dab1 PTB domain for binding to LUVs that contained PtdIns4,5P2, PtdIns4P, and PtdSer. Whereas GST remained in the high-speed supernatant in the presence or absence of LUVs, GST-Shc PTB domain and GST-Dab1 PTB domain fusion proteins appeared in the high-speed pellet fraction in the presence, but not in the absence, of LUVs (Fig. 5A). This suggests that the Dab1 PTB domain, like the Shc PTB domain, binds to LUVs containing mixed phospholipids.
FIG. 5.
The Dab1 PTB domain interacts with phosphoinositides in a stereospecific manner. (A) GST, GST-Shc PTB domain, and GST-Dab1 PTB domain proteins incubated in the presence (lanes 1 and 2) or absence (lanes 3 and 4) of LUVs containing 66% PtdSer, 17% PtdIns4P, and 17% PtdIns4,5P2. The LUVs were pelleted by high-speed centrifugation, and the proteins in the supernatant (S) and pellet (P) fractions were detected by Western blotting with anti-GST antibodies. (B) Dab1 PTB domain fusion protein incubated with LUVs (33% each PtdSer, PtdCho, and PtdEth) containing no phosphoinositides, 5% PtdIns4P, 5% PtdIns4,5P2, or 5% PtdIns3,4,5P3. Proteins in the supernatant (S) and pellet (P) fractions were detected as described for panel A. The fraction of the GST-PTB domain associated with LUVs was determined in each case. (C) Binding of the Dab1 PTB domain fusion protein to LUVs containing 0 or 5% PtdIns4,5P2 in the absence (lanes 1 to 4) or presence of various inositol phosphates at 100 μM (d-Ins1,3,4P3 [lanes 5 and 6], l-Ins1,4,5P3 [lanes 7 and 8], d-Ins1,4,5P3 [lanes 9 and 10], d-Ins1,4P2 [lanes 11 and 12], and d-Ins1P [lanes 13 and 14]). Only d-Ins1,4,5P3 was an effective competitor of binding. (D) Partial thrombin digestion of the GST-PTB domain fusion released the PTB domain and GST, which were then incubated with LUVs. The liberated PTB domain bound to LUVs containing PtdIns4,5P2, while the liberated GST did not. Molecular structures are depicted with inositol rings (hexagons), hydroxyl groups (open circles), phosphates (closed circles), and a diacylglycerol backbone (branch). Groups oriented above (large circles) and below (small circles) the plane of the paper are indicated.
To assess the phospholipid specificity of binding, LUVs were prepared containing equal quantities of PtdSer, PtdCho, and PtdEth and 0 or 5% (by weight) PtdIns4P, PtdIns4,5P2, or PtdIns3,4,5P3. This corresponds to approximately 7.5 μM phosphoinositide in the outer leaflet of the lipid bilayer. Whereas the GST-Dab1 PTB domain fusion protein bound to LUVs containing various phosphoinositides, it did not bind to LUVs lacking phosphoinositide (Fig. 5B). Binding to nonphosphorylated PtdIns or to PtdIns3,4,5P3 was less efficient than binding to PtdIns4P or PtdIns4,5P2 (data not shown). To further assess whether the binding to phosphoinositides is specific or simply reflects an affinity for strongly anionic phospholipids, binding to LUVs containing PtdIns4,5P2 was tested in the presence of various phosphorylated isomers of inositol (Fig. 5C). Binding was competed by 100 μM d-Ins1,4,5P3 but not by l-Ins1,4,5P3 or various other inositol phosphates. When tested at 30 μM, competition was again detected only with d-Ins1,4,5P3 (data not shown). The pattern of competition suggests that the Dab1 PTB domain specifically recognizes the phosphorylated inositol head group present in PtdIns4,5P2 and that binding is reduced by phosphate at the D3 position. The stereospecificity implies that binding is specific and that the PTB domain does not bind simply to highly charged molecules.
To determine if the dimerization of GST fusion proteins appreciably influenced the binding to the LUVs, we tested whether cleavage of the GST-Dab1 PTB fusion protein with thrombin significantly alters binding (Fig. 5D). While released GST did not bind the LUVs, the released Dab1 PTB domain and uncleaved fusion protein bound with comparable efficiencies. This suggests that GST-mediated dimerization does not artificially raise the apparent affinity of binding. However, the affinity of binding appears to be low. When various concentrations of the 32P-labelled Dab1 PTB domain were incubated with LUVs containing PtdIns4,5P2, binding increased approximately linearly to the maximum PTB domain concentration tested (2 μM) (data not shown). This suggests that the dissociation constant is in excess of 2 μM. For comparison, the dissociation constant for Shc PTB domain binding to similar LUVs was estimated to be 25 μM (41), and the Dab1 PTB domain appears to bind more efficiently than the Shc PTB domain (Fig. 5A).
Independent binding of the PTB domain to phosphoinositides and to peptides.
Phosphopeptides and phospholipids have been reported to mutually compete for binding to the Shc PTB domain (41, 60). To determine whether the Dab1 PTB domain binds to phosphoinositides and to peptides through the same binding site, we first created and tested a mutant PTB domain that does not bind to peptides. The C-terminal alpha helix of the PTB domain is highly conserved among Dab1, Shc, and other PTB domains (22). An invariant phenylalanine, F158 in Dab1, has been mutated to valine in Shc and X11 and found to be essential for peptide binding (2, 57). We replaced F158 with valine (F158V mutant) in Dab1 and tested its binding to peptides and lipids.
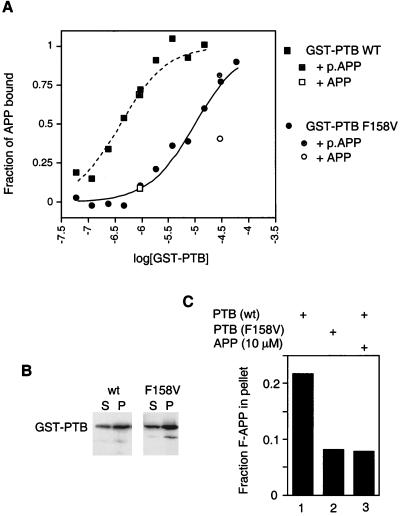
Full-length wild-type and F158V mutant Dab1 were expressed in 293 cells together with the epitope-tagged cytoplasmic domain of APP (mT-APP; Fig. 1B). Less mT-APP coprecipitated with F158V Dab1 p80 than with the wild type. The actual reduction in binding affinity caused by the F158V mutation was determined by using fluorescence depolarization (Fig. 6A). The wild-type Dab1 PTB domain fusion protein bound the fluorescent APP peptide with a dissociation constant of 0.4 μM (squares), while the mutant fusion protein bound it with an affinity of 10 μM (circles); i.e., there was a 25-fold decrease in affinity. Despite the reduced binding affinity, the mutant PTB domain retained specificity for the nonphosphorylated peptide (Fig. 6A, legend). Despite pronounced inhibition of peptide binding in vivo and in vitro, the F158V PTB domain fusion protein bound as well as the wild type to LUVs (Fig. 6B), indicating that a functional peptide binding site is not needed for binding to phosphoinositides.
FIG. 6.
Independence of peptide and phospholipid binding. (A) Reduced peptide binding of the F158V mutant PTB domain. Binding of wild-type (WT; squares) and mutant (circles) GST-PTB domains to a fluorescent APP peptide probe was determined by using fluorescence polarization. The hyperbolic curves were fitted with a Kd of 400 nM (wild type) or 10 μM (F158V mutant). Binding reactions were competed specifically by addition of the nonfluorescent APP peptide (open symbols) but not the phosphorylated APP (p.APP) peptide (shaded symbols). (B) Binding of wild-type (lanes 1 and 2) and F158V mutant (lanes 3 and 4) GST-PTB domains to LUVs containing 5% PtdIns4,5P2. Assays were performed as described in the legend to Fig. 4. S, supernatant; P, pellet. (C) Binding of fluorescent APP (F-APP) peptide to LUVs mediated by the GST-PTB domain. The fluorescent APP peptide was mixed with LUVs (containing 5% PtdIns4,5P2) and the wild-type (lanes 1 and 3) or F158V mutant (lane 2) GST-PTB domain in the absence (lanes 1 and 2) or presence (lane 3) of an excess of a nonfluorescent APP peptide competitor. The LUVs were isolated by centrifugation, and the associated fluorescent peptide was quantified.
Binding to membrane phosphoinositides could assist interaction between the PTB domain and a membrane protein, such as APP, LRP, or the LDL receptor, provided that peptide binding and phospholipid binding are not mutually competitive. Binding of the PTB to LUVs containing phosphoinositides was not inhibited by peptide concentrations exceeding the Kd, and binding of PTB to the peptide was not inhibited by LUVs (data not shown). Indeed, the fluorescent peptide efficiently bound to LUVs when the wild-type, but not a mutant, PTB domain was present (Fig. 6C). This shows that the PTB domain can bind simultaneously to peptides and to phosphoinositides and thus bridge between a peptide and a phospholipid membrane.
Subcellular distribution of Dab1.
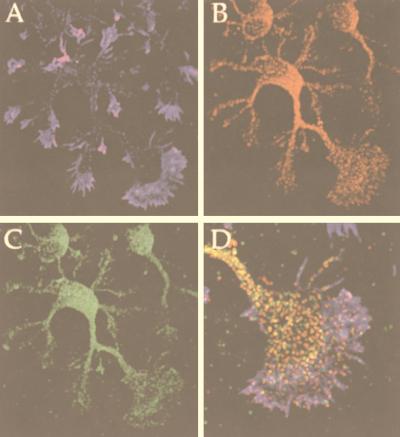
Dab1 function is important for neuronal placement in various parts of the brain, including the hippocampus (23, 48, 55). We investigated whether the protein-protein interactions detected here might be possible in hippocampal neurons. As a first step, we tested whether the Dab1 protein and APP are in the same compartment of the cell. We employed cultured embryonic hippocampal neurons, which develop a dominant axonal projection and several smaller dendritic processes and share many biological properties with their counterparts in the brain (17) (Fig. 7). Most of the Dab1 (Fig. 7B and D, red) and APP (Fig. 7C and D, green) was seen in the cell soma. Filamentous actin was detected with phalloidin (blue) and demarcates the leading edge of the growth cones. APP is reported to be predominantly localized in the endosomal compartment with a small, short-lived fraction at the cell surface (58). It is first delivered to the axon and then transported to the dendrites (49, 50, 58). Interestingly, Dab1 is also enriched in axons and both APP and Dab1 were in small particles that could represent intracellular vesicles. At high magnification, it is apparent that subsets of Dab1 and APP colocalize in the axonal shaft and the central region of the growth cone (Fig. 7D; yellow regions indicate overlap), suggesting that these molecules could interact under appropriate conditions.
FIG. 7.
Dab1 colocalizes with APP in primary neurons. (A to D) Triple labelling for F-actin (A; blue), Dab1 (B; red), and APP (C; green). D, merged image at higher magnification, showing the axonal growth cone. Overlap of Dab1 and APP appears yellow.
DISCUSSION
Evolutionary conservation of the Dab family PTB domain suggests that conserved aspects of Dab signaling involve regulated interactions of the PTB domain and other signaling molecules. In this study, we have identified binding partners for the Dab1 PTB domain as a step toward unravelling the Dab1 signaling pathway.
A comprehensive screen for protein ligands for the PTB domain showed that several proteins containing ΦXNPXY sequences were able to interact with the PTB domain in yeast. The binding is direct, since it occurs in vitro with the bacterially synthesized PTB domain and chemically synthesized peptides. The interactions may also be significant in cells and mediate Dab1 activity. The interacting proteins fell into two classes: the cytoplasmic signaling protein Ship and the transmembrane proteins of the APP and LDL receptor families. Recently, Trommsdorff et al. also found that the Dab1 PTB domain interacts with the cytoplasmic domains of APP, LRP, and the LDL receptor (50a). Ship has been characterized in hematopoietic cells (34), where certain splice forms of Dab1 are expressed (22). It is not known whether Ship and a new relative, Ship2 (40), are expressed in the brain. Of the transmembrane proteins identified as Dab1 PTB ligands, APP, APLP1, APLP2, and LRP are all expressed in the developing embryonic brain during the time when Dab1 function is important for neuronal positioning (35). Furthermore, both Dab1 and APP are present in one of the cell types whose position is abnormal in the absence of Dab1, the hippocampal neuron, and are found in overlapping regions within the cell, suggesting that they could interact with each other in vivo. Dab1 can bind to the cytoplasmic domain of APP in transfected cells, and the PTB domain of Dab1 binds to full-length APP in vitro. Binding is specific and has an affinity of approximately 0.5 μM. This is similar to the binding affinity of the X11 domain (59) and may be significant in the cell since the APP and LDL receptor family protein ligands are relatively abundant.
In contrast to other PTB domains, the PTB domain of Dab1 binds only to nonphosphorylated ligands. Tyrosine phosphorylation reduces the binding affinity approximately 250-fold. Such strong inhibition has not been reported for other PTB domains, but its significance is unclear since tyrosine phosphorylation of APP and LDL receptor family proteins has not been detected. On the other hand, Ship becomes tyrosine phosphorylated, probably at the Dab1 interaction site and additional tyrosines, when myeloid cells are treated with CSF-1 and some other cytokines (34). While tyrosine phosphorylation of Ship would promote interaction with the Shc PTB domain, it would also reduce interaction with the Dab1 PTB domain.
APP binds to other PTB domains besides Dab1. APP was detected in a two-hybrid screen for ligands for the PTB domain of FE65 (12), and two-hybrid screens with the APP cytoplasmic domain detected binding to the PTB domains of FE65 and X11 (5, 36). FE65 and X11 are brain proteins whose function(s) is unknown. The sequence requirements for binding of APP to the FE65 and X11 PTB domains were analyzed by Borg et al. (2). For both PTB domains, the Y of the NPXY sequence contributed little to binding affinity and could be replaced with alanine or phosphotyrosine with little change in affinity. This contrasts with the strong inhibitory effect of alanine or phosphotyrosine substitution on binding to the Dab1 PTB domain. Thus, different PTB domains recognize the APP sequence with high affinity but through different side chain contacts. Dab1, X11, and FE65 are expressed in the brain and may therefore compete for interaction with APP.
The function of Dab1 binding to LRP, APP, and their relatives could be to regulate trafficking or processing. The internalization signals of LRP and APP contain the NPXY sequence, which is bound by the Dab1 PTB domain (8, 9, 30). Nonetheless, it is unlikely that Dab1 functions in the internalization process per se. First, Dab1 is absent from many cell types that successfully internalize LDL receptors or APP. Second, the LDL receptor is thought to be clustered into coated pits by direct binding to clathrin (28). On the other hand, Dab1 may compete for internalization signals. By altering protein sorting into coated pits, Dab1 may affect membrane flow from the surface to intracellular membrane systems and hence influence membrane recycling, which is important for cell movement (6). APP is best known for its increased cleavage and the accumulation of a degradation product, beta amyloid, in Alzheimer’s disease (19). Interestingly, the NPXY motif appears to be important for proteolysis of APP to produce beta amyloid (29). Overexpression of X11 and FE65 affects APP processing in an opposing manner. While X11 overexpression leads to an increased half-life of APP, possibly by slowing the sorting of this protein into the endosomal compartment (3), overexpression of FE65 leads to increased translocation of APP to the cell surface and increased production of the proteolytic fragments (46). It remains to be determined what effect Dab1 might have on these processes.
It is also possible that Dab1 PTB domain signaling is mediated by proteins other than APP and LDL receptor family proteins. The high-affinity binding sequence found in APP family members, GYXNPXY, is found in approximately 10 other eukaryotic proteins listed in current (November 1998) databases, including the APP orthologs from drosophila and Caenorhabditis elegans. The high degree of conservation of this motif, 100% over the seven residues, suggests that selective pressures, possibly exerted by binding partners, act on it. The binding studies do not exclude the possibility that the Dab1 PTB domain has additional ligands with distinct sequences. Five cDNA clones were isolated that encode apparent Dab1 PTB domain binding partners that lack NPXY motifs in the interacting regions. They also lack other common sequence patterns. They may, therefore, bind to the PTB domain through novel interacting sequences.
PTB domains and PH domains are similar in structure. Many PH domains bind with 10−5 to 10−7 M affinity to phosphoinositides with characteristic stereospecificity. Similarly, the Shc PTB domain binds to PtdIns4P, PtdIns4,5P2, and PtdIns3,4,5P3 with affinities in the 10−4 to 10−5 M range (60). Making use of a triple-charge mutation of Shc that prevents lipid binding without preventing phosphopeptide binding, Ravichandran et al. (42) obtained evidence that lipid binding is important for Shc function. They suggested a model in which weak interaction between the Shc PTB domain and membrane phospholipids is a prerequisite for recruitment of Shc to activated growth factor receptors and Shc is released from phospholipids as it binds to the receptor. Using similar methods, we have found that the Dab1 PTB domain binds to PtdIns4P and PtdIns4,5P2 but less to PtdIns or PtdIns3,4,5P3. Since PtdIns4,5P2 is more abundant than PtdIns3,4,5P3, it is likely to be the major lipid bound to the Dab1 PTB domain in the cell. Unlike the Shc PTB domain (41, 60), the Dab1 PTB domain can bind simultaneously to synthetic peptides and phosphoinositides. Thus, binding to membrane phospholipids could reinforce binding to ΦXNPXY motifs in the cytoplasmic domains of transmembrane proteins. Unlike Shc, recruitment of Dab1 to a protein ligand would therefore not occur at the expense of binding to phospholipids.
The Dab1 protein acts within embryonic neurons and is required for appropriate neuronal placement within the brain. Recent findings suggest that it functions downstream of the extracellular matrix protein Reln that may define targets for the migrating neurons. Our findings show that the Dab1 PTB domain binds with high affinity to unphosphorylated targets and that binding is dramatically reduced by tyrosine phosphorylation. One of the targets identified in this study, Ship, is regulated by phosphorylation, while APP and LRP are not known to be tyrosine phosphorylated. However, Reln increases tyrosine phosphorylation of Dab1 itself (24), and Dab1 PTB domain function may be consequently regulated. Exposure of binding surfaces or changes in subcellular distribution could alter Dab1 PTB domain activity. It will be interesting to determine the Dab1 PTB domain functions required for Reln signaling and how the PTB domain ligands are involved in neuronal placement.
ACKNOWLEDGMENTS
We thank Steve Hahn for his encouragement in the use of the fluorescence polarization device, Wendy Thomas and John Glomsett for use of the LUV extrusion bomb, John Rasko for plasmid, Jürgen Wehland for helpful suggestions on the peptide arrays, Len Stevens and Lew Cantley for advice, Ching-Shih Chen for a kind gift of di-C16-PtdIns3,4,5P3, and Andrea Tiepold and Brigitte Kornak for help with the peptide synthesis.
This work was supported by R01-CA-41072 (J.A.C.) and NIH GM58801-01 (F.B.G.).
REFERENCES
- 1.Blaikie P, Immanuel D, Wu J, Li N, Yajnik V, Margolis B. A region in Shc distinct from the SH2 domain can bind tyrosine-phosphorylated growth factor receptors. J Biol Chem. 1994;269:32031–32034. [PubMed] [Google Scholar]
- 2.Borg J-P, Ooi J, Levy E, Margolis B. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borg J-P, Yang Y, De T B M, Margolis B, Turner R S. The X11alpha protein slows cellular amyloid precursor protein processing and reduces Abeta40 and Abeta42 secretion. J Biol Chem. 1998;273:14761–14766. doi: 10.1074/jbc.273.24.14761. [DOI] [PubMed] [Google Scholar]
- 4.Bork P, Margolis B. A phosphotyrosine interaction domain. Cell. 1995;80:693–694. doi: 10.1016/0092-8674(95)90347-x. [DOI] [PubMed] [Google Scholar]
- 5.Bressler S L, Gray M D, Sopher B L, Hu Q, Hearn M G, Pham D G, Dinulos M B, Fukuchi K, Sisodia S S, Miller M A, Disteche C M, Martin G M. cDNA cloning and chromosome mapping of the human Fe65 gene: interaction of the conserved cytoplasmic domains of the human beta-amyloid precursor protein and its homologues with the mouse Fe65 protein. Hum Mol Genet. 1996;5:1589–1598. doi: 10.1093/hmg/5.10.1589. [DOI] [PubMed] [Google Scholar]
- 6.Bretscher M S. Getting membrane flow and the cytoskeleton to cooperate in moving cells. Cell. 1996;87:601–606. doi: 10.1016/s0092-8674(00)81380-x. [DOI] [PubMed] [Google Scholar]
- 7.Chen R H, Corbalan-Garcia S, Bar-Sagi D. The role of the PH domain in the signal-dependent membrane targeting of Sos. EMBO J. 1997;16:1351–1359. doi: 10.1093/emboj/16.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W J, Goldstein J L, Brown M S. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J Biol Chem. 1990;265:3116–3123. [PubMed] [Google Scholar]
- 9.Davis C G, Van D I R, Russell D W, Brown M S, Goldstein J L. Low density lipoprotein receptor: identification of amino acids in the cytoplasmic domain required for rapid endocytosis. J Biol Chem. 1987;262:4075–4082. [PubMed] [Google Scholar]
- 10.Eck M J, Dhe-Paganon S, Trub T, Nolte R T, Shoelson S E. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell. 1996;85:695–705. doi: 10.1016/s0092-8674(00)81236-2. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson K M, Lemmon M A, Schlessinger J, Sigler P B. Structure of the high affinity complex of inositol triphosphate with a phospholipase C pleckstrin homology domain. Cell. 1995;83:1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- 12.Fiore F, Zambrano N, Minopoli G, Donini V, Duilio A, Russo T. The regions of the Fe65 protein homologous to the phosphotyrosine interaction/phosphotyrosine binding domain of Shc bind the intracellular domain of the Alzheimer’s amyloid precursor protein. J Biol Chem. 1995;270:30853–30856. doi: 10.1074/jbc.270.52.30853. [DOI] [PubMed] [Google Scholar]
- 13.Frank R. An easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron. 1992;48:9217–9232. [Google Scholar]
- 14.Frank R, Overwin H. SPOT synthesis. Epitope analysis with arrays of synthetic peptides prepared on cellulose membranes. Methods Mol Biol. 1996;66:149–169. doi: 10.1385/0-89603-375-9:149. [DOI] [PubMed] [Google Scholar]
- 15.Gertler F B, Bennett R L, Clark M J, Hoffmann F M. Drosophila abl tyrosine kinase in embryonic CNS axons: a role in axonogenesis is revealed through dosage-sensitive interactions with disabled. Cell. 1989;58:103–113. doi: 10.1016/0092-8674(89)90407-8. [DOI] [PubMed] [Google Scholar]
- 16.Gertler F B, Hill K K, Clark M J, Hoffmann F M. Dosage-sensitive modifiers of Drosophila abl tyrosine kinase function: prospero, a regulator of axonal outgrowth, and disabled, a novel tyrosine kinase substrate. Genes Dev. 1993;7:441–453. doi: 10.1101/gad.7.3.441. [DOI] [PubMed] [Google Scholar]
- 17.Goslin K, Banker G. Rat hippocampal neurons in low density culture. In: Banker G, Goslin K, editors. Culturing nerve cells. Cambridge, Mass: MIT Press; 1991. pp. 251–282. [Google Scholar]
- 18.Gustafson T A, He W, Craparo A, Schaub C D, O’Neill T J. Phosphotyrosine-dependent interaction of SHC and insulin receptor substrate 1 with the NPEY motif of the insulin receptor via a novel non-SH2 domain. Mol Cell Biol. 1995;15:2500–2508. doi: 10.1128/mcb.15.5.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy J. Amyloid, the presenilins and Alzheimer’s disease. Trends Neurosci. 1997;20:558–559. doi: 10.1016/s0166-2236(96)01030-2. [DOI] [PubMed] [Google Scholar]
- 20.Harlan J E, Hajduk P J, Yoon H S, Fesik S W. Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature. 1994;371:168–170. doi: 10.1038/371168a0. [DOI] [PubMed] [Google Scholar]
- 21.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howell B W, Gertler F B, Cooper J A. Mouse disabled (mDab1): a Src binding protein implicated in neuronal development. EMBO J. 1997;16:1165–1175. doi: 10.1093/emboj/16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howell B W, Hawkes R, Soriano P, Cooper J A. Neuronal position in the developing brain is regulated by mouse disabled-1. Nature. 1997;389:733–737. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- 24.Howell B W, Herrick T M, Cooper J A. Reelin-induced tyrosine phosphorylation of Disabled 1 during neuronal positioning. Genes Dev. 1999;13:643–648. doi: 10.1101/gad.13.6.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kavanaugh W M, Turck C W, Williams L T. PTB domain binding to signaling proteins through a sequence motif containing phosphotyrosine. Science. 1995;268:1177–1179. doi: 10.1126/science.7539155. [DOI] [PubMed] [Google Scholar]
- 26.Kavanaugh W M, Williams L T. An alternative to SH2 domains for binding tyrosine-phosphorylated proteins. Science. 1994;266:1862–1865. doi: 10.1126/science.7527937. [DOI] [PubMed] [Google Scholar]
- 27.Keegan K, Cooper J A. Use of the two-hybrid system to detect the association of the protein-tyrosine phosphatase, SHPTP2, with another SH2-containing protein, Grb7. Oncogene. 1996;12:1537–1544. [PubMed] [Google Scholar]
- 28.Kibbey R G, Rizo J, Gierasch L M, Anderson R G. The LDL receptor clustering motif interacts with the clathrin terminal domain in a reverse turn conformation. J Cell Biol. 1998;142:59–67. doi: 10.1083/jcb.142.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koo E H, Squazzo S L. Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J Biol Chem. 1994;269:17386–17389. [PubMed] [Google Scholar]
- 30.Lai A, Sisodia S S, Trowbridge I S. Characterization of sorting signals in the beta-amyloid precursor protein cytoplasmic domain. J Biol Chem. 1995;270:3565–3573. [PubMed] [Google Scholar]
- 31.Le N, Simon M A. Disabled is a putative adaptor protein that functions during signaling by the sevenless receptor tyrosine kinase. Mol Cell Biol. 1998;18:4844–4854. doi: 10.1128/mcb.18.8.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemmon M A, Ferguson K M, O’Brien R, Sigler P B, Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci USA. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S C, Songyang Z, Vincent S J, Zwahlen C, Wiley S, Cantley L, Kay L E, Forman K J, Pawson T. High-affinity binding of the Drosophila Numb phosphotyrosine-binding domain to peptides containing a Gly-Pro-(p)Tyr motif. Proc Natl Acad Sci USA. 1997;94:7204–7209. doi: 10.1073/pnas.94.14.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lioubin M N, Algate P A, Tsai S, Carlberg K, Aebersold R, Rohrschneider L R. p150Ship, a signal transduction molecule with inositol polyphosphate-5-phosphatase activity. Genes Dev. 1996;10:1084–1095. doi: 10.1101/gad.10.9.1084. [DOI] [PubMed] [Google Scholar]
- 35.Lorent K, Overbergh L, Moechars D, De Strooper B, Van Leuven F, Van Den Berghe H. Expression in mouse embryos and in adult mouse brain of three members of the amyloid precursor protein family, of the alpha-2-macroglobulin receptor/low density lipoprotein receptor-related protein and of its ligands apolipoprotein E, lipoprotein lipase, alpha-2-macroglobulin and the 40,000 molecular weight receptor-associated protein. Neuroscience. 1995;65:1009–1025. doi: 10.1016/0306-4522(94)00555-j. [DOI] [PubMed] [Google Scholar]
- 36.McLoughlin D M, Miller C C. The intracellular cytoplasmic domain of the Alzheimer’s disease amyloid precursor protein interacts with phosphotyrosine-binding domain proteins in the yeast two-hybrid system. FEBS Lett. 1996;397:197–200. doi: 10.1016/s0014-5793(96)01128-3. [DOI] [PubMed] [Google Scholar]
- 37.Mok S C, Wong K K, Chan R K, Lau C C, Tsao S W, Knapp R C, Berkowitz R S. Molecular cloning of differentially expressed genes in human epithelial ovarian cancer. Gynecol Oncol. 1994;52:247–252. doi: 10.1006/gyno.1994.1040. [DOI] [PubMed] [Google Scholar]
- 37a.National Center for Biotechnology Information. 13 April 1999, revision date. [On line.]. www.ncbi.nlm.nih.gov/BLAST. [11 May 1999, last date accessed].
- 38.O’Neill A, Craparo, Gustafson T A. Characterization of an interaction between insulin receptor substrate 1 and the insulin receptor by using the two-hybrid system. Mol Cell Biol. 1994;14:6433–6442. doi: 10.1128/mcb.14.10.6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 40.Pesesse X, Deleu S, De S F, Drayer L, Erneux C. Identification of a second SH2-domain-containing protein closely related to the phosphatidylinositol polyphosphate 5-phosphatase SHIP. Biochem Biophys Res Commun. 1997;239:697–700. doi: 10.1006/bbrc.1997.7538. [DOI] [PubMed] [Google Scholar]
- 41.Rameh L E, Arvidsson A K, Carraway K L, Couvillon A D, Rathbun G, Crompton A, VanRenterghem B, Czech M P, Ravichandran K S, Burakoff S J, Wang D S, Chen C S, Cantley L C. A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J Biol Chem. 1997;272:22059–22066. doi: 10.1074/jbc.272.35.22059. [DOI] [PubMed] [Google Scholar]
- 42.Ravichandran K S, Zhou M M, Pratt J C, Harlan J E, Walk S F, Fesik S W, Burakoff S J. Evidence for a requirement for both phospholipid and phosphotyrosine binding via the Shc phosphotyrosine-binding domain in vivo. Mol Cell Biol. 1997;17:5540–5549. doi: 10.1128/mcb.17.9.5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rebecchi M, Peterson A, McLaughlin S. Phosphoinositide-specific phospholipase C-delta 1 binds with high affinity to phospholipid vesicles containing phosphatidylinositol 4,5-bisphosphate. Biochemistry. 1992;31:12742–12747. doi: 10.1021/bi00166a005. [DOI] [PubMed] [Google Scholar]
- 44.Ron D, Dressler H. pGSTag-a versatile bacterial expression plasmid for enzymatic labeling of recombinant proteins. BioTechniques. 1992;13:866–869. [PubMed] [Google Scholar]
- 45.Rudnicki M A, McBurney M W. Cell culture methods and induction of differentiation of embryonal carcinoma cell lines. In: Robertson E, editor. Teratocarcinomas and embryonic stem cells: a practical approach. Oxford, England: IRL Press Ltd.; 1987. pp. 19–47. [Google Scholar]
- 46.Sabo S L, Lanier L M, Ikin A F, Khorkova O, Sahasrabudhe S, Greengard P, Buxbaum J D. Regulation of beta-amyloid secretion by FE65, an amyloid protein precursor-binding protein. J Biol Chem. 1999;274:7952–7957. doi: 10.1074/jbc.274.12.7952. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Sheldon M, Rice D S, D’Arcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell B W, Cooper J A, Goldowitz D, Curran T. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- 49.Simons M, Ikonen E, Tienari P J, Cid-Arregui A, Monning U, Beyreuther K, Dotti C G. Intracellular routing of human amyloid protein precursor: axonal delivery followed by transport to the dendrites. J Neurosci Res. 1995;41:121–128. doi: 10.1002/jnr.490410114. [DOI] [PubMed] [Google Scholar]
- 50.Tienari P J, De S B, Ikonen E, Simons M, Weidemann A, Czech C, Hartmann T, Ida N, Multhaup G, Masters C L, Van Leuven F, Beyreuther K, Dotti C G. The beta-amyloid domain is essential for axonal sorting of amyloid precursor protein. EMBO J. 1996;15:5218–5229. [PMC free article] [PubMed] [Google Scholar]
- 50a.Trommsdorff M, Borg J P, Margolis B, Herz J. Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein. J Biol Chem. 1998;273:33556–33560. doi: 10.1074/jbc.273.50.33556. [DOI] [PubMed] [Google Scholar]
- 51.van der Geer P, Pawson T. The PTB domain: a new protein module implicated in signal transduction. Trends Biochem Sci. 1995;20:277–280. doi: 10.1016/s0968-0004(00)89043-x. [DOI] [PubMed] [Google Scholar]
- 52.Vojtek A B, Hollenberg S M. Ras-Raf interaction: two-hybrid analysis. Methods Enzymol. 1995;255:331–342. doi: 10.1016/s0076-6879(95)55036-4. [DOI] [PubMed] [Google Scholar]
- 53.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 54.Wang D S, Chen C S. Synthesis of the D-3 series of phosphatidylinositol phosphates. J Org Chem. 1996;61:5905–5910. [Google Scholar]
- 55.Ware M L, Fox J W, Gonzalez J L, Davis N M, Lambert C, Russo C J, Chua Jr S C, Goffinet A M, Walsh C A. Aberrant splicing of a mouse disabled homolog, mdab1, in the scrambler mouse. Neuron. 1997;19:239–249. doi: 10.1016/s0896-6273(00)80936-8. [DOI] [PubMed] [Google Scholar]
- 56.Xu X X, Yang W N, Jackowski S, Rock C O. Cloning of a novel phosphoprotein regulated by colony-stimulating factor 1 shares a domain with the Drosophila disabled gene product. J Biol Chem. 1995;270:14184–14191. doi: 10.1074/jbc.270.23.14184. [DOI] [PubMed] [Google Scholar]
- 57.Yajnik V, Blaikie P, Bork P, Margolis B. Identification of residues within the SHC phosphotyrosine binding/phosphotyrosine interaction domain crucial for phosphopeptide interaction. J Biol Chem. 1996;271:1813–1816. doi: 10.1074/jbc.271.4.1813. [DOI] [PubMed] [Google Scholar]
- 58.Yamazaki T, Koo E H, Selkoe D J. Trafficking of cell-surface amyloid beta-protein precursor. II. Endocytosis, recycling and lysosomal targeting detected by immunolocalization. J Cell Sci. 1996;109:999–1008. doi: 10.1242/jcs.109.5.999. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Z, Lee C H, Mandiyan V, Borg J P, Margolis B, Schlessinger J, Kuriyan J. Sequence-specific recognition of the internalization motif of the Alzheimer’s amyloid precursor protein by the X11 PTB domain. EMBO J. 1997;16:6141–6150. doi: 10.1093/emboj/16.20.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou M M, Ravichandran K S, Olejniczak E T, Petros A M, Meadows R P, Sattler M, Harlan J E, Wade W S, Burakoff S J, Fesik S W. Structure and ligand recognition of the phosphotyrosine binding domain of Shc. Nature. 1995;378:584–592. doi: 10.1038/378584a0. [DOI] [PubMed] [Google Scholar]