Abstract
Background: MiRNAs are pivotal regulators involved in proliferation, apoptosis, invasion, metastasis, epithelial-mesenchymal transition (EMT), angiogenesis, drug resistance and autophagy in hepatocellular carcinoma (HCC). The aim of this study was to investigate the influence of miR-139-5p and its target genes on the outcomes of HCC.
Methods: Survival analysis of miR-139-5p in HCC was conducted in Kaplan-Meier plotter. Target genes of miR-139-5p were identified in TargetScan, miRTarBase and starBase. Gene Expression Omnibus (GEO) series were used for the validation of miR-139-5p target genes. Cox proportional regression model was also established.
Results: In Kaplan-Meier plotter, 163 HCC patients were included. MiR-139-5p downregulation was significantly associated with unfavorable overall survival (OS) and disease-free survival (DFS) in HCC patients (all P < 0.001). MiR-139-5p was significantly downregulated in HCC tumors and human hepatoma cell lines (all P < 0.05). As a target gene of miR-139-5p, CCT5 was overexpressed in HCC tumor tissues and peripheral blood mononuclear cells (all P < 0.05). A negative correlation between CCT5 and miR-139-5p was found in TCGA dataset. CCT5 overexpression was significantly associated with worse OS in HCC patients (P < 0.001), which was validated in the GSE14520 dataset (P = 0.017). CCT5 mRNA was significantly overexpressed in HCC patients with alpha-fetoprotein (AFP) > 300 ng/ml, BCLC staging B-C, TNM staging III and main tumor size > 5 cm (all P < 0.05). According to the Cox regression model of CCT5-interacting genes, HCC patients with high risk had poor OS compared to those with low risk in the TCGA dataset (P < 0.001), with the 1-year, 3-year, and 5-year ROC curves of an area under the curve (AUC) equal to 0.704, 0.662, and 0.631, respectively.
Conclusions: MiR-139-5p suppresses HCC tumor aggression and conversely correlated with CCT5. The miR-139-5p/CCT5 axis might perform crucial functions in the development of HCC.
Keywords: miR-139-5p, Chaperonin containing TCP1 subunit 5, CCT5, hepatocellular carcinoma, survival
Introduction
As a class of noncoding, highly conserved endogenous RNAs of approximately 18~25 nucleotides in length, microRNAs (miRNAs) can bind to specific mRNA sites and regulate mRNA expression for cleavage or translational repression 1, 2. Substantial evidence has revealed that miRNAs participate in various vital biological processes 3. Aberrant miRNA expression is implicated in numerous disorders, especially cancer. The important roles of miRNAs in malignant biological behavior display their potential as promising diagnostic and prognostic biomarkers 4.
As reviewed by Xu et al, miRNAs are pivotal regulators involved in proliferation, apoptosis, invasion, metastasis, epithelial-mesenchymal transition (EMT), angiogenesis, drug resistance and autophagy in hepatocellular carcinoma (HCC). Unfortunately, this group did not discuss the roles of miR-139 in the development of HCC 5. As a recognized tumor suppressor, miR-139-5p has been widely investigated in many malignancies, including breast cancer6, gastric cancer 7, colon cancer 8, bladder cancer 9, 10, lung cancer 11, prostate cancer 12, glioma 13, ovarian cancer 14, endometrial cancer 15, leukemia 16, 17, osteosarcoma 18, 19, esophageal cancer 20, 21 and oral squamous carcinoma 22. Conversely, a report by Pang C et al showed that miR-139-5p expression was significantly upregulated in prostate cancer patients compared with benign prostatic hyperplasia patients and healthy individuals 23. A bioinformatic analysis illustrated that miR-139-5p promotes the aggressiveness of adrenocortical cancer 24.
The interference of miR-139-5p could reverse the inhibitory effects of sevoflurane treatment on breast cancer cell migration, invasion, and EMT 25, and reverse the EMT of colon cancer stem cells 26. The miR-139-5p impeded the EMT in pancreatic cells27, cervical cancer cells 28, glioma 13, osteosarcoma 19, colorectal cancer cells 29, and in post-menopausal women with interstitial cystitis 30. In HCC, miR-139-5p functions to inhibit EMT and metastasis by targeting zinc finger E‑box binding to homeobox 1 (ZEB1) and ZEB2, and its expression is downregulated in HCC cells and tumor tissues 31, 32. MiR-139-5p inhibited HCC cell viability, migration and invasion and induced apoptosis by targeting kazal‐like domains proteoglycan 1 (SPOCK1), E26 transformation-specific 1 (ETS1) and NTRK like family member 4 (SLITRK4) 32-34. Given the controversial role of miR-139-5p in human malignancies and the importance of identifying target genes of miR-139-5p in HCC, we conducted an integrated bioinformatic analysis to investigate the roles and target genes of miR-139-5p in HCC patients.
Materials and methods
Data source
Survival analysis of miR-139-5p and its potential target genes was conducted in the Kaplan Meier plotter platform 35, 36. This database is processed by a PostgreSQL server, which integrates both gene expression and clinical data. To analyze the prognostic value of a particular gene, patient samples were divided into two groups by the proposed median cutoff of the biomarkers. Two patient groups were compared by Kaplan-Meier survival plot and the hazard ratio (HR) with 95% confidence intervals (CI) and log rank P value were calculated. When the miRNA ID hsa-miR-139-5p and non-commercial spotted platform were selected, 166 HCC patients were included in the analysis. Outcomes including overall survival (OS) and disease-free survival (DFS) were considered. No sex, tumor stage, cirrhosis status, metastasis, HBV infection, alcohol use or race restrictions.
Total RNA from 14 pairs of HCC tumors and adjacent tissues was isolated and purified in the GSE84402 dataset 22 for expression analysis of target genes of miR-139-5p. The GSE14520 dataset 37, 38 was used to validate relationships between miR-139-5p target genes and the survival and clinico-pathological characteristics of HCC patients. Data extraction methods used for the GSE14520 dataset have been described previously 39, 40.
Cell culture
The normal liver cells and hepatoma cell lines (HLF, HCCLM3, Hep3B, Huh6, and Huh7) were obtained from the American Type Culture Collection (ATCC, MD, USA). Cells were cultured in Dulbecco's modified Eagles' medium (DMEM/high glucose) supplemented with 10% fetal bovine serum (FBS) (Cellsera, NSW, Australia) containing 50 µg/ml penicillin and 50 µg/ml streptomycin at 37 °C in a 5% CO2 incubator. The medium was changed three times every week 41.
Quantitative Real time-PCR (qRT-PCR)
TaKaRa MiniBEST Universal RNA Extraction Kit (TaKaRa, Japan) was applied to extract the total RNA from samples according to the manufacture's recommendation. The reverse transcription of 500 ng total RNA was utilizing PrimeScript™ RT reagent Kit (TaKaRa, Japan) and quantitative real-time PCR with TB Green® Premix Ex Taq™ II (TaKaRa, Japan). The PCR was set at the initial denaturation of 2 min at 95 °C, following with 5 s at 95 °C, and 30 s at 60 °C in a total of 40 cycles, and was set at melt curve stage of 15 s at 95 °C, 1 min at 60 °C and 15s at 95 °C. All experiments were carried out in triplicate. The miR-139-5p expression was normalized to U6. The relative expression ratios of miR-139-5p were computed by the 2-ΔΔCT method. The primers involved in this assay were shown as follows: hsa-miR-139-5p, F: ACACTCCAGCTGGGTCTACAGTGCACGTG, R: GTGCAGGGTCCGAGGT; U6, F: AGAGAAGATTAGCATGGCCCCTG, R: ATCCAGTGCAGGGTCCGAGG.
MiR-139-5p target gene prediction
The TargetScan v7.2 42, miRTarBase 43, 44 and starBase v3.0 45, 46 databases were used for miR-139-5p target gene prediction. In both the TargetScan v7.2 and miRTarBase databases, human species were selected, and miR-139-5p was entered into the microRNA name dialog box. In the starBase database, the miRNA-target intersections function was used, and the following criteria were selected: human genome, hg19 assembly, miR-139-5p, CLIP-data ≥ 1, degradome-data ≥ 0, and program number ≥ 1. Common target genes of miR-139-5p in these three databases were calculated and drawn by the Venn diagram webtool (http://bioinformatics.psb.ugent.be/webtools/Venn/). The expression levels between tumor and nontumor tissues, including gene expression in peripheral blood mononuclear cells (PBMCs) between healthy individuals and HCC patients, were validated in the Gene Expression Omnibus (GEO) series with Raw.CEL files. Gene expression was calculated with the Affy 47 and Limma 48 packages in the R program. We have previously reported the detailed microarray processing approaches of gene expression in these GEO series 39.
Target gene expression and protein-protein-interaction analysis
The intersection of target genes of miR-139-5p and differentially expressed genes (DEGs) in the GSE84402 dataset from the GEO database was identified with the Venn diagram webtool (http://bioinformatics.psb.ugent.be/webtools/Venn/). Protein-protein-interaction analysis of miR-139-5p target genes of Homo sapiens organism was performed with the STRING v11.0 database (https://string-db.org/). Full network interacted proteins with interaction score more than 0.4 were selected.
Cox proportional hazard regression model establish
A Cox proportional hazard regression model was established based on The Cancer Genome Atlas (TCGA) database using the R program. The edgeR package 49 was used to identify DEGs in HCC tumor and adjacent tissues, the survival package 50 was used to conduct univariate and multivariate Cox regression analyses, and then the Cox regression formula was calculated. According to this formula, HCC patients were divided into two groups, the high- and low-risk groups 51. Survival analysis between these two groups was also performed by the survival package 50. The timeROC package was used to generate a receiver operating characteristic (ROC) curve of the model for the prediction of OS in HCC patients.
Statistical analysis
GraphPad Prism v7.0 (GraphPad Software, CA, US) was used. Student's t test or the Mann-Whitney U test were performed to analyze the differences in gene expression the two groups. Kaplan-Meier survival analysis was performed on the GSE14520 dataset. A two-tailed P < 0.05 was considered significant.
Results
Association between miR-139-5p and HCC survival
Using the miRpower liver cancer miRNA database in Kaplan-Meier plotter, 166 HCC patients were included in the non-commercial spotted platform. As shown in Figure 1, downregulation of miR-139-5p was significantly associated with poor OS in HCC patients (P = 7E-05, Figure 1A). Similarly, a low level of miR-139-5p in tumors significantly correlated with the 1-year, 3-year, and 5-year OS in HCC patients (all P < 0.001, Figure 1B-D).
Figure 1.
Low expression of miR-139-5p is associated with worse OS (A), 1-year (B), 3-year (C), and 5-year (D) OS in HCC patients.
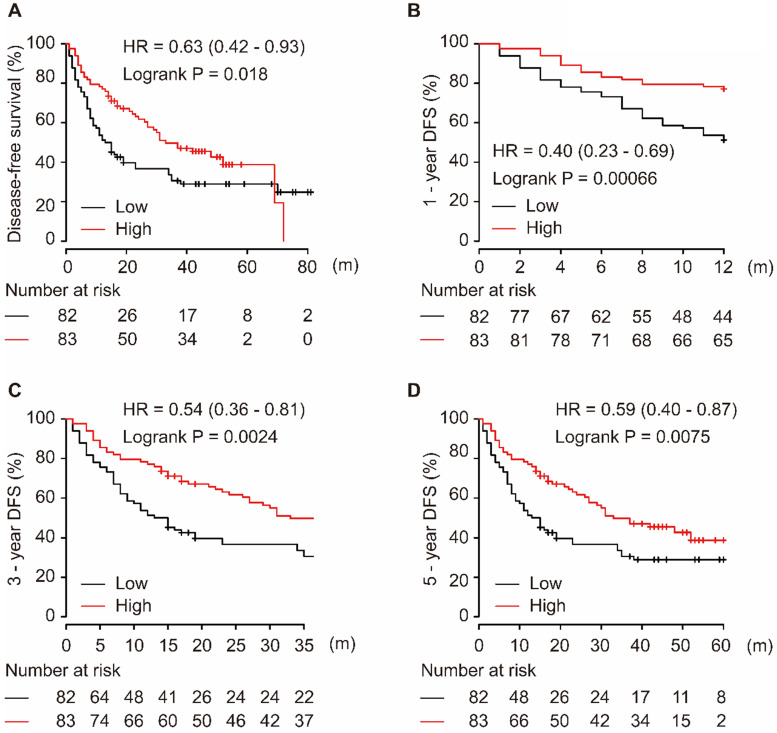
As shown in Figure 2, the downregulation of miR-139-5p in tumor tissues was significantly associated with worse DFS in patients with HCC (P = 0.018, Figure 2A). Additionally, low expression of miR-139-5p significantly contributed to worse 1-year, 3-year, and 5-year DFS in HCC patients (all P < 0.01, Figure 2B-D).
Figure 2.
Low expression of miR-139-5p is associated with worse DFS (A), 1-year (B), 3-year (C), and 5-year (D) DFS in HCC patients.
Target gene expression analysis
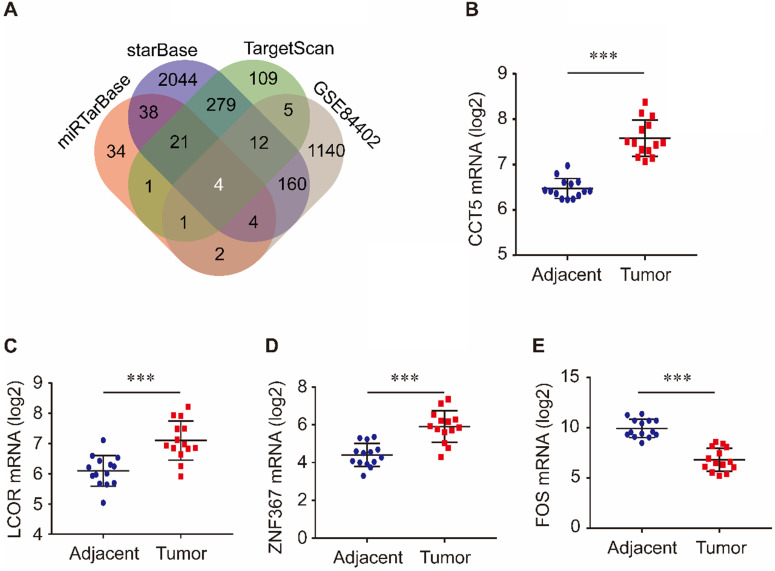
Target genes of miR-139-5p were identified in the TargetScan, miRTarBase, and starBase databases. As shown in Figure 3A, 432 target genes were identified in TargetScan, 105 were identified in miRTarBase and 2562 were identified in starBase. Furthermore, we investigated the common genes between these target genes and the DEGs in the GSE84402 dataset. Four genes, CCT5, FOS, LCOR, and ZNF367, were identified. As shown in Figure 3, CCT5, LCOR, and ZNF367 were evidently overexpressed in tumor tissues compared with adjacent tissues (all P < 0.001, Figure 3B-D), while FOS was significantly downregulated in tumor tissues compared with non-tumor tissues of HCC patients (P < 0.001, Figure 3E).
Figure 3.
Target genes of miR-139-5p and its common genes with differential expression genes in GSE84402 (A); CCT5 (B), LCOR (C), ZNF367 (D) and FOS mRNA (E) expression in tumor and adjacent tissues in GSE84402.
Association between CCT5 and overall survival in HCC
Among the four target genes (CCT5, FOS, LCOR, and ZNF367), only CCT5 showed significance for OS in HCC patients. As shown in Figure 4, CCT5 overexpression was significantly associated with worse OS in HCC patients (HR = 2.24, 95% CI = 1.57-3.20, P = 5.1E-06, Figure 4A). Moreover, a high level of CCT5 in tumor tissues was significantly associated with 1-year, 3-year, and 5-year OS in HCC patients (all HR > 2.0, P < 0.001, Figure 4B-D).
Figure 4.
CCT5 mRNA overexpression is associated with worse OS (A), 1-year (B), 3-year (C), and 5-year (D) OS in HCC patients.
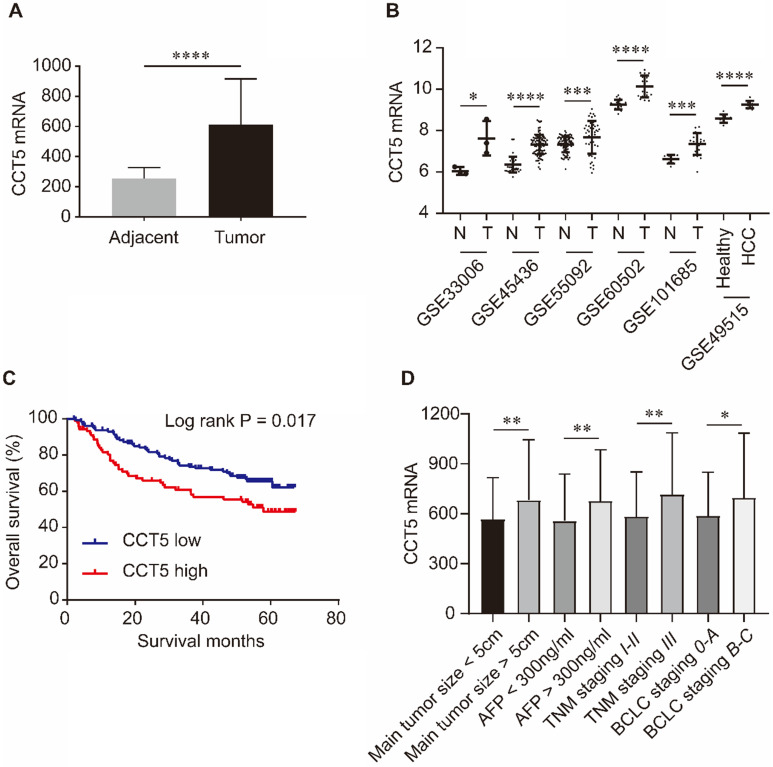
As shown in Figure 5, CCT5 mRNA was significantly upregulated in tumor tissues compared with adjacent tissues in the GSE14520, GSE33006, GSE45436, GSE55092, GSE60502 and GSE101685 datasets (all P < 0.05, Figure 5A,B). In addition, CCT5 mRNA was also significantly increased in the PBMCs of HCC patients compared to those of healthy individuals in the GSE49515 dataset (P < 0.0001, Figure 5B). High expression of CCT5 mRNA in tumors was a risk factor for OS in HCC patients (log-rank P = 0.017, Figure 5B). Additionally, CCT5 mRNA was significantly overexpressed in HCC patients with alpha-fetoprotein (AFP) > 300 ng/ml, BCLC staging B-C, TNM staging III and main tumor size > 5 cm (Figure 5C-F, respectively; all P < 0.05).
Figure 5.
CCT5 mRNA expression and its associations with OS and clinico-pathological features in HCC patients. CCT5 mRNA was upregulated in tumor tissues than that in adjacent tissues in GSE14520, GSE33006, GSE45436, GSE55092, GSE60502 and GSE101685 (A, B); CCT5 mRNA was significantly increased in PBMCs of HCC patients than that in healthy individuals in GSE49515 (B); high CCT5 mRNA was associated with worse OS (C), CCT5 mRNA was upregulated in HCC patients with main tumor size > 5cm, AFP > 300 ng/ml BCLC stage B-C and/or TNM stage III (D).
Correlation between miR-139-5p and CCT5
Human miR-139-5p expression was significantly downregulated in hepatoma cell lines including HLF, HCCLM3, Hep3B, Huh6, and Huh7 compared to that in normal liver cells (all P < 0.05, Figure 6A). We investigated the correlation between miR-139 and CCT5 in the TCGA dataset. As summarized in Figure 6, miR-139 expression was significantly downregulated in HCC tumors (P < 0.0001, Figure 6B), and CCT5 mRNA was significantly overexpressed in HCC tumors (P < 0.0001, Figure 6B). In addition, CCT5 mRNA was significantly downregulated in miR-139 high group (P < 0.0001, Figure 6C). As expected, a negative correlation was found between miR-139 and CCT5 mRNA (P < 0.001, Figure 6D).
Figure 6.
miR-139 was significantly downregulated in hepatoma cell lines compared to that in normal liver cells(A); miR-139 was also significantly downregulated in HCC tumors and CCT5 mRNA was significantly overexpressed in HCC tumors (B); CCT5 mRNA was significantly downregulated in miR-139 high samples (C); Negatively correlation was found between miR-139 and CCT5 mRNA in TCGA dataset (D).
Cox proportional hazard regression model
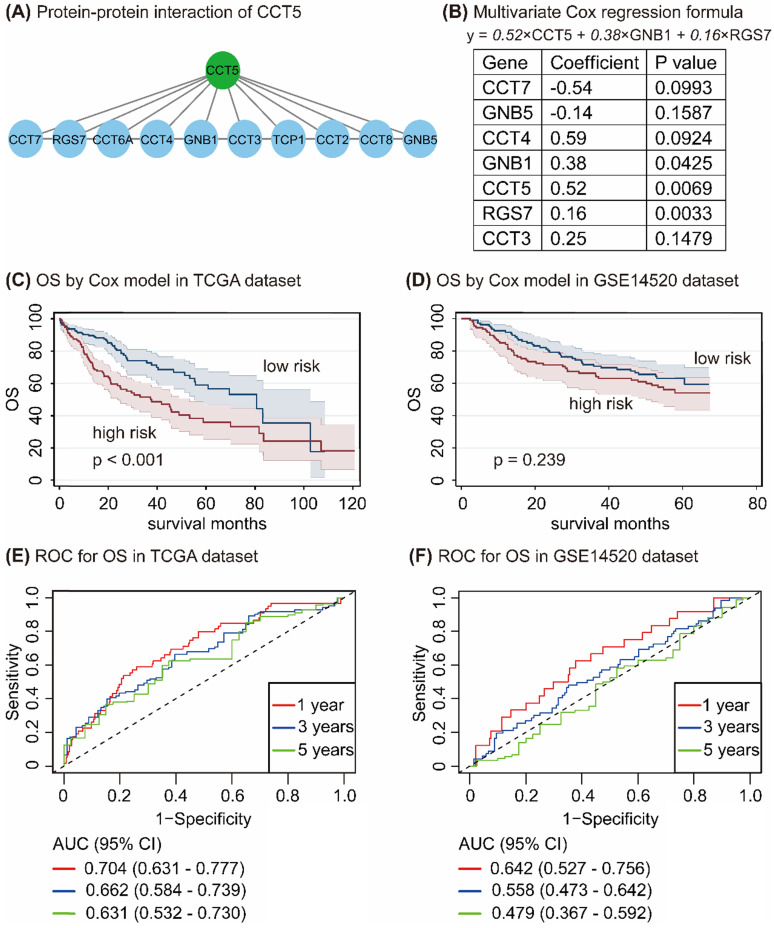
In the STRING database, 10 genes, namely, CCT7, RGS7, CCT6A, CCT4, GNB1, CCT3, TCP1, CCT2, CCT8, and GNB5, have interacted with CCT5 (Figure 7A). Univariate and multivariate Cox regression analyses in R program identified 7 genes, namely, CCT7, GNB5, CCT4, GNB1, CCT5, RGS7, and CCT3. CCT5, GNB1, and RGS7 were significantly associated with OS in HCC patients (all P < 0.05, Figure 7B), with the following formula: y = 0.52 × CCT5 + 0.38×GNB1 + 0.16 × RGS7 (Figure 7B). According to values obtained with the formula, HCC patients were divided into high-risk and low-risk groups in the TCGA and the GSE14520 datasets. As shown in Figure 7, HCC patients with high risk had poor OS compared with those of low-risk patients in the TCGA dataset (P < 0.001, Figure 7C), while no difference between this formula model and OS in HCC patients was found in the GSE14520 dataset (P = 0.239, Figure 7D). To determine the accuracy of this model for the prediction of the OS in HCC patients, the ROC curves with an area under the curve (AUC) in the TCGA and GSE14520 datasets were summarized in Figure 7E and Figure 7F, respectively.
Figure 7.
Protein-protein-interaction of CCT5 (A); Cox proportional hazard regression model of CCT5 interactive genes (B); Survival analysis based on Cox model grouped by risk scores in the TCGA dataset (C) and the GSE14520 dataset (D); and ROC curves of Cox model for OS in HCC patients in the TCGA dataset (E) and the GSE14520 dataset (F).
Discussion
Consistent with previous reports 32-34, our results revealed that the downregulation of miR-139-5p showed a significant association with worse survival in HCC patients. According to existing publications, miR-139-5p inhibits HCC progression by targeting several genes, such as ZEB1 and ZEB2 31, EZH2 52, SPOCK1 32, ETS1 33, XIST 53 and SLITRK4 34. MiR-139-5p was confirmed to negatively regulate ZEB1 and ZEB2 expression 31, which induces EMT and promotes the progression of malignant carcinoma 54, while overexpression of ZEB1 and ZEB2 abolishes the inhibitory effects of miR-139-5p on HCC cell migration and invasion 31. A previous report revealed that the zeste homolog 2 (EZH2) enhancer could promote HCC motility in vitro and pulmonary metastasis in a nude mouse model through epigenetically suppressing miR-139-5p by potent and extensive regulation of various signaling pathways involved in cell motility and metastasis 52. As an oncogene, SPOCK1 blocks apoptosis and promotes HCC metastasis 55. Overexpression of miR-139-5p suppressed HCC cell viability and invasion by targeting SPOCK1, promoted apoptosis, and inhibited tumor growth 32. A study by Hua et al. showed that miR-139-5p-induced aerobic glycolysis, proliferation, migration, and invasion were reversed by ETS1 overexpression, whereas ETS1 silencing induced the expression of miR-139-5p through posttranscriptional regulation 33. LncRNA X inactivate-specific transcript (XIST) induces the proliferation of HCC cells by promoting the cell cycle and protecting cells from apoptosis, and the mutual inhibitory action of XIST and miR-139-5p has also been investigated 53. Recently, SLITRK4 has been shown to be involved in the progression of HCC and to be targeted by miR-139-5p 34.
In addition to the target genes mentioned above, our analysis revealed that chaperonin-containing TCP1 subunit 5 (CCT5) might be a novel target gene of miR-139-5p linked to HCC survival and clinico-pathological features. CCT5 is a member of the chaperonin-containing TCP1 complex (CCT), also known as the TCP1 ring complex (TRiC). Its protein-folding activity is critical for cellular health. Misfolded proteins are associated with many human diseases, including cancer, making them potential therapeutic targets 56, 57. Highly expressed in cancer cell lines, CCT/TRiC is implicated in the folding of oncoproteins cyclin E, cyclin B, and p21(ras), which strongly indicates that it is involved in cell proliferation and tumorigenesis 58. CCT5 has mainly been investigated in neuropathology 59, 60. In HCC, CCT5 expression was upregulated in liver tumors, predicted shortened OS and DFS times. Gene enrichment analysis revealed that CCT5 was involved in the dysregulation of Myc target genes, hypoxia-inducible factor (HIF) target genes and the cell cycle, especially the G1/S transition, cell cycle arrest and apoptosis 61, 62. Additionally, CCT5 mutations might be associated with the accumulation of cytotoxic protein aggregates with tissue destruction and mitochondrial damage 63. In breast cancer, overexpression of CCT5 correlated with tumor purity and immune infiltration levels and was associated with poor survival rates 64. CCT5 mRNA was significantly upregulated in p53-mutated tumors and associated with a low response rate to docetaxel in breast cancer. Treatment of MCF-7 cells with CCT5-specific siRNA resulted in a significant enhancement of apoptosis induced by docetaxel 65. In non-small cell lung cancer (NSCLC), CCT5 was expressed at higher levels in tumor tissues and induced an autoantibody response in NSCLC sera 66. In addition, CCT5 was also overexpressed in sinonasal adenocarcinoma 67, esophageal squamous cell carcinoma 68 and multidrug-resistant gastric carcinoma cells 69. We suggest mechanistic insights into the role of CCT5 in tumorigenesis and tumor progression in the HCC population that should be considered in the future.
There are some limitations to our study. First, even though the suppressive impact of miR-139-5p on CCT5 was addressed in this study, our analysis did not involve experimental assays to verify the reciprocal repression between miR-139-5p and CCT5. Second, no experiments were performed to address the effects of the miR-139-5p/CCT5 axis on HCC cellular functions; Third, follow-up data of HCC patients from our own dataset were not available; And, the extension of Cox model in our research was relatively unfavorable in the GSE14520, further investigations are needed. However, according to our integrated analysis, we cautiously assumed that the miR-139-5p/CCT5 axis might be a novel mechanism in the progression of HCC. Since the hypothesis was based on a bioinformatic research, further investigations regarding the associations between miR-139-5p and CCT5 should be considered.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81803901, 81673788, and 81873136). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Data Availability
The data used to support the findings of this study are available from the corresponding author (Z. Yang) upon request.
Ethical Statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 3.Shah AA, Meese E, Blin N. Profiling of regulatory microRNA transcriptomes in various biological processes: a review. J Appl Genet. 2010;51:501–7. doi: 10.1007/BF03208880. [DOI] [PubMed] [Google Scholar]
- 4.Zaheer U, Faheem M, Qadri I. et al. Expression profile of MicroRNA: An Emerging Hallmark of Cancer. Curr Pharm Des. 2019;25:642–53. doi: 10.2174/1386207322666190325122821. [DOI] [PubMed] [Google Scholar]
- 5.Xu X, Tao Y, Shan L. et al. The Role of MicroRNAs in Hepatocellular Carcinoma. J Cancer. 2018;9:3557–69. doi: 10.7150/jca.26350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pajic M, Froio D, Daly S. et al. miR-139-5p Modulates Radiotherapy Resistance in Breast Cancer by Repressing Multiple Gene Networks of DNA Repair and ROS Defense. Cancer Res. 2018;78:501–15. doi: 10.1158/0008-5472.CAN-16-3105. [DOI] [PubMed] [Google Scholar]
- 7.Sun K, Hu P, Xu F. LINC00152/miR-139-5p regulates gastric cancer cell aerobic glycolysis by targeting PRKAA1. Biomed Pharmacother. 2018;97:1296–302. doi: 10.1016/j.biopha.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Ma X, Liu J, Li J. et al. miR-139-5p reverses stemness maintenance and metastasis of colon cancer stem-like cells by targeting E2-2. J Cell Physiol. 2019;234:22703–18. doi: 10.1002/jcp.28836. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Yu Y, Chen X. et al. MiR-139-5p is associated with poor prognosis and regulates glycolysis by repressing PKM2 in gallbladder carcinoma. Cell Prolif. 2018;51:e12510. doi: 10.1111/cpr.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y, Deng C, Zhang H. et al. Long non-coding RNA XIST promotes cell growth and metastasis through regulating miR-139-5p mediated Wnt/beta-catenin signaling pathway in bladder cancer. Oncotarget. 2017;8:94554–68. doi: 10.18632/oncotarget.21791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Yong-Hao Y, Xian-Guo W, Ming X. et al. Expression and clinical significance of miR-139-5p in non-small cell lung cancer. J Int Med Res. 2019;47:867–74. doi: 10.1177/0300060518815379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang B, Zhang W, Sun D. et al. Downregulation of miR-139-5p promotes prostate cancer progression through regulation of SOX5. Biomed Pharmacother. 2019;109:2128–35. doi: 10.1016/j.biopha.2018.09.029. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Li Q, Lin L. et al. Targeting the Notch1 oncogene by miR-139-5p inhibits glioma metastasis and epithelial-mesenchymal transition (EMT) BMC Neurol. 2018;18:133. doi: 10.1186/s12883-018-1139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Y, Jiang J, Jia H. et al. Recovery of miR-139-5p in Ovarian Cancer Reverses Cisplatin Resistance by Targeting C-Jun. Cell Physiol Biochem. 2018;51:129–41. doi: 10.1159/000495169. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Li C, Jiang Y. et al. Tumor-suppressor role of miR-139-5p in endometrial cancer. Cancer Cell Int. 2018;18:51. doi: 10.1186/s12935-018-0545-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R, Tang P, Wang F. et al. Tumor suppressor miR-139-5p targets Tspan3 and regulates the progression of acute myeloid leukemia through the PI3K/Akt pathway. J Cell Biochem. 2019;120:4423–32. doi: 10.1002/jcb.27728. [DOI] [PubMed] [Google Scholar]
- 17.Krowiorz K, Ruschmann J, Lai C. et al. MiR-139-5p is a potent tumor suppressor in adult acute myeloid leukemia. Blood Cancer J. 2016;6:e508. doi: 10.1038/bcj.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou L, Ma X, Yue J. et al. The diagnostic effect of serum miR-139-5p as an indicator in osteosarcoma. Cancer Biomark. 2018;23:561–7. doi: 10.3233/CBM-181744. [DOI] [PubMed] [Google Scholar]
- 19.Shi YK, Guo YH. MiR-139-5p suppresses osteosarcoma cell growth and invasion through regulating DNMT1. Biochem Biophys Res Commun. 2018;503:459–66. doi: 10.1016/j.bbrc.2018.04.124. [DOI] [PubMed] [Google Scholar]
- 20.Jiao W, Zhang J, Wei Y. et al. MiR-139-5p regulates VEGFR and downstream signaling pathways to inhibit the development of esophageal cancer. Dig Liver Dis. 2019;51:149–56. doi: 10.1016/j.dld.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 21.Liu R, Yang M, Meng Y. et al. Tumor-suppressive function of miR-139-5p in esophageal squamous cell carcinoma. PLoS One. 2013;8:e77068. doi: 10.1371/journal.pone.0077068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Huo X, Yang XR. et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pang C, Liu M, Fang W. et al. MiR-139-5p is Increased in the Peripheral Blood of Patients with Prostate Cancer. Cell Physiol Biochem. 2016;39:1111–7. doi: 10.1159/000447819. [DOI] [PubMed] [Google Scholar]
- 24.Agosta C, Laugier J, Guyon L. et al. MiR-483-5p and miR-139-5p promote aggressiveness by targeting N-myc downstream-regulated gene family members in adrenocortical cancer. Int J Cancer. 2018;143:944–57. doi: 10.1002/ijc.31363. [DOI] [PubMed] [Google Scholar]
- 25.Wu T, Sun L, Wang C. et al. Sevoflurane Suppresses the Migration, Invasion, and Epithelial-Mesenchymal Transition of Breast Cancer Cells Through the miR-139-5p/ARF6 Axis. J Surg Res. 2021;258:314–23. doi: 10.1016/j.jss.2020.08.051. [DOI] [PubMed] [Google Scholar]
- 26.Ma X, Liu J, Li J. et al. miR-139-5p reverses stemness maintenance and metastasis of colon cancer stem-like cells by targeting E2-2. J Cell Physiol. 2019;234:22703–18. doi: 10.1002/jcp.28836. [DOI] [PubMed] [Google Scholar]
- 27.Ma J, Zhang J, Weng YC. et al. EZH2-Mediated microRNA-139-5p Regulates Epithelial-Mesenchymal Transition and Lymph Node Metastasis of Pancreatic Cancer. Mol Cells. 2018;41:868–80. doi: 10.14348/molcells.2018.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J, Wang S, Liu P. et al. MiR-139-5p-ZEB1 is a Molecular Regulator of Growth, Invasion, and Epithelial-to-Mesenchymal Transition of Cervical Cancer. Cancer Manag Res. 2020;12:12723–33. doi: 10.2147/CMAR.S267634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, Liang X, Wang Y. et al. miR-139-5p Inhibits the Epithelial-Mesenchymal Transition and Enhances the Chemotherapeutic Sensitivity of Colorectal Cancer Cells by Downregulating BCL2. Sci Rep. 2016;6:27157. doi: 10.1038/srep27157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang C, Tong Z, Fang WL. et al. Microrna-139-5p inhibits epithelial-mesenchymal transition and fibrosis in post-menopausal women with interstitial cystitis by targeting LPAR4 via the PI3K/Akt signaling pathway. J Cell Biochem. 2018;119:6429–41. doi: 10.1002/jcb.26610. [DOI] [PubMed] [Google Scholar]
- 31.Qiu G, Lin Y, Zhang H. et al. miR-139-5p inhibits epithelial-mesenchymal transition, migration and invasion of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2. Biochem Biophys Res Commun. 2015;463:315–21. doi: 10.1016/j.bbrc.2015.05.062. [DOI] [PubMed] [Google Scholar]
- 32.Li P, Xiao Z, Luo J. et al. MiR-139-5p, miR-940 and miR-193a-5p inhibit the growth of hepatocellular carcinoma by targeting SPOCK1. J Cell Mol Med. 2019;23:2475–88. doi: 10.1111/jcmm.14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hua S, Lei L, Deng L. et al. miR-139-5p inhibits aerobic glycolysis, cell proliferation, migration, and invasion in hepatocellular carcinoma via a reciprocal regulatory interaction with ETS1. Oncogene. 2018;37:1624–36. doi: 10.1038/s41388-017-0057-3. [DOI] [PubMed] [Google Scholar]
- 34.Wu J, Zhang T, Chen Y. et al. MiR-139-5p influences hepatocellular carcinoma cell invasion and proliferation capacities via decreasing SLITRK4 expression. Biosci Rep. 2020;40:BSR20193295. doi: 10.1042/BSR20193295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy A, Lanczky A, Menyhart O. et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menyhart O, Nagy A, Gyorffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5:181006. doi: 10.1098/rsos.181006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roessler S, Jia HL, Budhu A. et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–12. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roessler S, Long EL, Budhu A. et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–66. doi: 10.1053/j.gastro.2011.12.039. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Y, Zhang J, Li L. et al. Carcinogenesis effects of E2F transcription factor 8 (E2F8) in hepatocellular carcinoma outcomes: an integrated bioinformatic report. Biosci Rep. 2020;40:BSR20193212. doi: 10.1042/BSR20193212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z, Zhuang L, Szatmary P. et al. Upregulation of heat shock proteins (HSPA12A, HSP90B1, HSPA4, HSPA5 and HSPA6) in tumour tissues is associated with poor outcomes from HBV-related early-stage hepatocellular carcinoma. Int J Med Sci. 2015;12:256–63. doi: 10.7150/ijms.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Z, Tao Y, Xu X. et al. Bufalin inhibits cell proliferation and migration of hepatocellular carcinoma cells via APOBEC3F induced intestinal immune network for IgA production signaling pathway. Biochem Biophys Res Commun. 2018;503:2124–31. doi: 10.1016/j.bbrc.2018.07.169. [DOI] [PubMed] [Google Scholar]
- 42.Agarwal V, Bell GW, Nam JW. et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou CH, Shrestha S, Yang CD. et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grimson A, Farh KK, Johnston WK. et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li JH, Liu S, Zhou H. et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang JH, Li JH, Shao P. et al. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–9. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gautier L, Cope L, Bolstad BM. et al. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 48.Ritchie ME, Phipson B, Wu D. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durisova M, Dedik L. SURVIVAL-an integrated software package for survival curve estimation and statistical comparison of survival rates of two groups of patients or experimental animals. Methods Find Exp Clin Pharmacol. 1993;15:535–40. [PubMed] [Google Scholar]
- 51.Yang S, Lin S, Liu K. et al. Identification of an immune-related RNA-binding protein signature to predict survival and targeted therapy responses in liver cancer. Genomics. 2021;113:795–804. doi: 10.1016/j.ygeno.2021.01.021. [DOI] [PubMed] [Google Scholar]
- 52.Au SL, Wong CC, Lee JM. et al. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology. 2012;56:622–31. doi: 10.1002/hep.25679. [DOI] [PubMed] [Google Scholar]
- 53.Mo Y, Lu Y, Wang P. et al. Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. Tumour Biol. 2017;39:1010428317690999. doi: 10.1177/1010428317690999. [DOI] [PubMed] [Google Scholar]
- 54.Gheldof A, Hulpiau P, van Roy F. et al. Evolutionary functional analysis and molecular regulation of the ZEB transcription factors. Cell Mol Life Sci. 2012;69:2527–41. doi: 10.1007/s00018-012-0935-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Chen L, Chan TH. et al. SPOCK1 is regulated by CHD1L and blocks apoptosis and promotes HCC cell invasiveness and metastasis in mice. Gastroenterology. 2013;144:179–91. doi: 10.1053/j.gastro.2012.09.042. e4. [DOI] [PubMed] [Google Scholar]
- 56.Pereira JH, McAndrew RP, Sergeeva OA. et al. Structure of the human TRiC/CCT Subunit 5 associated with hereditary sensory neuropathy. Sci Rep. 2017;7:3673. doi: 10.1038/s41598-017-03825-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roh SH, Kasembeli M, Bakthavatsalam D. et al. Contribution of the Type II Chaperonin, TRiC/CCT, to Oncogenesis. Int J Mol Sci. 2015;16:26706–20. doi: 10.3390/ijms161125975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boudiaf-Benmammar C, Cresteil T, Melki R. The cytosolic chaperonin CCT/TRiC and cancer cell proliferation. PLoS One. 2013;8:e60895. doi: 10.1371/journal.pone.0060895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Min W, Angileri F, Luo H. et al. A human CCT5 gene mutation causing distal neuropathy impairs hexadecamer assembly in an archaeal model. Sci Rep. 2014;4:6688. doi: 10.1038/srep06688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sergeeva OA, Tran MT, Haase-Pettingell C. et al. Biochemical characterization of mutants in chaperonin proteins CCT4 and CCT5 associated with hereditary sensory neuropathy. J Biol Chem. 2014;289:27470–80. doi: 10.1074/jbc.M114.576033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao L, Zou X, Liu L. The TCP1 ring complex is associated with malignancy and poor prognosis in hepatocellular carcinoma. Int J Clin Exp Pathol. 2019;12:3329–43. [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Q, Huang WR, Chih WY. et al. Cdc20 and molecular chaperone CCT2 and CCT5 are required for the Muscovy duck reovirus p10.8-induced cell cycle arrest and apoptosis. Vet Microbiol. 2019;235:151–63. doi: 10.1016/j.vetmic.2019.06.017. [DOI] [PubMed] [Google Scholar]
- 63.Macario AJ, de Macario EC. Molecular mechanisms in chaperonopathies: clues to understanding the histopathological abnormalities and developing novel therapies. J Pathol. 2020;250:9–18. doi: 10.1002/path.5349. [DOI] [PubMed] [Google Scholar]
- 64.Xu WX, Song W, Jiang MP. et al. Systematic Characterization of Expression Profiles and Prognostic Values of the Eight Subunits of the Chaperonin TRiC in Breast Cancer. Front Genet. 2021;12:637887. doi: 10.3389/fgene.2021.637887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ooe A, Kato K, Noguchi S. Possible involvement of CCT5, RGS3, and YKT6 genes up-regulated in p53-mutated tumors in resistance to docetaxel in human breast cancers. Breast Cancer Res Treat. 2007;101:305–15. doi: 10.1007/s10549-006-9293-x. [DOI] [PubMed] [Google Scholar]
- 66.Gao H, Zheng M, Sun S. et al. Chaperonin containing TCP1 subunit 5 is a tumor associated antigen of non-small cell lung cancer. Oncotarget. 2017;8:64170–9. doi: 10.18632/oncotarget.19369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tripodi D, Quemener S, Renaudin K. et al. Gene expression profiling in sinonasal adenocarcinoma. BMC Med Genomics. 2009;2:65. doi: 10.1186/1755-8794-2-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uchikado Y, Inoue H, Haraguchi N. et al. Gene expression profiling of lymph node metastasis by oligomicroarray analysis using laser microdissection in esophageal squamous cell carcinoma. Int J Oncol. 2006;29:1337–47. [PubMed] [Google Scholar]
- 69.Ludwig A, Dietel M, Lage H. Identification of differentially expressed genes in classical and atypical multidrug-resistant gastric carcinoma cells. Anticancer Res. 2002;22:3213–21. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author (Z. Yang) upon request.