Abstract
In Drosophila, two classes of genes, the trithorax group and the Polycomb group, are required in concert to maintain gene expression by regulating chromatin structure. We have identified Trithorax protein (TRX) binding elements within the bithorax complex and have found that within the bxd/pbx regulatory region these elements are functionally relevant for normal expression patterns in embryos and confer TRX binding in vivo. TRX was localized to three closely situated sites within a 3-kb chromatin maintenance unit with a modular structure. Results of an in vivo analysis showed that these DNA fragments (each ∼400 bp) contain both TRX- and Polycomb-group response elements (TREs and PREs) and that in the context of the endogenous Ultrabithorax gene, all of these elements are essential for proper maintenance of expression in embryos. Dissection of one of these maintenance modules showed that TRX- and Polycomb-group responsiveness is conferred by neighboring but separable DNA sequences, suggesting that independent protein complexes are formed at their respective response elements. Furthermore, we have found that the activity of this TRE requires a sequence (∼90 bp) which maps to within several tens of base pairs from the closest neighboring PRE and that the PRE activity in one of the elements may require a binding site for PHO, the protein product of the Polycomb-group gene pleiohomeotic. Our results show that long-range maintenance of Ultrabithorax expression requires a complex element composed of cooperating modules, each capable of interacting with both positive and negative chromatin regulators.
Body segment identity in many organisms is achieved, in large part, through the activities of homeotic genes during development. In Drosophila, the establishment and maintenance of their patterns of expression are critical for the determination of the fates of embryonic cells. Two groups of genes, the trithorax group (trxG) (reviewed in reference 19) and the Polycomb group (PcG) (reviewed in references 3, 25, 29, and 38), play a predominant role in maintenance of the on and off states, respectively, of homeotic gene expression during development. It has been proposed that the products of different PcG genes assemble in a multimeric complex only at target genes that are not actively being transcribed, ostensibly locking these genes in an inactive state. This presumably imprints a determined state of the chromatin which could be inherited by the cellular progeny (25). Indeed, several Polycomb-group (PcG) proteins analyzed thus far colocalized at a large number of sites on salivary gland polytene chromosomes, suggesting that they often function together (11, 23, 33). Moreover, it was shown that the Polycomb (Pc) and polyhomeotic products are constituents of a large multimeric protein complex (15). Contrasting with PcG repression is activation by trxG genes. The trxG includes trithorax (trx), brahma (brm), Trithorax-like (Trl), ash1, ash2, and more than 10 additional members, many of which are only minimally characterized. The products of these genes function as transcriptional activators that sustain particular patterns of homeotic gene expression which act antagonistically to those of the PcG. It has been shown that in trx mutant embryos, expression of all bithorax complex (BX-C) genes and several Antennapedia complex (ANT-C) genes are affected in a tissue-, parasegment (PS)-, and promoter-specific fashion (4, 24, 36). Like PcG gene products, those of the trxG have been found at multiple sites on polytene chromosomes, suggesting that targets of these proteins are not limited to the genes of the homeotic complexes. Indeed, it was shown that the region-specific homeotic gene fork head is a direct target gene of trx based on Trithorax protein (TRX) binding on polytene chromosomes (21).
It is thought that genes of both the trxG and PcG encode chromatin-associated regulatory proteins, because both Polycomb protein (Pc) and TRX have homologies to modifiers of position-effect variegation, which are believed to affect transcription through changes in chromatin structure. It has been suggested that, like PcG proteins, trxG proteins (trxG) act in multimeric complexes, because mutations in several members of the trxG produce dose-dependent effects with trx and with each other (37). Binding of TRX to salivary gland polytene chromosomes depends on the presence of the products of ash-1, a trxG gene, and E(z), a PcG gene (21). Interestingly, binding of two other PcG proteins has also been shown to depend on the presence of E(z) (33), suggesting that the protein products of these two genetically antagonistic groups may interact within a similar “core complex.” In transient expression experiments using a Drosophila haploid cell line, Chang et al. (8) have defined TRX and Pc response elements (TRE and PRE) upstream of the Ultrabithorax (Ubx) gene promoter and have shown that trx-dependent activity can be abrogated by increasing the amount of Pc protein.
There is evidence that some trxG genes are involved in chromatin remodeling. The gene product of brm is strikingly similar to the Saccharomyces cerevisiae global transcriptional activator SNF2/SWI2 (45), including a nucleotide-dependent ATPase-presumptive helicase domain that is essential for SNF2 activity (reviewed in references 28 and 44). Genetic and biochemical studies of SNF2, BRM, and related human proteins have suggested that these proteins are components of large protein complexes that help DNA binding regulatory proteins overcome the repressive effects of chromatin on transcription. The yeast SWI/SNF and analogous human complexes both use the energy of ATP hydrolysis to disrupt nucleosome structure in the promoter region of model target genes. A recent biochemical analysis of the ATP-dependent nucleosome remodeling factor NURF, which is required in concert with GAGA factor (a product of the trxG gene Trl) to generate an accessible heat shock promoter, showed that the NURF complex is biochemically distinct from the SWI/SNF complex (48). The failure to detect significant sequence specificity in the binding of the SWI/SNF complex to DNA (20) underscores the fact that the mechanism by which these complexes are recruited to particular target genes is still largely unknown. One possibility might be through an interaction of remodeling complexes with other trxG proteins. Indeed, SNR1, a member of the Drosophila SWI/SNF complex, and INI1, a homologous component of the mammalian SWI/SNF complexes (13, 50), interact with conserved C-terminal domains of TRX and its human homologue, ALL-1/HRX, respectively (34). It is not known whether TRX and ALL-1/HRX are components of these complexes. ALL-1/HRX was not detected in the mammalian SWI/SNF complex, purified to homogeneity, from mammalian cells (50). It is possible, therefore, that the detected interactions between TRX and SNR1 and between ALL-1 and INI1 are transient or might occur only in specific cell types.
Although trx and Pc have a well established set of target genes, their mechanism of action is still poorly understood. The primary difficulty lies in the absence of a system to directly identify target sequences. This is partly due to the absence of specific DNA binding properties for most of the trxG and PcG proteins that have been cloned to date (see Discussion). Given the limited number of binding sites for these proteins on salivary gland polytene chromosomes, it is clear that there is some mechanism which brings them to specific target genes. In the absence of direct recognition of DNA by some members of these groups, this mechanism presumably involves protein-protein interactions. It is, therefore, important to localize the DNA elements on which these proteins reside. This would provide an assay to test directly the properties of these elements both in vitro and in vivo. In the experiments described here, we demonstrate that TRX is localized to several discrete, functionally relevant regulatory regions within the 300-kb BX-C and that each of these regions coincides with genetically defined PREs and/or with regions where Pc has been localized. In the Ubx gene, we have further localized TRX binding regions to three neighboring 300- to 400-bp DNA fragments located roughly 25 kb upstream of the promoter. Each of these TRX binding elements is functionally important, and each element also contains essential PREs. Furthermore, we mapped TRE and PRE sequences within one of these elements (module C) and showed that they are separable. Thus, upstream of the Ubx promoter there is a complex 3-kb chromatin maintenance unit which consists of multiple discrete modules. Each of these modules is essential for the function of the unit, and at least one contains separable TREs and PREs.
MATERIALS AND METHODS
PCR-linked immunoprecipitation.
Plasmid, phage, or genomic DNA was digested independently with Sau3A, MspI, or both. The corresponding adapters for PCR, similar to those described by Saunders et al. (35), were ligated to the resulting DNA fragments. Nuclei from 2- to 20-h-old Drosophila embryos were isolated as previously described (41). Protein extracts were obtained as previously described (12). All DNA binding reactions were carried out for 30 min at 25°C in 50-μl volumes containing 9 μg of nuclear extract and 0.01 μg of cloned target DNA or 0.5 μg of genomic DNA previously ligated with PCR adapters. Two different binding buffers were used. Buffer 1 consisted of 10 mM Tris-HCl (pH 7.5), 80 mM NaCl, 10 mM KCl, 0.01 mM ZnCl2, 1 mM MgCl2, 10 μg of bovine serum albumin/ml, 5% glycerol, 0.025% Nonidet P-40, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, 10 μg of pepstatin/ml, 1 μg of aprotinin/ml, and 0.5 μg of λ phage DNA digested with MspI and Sau3A. Buffer 2 consisted of 10 mM Tris-HCl (pH 7.9), 17 mM NaCl, 100 mM KCl, and 2 μg of poly(dI-dC) · poly(dI-dC), and concentrations of the other components were the same as for buffer 1. The subsequent immunoprecipitations were carried out on a rocking platform in 400 μl of either buffer 1 or 2 at room temperature. Samples were precleared during a 30-min incubation period with 5 mg of protein A-sepharose and then centrifuged for 1 min at 12,000 rpm. The solution was then removed from the beads and placed in a new tube, and 4 μl of either purified antibodies (or immune serum) or preimmune serum (as a control) was added to the supernatant. Following a 1- to 2-h incubation, 5 mg of protein A-sepharose was added, and incubation was continued for 1 h. After centrifugation, the resulting pellet was washed three times for 20 min, and the DNA was eluted from the pellet at 60°C for 5 min in a buffer containing 10 mM Tris-HCl (pH 8.0), 0.4 M NaCl, 10 mM EDTA, 0.5% sodium dodecyl sulfate, and 0.04 mg of tRNA/ml and was then purified for PCR amplification, which was performed in a Perkin-Elmer Cetus apparatus for 15 to 25 cycles (95°C for 45 s, 56°C for 1 min, and 72°C for 3 min). An aliquot of the amplified DNA was labeled with 32P by random priming and used for Southern blot analysis. When genomic DNA was used for binding, the PCR-amplified material was subjected to a second round of immunoprecipitation and PCR amplification. In the experiments with deletion mutants (see Fig. 7), DNA after immunoprecipitation and PCR amplification was analyzed by Southern blot hybridization with the labeled C and D DNA fragments as probes.
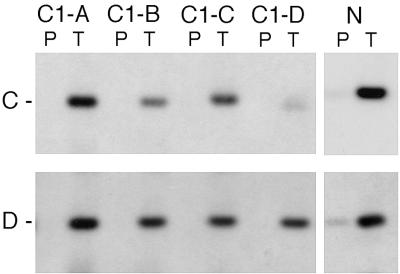
FIG. 7.
Binding of the TRX protein is affected by mutations in the C TRE. DNA fragments obtained after one round of immunoprecipitation of pCaSpeR3 DNA (containing either intact 4-kb BamHI-KpnI fragment, N, or the same fragment with the ΔC1-B, ΔC1-C, and ΔC1-D deletions) with TRX (T) antibody or with preimmune serum (P), following incubation with nuclear extracts (see Materials and Methods), were run on the agarose gel and transferred onto a nylon membrane. The filters were probed separately with the 32P-labeled C (upper panel) and D (lower panel) DNA fragments.
Construction of transposons.
All 13-kb constructs (see Fig. 3A) were made by inserting a Ubx-lacZ fusion gene from pMBO141 plasmid (40) and the following three fragments from the bxd/pbx region into a pCaSpeR3 vector (reference 46; see also Fig. 3): SspI-BamHI (nucleotides [nt] 209,575 to 210,192); HindIII-BglII fragment with or without deletions (nt 214,871 to 221,674); BglII-EcoRI (nt 226,706 to 232,519) (GenBank accession no. U31961). All 4-kb constructs were made by inserting a BamHI-KpnI fragment (nt 216,487 to 220,533) into a pCaSpeR3 vector. The orientation of the insert with respect to the mini-white gene was the same in both types of vectors. Deletions were made by conventional techniques and were confirmed by sequencing.
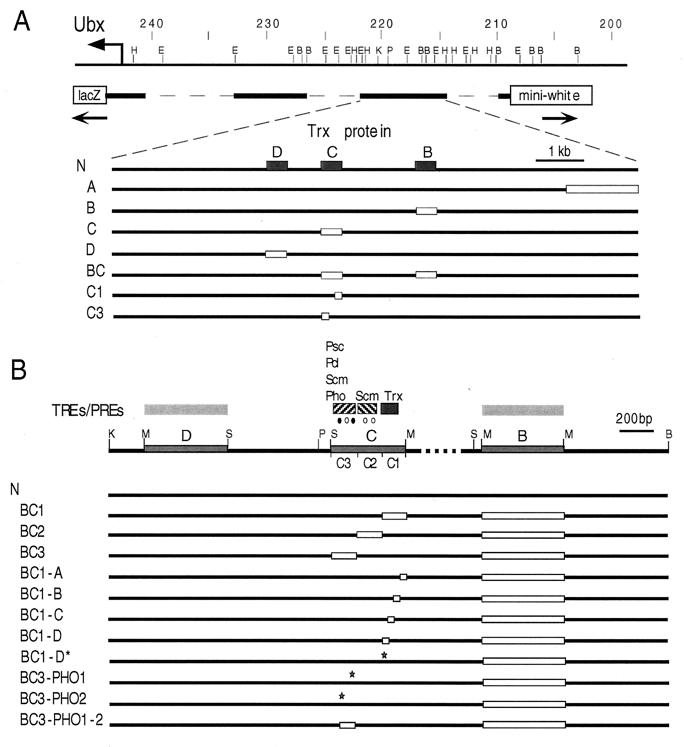
FIG. 3.
Map of constructs used to detect TREs and PREs in the bxd region of Ubx. (A) Partial map of the BX-C including the bxd/pbx regulatory region. Map coordinates are as in Fig. 1. A basal 13-kb construct is shown as a solid bar beneath the DNA line. Deletions within TRX binding elements used to generate transgenic lines are indicated as open boxes beneath the basal construct. The coordinates of the deleted regions in the constructs are as follows: ΔA, HindIII-BamHI (nt 214,875 to 216,285); ΔB, MspI-MspI (nt 217,111 to 217,626); ΔC, MspI-Sau3A (nt 218,835 to 219,249); ΔD, Sau3A-MspI (nt 219,700 to 220,118); ΔC1 (nt 218,835 to 218,959). (B) Map of the multiple TRE-PRE-containing expression maintenance modules. The basal 4-kb construct with a mini-white reporter gene contains the C and D TRX binding elements, approximately a 1-kb fragment which separates the B and C elements, and approximately 0.8 kb of flanking sequences. The coordinates of the deletions in the constructs are as follows: ΔB and ΔC1 are as in panel A; ΔC2 (nt 218,960 to 219,088); ΔC3 (nt 219,089 to 219,249). ΔBC1-A, ΔBC1-B, and ΔBC1-C are deletions of nt 1 to 27, 28 to 61, and 86 to 122, respectively, in the C1 fragment indicated above. The mutation AC to TG in the C1-D fragment and the deletion of the PHO binding site in the C3 fragment are indicated by stars. Consensus binding sites for PHO (filled circles) and GAGA (open circles) in the C fragment are indicated above the map. H, HindIII; E, EcoRI; B, BamHI; P, PstI; K, KpnI; S, Sau3A; M, MspI.
Generation and analysis of transgenic lines.
Injections were performed by standard procedures (42) into a homozygous yw; +/+; +/+ strain. In some cases, P-element insertions were mobilized by using the endogenous transposase insertion P[ry+ Δ2-3]99B, and new transformant lines were selected based on a change in eye color. To test the effects of the mutations of PcG and trx on white gene expression, transformants from each tested line were crossed to flies from balancer stocks containing mutant loci. The effects of the following mutations were tested: trxB11, Psc1, Pcl11, ScmD1, and phob/phocv. For all comparisons, flies of the same sex and age were compared, and to avoid the potential effects from balancer chromosomes on eye color, comparisons were made with unbalanced heterozygotes for each transgenic line.
In situ hybridization and immunostaining of embryos.
A digoxigenin-labeled antisense RNA probe specific for lacZ RNA was used for in situ hybridization to whole-mount embryos (26). Double labeling of embryos to distinguish mutants of trx was carried out by using a probe specific for Ubx RNA and anti-β-galactosidase antibodies (1:100 dilution; Cappel) as described by Mullen and DiNardo (26).
In situ hybridization and immunostaining of polytene chromosomes.
Drosophila polytene chromosome spreads were prepared from salivary glands of third-instar larvae as previously described (14). The pCaSpeR-mini-white DNA was labeled with digoxigenin-11-dUTP (Boehringer Mannheim) by using a random-primed DNA labeling kit (Boehringer Mannheim) and was used as a probe for in situ hybridization. Hybridization was performed for approximately 20 h at 37°C in a solution containing 50% formamide, 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 10% dextran sulfate, and 400 μg of salmon sperm DNA/ml. Anti-digoxigenin-fluorescein antibody (Boehringer Mannheim) was used for detection. Fluorescent labeling of TRX on polytene chromosomes was carried out essentially as described previously (21) by using N1 anti-TRX antibody. Texas-red-conjugated goat anti-rabbit immunoglobulin G (Jackson Immunoresearch Laboratory) was used as secondary antibody at a 1:200 dilution. Chromosomes were counterstained with Hoechst 33258 (Sigma). The slides were mounted in Vectashield mounting medium for fluorescence (Vector). Images of labeled chromosomes were acquired with a Zeiss microscope equipped with a digital camera and were processed with the Adobe Photoshop program.
RESULTS
In this work, we attempted to localize TRX protein to the regulatory regions of the BX-C, which contains three homeotic genes, the expression of which is maintained by the activity of the trx gene. Little is known about the structure of TREs and PREs, although some have been localized genetically and in cell culture to within several kilobases of DNA (7, 8). The structure of these response elements remains elusive, in large part because of the failure of the gene products involved to show DNA binding specificity. Our own attempts to detect direct DNA binding by portions of the TRX protein expressed in bacteria were unsuccessful, suggesting that it binds to DNA through interactions with unknown DNA binding proteins. Also, immunoprecipitation from embryonic nuclear extracts with TRX antibodies was not sensitive enough to detect TRX-DNA complexes (our unpublished results).
TRX protein is localized to multiple discrete binding regions of the BX-C.
We developed a PCR-linked immunoprecipitation procedure, which involves PCR amplification of DNA fragments retained in a pellet after immunoprecipitation of TRX-DNA complexes from embryonic nuclear extracts by using TRX-specific antibody (see Materials and Methods). The amplified material was subsequently used as a probe for Southern blot analysis. We found that two rounds of precipitation and amplification were essential for the successful application of this technique. First, target DNA was digested with two restriction enzymes before incubation with nuclear extracts to ensure that DNA fragments were all of an efficiently amplifiable size (<1 kb). This is important because large DNA fragments are not amplified as efficiently. Since it is possible that binding sites may be cleaved by a particular enzyme, we separately analyzed digests with more than one enzyme. Second, different numbers of PCR cycles for amplification of the final pellet material were tested in order to minimize the background signal. This was monitored by using pellets obtained with both preimmune serum and with an unrelated antibody as controls. Once conditions were optimized, this technique was very sensitive and gave quite reproducible results. One round of immunoprecipitation was sufficient to detect TRX binding fragments when this procedure was applied to several phage clones containing approximately 50 kb of Ubx upstream regulatory DNA (see below). We then extended this approach to use genomic DNA as starting material. Following two rounds of immunoprecipitation and PCR amplification, the DNA was used to probe overlapping phage clones which cover the entire 300 kb of the BX-C.
Figure 1 shows that TRX protein is localized to 10 discrete fragments of the BX-C. The identification of TRX binding regions within the regulatory DNA of all three genes of the BX-C, (Ubx, abdominal-A [abd-A], and Abdominal-B [Abd-B]), is striking since trx is required to maintain the expression levels of all three genes in embryos (4, 24, 36). Next, we proceeded with high-resolution mapping of the TRX binding sites, an example of which is shown in Fig. 2 for the bxd/pbx region of Ubx. Since the sequence of the entire BX-C is now available (22), we have mapped, with one exception, the identified TRX binding sites to DNA fragments of between 200 and 2,000 bp (Table 1). TRX protein was found in several regulatory regions of the BX-C: abx, bxd/pbx, iab-2, iab-3, iab-4, iab-7, and iab-8 (Fig. 1). A number of studies have defined PREs (6, 7, 16, 32, 39) and Pc protein binding sites in the BX-C (9, 43). Comparison of our data with those results showed that six TRX binding sites in bxd/pbx, iab-3, iab-4, and iab-7 regulatory regions are either within or very close to minimal PREs or PC binding sites identified previously (Fig. 1). Interestingly, we detected several signals in the bxd/pbx region, suggesting that there are multiple TRX binding sequences within this regulatory region.
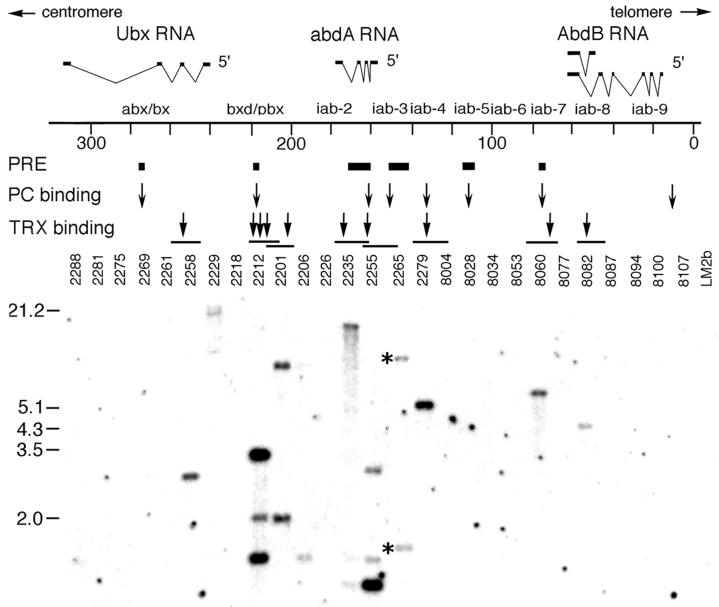
FIG. 1.
Localization of the TRX protein in the BX-C. (Top) A diagram of the molecular organization of the BX-C. Map coordinates (in kilobases) are based on the complete sequence of the BX-C (22). The Ubx, abdA, and AbdB transcription units and regulatory regions are shown above the DNA. PREs (7, 16, 32, 39) and Pc protein binding regions (43) are shown beneath the DNA as filled bars and arrows, respectively. The TRX protein binding regions are indicated by arrows above phage clones that contain TRX binding fragments, which are shown as horizontal lines. (Bottom) Phage clones are indicated by numbers. Equal amounts (∼1 μg) of each of the 27 overlapping λ Charon clones (2) were digested with EcoRI and transferred onto a nylon membrane. The membrane was hybridized with 32P-labeled PCR-amplified fragments of genomic DNA obtained after two rounds of amplification-immunoprecipitation with TRX antibody (see Materials and Methods). Positions of molecular weight markers are indicated on the left. Asterisks indicate two bands also seen in control experiments with preimmune serum that were probably due to repeats in genomic DNA (these bands were not seen in the preimmune serum control lane when cloned phage DNA was used for immunoprecipitation, as in Fig. 2).
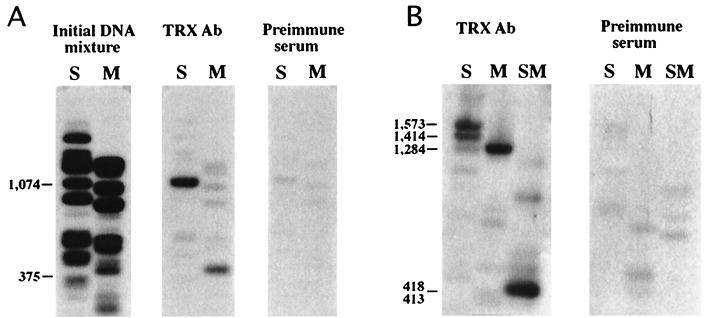
FIG. 2.
Localization of TRX protein in the bxd region of Ubx. DNA fragments obtained after one round of immunoprecipitation of pMBO1253 DNA (containing a 14.4-kb SalI-HindIII fragment with the map coordinates −18.1 to 3.7 [40] with TRX antibody or with preimmune serum from the same rabbit, following incubation with nuclear extracts (see Materials and Methods) were 32P labeled and used to probe filters containing the same DNA digested with Sau3A (S), MspI (M), or both (SM). Fragment B was detected following incubation of DNA with nuclear extract in binding buffer 1 (A), while fragments C and D were detected by using binding buffer 2 (B). Lengths of binding fragments are shown on the left. Gels in panel B were run for a longer time to resolve a doublet in the S lane. The initial DNA mixture was obtained by end labeling λ1253 DNA digested with Sau3A and MspI. The coordinates of the TRX binding fragments in the complete sequence of the BX-C (accession no. U31961) are as follows: B, 217,111 to 217,626; C, 218,834 to 219,314; D, 219,701 to 220,118. (C) TRX binding to the 4-kb N transgene (Fig. 3B) on polytene chromosomes. Localization of the N18 transgene to the 100F region of 3R by in situ hybridization (top). TRX is not localized at 100F in the wild-type larva (middle). The new TRX binding site appears at 100F in the N18 transformant larva (bottom). Arrows indicate the site of insertion of the N transgene.
TABLE 1.
Coordinates of TRX binding sites in the BX-C
| Phage | Coordinates (nt) |
|---|---|
| 8082 | 48,720–59,209 |
| 8060 | 72,602–72,824 |
| 2279 | 133,282–133,605 |
| 2255 | 161,454–163,515 |
| 2235 | 174,020–175,162 |
| 2201, 2212 | 202,535–202,817 |
| 2212 | 217,111–217,626 |
| 2212 | 218,834–219,314 |
| 2212 | 219,701–220,118 |
| 2258 | 253,192–253,583 |
High-resolution mapping of TRX protein in the bxd region of Ubx.
We then concentrated on a more detailed localization of TRX binding elements in the bxd/pbx region, since this region contains a TRE detected in cell culture experiments (8) as well as the best-studied PRE (7). In the bxd/pbx region, TRX is localized to three DNA fragments (each ∼400 bp), which we termed B, C, and D. These elements are separated by approximately 0.5 kb (C and D) and 1 kb (B and C) of DNA (Fig. 2 and 3). Fragment B was immunoprecipitated with TRX antibody after incubation in a buffer which was different from that used to localize the C and D fragments (see Materials and Methods), suggesting that there might be indirect binding via interactions between TRX and different DNA binding proteins. This is also suggested by the absence of extended common sequence motifs in the three fragments, which is also consistent with our inability to detect binding of bacterially expressed portions of TRX to this region of DNA (our unpublished data). Mapping of TRX binding fragments (Fig. 3) showed that all three of these sites are within or close to the smallest regions to which a TRE and PRE were previously mapped (7, 8).
TRX binds in vivo to the bxd region of Ubx.
To confirm the results of the in vitro immunoprecipitation experiments, we needed to address whether TRX protein binds in vivo to the detected TRX binding DNA fragments. To this end, we used transgenic fly lines carrying a 4-kb fragment of the bxd region of Ubx. This DNA fragment (construct N in Fig. 3B) includes all three detected TRX binding fragments, B, C, and D. Once the cytological localization of the transgene was detected by in situ hybridization (100F in line N18; Fig. 2C, top), we immunostained polytene chromosomes prepared from the wild-type and transgenic larvae with TRX antibody. Figure 2C shows that a new TRX signal is observed at the site of insertion of the N transgene in the N18 line, and this signal is absent in wild-type chromosomes. The same results were obtained with other transgenic N lines (not shown). An additional signal was also observed in similar experiments by using a larger (14-kb) transgene which also contained this same bxd region of Ubx (10). These results provide additional in vivo evidence that TRX is physically associated with the bxd regulatory element of Ubx.
TRX binding fragments of the bxd region contain both TREs and PREs.
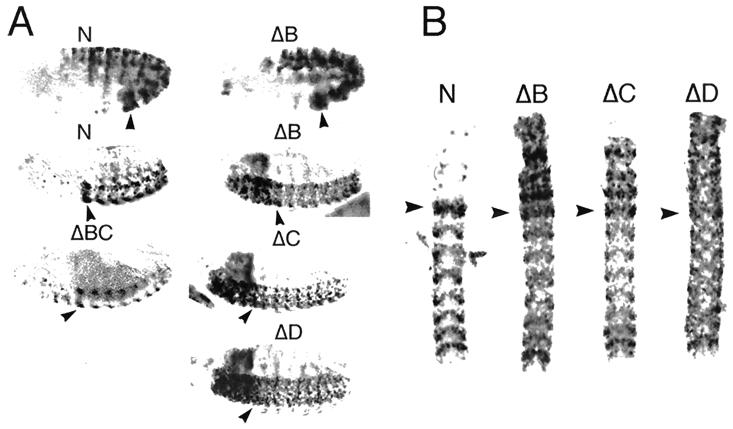
To understand the functional significance of the TRX binding fragments, we constructed a number of lacZ reporter plasmids (Fig. 3) which were introduced into Drosophila embryos via P-element-mediated transformation, and several transgenic fly lines were established for each construct. These 13-kb constructs contained the entire bxd regulatory region, including TRX binding elements as well as multiple embryonic and larval enhancers (30), in an attempt to mimic the regulation of the endogenous gene. The domain of expression of the endogenous Ubx gene is restricted to the abdominal segments with its highest levels of expression in PS6. Ubx expression, which is initiated by the products of the gap and pair-rule genes, is then maintained by the trxG and PcG products beginning from embryonic stages 10 and 11. Expression of lacZ in embryos carrying the wild-type N construct (Fig. 4A) and the shorter ΔA construct (not shown) closely mimics the expression pattern of endogenous Ubx at all developmental stages. These results show that none of the regulatory elements in the bxd region required for proper initiation of Ubx expression was deleted in our constructs and that our basal transgene contains all TREs and PREs required for proper maintenance of Ubx expression during embryogenesis. Deletion of 1.4 kb of DNA (ΔA lines) in the distal portion has no effect on the embryonic expression pattern.
FIG. 4.
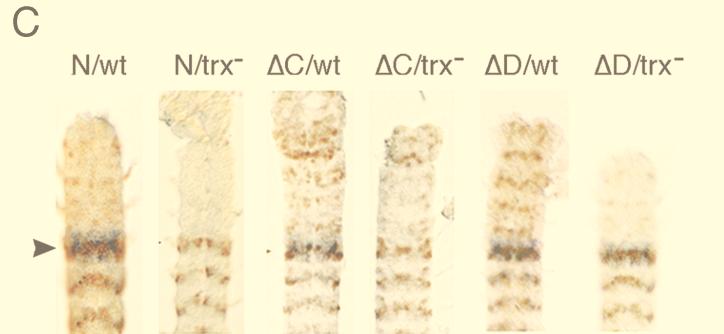
Expression of lacZ in transformant lines in wild-type and mutant trx embryos. (A) lacZ RNA, detected by in situ hybridization to whole-mount embryos, in the N, ΔB (upper row), and all the other tested lines (not shown) is expressed in PS6 to 13 at embryonic stage 10. At embryonic stages 15 and 16, expression of the N construct is still restricted to the region of the embryo posterior to PS6, while in ΔB, ΔC, and ΔD embryos, lacZ is strongly expressed in the anterior neuromeres and in the supraesophageal ganglia (right). At this stage in ΔBC embryos expression of lacZ in the anterior is strongly decreased compared to expression in ΔB, ΔC, and ΔD embryos. In all embryos with the deletion constructs, expression of lacZ in the posterior parasegments is weaker than in N embryos. The anterior region is to the left. (B) Expression of lacZ RNA in the CNS of N, ΔB, ΔC, and ΔD lines. Expression in neuromeres posterior to PS5 is visibly reduced in the deletion lines. The extent and pattern of anteriorly expressed lacZ are different in each of the ΔB, ΔC, and ΔD lines. The anterior end is at the top. (C) The effect of trxB11 mutation on the expression of N, ΔC, and ΔD transgenes. Expression of lacZ RNA in the CNS of N, ΔC, and ΔD transgenes in wild-type and mutant trx embryos. Embryos were doubly stained with antibodies against β-galactosidase (brown) and lightly stained by in situ hybridization with a probe specific to endogenous Ubx RNA (blue). trxB11 mutant embryos were identified by a decrease in Ubx expression primarily in PS6. Expression in neuromeres posterior to PS5 is visibly reduced in the trx mutant in the N line. In the ΔC and ΔD lines, no reduction of β-galactosidase expression in the trxB11 mutant is seen in cells at the periphery of the CNS, where expression of endogenous Ubx is strongly affected by trxB11 mutations (8, 24). In the trxB11 mutant embryos, expression is severely reduced in the anterior neuromeres of the ΔC and ΔD embryos. In the ΔC and ΔD lines, trxB11 mutation also causes an increase of expression in the cells along the midline of the CNS posterior to PS6. This effect is likely to be indirect, due to decreased repression by Ubx and abd-A proteins, since in a trxB11 mutant, expression of all BX-C genes is decreased. Consistent with this interpretation, it has been shown previously that several transgenes, including one similar to the N construct, contain elements that mediate partial repression by the Ubx and abd-A genes (39). Arrowheads indicate PS6.
We also constructed plasmids in which each of the TRX binding fragments (constructs ΔB, ΔC, and ΔD) or one of the fragments in the neighboring region (ΔA) was deleted. For further analysis, we used only those lines in which initiation of expression of the lacZ reporter resembled that of the endogenous Ubx gene. In embryos of each of these chosen lines (including ΔC1 and ΔC3 [see below]), expression was restricted (at least until embryonic stage 10) to the region posterior to PS5, as is expression of the endogenous Ubx gene (Fig. 4A). Analysis of the expression patterns of the three deleted transgenes (ΔB, ΔC, and ΔD) revealed quite striking results. Although in all three cases both the initiation and maintenance of expression at early stages were indistinguishable from those of the N and ΔA constructs, beginning at stage 11, lacZ RNA began to be ectopically expressed in head structures and anterior regions of the central nervous system (CNS) (Fig. 4). Both the timing and the level of this ectopic expression in the anterior portion of the embryo can be explained by the deletion of a strong PRE, leading to a loss of the anterior boundary of expression of all three transgenes. This was true for all of the transgenic lines with these deletion constructs. Since patterns and levels of expression are to some extent different in each of the three constructs (Fig. 4B), each deletion may have removed PREs with somewhat different properties, perhaps ones that respond to a particular subset of PcG proteins. This is consistent with the previous finding that mutations in several PcG proteins produce similar but distinct effects on the expression of a bxd transgene that is roughly equivalent to the one used here (bxd14) (39).
At late embryonic stages, we also observed, for each of the deletion constructs, reduced levels of expression in the periphery of the neuromeres of regions posterior to PS5 compared to those of the N and ΔA lines (Fig. 4A and B). This decrease in expression is reminiscent of the decreased Ubx expression observed in embryos homozygous for the trxB11 null allele (4, 24, 36) and of the decreased expression of a bxd14 construct similar to our constructs (8). The effect of the homozygous trxB11 null allele has been described previously (4, 24, 36). Although the null trx mutation does not completely abolish Ubx and bxd14 expression, it is manifested by a visible decrease of expression in cells at the periphery of the CNS in all of the abdominal neuromeres, with the strongest reduction in PS6. Reduction of lacZ expression in the ΔB, ΔC, and ΔD lines in the region posterior to PS5 is similar to this effect, suggesting that deletion of any of these elements gives an expression pattern similar to that given when trx function is removed. Since these findings suggest that essential TREs have been deleted in these transgenes carrying deletions in the TRX binding elements, we analyzed expression of the N, ΔB, ΔC, and ΔD transgenes in a trxB11 mutant background to see whether further reduction of expression would be observed. Compared to the effect of trxB11 on the N construct, no further reduction of expression in the posterior region of the embryo was detected in trx mutant embryos (ΔC and ΔD in Fig. 4C). Similar results were obtained with the ΔB lines (not shown). These results suggest that all three of the TRX binding elements detected in our immunoprecipitation experiments contain functional TREs and that, at least in the context of our transgene, these elements are all essential for full expression in embryos.
Interestingly, in trx mutant embryos, expression anterior to PS6 is severely decreased in each of the deletion constructs (Fig. 4C). This unexpected result shows that trx products are also required for the expression of the transgene in the anterior portion of the embryo. We constructed transgenic lines carrying a deletion of two TREs/PREs, B and C (ΔBC in Fig. 3). Expression of the lacZ reporter gene in the ΔBC line is very weak in the anterior portion of the embryo (Fig. 4A) compared to that of the ΔB and ΔC lines, and it is reminiscent of the expression pattern observed in ΔB, ΔC, and ΔD in trx mutant embryos (Fig. 4C). This suggests that the simultaneous deletion of two TREs leads to the same effect as complete removal of TRX function in the anterior portion of the embryo. This result confirms our previous conclusion that the TRX binding B and C fragments each contain functional TREs.
Properties of individual TREs/PREs.
Our results suggest that multiple elements within the bxd region of Ubx are crucial for proper expression in embryos. We also analyzed the expression of the mini-white gene in our transgenic lines by examining the eye color of adult flies. We were particularly interested in comparing the effects of a trx mutation on the expression of the lacZ and white reporter genes in our constructs with deletions. Although we saw no effect of a trx null mutation on the expression of lacZ in the posterior region of embryos in the ΔB, ΔC, and ΔD lines (see above), the expression of the white gene was decreased in the eyes of flies heterozygous for trxB11 (Table 2). Quite strikingly, no effect of the trxB11 mutation was observed when both elements B and C were deleted (ΔBC construct). This suggests that although two TREs together retain some activity in both tests (lacZ expression in embryos and white expression), one element is incapable of providing a substantial level of trx responsiveness even in the apparently more sensitive white expression assay. This is in contrast to the results obtained in the background of PcG mutations: deletion of two elements, B and C in ΔBC, did not abolish the responsiveness of the white gene in this construct to the two PcG mutants tested (Psc1 and Pcl11; results not shown). Since ΔBC in the context of a truncated form of the 13-kb construct (4-kb constructs; Fig. 3 and below) completely abrogates responsiveness of the white gene to the same PcG alleles (see below), this suggests that the 13-kb DNA fragment might contain more than the three PREs detected in these experiments. This would be consistent with a requirement for multiple PREs to generate a strong response to PcG function.
TABLE 2.
Effect of the trxB11 null mutation on white gene expression in the 13-kb transgenic lines
| Construct | No. of lines showing decreased expression/no. of lines testeda |
|---|---|
| N | 2/2 |
| ΔB | 2/2 |
| ΔC | 2/2 |
| ΔD | 2/2 |
| ΔBC | 0/3 |
The ratio represents the number of lines showing a decrease in white gene expression in response to the heterozygous trxB11 allele to the number of transgenic lines tested.
TREs and PREs in the C module are conferred by separable DNA sequences.
The finding that TREs and PREs are closely situated suggests that these elements might in fact be the same or overlapping DNA sequences. To test this, we first pursued dissection of the central 412-bp C element by using the mini-white gene as a reporter. We constructed transgenic lines carrying vectors with a 4-kb DNA fragment in which the B element was deleted entirely and the C element carried partial deletions. As a control, we generated a number of lines carrying a deletion of the B element alone. The design of these experiments was based on the previous result which showed that no effect of the trx mutation on white gene expression was observed when both elements B and C were deleted but that responsiveness remained when either one alone was deleted (Table 2). These 4-kb transgenic lines (Fig. 3B) were tested for the loss of TRE and/or PRE activities by examining changes in eye color in the background of trxB11 and each of three PcG mutants, Psc1, Pcl11, and ScmD1. In the majority of the control ΔB lines, the responsiveness of the white gene expression to trxB11 mutation remained unchanged (Table 3). The results of experiments with deletion constructs, which are summarized in Table 3 and Fig. 3B, show that deletion of the C1 fragment but not the C2 and C3 fragments eliminates responsiveness of this construct to the heterozygous trxB11 mutation in all lines tested. This suggests that a functional TRE is located only in the C1 element. We also observed a clear difference in the responsiveness of these 4-kb transgenes to the PcG mutants (Table 3). One half or more of the ΔBC1 lines tested showed an increase in eye color, when heterozygous for any of the three PcG mutations, indicating the presence of PREs in this construct. The fact that not all lines were responsive to the PcG mutations was expected, since the effects of various PcG mutations on the same transgene depend on the chromosomal insertion site (14, 18), and therefore many lines were tested. Thus, deletion of the TRE-containing 124-bp DNA fragment C1 does not remove PRE activity, suggesting that TRE and PRE activities are conferred by different sequences. Further analysis showed that the responsiveness to all three PcG alleles was lost in the ΔBC3 lines. Interestingly, although the responsiveness to Psc and Pcl mutations was unaffected by deletion of the C2 fragment, the responsiveness to the Scm mutation was lost. Although this analysis is based on the results of the white gene expression assay only, these results indicate that this module may contain two distinct PREs in the neighboring 128-bp C2 and 160-bp C3 fragments. They also suggested that both putative PREs depend on the activity of the Scm gene product and that the activity of the C3 PRE, in addition, depends on the presence of the Psc and Pcl gene products. These results were also confirmed by another test based on an intrinsic feature of PRE activity, its ability to cause pairing-sensitive repression of white gene expression. Normally, animals homozygous for the mini-white transgene have darker eyes than their heterozygous siblings. However, when PREs are included in a particular transgene, homozygotes typically have a lighter eye color than heterozygotes. Consistent with this and with the fact that both C2 and C3 contain PREs, we observed pairing-sensitive repression in homozygotes of most of the ΔB and ΔBC1 lines tested (Table 3). (Since the control ΔB lines retained the property of pairing-sensitive repression, they were not tested for the effects of the individual PcG mutations.) No pairing-sensitive repression was detected, however, in any of the ΔBC2 and ΔBC3 lines tested. These results are in agreement with the results of our previous analysis, which suggested that the C2 and C3 fragments each contain functional PREs, and they further indicate that these PREs are functionally nonredundant in this assay.
TABLE 3.
Effects of trx and PcG mutations on white gene expression and on pairing-sensitive repression in ΔBC 4-kb transgenic lines
| Construct | No. of lines showing a change in gene expression/no. of lines tested
|
|||||
|---|---|---|---|---|---|---|
| trxB11 | Psc1 | Pcl11 | ScmD1 | phob/phocv | Pairing-sensitive repressionb | |
| ΔB | 6/9a | NT | NT | NT | NT | 5/7b |
| ΔBC1 | 0/11 | 7/10 | 6/10 | 5/10 | NT | 3/3 |
| ΔBC2 | 4/5 | 4/7 | 7/7 | 0/7 | NT | 0/5 |
| ΔBC3 | 4/6 | 0/5 | 0/5 | 0/5 | NT | 0/5 |
| ΔBC3-PHO1 | NTc | 4/8 | 3/8 | 2/8 | 0/3 | 0/6 |
| ΔBC3-PHO2 | NT | 7/10 | 6/10 | 5/10 | 3/4 | 5/8 |
| ΔBC3-PHO1-2 | NT | 2/8 | 2/8 | 2/8 | 1/4 | 1/7 |
Ratios in columns trxB11 to phob/phocv represent the number of transgenic lines which showed either a decrease (in trx mutants) or an increase (in PcG mutants) in eye pigmentation in response to heterozygous mutant alleles to the total number of lines tested. For pho mutants, the effect was tested in the phob/phocv heteroallelic combination.
Ratios in this column represent the number of lines with decreased white gene expression in homozygotes versus heterozygotes to the number of lines tested for pairing-sensitive repression.
NT, not tested.
Interestingly, the putative C3 PRE that interacts with all three PcG genes tested contains two consensus binding sites for the PHO, the protein product of the gene pleiohomeotic (Fig. 3B). PHO is a PcG protein that binds specifically to a PRE in the engrailed gene and is the only sequence-specific DNA binding protein of the PcG identified thus far (5). To assess the functional significance of these consensus binding sites, we constructed transgenic lines carrying deletions in each of these sites, as well as a deletion of 112 bp in the C3 region (ΔBC3-PHO1-2) which includes both putative PHO binding sites and a consensus binding site for the GAGA factor. A deletion of one of the putative binding sites had no discernible effect on the responsiveness of the expression of the transgene to four PcG mutations tested (compare ΔBC1 to ΔBC3-PHO2), including a pho mutation (in a phob/phocv heteroallelic combination). This suggests that this site may be either nonfunctional or functionally redundant (Table 3). However, in most of the lines tested, a deletion of another PHO binding site (ΔBC3-PHO1) as well as a deletion which removes both putative PHO binding sites decrease the responsiveness of white gene expression to mutations in three PcG genes, pho (phob/phocv), Pcl11 and ScmD1 (Table 3). The results with the Psc1 allele are less conclusive since one-half of the ΔBC3-PHO1 lines remained sensitive to the dosage of the Psc protein product. Further, the frequency of pairing-sensitive repression was reduced in most of the ΔBC3-PHO1 and ΔBC3-PHO1-2 lines (compare to ΔB, ΔBC1, and ΔBC3-PHO2), confirming the importance of this PHO1 binding site for PcG-mediated repression. The results of these experiments suggest that the protein products of four PcG genes, pho, Psc, Pcl, and Scm, are required for the activity of the C3 PRE. The involvement of multiple PcG products in the activity of this relatively small region (160 nt), as well as the requirement for the PHO site for most or all of the PcG responsiveness, suggests the possibility that a multiprotein complex containing these products is associated with this element.
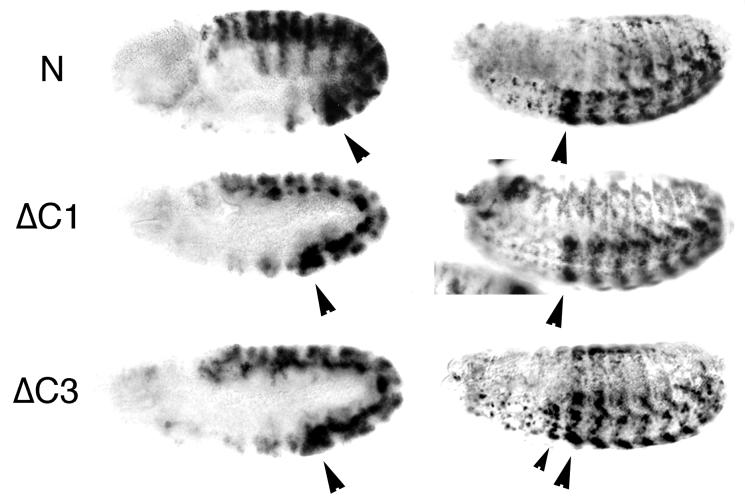
Since these results are of general significance and suggest that the TRE and PRE activities of region C are conferred by different sequences, and since some PRE activity could have gone undetected in the experiments described above, it was necessary to confirm these findings in embryos. To this end, we constructed transgenic fly lines carrying a 13-kb transgene in which either the C1 TRE-containing fragment or the C3 PRE-containing fragment was deleted (ΔC1 and ΔC3; Fig. 5). Analysis of the embryonic expression pattern of the lacZ reporter gene in several ΔC1 transgenic lines showed that the anterior boundary of lacZ expression is properly maintained at PS6, as it is in the wild-type N construct (Fig. 5). This suggests that no functionally important PRE is present in the C1 DNA fragment. The expression level of the reporter gene in these lines is decreased in the posterior PSs, especially in PS6, which is reminiscent of the effect of a trx mutation on lacZ expression (Fig. 4C), confirming the loss of an essential TRE. In the transgenic lines carrying the ΔC3 construct, we observed anterior overexpression of lacZ in PS5, suggesting that some PRE activity has been lost (Fig. 5). However, the level of anterior overexpression is significantly lower than in the ΔC lines, where both PREs have been deleted. This indicates that each of the PREs within fragment C are essential for maintenance of the anterior boundary of expression but that the C1 fragment is dispensable for the PcG responsiveness. Thus, the results of our experiments clearly show that the primary TRE and PRE activities within the C element are conferred by different DNA sequences.
FIG. 5.
Expression of lacZ in ΔC1 and ΔC3 transformant lines. lacZ RNA in the N, ΔC1, and ΔC3 embryos (left column) is expressed in PS6 to 13 at embryonic stage 10. In ΔC1 embryos, at embryonic stage 15 and 16 (right column), there is no expression anterior to PS6. At this stage in ΔC1 embryos, expression of lacZ is decreased compared to expression in N. In ΔC3 embryos, the anterior boundary is shifted to PS5 (small arrowhead). Large arrowheads indicate PS6.
TRE activity and TRX binding require a sequence which is juxtaposed to the C2 PRE.
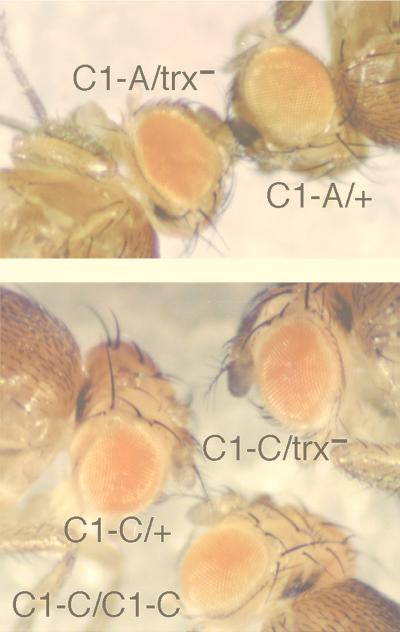
In order to further localize the sequences responsible for TRE activity, we generated transgenic lines which contained four nonoverlapping 28-, 34-, 24-, and 36-bp deletions in the C1 sequence (Fig. 3B). Analysis of the effect of the trx null mutation on the expression of the white reporter gene showed that each of the three small deletions, ΔBC1-B, ΔBC1-C, and ΔBC1-D, abolished responsiveness to a trx mutation in a majority of transgenic lines tested (Table 4). We found short homologous sequences AACAA between the detected TRE-containing fragment C1 (three repeats) and the TRE-PRE-containing module D (two repeats). To test whether these homologous sequences are required for the TRE activity, we constructed transgenic lines carrying an AC-to-TG mutation in the central AACAA repeat in the TRE C1-D subsequence. Strikingly, this mutation had the same deleterious effect on the responsiveness of the transgene to the trx mutation as did deletion of the whole C1 fragment (Table 4). The expression of the white gene in the lines carrying the other deletion, ΔBC1-A, remained sensitive to a change in the dose of trx (Fig. 6), suggesting that this sequence is dispensable for TRE activity. We suggest that in the C1 fragment, there is a single functional TRE which is represented by a sequence approximately 90 bp in length. Since we were not able to demonstrate that TRX binds directly to DNA, we believe that it is likely to bind to the identified TREs through interactions with other primary DNA binding proteins.
TABLE 4.
Effect of trx mutation on white gene expression in ΔBC1 transgenic lines
| Construct | No. of lines showing a change in gene expression/no. of lines testeda |
|---|---|
| ΔBC1-A | 6/6 |
| ΔBC1-B | 1/4 |
| ΔBC1-C | 2/8 |
| ΔBC1-D | 1/5 |
| ΔBC1-D* | 1/5 |
Ratios represent the number of lines which showed a decrease in eye color in response to heterozygous trxB11 allele to the number of lines tested.
FIG. 6.
The effect of the trxB11 mutation in heterozygotes on expression of the white gene, and pairing-sensitive repression in ΔBC1-A and ΔBC1-C transgenic flies. Expression of white in the eyes of the ΔBC1-A heterozygous line is strongly decreased in trxB11 heterozygotes (top) (C1-A/+ appears to the left of the label, and C1-A/trx− is shown to the right of the label), but expression of white in the ΔBC1-C heterozygous line is unaffected by the trxB11 mutation (bottom). Expression of white is significantly decreased in the eyes of homozygotes compared to that in heterozygotes in the ΔBC1-C line.
In order to test the correspondence between TRE function in vivo and TRX binding in vitro, we asked whether the deletion of our C1 subregions significantly affected the ability of TRX protein to associate with the C fragment. The design of these experiments was similar to those described above, except that P-element vectors containing the 4-kb bxd inserts, N, ΔBC1-A, ΔBC1-B, ΔBC1-C, and ΔBC1-D, were used for binding to TRX protein in nuclear extracts. Following immunoprecipitation, material was PCR amplified and tested for the presence of the C (or ΔC deletions) and D fragments (as a control) by Southern hybridization. The results of these experiments, shown in Fig. 7, show that removing either C1-B or C1-D subfragments clearly reduce TRX binding, with ΔC1-D having the strongest effect, while removing C1-A or C1-C had little or no effect. These results correlate well with the requirement for both the C1-B and C1-D subregions for TRE activity in vivo. However, the fact that C1-C is also required for TRE activity but shows little effect on TRX binding in vitro suggests either that TRX binding in vivo has somewhat more stringent requirements or that another activity, in addition to TRX recruitment, is required for TRE activity in vivo.
DISCUSSION
The common feature of trxG and PcG proteins, which are required for the activation and repression of their target genes, respectively, is to “lock in” a particular state of gene activity early in embryogenesis and to maintain this state through many cell generations. The mechanism of their functioning is poorly understood but is believed to involve specific alterations of chromatin structure. There are two major criteria which have been used to identify functional PREs: pairing sensitivity of white gene expression in the eye and PcG-dependent repression of reporter genes in embryos. Most studies aimed at fine mapping of PREs using these criteria have used minimal regulatory regions consisting of relatively short DNA fragments. However, this approach may be limited if maintenance of the chromatin states involves cooperative interaction within larger regulatory regions. Other approaches have included mapping of PcG protein binding sites, either by the direct immunoprecipitation of chromatin or by analyzing the appearance of a new binding site on polytene chromosomes at the site of insertion of a transgene that contains a PRE. These studies suggest that there are multiple binding sites for Pc, as well as for other PcG proteins, in the BX-C and in other target genes and that some of these sites overlap well-characterized PREs (6, 7, 16, 32, 39, 43). Some of these sites were mapped close to binding sites for the GAGA factor, a product of the trxG gene Trl (43). Paradoxically, in the case of the iab-7 PRE, which contains GAGA binding sites, a Trl mutation caused suppression of PcG-mediated silencing, an effect that is expected from a PcG mutation (17). In the case of the Mcp PRE, which also contains consensus GAGA binding sites, Trl mutations had no apparent effect on silencing (17). A TRE, situated closely to a PRE, has also been mapped in the bxd region of the BX-C (7, 8). However, since this mapping of trxG and PcG binding sites and response elements was performed at low resolution (from several hundred base pairs to several kilobases), the question of whether TREs and PREs are separable functional elements remained unanswered. In addition, the existing information suggests that there can be multiple PREs contained within several kilobases, which can function independently in regulating white gene expression. This lack of resolution of individual response elements has prevented their functional characterization, including an analysis of the proteins which may interact with these elements.
Following our mapping of TRX protein binding regions within the BX-C, we chose a different approach to analyze the significance of the binding sites. Rather than attempting to analyze small fragments in isolation, we created a transgene that included the TRX binding sites in a more normal context, within a 13-kb region of Ubx regulatory DNA. This region is sufficient to emulate most aspects of the normal embryonic expression patterns of the endogenous Ubx gene. This analysis was complemented by examining the effects on a second reporter within the same transgene, the white gene, and by studying the responsiveness of the various transgenes to mutations in trx and PcG genes. The results provide several important insights into the functioning of both TREs and PREs in the BX-C. (i) There are multiple regulatory “maintenance modules” in which TREs and PREs are located in close proximity. (ii) Each of the modules tested is essential for the proper maintenance of the embryonic expression patterns. (iii) The TRE and PREs within the C module are represented by separable DNA sequences.
Discrete DNA sequences of TREs and PREs.
We used two criteria to identify TRE and PRE activities within our reporter constructs: maintenance of lacZ reporter expression patterns in embryos, and trx- and PcG-dependent maintenance of white gene expression in the eyes of adult flies, including their effects on pairing-sensitive repression of white. Ultimately the results obtained with both reporter genes led to similar conclusions, and the TRE and PRE activities of module C were shown to be conferred by neighboring but separable DNA sequences (Fig. 3, Table 3). An essential TRE and two distinct PREs were detected in this central C module. Further analysis has shown that the TRE activity and TRX binding require a 90-bp region, which, based on its length, is likely to bind more than just a single protein. This suggests that a number of primary DNA proteins may be associated with the TRE in the C1 fragment. A gel-shift analysis of the protein binding properties of this 90-bp TRE fragment suggests that this fragment contains two core binding sequences which are required for apparent cooperative binding by several nuclear proteins (20a). These core sequences, which are located on the boundary of the C1-B and C1-C fragments and in the C1-D fragment appear to contribute to the formation of a large multiprotein complex. Therefore, there is a direct correlation between the sequences which are required to form this protein complex and those which are required for the TRX binding and TRE activity. Most strikingly, the AACAA repeat in the C1-D fragment seems to be crucial for forming this complex, as it was shown to be crucial for the TRE activity, since complex formation is virtually abolished when the AC residues are changed to TG (our unpublished results). Since direct TRX protein binding to DNA has not been established by using a number of DNA binding assays and since the DNA binding protein complex in the C1 fragment does not contain TRX (20a), we suggest that TRX binds to this TRE through interactions with a complex of primary DNA binding proteins. At present, we do not know the identity of the proteins that associate with these TREs. While it would not be surprising to find that some are products of the trxG, it is unlikely that the GAGA factor or the ZESTE protein are primary binding proteins in this case, since the C1 element does not contain consensus binding sites for either of these proteins.
Our analysis suggested that the C2 and C3 fragments each contain a PRE. Each of these elements, C2 and C3, is required to confer pairing-sensitive repression on a white reporter gene. These elements may be functionally different because their activities require different sets of PcG proteins (Table 3) and because there is no significant sequence similarity between them. Both PREs are apparently also required, in concert, to provide full maintenance function in embryos. This follows from the fact that while very strong anterior overexpression occurs in embryos when the entire C fragment is deleted, only moderate overexpression in PS5 results from the deletion of a single PRE (C3; Fig. 4A). In addition, one of these PREs, C3, may contain a functionally important binding site for the PcG protein PHO (5), since deletion of this binding site abrogates both pairing-sensitive repression and responsiveness of the white reporter gene to three PcG mutations, pho, Pcl, and Scm. (Table 3). Therefore, we suggest that the protein products of these three PcG genes may be components of a putative PcG protein complex that is formed at the C3 PRE. In addition to the PHO binding sites, the C2 and C3 DNA fragments contain three consensus binding sites for the GAGA protein (Fig. 3). Further analysis is required to determine whether the deletion of GAGA sites has an effect on PRE function. It is likely, however, that PHO and GAGA are not the only primary DNA binding proteins in these PREs, since the C2 PRE does not contain consensus PHO binding sites. These proteins may be DNA-interacting components of particular subsets of PcG complexes, as has been suggested by Brown et al. (5) for the engrailed PRE.
Multiple TREs and PREs are essential in embryos.
We have shown that the TREs and PREs in the bxd region of Ubx are clustered in three closely situated maintenance modules, each approximately 400 bp. Each module contains elements for both of these opposing activities. Our analysis is focused on the TRX protein, although it is possible that there are other positive maintenance elements in this region which require the products of other trxG genes. Similarly, since we analyzed PRE function only in TRX binding regions, some PREs may have gone undetected. Despite these limitations, we discovered several TRE- and PRE-containing modules in the bxd region of Ubx which are all essential for proper Ubx expression, since deletion of any one of the three modules leads to a significant loss of maintenance activity in embryos. In the context of a natural Ubx promoter and a large part of its regulatory region, these modules were all essential in embryos with respect to PRE and TRE function. However, when tested for effects on white gene expression, deletion of individual elements did not completely abolish either eye color variegation or the responsiveness to trx and PcG mutations. These differences suggest that repression of white expression in adults may not accurately reflect the function of these elements in the context of the entire Ubx gene. Thus, cooperative interactions among multiple PREs and TREs are required for proper function of the Ubx gene, and these interactions may involve activities not reflected in assays with reporters unrelated to Ubx expression.
Interestingly, we found that trx function is required for Ubx expression not only in its normal domain of expression in the posterior region of the embryo but also in the anterior region, when Ubx is overexpressed due to a deletion of PREs. There are clear differences between the anterior and posterior regions of the embryo with respect to both the effect of a trx mutation and the requirements of TREs for the expression patterns of our transgenes. First, in trx mutants, the loss of expression in the anterior is much more severe than it is in the posterior. Second, anterior expression is very strong when one of the three TREs is deleted, and only a simultaneous deletion of two TREs leads to a decrease of this expression which is comparable with the effect of a trxB11 null mutation. In the posterior, however, deletion of only one element mimics an almost complete loss of trx-related activity, and deletion of two elements has no further effect. This might be explained by a different mode of functioning in the anterior versus posterior regions of the embryo. Such a functional difference is also suggested by our previous observation that different trx protein products, which result from alternatively spliced mRNAs, may be required for the maintenance of expression of the ANT-C genes (in the anterior region) and not for maintenance of BX-C gene expression (in the posterior regions). This is based on an analysis of the effects of different trx alleles on the two homeotic complexes and on the finding that the expression of one of the early trx RNAs is spatially restricted to the posterior region where the BX-C genes are expressed (36). Based on these observations, we conclude that there are quite complex requirements for the activities of the three maintenance modules in different cells. Functionally similar maintenance units may regulate other genes in the BX-C as well, since the other TRX binding regions we detected are associated with either PRE function, Pc protein binding sites, or both.
Functional relationships between TRX and PcG proteins within a TRE-PRE module.
Our finding of discrete TRE and PRE sequences argues against a direct competition between the proteins of these opposing groups for binding sites on DNA, although the question of whether they normally occupy their response elements simultaneously within a given module remains open. In addition, some data suggest that the association of trxG and PcG proteins with a particular gene depends on its transcriptional status. First, the strength of TRX binding to salivary gland polytene chromosomes at the site of a transcriptionally active gene, such as fork head, is much higher than it is at the location of the BX-C, which is silent in the salivary glands (21). Second, immunoprecipitation of Pc protein from Drosophila cells was found to be more abundant at silenced genes than at activated genes of the BX-C (27). Third, when transcription of a reporter gene is induced by GAL4, several PcG proteins are displaced from the chromosomal site of insertion of a Fab-7 transgene (52). Although this is not directly related to trxG functioning, it indicates that PcG proteins are not bound abundantly to actively transcribed genes, suggesting that there might be quantitative or qualitative differences in bound trxG and PcG protein complexes depending on the transcriptional activity of a particular gene. Our work suggests that the occupancy of TREs and PREs may be independent rather than mutually exclusive. Since the formation of functional activating or silencing complexes may depend on and in turn lead to the maintenance of the on-off state of Ubx expression in a particular tissue, we suggest that the occupancy of the TRE by a functional trxG complex alters, directly or indirectly, the composition of nearby PRE complexes without necessarily abolishing binding by PcG proteins.
How do active trxG and PcG protein complexes function? There have as yet been no specific biochemical activities associated with PcG proteins. Most of the PcG proteins are associated with chromosomes, and it is assumed that they act by forming repressive multiprotein complexes that prevent active transcription. One of the functions of trxG proteins may be simply to counteract the formation of these repressive PcG complexes and thus to increase the accessibility of enhancers to the neighboring regions of DNA. However, there is growing evidence that the trxG represents a heterogeneous family of proteins with diverse functions. Some of them, such as TRX, ASH1, ASH2, GAGA, and ZESTE, are associated with particular sites on polytene chromosomes (1, 10, 21, 33, 47, 49), while others, such as BRM and SNR1, are found in chromatin remodeling complexes that may not be associated with specific chromosomal regions. There is some evidence that one of the functions of trxG proteins may be to recruit chromatin remodeling complexes to DNA. GAGA factor is required for the function of one chromatin remodeling complex, the Drosophila NURF complex (49), and TRX was shown to physically interact with SNR1, a component of the Drosophila SWI/SNF complex (34). However, there is no evidence thus far that these interactions are mediated through particular TREs. In addition, there is evidence that TRX and its human homologue, ALL-1/HRX, may be involved directly in the activation of promoters, since both of these proteins possess transactivation activity in cells (8, 31, 51). Therefore, it is likely that trxG proteins not only can counteract formation of PcG-mediated repressive chromatin structure but may also play a more direct role in maintaining transcription.
In conclusion, we have identified in the Ubx regulatory region three discrete TRE/PRE modules. These modules are contained within a complex, 3-kb maintenance unit in which each detected element is essential with respect to both PRE and TRE function. Furthermore, we found that TRX binds sequences in other regulatory regions of the BX-C that are consistently associated with either PRE function, Pc protein binding, or both, suggesting the possibility that similar maintenance units are employed for regulation of other genes in the complex. Functional dissection of one of these modules showed that the TRE and PRE activities can be ascribed to separable DNA elements, even though they are located within tens of nucleotides of each other. This proximity suggests that there may be some direct interaction between protein complexes formed at these elements. In addition, the TREs and PREs that we have identified do not contain extensive sequence similarities, suggesting that they are bound by protein complexes of different composition.
ACKNOWLEDGMENTS
We thank W. Bender for clones of the BX-C; V. Pirrotta, J. Jaynes, R. Jones and W. Bender for discussions; and J. Jaynes and S. Smith for critically reading the manuscript.
This work was supported by a grant from the National Cancer Institute to A.M.
Footnotes
This work is dedicated to the memory of Tadaatsu Goto.
REFERENCES
- 1.Adamson A L, Shearn A. Molecular genetic analysis of Drosophila ash2, a member of the trithorax group required for imaginal disc pattern formation. Genetics. 1996;144:621–633. doi: 10.1093/genetics/144.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender W, Akam M, Karch F, Beachy P A, Peifer M, Spierer P, Lewis I B, Hogness D S. Molecular genetics of the bithorax complex in Drosophila melanogaster. Science. 1983;221:23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- 3.Bienz M, Müller J. Transcriptional silencing of homeotic genes in Drosophila. BioEssays. 1995;17:775–784. doi: 10.1002/bies.950170907. [DOI] [PubMed] [Google Scholar]
- 4.Breen T R, Harte P J. trithorax regulates multiple homeotic genes in the bithorax and Antennapedia complexes and exerts different tissue-specific, parasegment-specific and promoter-specific effects on each. Development. 1993;117:119–134. doi: 10.1242/dev.117.1.119. [DOI] [PubMed] [Google Scholar]
- 5.Brown J L, Mucci D, Whiteley M, Dirksen M L, Kassis J A. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 6.Busturia A, Bienz M. Silencers in Abdominal-B, a homeotic Drosophila gene. EMBO J. 1993;12:1415–1425. doi: 10.1002/j.1460-2075.1993.tb05785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan C-S, Rastelli L, Pirrotta V A. Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang Y-L, King B O, O’Connor M, Mazo A, Huang D-H. Functional reconstruction of trans regulation of the Ultrabithorax promoter by the products of two antagonistic genes, trithorax and Polycomb. Mol Cell Biol. 1995;15:6601–6612. doi: 10.1128/mcb.15.12.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang A, O’Connor M, Paro R, Simon J, Bender W. Discrete Polycomb-binding site in each parasegmental domain of the bithorax complex. Development. 1995;121:1681–1689. doi: 10.1242/dev.121.6.1681. [DOI] [PubMed] [Google Scholar]
- 10.Chinwalla V, Jane E P, Harte P J. The Drosophila trithorax protein binds to specific chromosomal sites and is co-localized with Polycomb at many sites. EMBO J. 1995;14:2056–2065. doi: 10.1002/j.1460-2075.1995.tb07197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeCamillis M, Cheng N S, Pierre D, Brock H W. The polyhomeotic gene of Drosophila encodes a chromatin protein that shares polytene chromosome-binding sites with Polycomb. Genes Dev. 1992;6:223–232. doi: 10.1101/gad.6.2.223. [DOI] [PubMed] [Google Scholar]
- 12.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dingwall A K, Beek S J, McCallum C M, Tamkun J W, Kalpana G V, Goff S P, Scott M P. The Drosophila SNR1 and Brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol Biol Cell. 1995;6:777–791. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fauvarque M-O, Dura J-M. polyhomeotic regulatory sequences induce developmental regulator-dependent variegation and targeted P-element insertions in Drosophila. Genes Dev. 1993;7:1508–1520. doi: 10.1101/gad.7.8.1508. [DOI] [PubMed] [Google Scholar]
- 15.Franke A, DeCamillis M, Zink D, Cheng N, Brock H W, Paro R. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 1992;11:2941–2950. doi: 10.1002/j.1460-2075.1992.tb05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 17.Hagstrom K, Muller M, Schedl P. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics. 1997;146:1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kassis J A. Unusual properties of regulatory DNA from the Drosophila engrailed gene: three “pairing-sensitive” sites within a 1.6 kb region. Genetics. 1994;136:1025–1038. doi: 10.1093/genetics/136.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennison J A. Transcriptional activation of Drosophila homeotic genes from distant regulatory elements. Trends Genet. 1993;9:75–79. doi: 10.1016/0168-9525(93)90227-9. [DOI] [PubMed] [Google Scholar]
- 20.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 20a.Kuzin, A. Unpublished results.
- 21.Kuzin B, Tillib S, Sedkov Y, Mizrokhi L, Mazo A. The Drosophila trithorax gene encodes a chromosomal protein and directly regulates the region-specific homeotic gene fork head. Genes Dev. 1994;8:2478–2490. doi: 10.1101/gad.8.20.2478. [DOI] [PubMed] [Google Scholar]
- 22.Martin C H, Mayeda C A, Davis C A, Ericsson C L, Knafels J D, Mathog D R, Celniker S E, Lewis E B, Palazzolo M J. Complete sequence of the bithorax complex of Drosophila. Proc Natl Acad Sci USA. 1995;92:8398–8402. doi: 10.1073/pnas.92.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin E C, Adler P N. The Polycomb group gene Posterior Sex Combs encodes a chromosomal protein. Development. 1993;117:641–655. doi: 10.1242/dev.117.2.641. [DOI] [PubMed] [Google Scholar]
- 24.Mazo A M, Huang D H, Mozer B A, Dawid I B. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc Natl Acad Sci USA. 1990;87:2112–2116. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moehrle A, Paro R. Spreading the silence: epigenetic transcriptional regulation during Drosophila development. Dev Genet. 1994;15:478–484. doi: 10.1002/dvg.1020150606. [DOI] [PubMed] [Google Scholar]
- 26.Mullen J R, DiNardo S. Establishing parasegments in Drosophila embryos: roles of the odd-skipped and naked genes. Dev Biol. 1995;169:295–308. doi: 10.1006/dbio.1995.1145. [DOI] [PubMed] [Google Scholar]
- 27.Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- 28.Peterson C L, Tamkun J W. The SWI/SNF complex: a chromatin remodeling machine. Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 29.Pirrotta V. PcG complexes and chromatin silencing. Curr Opin Genet Dev. 1997;7:249–258. doi: 10.1016/s0959-437x(97)80135-9. [DOI] [PubMed] [Google Scholar]
- 30.Poux S, Kostic C, Pirrotta V. Hunchback-independent silencing of late Ubx enhancers by a Polycomb group response element. EMBO J. 1996;17:4713–4722. [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad R, Yano T, Sorio C, Nakamura T, Rallapalli R, Gu Y, Leshkowitz D, Croce C M, Canaani E. Domains with transcriptional regulatory activity within the ALL1 and AF4 proteins involved in acute leukemia. Proc Natl Acad Sci USA. 1995;92:12160–12164. doi: 10.1073/pnas.92.26.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quin S, Capovilla M, Pirrotta V. Molecular mechanisms of pattern formation by the BRE enhancer of the Ubx gene. EMBO J. 1993;12:3865–3877. doi: 10.1002/j.1460-2075.1993.tb06065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rastelli L, Chan C S, Pirotta V. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on enhancer of zeste function. EMBO J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozenblatt-Rosen O, Rozovskaia T, Buracov D, Sedkov Y, Tillib S, Blechman J, Croce C, Mazo A, Canaani E. The C-terminal SET domains of ALL-1 and Trithorax interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci USA. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saunders R D C, Glover D M, Ashburner M, Siden-Kiamos I, Louis C, Monastirioti M, Savakis C, Kafatos F. PCR amplification of DNA microdissected from a single polytene chromosome band: a comparison with conventional microcloning. Nucleic Acids Res. 1989;17:9027–9037. doi: 10.1093/nar/17.22.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sedkov Y, Tillib S, Mizrokhi L, Mazo A. The bithorax complex is regulated by trithorax earlier during Drosophila embryogenesis than is the Antennapedia complex, correlating with a bithorax-like expression pattern of distinct early trithorax transcripts. Development. 1994;120:1907–1917. doi: 10.1242/dev.120.7.1907. [DOI] [PubMed] [Google Scholar]
- 37.Shearn A. The ash-1, ash-2 and trithorax genes of Drosophila melanogaster are functionally related. Genetics. 1989;121:517–525. doi: 10.1093/genetics/121.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 39.Simon J, Chiang A, Bender W, Shimmel M J, O’Connor M. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev Biol. 1993;158:131–144. doi: 10.1006/dbio.1993.1174. [DOI] [PubMed] [Google Scholar]
- 40.Simon J, Peifer M, Bender W, O’Connor M. Regulatory elements of the bithorax complex that control expression along the anterior-posterior axis. EMBO J. 1990;9:3945–3956. doi: 10.1002/j.1460-2075.1990.tb07615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soeller W C, Poole S J, Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 1988;2:68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- 42.Spradling A C. P element-mediated transformation. In: Roberts D B, editor. Drosophila: a practical approach. Washington, D.C: IRL Press; 1986. pp. 175–197. [Google Scholar]
- 43.Strutt H, Cavalli G, Paro R. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamkun J W. The role of brahma and related proteins in transcription and development. Curr Opin Genet Dev. 1995;5:473–477. doi: 10.1016/0959-437x(95)90051-h. [DOI] [PubMed] [Google Scholar]
- 45.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 46.Thummel C S, Boulet A M, Lipshitz H D. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- 47.Tripoulas N, LaJeunesse D, Gildea J, Shearn A. The Drosophila ash1 gene product, which is localized at specific sites on chromosomes, contains a SET domain and a PHD finger. Genetics. 1996;143:913–928. doi: 10.1093/genetics/143.2.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 KDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 49.Tsukiyama T, Wu C. Chromatin remodeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 50.Wang W, Côte J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Gaff S P, Yaniv M, Workman J L, Crabtree G R. Purification to biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5380. [PMC free article] [PubMed] [Google Scholar]
- 51.Zeleznik-Le N J, Harden A M, Rowley J D. 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc Natl Acad Sci USA. 1994;91:10610–10614. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zink D, Paro R. Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J. 1995;14:5660–5671. doi: 10.1002/j.1460-2075.1995.tb00253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]