Abstract
The importance of long noncoding RNAs (lncRNAs) in the oncogenicity of hepatocellular carcinoma (HCC) has been widely studied. However, the detailed functions of ZSCAN16 antisense RNA 1 (ZSCAN16-AS1) have seldom been explored in HCC until the present study. In the present study, experiments were performed to clarify whether ZSCAN16-AS1 is implicated in the oncogenesis and progression of HCC and to explore the possible underlying mechanisms. ZSCAN16-AS1 expression was analyzed using reverse transcription-quantitative PCR. The effects of ZSCAN16-AS1 on the biological behavior of HCC cells were demonstrated by functional experiments. The direct binding capacity of ZSCAN16-AS1 with microRNA-451a (miR-451a) was indicated by the luciferase reporter assay and RNA immunoprecipitation. The high expression of ZSCAN16-AS1 was confirmed in HCC by The Cancer Genome Atlas database and the cohort of the present study. Survival data revealed that patients with a high ZSCAN16-AS1 level had worse prognosis compared with those with a low ZSCAN16-AS1 level. Following ZSCAN16-AS1 knockdown, HCC cell proliferation, migration and invasion were curbed, whereas cell apoptosis was promoted in vitro. The absence of ZSCAN16-AS1 restricted tumor growth of HCC cells in vivo. Mechanistically, ZSCAN16-AS1 acted as a competing endogenous RNA by decoying miR-451a in HCC cells. Furthermore, activating transcription factor 2 (ATF2), a direct target of miR-451a, was under the regulation of ZSCAN16-AS1, which was exerted by sequestering miR-451a. In addition, miR-451a knockdown or ATF2 resumption reversed the proliferation suppression, apoptosis promotion and migration and invasion inhibition triggered by ZSCAN16-AS1 silencing. In conclusion, ZSCAN16-AS1, a pro-oncogenic lncRNA, aggravated the malignancy of HCC by controlling the miR-451a/ATF2 axis. An understanding of the competing endogenous RNA network of ZSCAN16-AS1/miR-451a/ATF2 in HCC might be instrumental in the development of attractive targets for molecular therapy.
Keywords: competing endogenous RNA, long noncoding RNA, hepatocellular carcinoma, microRNA
Introduction
Hepatocellular carcinoma (HCC), which is the primary subtype of liver cancer, is the sixth most frequent malignant tumor and the second deadliest of all types of cancer globally (1). HCC is characterized by aggressive behavior, high morbidity, easily occurring metastasis and resistance to currently available chemotherapeutic drugs (2). In 2015, 466,000 new HCC cases were diagnosed in China, of which 422,000 patients succumbed to fatal tumors (3). Currently, surgical excision is a treatment option for patients with early-stage HCC, but the vast majority of cases are diagnosed at an advanced stage, at which point the curative effect of radiochemotherapy is poor (4). Over the last decade, the clinical outcomes of HCC have substantially improved due to the significant progress in diagnostic and therapeutic techniques (5). However, even when receiving first-line anticancer therapies, >50% of patients with HCC will experience tumor relapse, including intrahepatic and distant metastasis, which results in poor prognosis (6). Thus, research focusing on the mechanisms modulating HCC metastasis and aggressiveness are urgently needed for the development of attractive therapeutic targets.
In the human genome, only 2% of the RNA can be translated into protein and the residual RNAs lack protein-coding ability and are thus termed noncoding RNAs (7). Long noncoding RNAs (lncRNAs) are defined as a form of noncoding RNA transcripts that are >200 nucleotides in length (8). Studies have focused on the important functions of lncRNAs in cancer research (9–11). Differentially expressed lncRNAs have been observed in human cancers and contribute to various aspects of the biological activity of tumors (12,13). In HCC, a number of lncRNAs are reported to be dysregulated and regulate anti- or pro-oncogenic activities during hepatocarcinogenesis and cancer progression (14,15).
MicroRNAs (miRNAs) are another type of noncoding RNA transcript that also exerts nonprotein-coding capacity (16). miRNAs can bind directly to the 3′-untranslated region (UTR) of downstream target genes and consequently inhibit translation or mRNA degradation, thereby controlling gene expression at the posttranslational level (17). Notably, lncRNAs are capable of interacting with miRNAs and thus implementing their functions (18). As gene regulators, lncRNAs can work as competing endogenous RNAs (ceRNAs) by decoying or competing with shared miRNAs, thereby lowering the miRNA-induced inhibition of gene expression (19). Therefore, studying lncRNAs and miRNAs may facilitate research on HCC diagnosis and anticancer treatments.
The association of lncRNAs with HCC has been widely reported (20–22), yet detailed functions of ZSCAN16-AS1 have seldom been unraveled in HCC. The present study examined the expression pattern of ZSCAN16-AS1 in HCC. To address the functions of ZSCAN16-AS1 in HCC, loss-of-function assays were implemented in vitro and in vivo. In addition, the possible molecular events through which ZSCAN16-AS1 exerts its tumorigenic actions in HCC were demonstrated.
Materials and methods
Clinical specimens and cell lines
The present study was approved by the Ethics Committee of The People's Hospital of Tongliang District (approval no. EC.2015-39). All patients provided informed written consent for the use of their tissue samples. A total of 47 pairs of HCC tissues and adjacent normal tissues (distance, 2 cm) were obtained from patients (31 males and 16 females; age range, 41–68 years) in The People's Hospital of Tongliang District between May 2015 and November 2016. Following tumor excision, the tumors were quickly stored in liquid nitrogen. The patients had not received chemotherapy, radiotherapy, or other anticancer treatments.
The transformed human liver epithelial-3 (THLE-3) cell line and two HCC cell lines (Hep3B and HuH7) were acquired from the Cell Bank of the Chinese Academy of Sciences. THLE-3 was cultured in bronchial epithelial basal medium (BEGM; Clonetics Corporation), to which 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 5 ng/ml EGF and 70 ng/ml phosphoethanolamine were added. Hep3B and HuH7 cells were grown in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS, 1% GlutaMAX, 1% nonessential amino acids and 1% penicillin/streptomycin. An additional 100 mM 1% sodium pyruvate solution was added to the Hep3B cell culture.
In addition, two other HCC cell lines, SNU-398 and SNU-182, were purchased from the ATCC. SNU-398 cells were maintained in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS and 1% penicillin/streptomycin. RPMI-1640 with 10% FBS, 1% GlutaMAX, 1% nonessential amino acids and 1% penicillin/streptomycin were applied to SNU-182 cells. All cells were kept in a humidified atmosphere equipped with 5% CO2 at 37°C.
Cell transfection
Small interfering RNAs (siRNAs) targeted to silence ZSCAN16-AS1 (si-ZSCAN16-AS1s) and nontargeted siRNA (si-NC) were designed and synthesized by Shanghai GenePharma Co., Ltd. The si-ZSCAN16-AS1#1 sequence was 5′-TTGTAAAATTGAAATATTTGAAT-3′; the si-ZSCAN16-AS1#2 sequence was 5′-TACCAAAAAATAAAAATATGAAC-3′; the si-ZSCAN16-AS1#3 sequence was 5′-GGCATACTTAGTTTTACATTTTT-3′; and the si-NC sequence was 5′-CACGATAAGACAATGTATTT-3′. miR-451a mimic and miR-451a inhibitors were obtained from Guangzhou RiboBio Co., Ltd., with miRNA mimic control (NC mimic) and miRNA inhibitor control (NC inhibitor) as the controls. The miR-451a mimic sequence was 5′-UUGAGUCAUUACCAUUGCCAAA-3′ and the NC mimic sequence was 5′-UUGUACUACACAAAAGUACUG-3′. The miR-451a inhibitor sequence was 5′-AACUCAGUAAUGGUAACGGUUU-3′ and the NC inhibitor sequence was 5′-ACUACUGAGUGACAGUAGA-3′. The cDNA encoding the ATF2 coding sequence was amplified by Shanghai GenePharma Co., Ltd., and was inserted into the pcDNA3.1 plasmid, thereby generating the pcDNA3.1-ATF2 plasmid. The empty pcDNA3.1 plasmid was applied as the control. Cells were transfected with siRNAs (100 pmol), miRNA mimic (100 pmol), miRNA inhibitor (100 pmol) or plasmid (4 µg) utilizing Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). After 6 h incubation with transfection reagent at 37°C, the medium was replaced with fresh medium. Then, 48 h after transfection, reverse transcription-quantitative (RT-q) PCR, cell apoptosis detection by flow cytometry, Transwell cell migration and invasion assays and western blotting were performed. A Cell Counting Kit-8 (CCK-8) assay was implemented at 24 h post-transfection.
RT-qPCR
Tissues or cells (2×106) were immersed in Beyozol (Beyotime Institute of Biotechnology) for total RNA extraction. To quantify miRNA expression, reverse transcription was performed according to the manufacturer's instructions, using a miRcute miRNA First-Strand cDNA Synthesis kit followed by PCR amplification using the miRcute miRNA qPCR Detection kit SYBR-Green (both from Tiangen Biotech Co., Ltd.). U6 small nuclear RNA served as an endogenous control to analyze miRNA expression. To analyze ZSCAN16-AS1 and ATF2 expression, a PrimeScript RT Reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd.) was used to carry out reverse transcription. The obtained cDNA was subjected to TB Green Premix Ex Taq™ II (Takara Biotechnology Co., Ltd.) to perform qPCR. The thermocycling conditions were as follows: Initial denaturation at 95°C for 30 sec, followed by 40 cycles at 95°C for 3 sec, 60°C for 30 sec and 72°C for 30 sec. Relative ZSCAN16-AS1 and ATF2 expression was normalized to that of GAPDH. The 2−ΔΔCq method (23) was used to process all data. RNA extraction, cDNA synthesis, and qPCR were performed according to the manufacturer's protocols. These experiments were repeated three times.
The primers were as follows: ZSCAN16-AS1, 5′-GGGCTGCAATAAAACAGCAAA-3′ (forward) and 5′-CAATTTCCTATCCCGACCCTCT-3′ (reverse); ATF2, 5′-CAGGAACTGTTCTAGCACCAGC-3′ (forward) and 5′-CAGGAGTTTCAGGCTGCAGTAA-3′ (reverse); GAPDH, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ (forward) and 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ (forward); miR-23c, 5′-TCGGCAGGGGGTAATCACTGG-3′ (forward) and 5′-CACTCAACTGGTGTCGTGGA-3′ (reverse); miR-23b-3p, 5′-TCGGCAGGGGAAATCCCTGG-3′ (forward) and 5′-CACTCAACTGGTGTCGTGGA-3′ (reverse); miR-130a-5p, 5′-TCGGCAGGGCUCUUUUCACAUU-3′ (forward) and 5′-CACTCAACTGGTGTCGTGGA-3′ (reverse); miR-181a-5p, 5′-TCGGCAGGAACAUUCAACGCUG-3′ (forward) and 5′-CACTCAACTGGTGTCGTGGA-3′ (reverse); miR-181c-5p, 5′-TCGGCAGGAACAUUCAACCUG-3′ (forward) and 5′-CACTCAACTGGTGTCGTGGA-3′ (reverse); miR-4524a-5p, 5′-TCGGCAGGAUAGCAGCAUGAAC-3′ (forward) and 5′-CACTCAACTGGTGTCGTGGA-3′ (reverse); miR-451a, 5′-TCGGCAGGAAACCGUUACCAUU-3′ (forward) and 5′-CACTCAACTGGTGTCGTGGA-3′ (reverse); miR-146a-5p, 5′-TCGGCAGGUGAGAACUGAAUUC-3′ (forward) and 5′-CACTCAACTGGTGTCGTGGA-3′ (reverse); miR-22-3p, 5′-TCGGCAGGACAGTTCTTCAACT-3′ (forward) and 5′-CACTCAACTGGTGTCGTGGA-3′ (reverse); and U6, 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse).
CCK-8 assay
Transfected cells were harvested to prepare a cell suspension. A 100 µl volume of cell suspension carrying 2×103 cells was then transferred to 96-well plates, followed by cultivation for different time points (0, 24, 48 and 72 h) in an incubator. At different time points (0, 24, 48 and 72 h), 10 µl of CCK-8 reagent (Dojindo Molecular Technologies, Inc.) was employed to treat the cells at 37°C for another 2 h. Finally, the absorbance at 450 nm was measured using a microplate reader.
Cell apoptosis detection by flow cytometry
The collected cells were transferred to flow tubes. Following centrifugation at 4°C at 1,000 × g for 5 min, the supernatant fluid was removed and transfected cells were resuspended in 195 µl of Annexin V-FITC binding buffer from the Annexin V-FITC Apoptosis Detection kit (Beyotime Institute of Biotechnology). Immediately, 5 µl of Annexin V-FITC and 10 µl of PI were added to the cell suspension, followed by 15 min of cultivation without light. A flow cytometer (FACScan, BD Biosciences) was used to analyze cell apoptosis. Data were analyzed using CellQuest software v.2.9 (BD Biosciences). The percentage of early + late apoptotic cells was calculated.
Transwell cell migration and invasion assays
The harvested cells were centrifuged at room temperature at 1,000 × g for 15 min and resuspended in culture medium without serum. The cell concentration was adjusted to 2.5×105 cells/ml. Transwell inserts (8-µm filter; BD Biosciences) that were precoated with Matrigel (BD Biosciences) were used for the invasion test, whereas the migration test was performed without Matrigel. Precoating was performed at 37°C for 2 h. A total of 200 µl of cell suspension was seeded into the upper chambers, while 20% FBS-contained 600 µl of culture medium was applied to the lower chambers as a chemoattractant. One day later, the cells remaining in the upper chambers were cleaned with a cotton bud. The cells that passed through the pores were fixed in 4% paraformaldehyde at room temperature for 30 min and stained with 0.1% crystal violet at room temperature for 30 min. Images of migrated or invaded cells were obtained and quantified under an optical light microscope (magnification, ×200). A total of five fields were randomly selected and the number of migrated/invaded cells was counted.
Tumor xenograft model
Experiments involving animals were implemented under the approval of the Institutional Animal Care and Use Committee of The People's Hospital of Tongliang District. A 2nd lentiviral system was used in the production of lentiviruses. Short hairpin RNA (shRNA) targeting ZSCAN16-AS1 (sh-ZSCAN16-AS1) and the negative control shRNA (sh-NC) were acquired from GenePharma Inc. The sh-ZSCAN16-AS1 sequence was 5′-CCGGTTGTAAAATTGAAATATTTGAATCTCGAGATTCAAATATTTCAATTTTACAATTTTTG-3′ and sh-NC sequence was 5′-CCGGCACGATAAGACAATGTATTTCTCGAGAAATACATTGTCTTATCGTGTTTTTG-3′. Following insertion into the pLKO.1 vector (Addgene Inc.), they were transfected into 239T cells (Cell Bank of the Chinese Academy of Sciences) in the presence of lentiviral packaging plasmid psPAX2 and envelope expression plasmid pMD2.G (both from Addgene Inc.). The transfection was implemented using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The proportion of lentiviral plasmid: pLKO.1: psPAX2: pMD2.G was 2:1:1 and 30 µg plasmids were employed in lentivirus packaging. Following 5 h incubation at 37°C with 5% CO2, the culture medium was discarded and cells were maintained in fresh Dulbecco's modified Eagle medium that was supplemented with 10% FBS, 1% glutamax, 1% non-essential amino acids and 1% sodium pyruvate solution (all from Gibco; Thermo Fisher Scientific, Inc.). Lentiviruses expressing sh-ZSCAN16-AS1 or sh-NC were harvested via ultracentrifugation at 4°C at 1,000 × g for 2 h and then mixed with polybrene (5 µg/ml; Sigma-Aldrich; Merck KGaA) and Dulbecco's modified Eagle's medium. Following injection into HuH7 cells with a multiplicity of infection 5, puromycin was applied to select HuH7 cells with stable ZSCAN16-AS1 ablation.
For subcutaneous injection, a total of 2×106 sh-ZSCAN16-AS1- or sh-NC-transfected HuH7 cells were resuspended in 100 µl phosphate buffer saline and implanted into the flank of 4- to 5-week-old male BALB/c nude mice (n=6; mean weight, 20.4 g), which were obtained from the Shanghai SLAC Laboratory Animal Co., Ltd. All mice were housed under specific pathogen-free conditions at 25°C and 50% humidity, with a 10:14 light/dark cycle and ad libitum access to food and water. The tumor size was examined weekly. After four weeks, all mice were euthanized by cervical dislocation and the tumor xenografts were excised, imaged and weighed. The volume of the tumor xenografts was calculated using the following formula: volume=(length × width2)/2.
Subcellular fractionation assay
Cytoplasmic and nuclear fractions of HCC cells were separated via a Cytoplasmic and Nuclear RNA Purification kit (Norgen Biotek Corp.). RNA was then analyzed with RT-qPCR to calculate the relative distribution of ZSCAN16-AS1 in HCC cells.
Bioinformatics prediction
StarBase 3.0 (http://starbase.sysu.edu.cn/) was applied for the prediction of direct binding between ZSCAN16-AS1 and miR-451a. TargetScan (http://www.targetscan.org), miRDB (http://mirdb.org/) and StarBase 3.0 were used to determine the target genes of miR-451a.
Luciferase reporter assay
ZSCAN16-AS1 and ATF2 fragments carrying the miR-451a binding site were amplified before they were inserted into the pMIR-luciferase reporter plasmid (Promega Corporation). The resulting luciferase reporter plasmids were labeled as ZSCAN16-AS1-wild-type (wt) and ATF2-wt. The luciferase reporter plasmids that contained a mutant (mut) miR-451a binding site were synthesized following the same experimental steps and the resulting plasmids were labeled ZSCAN16-AS1-mut and ATF2-mut. HCC cells were transfected with miR-451a mimic or NC mimic in parallel with the luciferase reporter plasmids utilizing Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h of cultivation, a dual-luciferase reporter assay system (Promega Corporation) was adopted for the assessment of luciferase activity.
RNA immunoprecipitation (RIP)
The Magna RIP RNA-Binding Protein Immunoprecipitation kit (EMD Millipore) was applied in the assay. In brief, 1×106 HCC cells were harvested and cultivated in complete RIP lysis buffer to obtain whole-cell extracts. Then, RIP buffer supplemented with magnetic beads that were conjugated with human anti-Ago2 or anti-IgG antibodies (Millipore) was utilized to incubate the cell extract overnight at 4°C. An input control was assayed simultaneously to function as the positive control, while IgG served as the negative control. Magnetic beads were collected via centrifugation at 4°C at 1,000 × g for 5 min and probed with proteinase K to remove the protein. After extracting the immunoprecipitated RNA, the relative enrichment of ZSCAN16-AS1, miR-451a and ATF2 was detected by RT-qPCR.
Western blot analysis
Total protein from the cultured cells was extracted by lysis in cell lysis buffer for western blotting and immunoprecipitation (IP; Beyotime Institute of Biotechnology) and quantified using an Enhanced BCA Protein Assay kit (Beyotime Institute of Biotechnology). Protein was added to the loading buffer and each well was loaded with equal amounts of protein (30 µg/lane). The separated proteins were transferred to PVDF membranes and then subjected to 10% SDS-PAGE electrophoresis. After that, the membranes were sealed with 5% defatted milk at room temperature for 2 h, after which the membranes underwent 12 h incubation at 4°C with primary antibodies specifically binding to ATF2 (cat. no. ab239361; 1:1,000) or GAPDH (ab181602; 1:1,000; Abcam). After 2 h of cultivation at room temperature with horseradish peroxidase-labelled secondary antibody (cat. no. ab205718; 1:5,000; Abcam), the signals were detected with a BeyoECL Plus kit (Beyotime Institute of Biotechnology). Quantity One software version 4.62 (Bio-Rad Laboratories, Inc.) was adopted for densitometry.
Statistical analysis
Data were calculated from three biological repeats of each assay and presented as the mean ± standard deviation. Paired Student's t-test was used to compare ZSCAN16-AS1, miR-451a and ATF2 expression between HCC tumor tissues and normal tissues. Significant differences between other two groups were detected by unpaired Student's t-test. One-way analysis of variance followed by Tukey's test was used to compare the differences among multiple groups. Pearson's correlation test was used to determine the expression correlations among ZSCAN16-AS1, miR-451a and ATF2. The overall survival curves were plotted utilizing the Kaplan-Meier method and were compared using the log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
The upregulation of ZSCAN16-AS1 indicates a poor prognosis for HCC
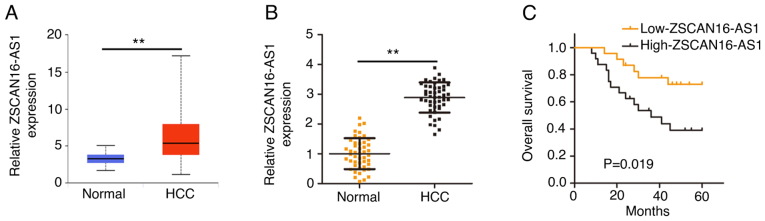
To comprehensively disclose the expression of ZSCAN16-AS1, its expression in HCC was first analyzed using The Cancer Genome Atlas (TCGA) database. ZSCAN16-AS1 level in HCC tumor tissues was clearly increased compared with normal tissues (Fig. 1A). RT-qPCR was implemented to further confirm ZSCAN16-AS1 expression in HCC tissues and matched adjacent normal tissues (n=47). Notably high ZSCAN16-AS1 level was validated in HCC tissues (Fig. 1B). In addition, all individuals were divided into low-ZSCAN16-AS1 or high-ZSCAN16-AS1 expression groups according to the median value of ZSCAN16-AS1 expression in the 47 HCC tissues. Patients in the high-ZSCAN16-AS1 expression group displayed shorter overall survival rates than patients in the low-ZSCAN16-AS1 expression group (Fig. 1C).
Figure 1.
Overexpressed ZSCAN16-AS1 indicates poor prognosis for patients with HCC. (A) ZSCAN16-AS1 expression in HCC was analyzed based on The Cancer Genome Atlas database. (B) The measurement of ZSCAN16-AS1 expression in HCC tissues was conducted applying reverse transcription-quantitative PCR. (C) Kaplan-Meier method was used to examine the relationship between ZSCAN16-AS1 expression and overall survival in patients with HCC. **P<0.01. HCC, hepatocellular carcinoma.
ZSCAN16-AS1 knockdown exerts inhibitory effects on the malignant processes of HCC cells
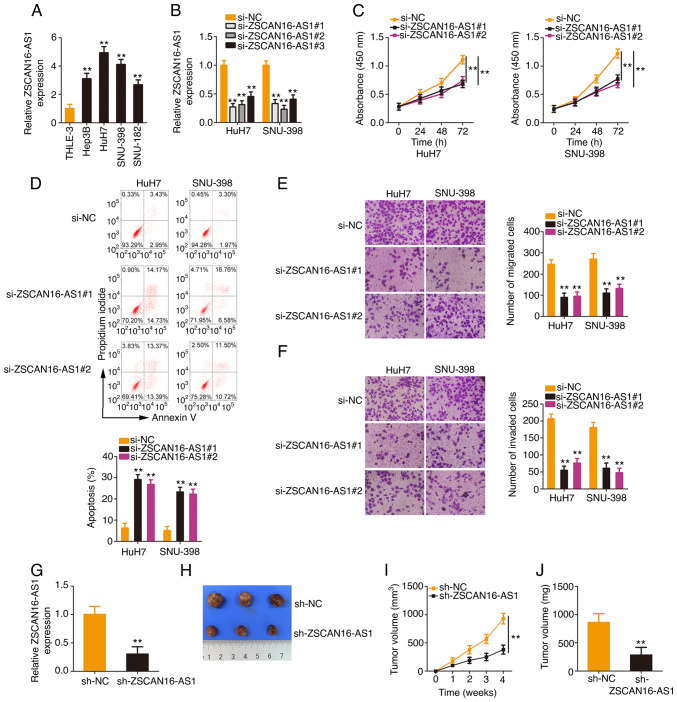
To explore the biological actions of ZSCAN16-AS1, ZSCAN16-AS1 expression in HCC cell lines was measured. All four HCC cell lines presented higher ZSCAN16-AS1 expression levels, especially in the HuH7 and SNU-398 cell lines (Fig. 2A). Therefore, they were selected for use in following experiments. ZSCAN16-AS1 expression was effectively silenced in HuH7 and SNU-398 cells after transfection with si-ZSCAN16-AS1 (Fig. 2B). To avoid off-target effects, two siRNAs, si-ZSCAN16-AS1#1 and si-ZSCAN16-AS1#2, were employed in the loss-of-function assays. Functionally, the CCK-8 assay confirmed that the proliferation of HCC cells was clearly hindered after ZSCAN16-AS1 depletion (Fig. 2C). Additionally, ZSCAN16-AS1-silenced HCC cells showed more apoptosis than the si-NC group (Fig. 2D). Furthermore, the number of migrated (Fig. 2E) and invaded (Fig. 2F) cells in si-ZSCAN16-AS1-transfected HCC cells was clearly less than that in the si-NC-transfected cells, which indicated that ZSCAN16-AS1 depletion impaired the migratory and invasive capacities of HCC cells. In addition, data obtained from the tumor xenograft model revealed that the tumor xenografts originating from HuH7 cells with stable ZSCAN16-AS1 depletion (Fig. 2G) were smaller (Fig. 2H and I) and lighter (Fig. 2J) compared with those of the sh-NC group. Altogether, ZSCAN16-AS1 exhibited oncogenic properties that promote malignant behaviors in HCC cells.
Figure 2.
The loss of ZSCAN16-AS1 impedes the malignant properties of HCC cells. (A) ZSCAN16-AS1 level in different HCC cell lines was measured by RT-qPCR. (B) HCC cells were silenced by transfection with si-ZSCAN16-AS1, which was confirmed by RT-qPCR. (C) A CCK-8 assay was used to measure the proliferation of ZSCAN16-AS1-silenced HCC cells. (D) Apoptosis of si-ZSCAN16-AS1-transfected HCC cells. Transwell cell (E) migration and (F) invasion assays presented a change in the motility of HCC cells after ZSCAN16-AS1 knockdown (magnification, ×200). (G) HuH7 cells were stably transfected with sh-ZSCAN16-AS1 and then subjected to RT-qPCR analysis to detect the transfection efficacy. (H) Representative pictures show the tumor xenografts. (I) Nude mice were injected with HuH7 cells with stable sh-ZSCAN16-AS1 or sh-NC expression. The growth curves were plotted utilizing the monitored tumor width and length. (J) Weight of the tumor xenografts. **P<0.01. HCC, hepatocellular carcinoma; RT-qPCR, reverse transcription-quantitative PCR; sh, short hairpin; NC, negative control.
ZSCAN16-AS1 is a ceRNA and operates as a molecular sponge for miR-451a in HCC
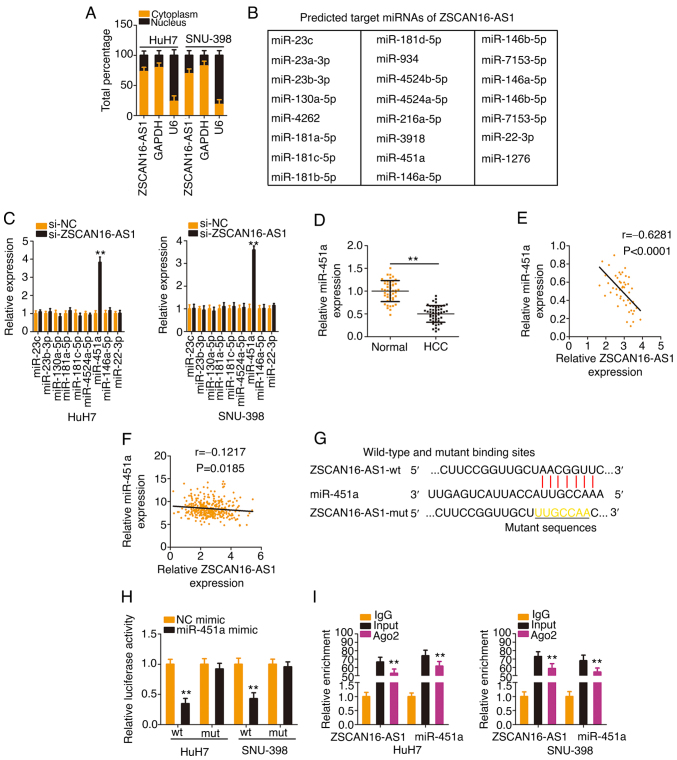
To investigate the mechanisms underlying the oncogenic actions of ZSCAN16-AS1, a subcellular fractionation assay was applied to separate the cytoplasmic and nuclear fractions of HCC cells and RT-qPCR was then performed to assess the distribution characteristics of ZSCAN16-AS1, which was validated to be located mostly in the cytoplasm of HCC cells (Fig. 3A). Given the localization of ZSCAN16-AS1 in HCC, it was inferred that ZSCAN16-AS1 may perform tumor-promoting roles via a ceRNA. Using StarBase 3.0, 23 miRNAs (Fig. 3B) were predicted to be targets of ZSCAN16-AS1. Notably, by comparing the 23 miRNAs with those dysregulated miRNAs in the TCGA-HCC database, miR-23c, miR-23b-3p, miR-130a-5p, miR-181a-5p, miR-181c-5p, miR-4524a-5p, miR-451a, miR-146a-5p and miR-22-3p were found to be decreased in HCC. RT-qPCR was then performed to examine the regulatory actions of ZSCAN16-AS1 on these candidates in HCC cells. As Fig. 3C shows, downregulation of ZSCAN16-AS1 evidently increased miR-451a expression, whereas the other miRNAs were unaffected. Decreased miR-451a expression was identified in HCC (Fig. 3D) and presented an inverse correlation with ZSCAN16-AS1 expression (Fig. 3E). Using TCGA database, an inverse correlation was observed between ZSCAN16-AS1 and miR-451a in HCC (Fig. 3F). Furthermore, the direct binding between ZSCAN16-AS1 and miR-451a (Fig. 3G) was corroborated by RIP and luciferase reporter assays. Overexpressed miR-451a reduced the luciferase activity of the ZSCAN16-AS1-wt reporter plasmid; nevertheless, mutation of the binding site abolished the inhibitory action (Fig. 3H). The outcomes of the RIP assay showed that ZSCAN16-AS1 and miR-451a were enriched in Ago2-containing immunoprecipitated RNA in contrast to that of the IgG control (Fig. 3I). These results suggest that ZSCAN16-AS1 worked as a miR-451a sponge in HCC cells.
Figure 3.
ZSCAN16-AS1 acts as a natural miR-451a sponge in HCC. (A) Relative abundances of ZSCAN16-AS1 in the cytoplasmic and nuclear fractions of HCC cells. (B) The putative targets of ZSCAN16-AS1 predicted by StarBase 3.0. (C) The levels of miR-23c, miR-23b-3p, miR-130a-5p, miR-181a-5p, miR-181c-5p, miR-4524a-5p, miR-451a, miR-146a-5p and miR-22-3p were detected in HCC cells following ZSCAN16-AS1 knockdown. (D) miR-451a expression was measured by reverse transcription-quantitative PCR in HCC tissues. (E) The relation between ZSCAN16-AS1 and miR-451a levels in 47 HCC tissues. (F) The expression correlation between ZSCAN16-AS1 and miR-451a in HCC was examined using The Cancer Genome Atlas database. (G) The predicted complementary sequence between ZSCAN16-AS1 and miR-451a. (H) Luciferase activities of ZSCAN16-AS1-wt or ZSCAN16-AS1-mut were determined in the presence of miR-451a mimic or NC mimic. (I) RNA immunoprecipitation assay was implemented to evaluate ZSCAN16-AS1 and miR-451a enrichment in immunoprecipitants of HCC cells. **P<0.01. miR, microRNA; HCC, hepatocellular carcinoma; wt, wild-type; mut, mutant; NC, negative control.
miR-451a exerts anti-oncogenic actions in HCC
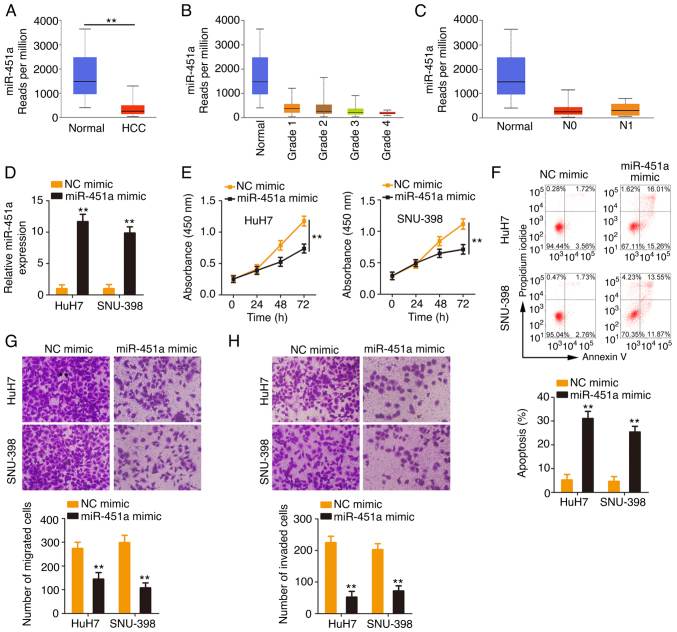
Considering the downregulation of miR-451a in HCC, the present study next addressed its clinical value and detailed its roles in regulating the malignant behaviors of HCC cells. As shown in the TCGA database, low miR-451a expression (Fig. 4A) was closely correlated with the tumor grade and lymph node metastasis (Fig. 4B and C) in HCC. miR-451a was overexpressed in HCC cells after miR-451a mimic transfection (Fig. 4D). Ectopic miR-451a expression inhibited the proliferative ability of HCC cells (Fig. 4E). In addition, the percentage of apoptotic HCC cells was significantly increased by miR-451a mimic transfection (Fig. 4F). Furthermore, HCC cells transfected with miR-451a mimic manifested lower migratory and invasive (Fig. 4G and H) abilities than the NC mimic control group. Overall, miR-451a exerted anti-oncogenic roles in HCC.
Figure 4.
Enforced miR-451a expression exerts tumor-inhibiting roles in HCC cells. (A) miR-451a expression in HCC was analyzed based on TCGA database. (B) The correlation between miR-451a expression and tumor grade in the TCGA-HCC database. (C) Correlation between miR-451a expression and lymph node metastasis in the TCGA-HCC database. (D) Following transfection of HCC cells with miR-451a mimic or NC mimic, reverse transcription-quantitative PCR was conducted to evaluate the transfection efficacy. (E) Proliferation and (F) apoptosis of miR-451a-upregulated HCC cells. (G and H) The motility (magnification, ×200) of miR-451a mimic-transfected HCC cells was examined. **P<0.01. miR, microRNA; HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; NC, negative control.
ATF2 is under the regulation of the ZSCAN16-AS1/miR-451a axis in HCC
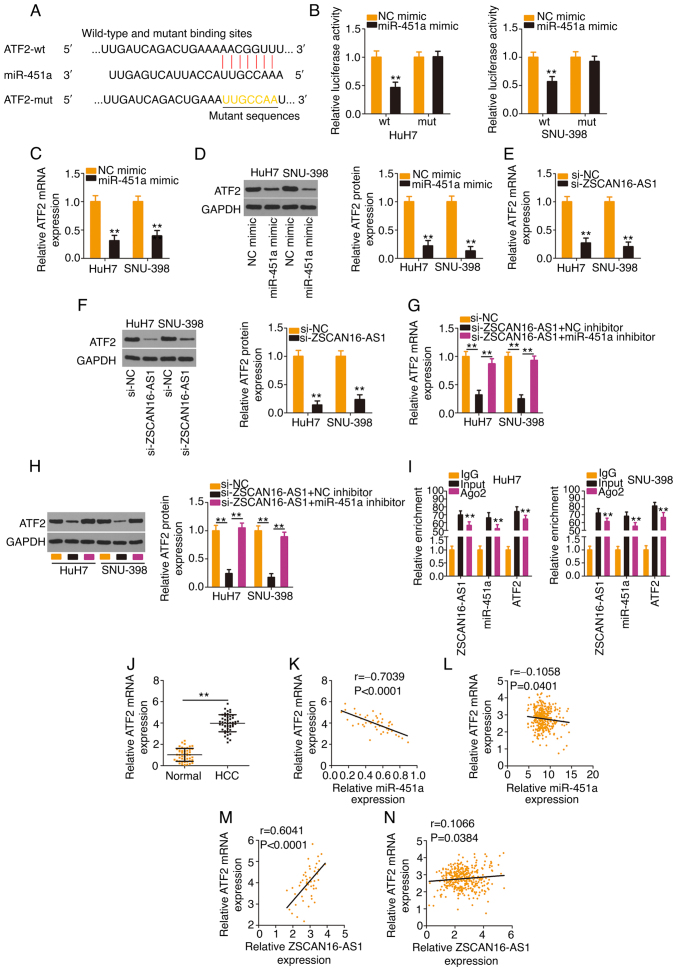
Using bioinformatics analysis, a potential binding site of miR-451a was identified in the ATF2 3′-UTR (Fig. 5A). The results of the luciferase reporter assay ascertained that transfection with the miR-451a mimic weakened the activity of the ZSCAN16-AS1-wt reporter plasmid but not that of the reporter plasmid ZSCAN16-AS1-mut in HCC cells (Fig. 5B). In addition, enforced miR-451a expression appeared to lower ATF2 levels (Fig. 5C and D) in HCC cells. After identifying ATF2 as a direct target of miR-451a, subsequent experiments were implemented to explore whether a ceRNA pathway consisting of ZSCAN16-AS1, miR-451a and ATF2 exists in HCC. The data revealed that ATF2 expression (Fig. 5E and F) was decreased in ZSCAN16-AS1-deficient HCC cells but was restored by cotransfection with the miR-451a inhibitor (Fig. 5G and H). Furthermore, ZSCAN16-AS1, miR-451a and ATF2 were all enriched in Ago2-containing immunoprecipitated RNA compared with those of the IgG control (Fig. 5I). A higher ATF2 level was detected in HCC tissues compared with adjacent normal tissues (Fig. 5J). In addition, a negative correlation between ATF2 and miR-451a was confirmed in the cohort of the present study (Fig. 5K) and TCGA database (Fig. 5L). A positive correlation between ATF2 and ZSCAN16-AS1 was identified in the 47 HCC tissues (Fig. 5M) and TCGA database (Fig. 5N). Thus, ZSCAN16-AS1 is a ceRNA that sequesters miR-451a and consequently positively regulates ATF2 expression in HCC.
Figure 5.
The ZSCAN16-AS1/miR-451a axis regulates ATF2 expression in HCC cells. (A) The predicted binding site of miR-451a within the 3′-UTR of ATF2. The mutated binding site is also shown. (B) HCC cells were transfected with ATF2-wt or ATF2-mut in combination with miR-451a mimic or NC mimic; 48 h later, the luciferase activity was quantified. (C and D) ATF2 expression was determined in HCC cells when miR-451a was overexpressed. (E) mRNA and (F) protein expression of ATF2 in HCC cells following ZSCAN16-AS1 ablation. si-ZSCAN16-AS1 together with miR-451a inhibitor or NC inhibitor was transfected into HCC cells, followed by quantification of ATF2 (G) mRNA and (H) protein levels. (I) RNA immunoprecipitation was used to assess ZSCAN16-AS1, miR-451a and ATF2 enrichment in immunoprecipitants in HCC cells. (J) ATF2 level was determined in HCC tissues using reverse transcription-quantitative PCR. (K) The correlation of ATF2 mRNA and miR-451a expression in 47 HCC tissues. (L) The expression correlation between ATF2 and miR-451a in HCC was examined using TCGA database. (M) The relation between ATF2 mRNA and ZSCAN16-AS1 in 47 HCC tissues. (N) The expression correlation between ZSCAN16-AS1 and ATF2 in HCC was examined using TCGA database. **P<0.01. miR, microRNA; ATF2, activating transcription factor 2; HCC, hepatocellular carcinoma; UTR, untranslated region; wt, wild-type; mut, mutant; NC, negative control; TCGA, The Cancer Genome Atlas; si, small interfering.
miR-451a/ATF2 axis mediates the pro-oncogenic actions of ZSCAN16-AS1 in HCC
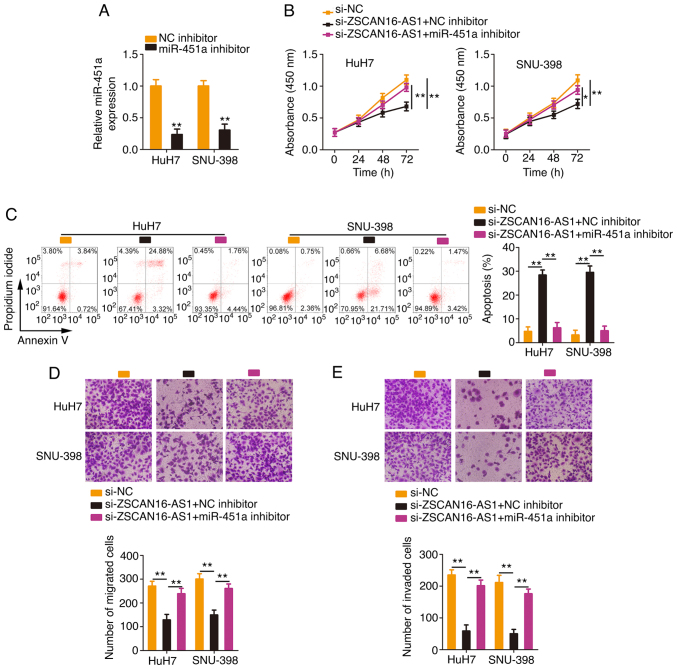
Rescue experiments were conducted with the aim of illustrating whether the miR-451a/ATF2 axis is required for the cancer-repressing actions of si-ZSCAN16-AS1 in HCC cells. The efficiency of the miR-451a inhibitor transfection was tested via RT-qPCR (Fig. 6A). The miR-451a inhibitor or NC inhibitor together with si-ZSCAN16-AS1 were introduced into HCC cells. The inhibition of miR-451a ameliorated the antiproliferative roles of si-ZSCAN16-AS1 in HCC cells (Fig. 6B). Flow cytometry demonstrated that miR-451a inhibitor cotransfection eliminated the promotive effect of ZSCAN16-AS1 depletion on HCC cell apoptosis (Fig. 6C). Similarly, Transwell cell migration and invasion assays showed the same tendency (Fig. 6D and E).
Figure 6.
miR-451a inhibition counteracts the tumor-inhibiting actions of ZSCAN16-AS1 knockdown in HCC cells. (A) Reverse transcription-quantitative PCR analysis detected the efficacy of the miR-451a inhibitor in silencing endogenous miR-451a expression. HCC cells were cotransfected with miR-451a inhibitor or NC inhibitor and si-ZSCAN16-AS1, followed by the determination of cell (B) proliferation and (C) apoptosis. (D) Migratory and (E) invasive (magnification, ×200) capabilities of the aforementioned cells. *P<0.05 and **P<0.01. miR, microRNA; HCC, hepatocellular carcinoma; NC, negative control; si, small interfering.
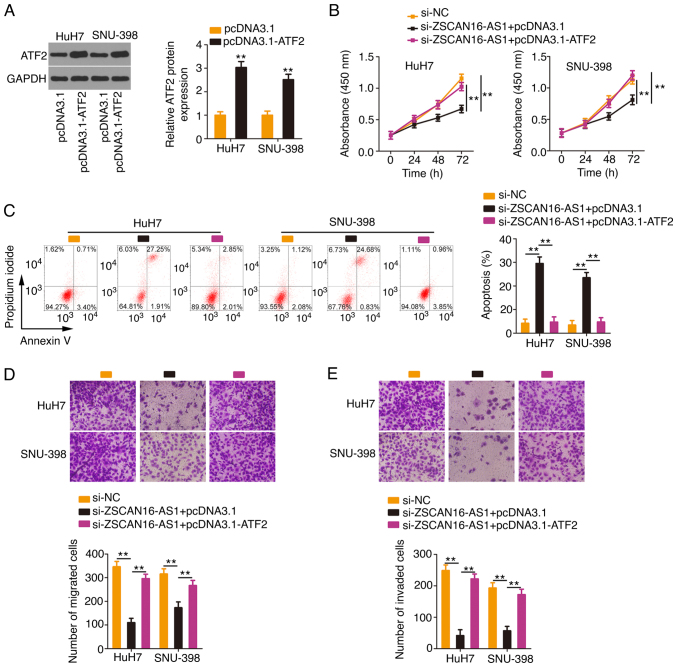
HCC cells were transfected with the ATF2-overexpressing plasmid pcDNA 3.1-ATF2 (Fig. 7A) or empty pcDNA 3.1 plasmid in the presence of si-ZSCAN16-AS1. Proliferation inhibition and apoptosis promotion (Fig. 7B and C) by ZSCAN16-AS1 silencing could be rescued in HCC cells after cotransfection with pcDNA3.1-ATF2. In addition, ATF2 overexpression restored the motility (Fig. 7D and E) of HCC cells, which were hindered by si-ZSCAN16-AS1. The above results affirmed that ZSCAN16-AS1 exerted tumorigenic roles in HCC via the miR-451a/ATF2 axis.
Figure 7.
Overexpressed ATF2 reverses the suppression of malignant properties in HCC cells caused by ZSCAN16-AS1 depletion. (A) ATF2 was overexpressed in HCC cells following transfection with pcDNA3.1-ATF2, which was corroborated by western blotting. ZSCAN16-AS1-depleted HCC cells were further transfected with pcDNA3.1-ATF2 or pcDNA3.1. Cellular (B) proliferation, (C) apoptosis, (D) migration and (E) invasion (magnification, ×200) were investigated. **P<0.01. ATF2, activating transcription factor 2; HCC, hepatocellular carcinoma.
Discussion
The importance of lncRNAs in the oncogenicity of HCC has been widely studied (15,24). Studies have demonstrated the abnormal expression of lncRNAs in HCC and confirm it to be an important driving force for HCC malignancy (25–27). Thus, lncRNAs may be effective targets for HCC diagnosis, prognosis and therapy. At present, numerous lncRNAs have been validated in the human genome by transcriptome sequencing, but the detailed roles of various lncRNAs in HCC genesis and development have not been thoroughly characterized and remain to be determined. The present study clarified whether ZSCAN16-AS1 was implicated in the aggressive properties of HCC and the possible mechanisms involved were explored.
A number of lncRNAs are reported to be involved in HCC etiology and progression. For example, ANCR (28), LINC02580 (29) and MT1JP (30) are underexpressed in HCC and are confirmed to be tumor suppressors. By contrast, TMPO-AS1 (31), ST8SIA6-AS1 (32) and ADAMTS9-AS1 (33) are overexpressed in HCC and aggravate malignant characteristics. However, until now, there has been no relevant study regarding the relationship between ZSCAN16-AS1 and HCC progression. In the present study, the data confirmed the high expression of ZSCAN16-AS1 in HCC by the TCGA database and the cohort of the present study. Analysis of the survival data revealed that overexpressed ZSCAN16-AS1 exhibited a prominent correlation with poor clinical outcomes of HCC patients. Following ZSCAN16-AS1 silencing, HCC cell proliferation, migration and invasion were inhibited, whereas cell apoptosis was promoted in vitro. Furthermore, the absence of ZSCAN16-AS1 restricted the tumor growth of HCC cells in vivo. Accordingly, the present study identified a novel oncogenic lncRNA, ZSCAN16-AS1, in HCC.
An understanding of the detailed mechanisms by which ZSCAN16-AS1 regulates HCC malignancy may offer novel insights into gene regulatory networks, which may lead to important implications in cancer. As gene regulators, lncRNAs take part in gene regulation by employing different mechanisms. Generally, lncRNAs located in the nucleus directly bind to proteins and control gene expression at the transcriptional level (34). By contrast, cytoplasmic lncRNAs serve as endogenous decoys for miRNAs through sequence complementarity, thereby influencing gene expression and signal transduction pathways at the posttranscriptional level (35). Consequently, the actions of lncRNAs are determined by their unique subcellular localizations. The present study demonstrated that ZSCAN16-AS1 was prominently distributed in the cytoplasm of HCC cells, which provided evidence for ZSCAN16-AS1 as a ceRNA.
Using StarBase 3.0, a potential interaction between miR-451a and ZSCAN16-AS1 was predicted. RT-qPCR analysis demonstrated that the downregulation of ZSCAN16-AS1 increased miR-451a expression in HCC cells. A luciferase reporter assay combined with RIP and correlation analysis identified ZSCAN16-AS1 as an endogenous molecular sponge for miR-451a. miR-451a was further demonstrated to directly target ATF2 in HCC cells through mechanistic experiments. In addition, ATF2 was shown to be positively controlled by ZSCAN16-AS1 in HCC cells, via miR-451a sequestration. Together these results established a new ceRNA pathway in HCC that consists of ZSCAN16-AS1, miR-451a and ATF2.
Low expression of miR-451a was previously reported in HCC (36,37), which was in accordance with the present study. In addition, the data revealed that miR-451a showed a significant correlation with the tumor grade and lymph node metastasis of HCC patients and served a cancer-inhibiting role in HCC cells. Mechanistically, ATF2 is a downstream effector of miR-451a in HCC, as verified by bioinformatics prediction, the luciferase reporter assay and molecular analysis. ATF2, which is a member of the cAMP response element-binding family, performs important regulatory activities in the genesis and progression of various human cancers (38,39). In the present study, ATF2 was found to be in control of the ZSCAN16-AS1/miR-451a axis. Notably, rescue experiments further confirmed that miR-451a knockdown or ATF2 resumption rescued the proliferation suppression, apoptosis promotion and migration and invasion inhibition caused by ZSCAN16-AS1 depletion. Therefore, the oncogenicity of HCC was weakened by ZSCAN16-AS1 due to its nature as a ceRNA and sponge of miR-451a, which thus alleviated miR-451a-mediated ATF2 downregulation.
The present study identified a novel lncRNA, ZSCAN16-AS1, which was overexpressed in HCC and related to poor prognosis in HCC patients. ZSCAN16-AS1 aggravated the malignancy of HCC cells by regulating the miR-451a/ATF2 axis. An in-depth understanding of the ceRNA network of ZSCAN16-AS1/miR-451a/ATF2 in HCC might be instrumental in the development of attractive targets for molecular therapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
WW and CL designed the study and wrote the manuscript. CL, QW, CS, HW and BZ conducted all experiments. QW analyzed the data. CL and WW confirm the authenticity of all the raw data. All authors have read and approved the final draft.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The People's Hospital of Tongliang District and was conducted in accordance with the principles of the Declaration of Helsinki. All patients provided informed written consent for the use of their tissue samples. Experiments involving animals were implemented under the approval of the Institutional Animal Care and Use Committee of The People's Hospital of Tongliang District.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 2.Helal Tel A, Radwan NA, Shaker M. Extrahepatic metastases as initial manifestations of hepatocellular carcinoma: An Egyptian experience. Diagn Pathol. 2015;10:82. doi: 10.1186/s13000-015-0313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Inchingolo R, Posa A, Mariappan M, Spiliopoulos S. Locoregional treatments for hepatocellular carcinoma: Current evidence and future directions. World J Gastroenterol. 2019;25:4614–4628. doi: 10.3748/wjg.v25.i32.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156:477–491.e1. doi: 10.1053/j.gastro.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gelli M, Sebagh M, Porcher R, Romanelli E, Vibert E, Sa Cunha A, Castaing D, Rosmorduc O, Samuel D, Adam R, Cherqui D. Liver resection for early hepatocellular carcinoma: Preoperative predictors of non transplantable recurrence and implications for treatment allocation. Ann Surg. 2020;272:820–826. doi: 10.1097/SLA.0000000000004259. [DOI] [PubMed] [Google Scholar]
- 7.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlsson O, Baccarelli AA. Environmental health and long non-coding RNAs. Curr Environ Health Rep. 2016;3:178–187. doi: 10.1007/s40572-016-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Meng H, Bai Y, Wang K. Regulation of lncRNA and its role in cancer metastasis. Oncol Res. 2016;23:205–217. doi: 10.3727/096504016X14549667334007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolha L, Ravnik-Glavac M, Glavac D. Long noncoding RNAs as biomarkers in cancer. Dis Markers. 2017;2017:7243968. doi: 10.1155/2017/7243968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Botti G, Marra L, Malzone MG, Anniciello A, Botti C, Franco R, Cantile M. LncRNA HOTAIR as prognostic circulating marker and potential therapeutic target in patients with tumor diseases. Curr Drug Targets. 2017;18:27–34. doi: 10.2174/1389450117666151209122950. [DOI] [PubMed] [Google Scholar]
- 14.Heo MJ, Yun J, Kim SG. Role of non-coding RNAs in liver disease progression to hepatocellular carcinoma. Arch Pharm Res. 2019;42:48–62. doi: 10.1007/s12272-018-01104-x. [DOI] [PubMed] [Google Scholar]
- 15.Abbastabar M, Sarfi M, Golestani A, Khalili E. lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI J. 2018;17:900–913. doi: 10.17179/excli2018-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Fan S, Song E. Noncoding RNAs: New players in cancers. Adv Exp Med Biol. 2016;927:1–47. doi: 10.1007/978-3-319-32189-9_22. [DOI] [PubMed] [Google Scholar]
- 17.Abba M, Mudduluru G, Allgayer H. MicroRNAs in cancer: Small molecules, big chances. Anticancer Agents Med Chem. 2012;12:733–743. doi: 10.2174/187152012802650273. [DOI] [PubMed] [Google Scholar]
- 18.Yang C, Wang Y, Xue W, Xie Y, Dong Q, Zhu C. Competing endogenous RNA (ceRNA) network analysis of autophagy-related genes in hepatocellular carcinoma. Pharmgenomics Pers Med. 2020;13:445–462. doi: 10.2147/PGPM.S267563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niu ZS, Wang WH, Dong XN, Tian LM. Role of long noncoding RNA-mediated competing endogenous RNA regulatory network in hepatocellular carcinoma. World J Gastroenterol. 2020;26:4240–4260. doi: 10.3748/wjg.v26.i29.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaes E, Khan M, Mombaerts P. Statistical analysis of differential gene expression relative to a fold change threshold on NanoString data of mouse odorant receptor genes. BMC bioinformatics. 2014;15:39. doi: 10.1186/1471-2105-15-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ballester M, Cordon R, Folch JM. DAG expression: High-throughput gene expression analysis of real-time PCR data using standard curves for relative quantification. PLoS One. 2013;8:e80385. doi: 10.1371/journal.pone.0080385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Iterson M, t Hoen PA, Pedotti P, Hooiveld GJ, den Dunnen JT, van Ommen GJ, Boer JM, Menezes RX. Relative power and sample size analysis on gene expression profiling data. BMC Genom. 2009;10:439. doi: 10.1186/1471-2164-10-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Peng L, Yuan XQ, Zhang CY, Peng JY, Zhang YQ, Pan X, Li GC. The emergence of long non-coding RNAs in hepatocellular carcinoma: An update. J Cancer. 2018;9:2549–2558. doi: 10.7150/jca.24560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mai H, Zhou B, Liu L, Yang F, Conran C, Ji Y, Hou J, Jiang D. Molecular pattern of lncRNAs in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:198. doi: 10.1186/s13046-019-1213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim LJ, Wong SYS, Huang F, Lim S, Chong SS, Ooi LL, Kon OL, Lee CG. Roles and regulation of long noncoding RNAs in hepatocellular carcinoma. Cancer Res. 2019;79:5131–5139. doi: 10.1158/0008-5472.CAN-19-0255. [DOI] [PubMed] [Google Scholar]
- 27.Wei L, Wang X, Lv L, Liu J, Xing H, Song Y, Xie M, Lei T, Zhang N, Yang M. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer. 2019;18:147. doi: 10.1186/s12943-019-1086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song XZ, Xu XJ, Ren XN, Ruan XX, Wang YL, Yao TT. LncRNA ANCR suppresses the progression of hepatocellular carcinoma through the inhibition of Wnt/β-catenin signaling pathway. OncoTargets Ther. 2020;13:8907–8917. doi: 10.2147/OTT.S260556. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Xu L, Wang Z, Yin C, Pan F, Shi T, Tian Y. Long noncoding RNA LINC02580 suppresses the invasion-metastasis cascade in hepatocellular carcinoma by targeting SRSF1. Biochem Biophys Res Commun. 2020;533:685–691. doi: 10.1016/j.bbrc.2020.10.061. [DOI] [PubMed] [Google Scholar]
- 30.Mo W, Dai Y, Chen J, Liang L, Xu S, Xu X. Long Noncoding RNA (lncRNA) MT1JP Suppresses Hepatocellular Carcinoma (HCC) in vitro. Cancer Manag Res. 2020;12:7949–7960. doi: 10.2147/CMAR.S253496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Shen Z. LncRNA TMPO-AS1 aggravates the development of hepatocellular carcinoma via miR-429/GOT1 axis. Am J Med Sci. 2020;360:711–720. doi: 10.1016/j.amjms.2020.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Xu S, Hu C, Fang K, Zhou J, Guo Z, Zhu G, Li L. LncRNA ST8SIA6-AS1 promotes hepatocellular carcinoma progression by regulating MAGEA3 and DCAF4L2 expression. Biochem Biophys Res Commun. 2020;533:1039–1047. doi: 10.1016/j.bbrc.2020.09.115. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Li H, Hu Y, Wang F. Long non-coding RNA ADAMTS9-AS1 exacerbates cell proliferation, migration, and invasion via triggering of the PI3K/AKT/mTOR pathway in hepatocellular carcinoma cells. Am J Transl Res. 2020;12:5696–5707. [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang XZ, Liu H, Chen SR. Mechanisms of long non-coding RNAs in cancers and their dynamic regulations. Cancers (Basel) 2020;12:1245. doi: 10.3390/cancers12051245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Lu B. The roles of ceRNAs-mediated autophagy in cancer chemoresistance and metastasis. Cancers (Basel) 2020;12:2926. doi: 10.3390/cancers12102926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei GY, Hu M, Zhao L, Guo WS. miR-451a suppresses cell proliferation, metastasis and EMT via targeting YWHAZ in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:5158–5167. doi: 10.26355/eurrev_201906_18180. [DOI] [PubMed] [Google Scholar]
- 37.Zhao S, Li J, Zhang G, Wang Q, Wu C, Zhang Q, Wang H, Sun P, Xiang R, Yang S. Exosomal miR-451a functions as a tumor suppressor in hepatocellular carcinoma by targeting LPIN1. Cell Physiol Biochem. 2019;53:19–35. doi: 10.33594/000000118. [DOI] [PubMed] [Google Scholar]
- 38.Huebner K, Procházka J, Monteiro AC, Mahadevan V, Schneider-Stock R. The activating transcription factor 2: An influencer of cancer progression. Mutagenesis. 2019;34:375–389. doi: 10.1093/mutage/gez041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watson G, Ronai ZA, Lau E. ATF2, a paradigm of the multifaceted regulation of transcription factors in biology and disease. Pharmacol Res. 2017;119:347–357. doi: 10.1016/j.phrs.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.