Abstract
During the terminal differentiation of skeletal myoblasts, the activities of myogenic factors regulate not only tissue-specific gene expressions but also the exit from the cell cycle. The induction of cell cycle inhibitors such as p21 and pRb has been shown to play a prominent role in the growth arrest of differentiating myoblasts. Here we report that, at the onset of differentiation, activation by MyoD of the Rb, p21, and cyclin D3 genes occurs in the absence of new protein synthesis and with the requirement of the p300 transcriptional coactivator. In differentiated myocytes, cyclin D3 also becomes stabilized and is found nearly totally complexed with unphosphorylated pRb. The detection of complexes containing cyclin D3, cdk4, p21, and PCNA suggests that cdk4, along with PCNA, may get sequestered into high-order structures held together by pRb and cyclin D3. Cyclin D3 up-regulation and stabilization is inhibited by adenovirus E1A, and this correlates with the ability of E1A to promote pRb phosphorylation; conversely, the overexpression of cyclin D3 in differentiated myotubes counteracts the E1A-mediated reactivation of DNA synthesis. These results indicate that cyclin D3 critically contributes to the irreversible exit of differentiating myoblasts from the cell cycle.
Skeletal muscle differentiation is characterized by terminal withdrawal from the cell cycle, the coordinated activation of muscle-specific gene expression, and the fusion of myoblasts into multinucleated myotubes. As in most cell types, proliferation and differentiation of skeletal myoblasts are mutually exclusive events. Established mouse myogenic cell lines have allowed the identification of muscle-specific transcription factors, belonging to the MyoD family, which determine the initiation and the maintenance of the myogenic program (reviewed in references 8, 18, 60, and 94). Muscle regulatory factors (MRFs) are basic helix-loop-helix transcription factors, which promote skeletal muscle differentiation by binding to a consensus sequence, termed E-box, present in the regulatory region of many muscle-specific genes (12, 95).
Besides regulating tissue-specific gene expression, the activity of MRFs is also involved in promoting cell cycle arrest (37, 42). Although the molecular mechanisms responsible for the coupling of cell cycle arrest with terminal differentiation of muscle cells have not been completely elucidated, several functional interactions between myogenic factors and cell cycle regulatory proteins have now been clarified. We have previously reported that MyoD induces transcription of the retinoblastoma growth suppressor gene (pRb) by a mechanism independent of direct binding of MyoD to the Rb gene promoter (46). It has also been shown that MyoD can mediate the transcriptional induction of the cell cycle inhibitor p21 (28).
A critical role for pRb activity in muscle cells was first suggested by the finding that the ability of DNA tumor virus oncoproteins, such as adenovirus E1A, simian virus 40 (SV40) large T antigen, and polyomavirus large T antigen, to inhibit myogenic differentiation is related to their ability to bind (and hence inactivate) the pRb family of proteins (10, 25, 43, 84). The importance of pRb in myogenesis is also indicated by the observation that muscle differentiation is associated with induced expression of pRb (19), which shows enhanced nuclear affinity and a hypophosphorylated, active state (25, 86). Further studies have more directly demonstrated that pRb function is required for myoblast differentiation; in fact, by using cells derived from mouse embryos specifically deficient for pRb, it has been demonstrated that pRb activity is required both for MyoD-mediated activation of muscle structural genes and irreversible cell cycle withdrawal (58, 78). Moreover, physical interaction between pRb and MyoD has been found both in vitro and in vivo (25), though the question of how such interaction regulates MyoD or pRb activity remains unanswered.
The function of pRb is known to be inactivated through phosphorylation by cyclin-dependent kinases (cdk’s), which act in conjunction with their regulatory partners, the cyclins (reviewed in references 54, 57, 72, 80). As expected, the expression in muscle cells of most cyclins is down-regulated at the onset of terminal differentiation, as cells arrest in the G0/G1 phase of the cell cycle (33, 70, 91), with the notable exception of cyclin D3, whose expression is actually induced during terminal differentiation (36, 71). The activity of cdk’s is negatively regulated by cdk inhibitors, which bind to either cdk or cyclin-cdk complexes, inhibiting their activity and blocking cell cycle progression (reviewed in references 30 and 81). In addition, the cdk inhibitor p21 can also function as a direct inhibitor of DNA polymerase by binding to the proliferating cell nuclear antigen (PCNA) subunit (89). Increased expression of the p21 and p18 cdk inhibitors has been associated with the process of terminal muscle differentiation (1, 27, 28, 52, 65, 66).
In addition to pRb, another important cellular protein, p300, targeted by viral oncoproteins and involved in cell cycle control, was first suggested as a regulator of myogenic differentiation based on E1A’s requirement for p300 binding to inhibit differentiation (10, 56). Subsequently, p300 has been identified as a transcriptional adapter which assists the function of several transcriptional activators by mediating their communication with the basal transcription machinery (2, 16, 39). Recently, we and others have demonstrated that p300 enhances the transcriptional activity of MyoD and is involved in both the cell growth arrest and the myogenic activity of MyoD (17, 67, 77, 102).
To further clarify the functional links between myogenic factors and inhibitors of cell proliferation, in this study we have addressed two questions. The first question is whether the mechanism by which MyoD induces Rb, p21, and cyclin D3 during muscle differentiation is common to these three genes, and in particular, whether they are direct targets of MyoD transactivation or whether a new synthesis of factors is needed. Furthermore, we wanted to determine whether p300 and/or pRb function is implicated in this induction by MyoD. The second question concerns the functional role of a normally proliferative factor such as cyclin D3 in terminally differentiated myotubes. To investigate these problems, we used cells expressing a hormone-inducible MyoD protein and cells in which myogenic differentiation is blocked at different stages by adenovirus E1A mutants.
MATERIALS AND METHODS
Plasmids and probes.
Many of the probes used in this study have been described previously (46). The mouse p21 probe was prepared by EcoRI plus HindIII digestion from the pGDSV7S-mwaf1 plasmid, obtained from C. Schneider (Trieste, Italy). The mouse cyclin D3 probe was obtained by EcoRI digestion from the pcN2.cyl3 plasmid (50).
The puromycin resistance gene vector pBABE-puro (55) was kindly provided by H. Land (Imperial Cancer Research Fund, London, United Kingdom). The MyoD-ER chimerical construct, obtained by fusing the hormone binding domain of the estrogen receptor in frame to the MyoD gene, was supplied by H. Weintraub and previously characterized (32).
In the transient expression experiments, the muscle creatine kinase (MCK) promoter luciferase reporter plasmid was −1256 MCK-luc; the human cyclin D3 was expressed from a cytomegalovirus expression vehicle (Rc/CMV; Invitrogen) and was kindly provided by J. Pines (Cambridge, United Kingdom); the β-galactosidase expression plasmid, CMV-β, was purchased from Clontech.
In the microinjection experiments, human cyclin D3 was expressed from a CMV expression vehicle (Rc/CMV; Invitrogen). The E1A wild-type (wt) and the E1A N-terminal mutant (RG2) proteins (kindly provided by E. Moran, Philadelphia, Pa.) were both in the 12S context and expressed under the control of the E1A gene promoter (90). The E1A N-terminal mutant, RG2, used in this study carries the same amino acid substitution as the E1A pm563 mutant (96) that we used previously to generate the stable E1A N-terminal mutant-expressing C2 cell line (10).
Cells and DNA transfections and luciferase assays.
Clone 7 of the C2 line of mouse myoblasts (101) was obtained from M. Buckingham (Institut Pasteur, Paris, France). C2 myoblasts were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 20% fetal bovine serum (HyClone). Myoblasts were carefully passaged before cell-cell contact, to avoid selection. To induce differentiation, 2 × 105 cells, seeded in 100-cm-diameter dishes and allowed to grow for 3 days (until they reached 80 to 90% confluence), were exposed for 2 days to DMEM containing 2% fetal bovine serum. E1A-expressing C2 cell lines, generated by stable transfection of wt and E1A mutant derivatives, have been previously described and characterized (10).
C3H10T1/2 fibroblasts were grown in DMEM plus 20% defined and supplemented calf serum (HyClone).
CC42 Rb−/− myogenic cells (78) were grown in DMEM supplemented with 20% fetal bovine serum (HyClone).
Stable cell lines in C3H10T1/2 fibroblasts were generated by cotransfecting 0.5 μg of the puromycin resistance gene vector pBABEpuro together with 5 μg of the MyoD-ER construct and 4.5 μg of mouse genomic DNA, as a carrier, by using the calcium-phosphate precipitation method (97). Transfected cells were maintained for 10 days in puromycin at 2 μg/ml, and then resistant clones were selected to duplicate multiwell dishes. Cells were kept in DMEM without phenol red containing 20% double-stripped defined and supplemented calf serum (75) throughout this period. Clones that exhibited morphological differentiation after 48 h in the presence of estradiol (10−7 M) and 2% calf serum were expanded from the undifferentiated duplicate plate.
For transient transfections, 105 C2 myoblasts were seeded onto 35-mm-diameter dishes the day before transfection. The MCK luciferase reporter (0.5 μg) was mixed with 0.75, 1, or 2 μg of the Rc/CMV-cyclin D3 expression construct and 0.2 μg of CMV-β; total transfected plasmid in each transfection was held constant by the addition of an empty Rc/CMV. Plasmids were mixed in OPTIMEM with 12 μl of Lipofectamine reagent (GIBCO BRL) and incubated with the cells for 5 h, according to the manufacturer’s instructions. All transfections were done in duplicate and repeated three times. After 12 h, cells were trypsinized, one half of these cells were plated onto 90-mm-diameter dishes, and the other half were plated onto 60-mm-diameter dishes and maintained in growth medium for an additional 48 h. To monitor MCK expression under differentiating conditions, transfected cells were transferred to differentiation medium 5 h after the addition of DNA and maintained under these conditions for an additional 48 h. Cells were then lysed in 100 μl of extraction buffer (100 mM potassium phosphate [pH 7.8], 1% Triton X-100, 1 mM dithiothreitol [DTT]) for 10 min at 4°C, and cell debris was pelleted by centrifugation in a microcentrifuge for 5 min at 4°C. Total protein in the extracts was determined by the Bradford assay (6), and the luciferase activity of equal amounts of protein was determined exactly as described by de Wet et al. (14). Luciferase activity was normalized for β-galactosidase activity, as determined according to the method described by Rosenthal et al. (74).
Microinjection experiments and immunofluorescence.
Microinjection experiments were performed as previously described (24, 67). However, 3-day-old C2 myotubes, cultured in 2% fetal calf serum (FCS)-enriched medium, were injected into the nuclei with 0.2 μg of the E1A N-terminal mutant (12S E1A RG2) and either 0.6 μg of the cyclin D3 expression vector Rc/CMV-CycD3 or 0.6 μg of the Rc/CMV empty vector; all the DNA preparations were in Tris-EDTA (TE; pH 7.6). Twelve hours before the injection, cells were shifted to 0.1% FCS, and 4 h after the injection, bromodeoxyuridine (BrdU) was added for an additional 18 h. Cells were then washed in phosphate-buffered saline (PBS), fixed, and processed for immunofluorescence. To detect E1A expression, cells were fixed in a 1:2 methanol-acetone solution, dried, preincubated with 5% bovine serum albumin (BSA) in PBS, and incubated for 30 min at 37°C with a 1:5 dilution of hybridoma-conditioned medium containing the M73 mouse monoclonal anti-E1A antibody (29). Specifically bound antibody was visualized by incubation with rhodamine-conjugated second-step anti-mouse immunoglobulin (Ig) antibody (Cappel). Immunofluorescence for detection of BrdU, as DNA synthesis indicator, was performed with an anti-BrdU antibody directly conjugated with fluorescein (Boehringer), according to the manufacturer’s instructions. After immunofluorescence treatment, nuclei were stained by a 3-min incubation in a 1-μg/ml solution of 4′,6-diamidino-2-phenylindole (DAPI) in PBS.
Northern blot analysis.
Total cellular RNA was isolated by using the method of Chomczynski and Sacchi (11). Ten micrograms of each RNA sample was size fractionated on 1.2% agarose-30% formaldehyde gels and transferred to Qiabrane nylon filters (Qiagen), as described by Thomas (85). The integrity and the amount of RNA were checked by ethidium bromide staining of ribosomal RNA. DNA fragments, purified by low-melting agarose gels, were 32P labelled by random priming and used to probe the filters. Hybridization was carried out for 24 h at 42°C in 50% formamide, 5× SSPE (1× SSPE is 0.15 M NaCl, 0.01 M sodium phosphate, 0.001 M EDTA [pH 7.7]), 5× Denhardt’s solution (1× Denhardt’s is 0.02% Ficoll, 0.02% polyvinyl-pyrrolidone, 0.02% BSA), 0.5% sodium dodecyl sulfate (SDS), 20 μg of salmon sperm DNA, and 2 × 106 cpm of 32P-labelled probes per ml. Filters were washed three times in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M Na-citrate), 0.2% SDS, for 30 min at 58°C, followed by exposition to phosphorstorage screens (Molecular Dynamics).
Immunoprecipitation and Western blot analyses.
For immunoprecipitation experiments, exponentially growing or differentiated C2 cells were rinsed two times with PBS, and cell lysates were prepared by the addition of ice-cold Nonidet P-40 (NP-40) lysis buffer (Tris-HCl [pH 7.4], 50 mM; NaCl, 250 mM; NP-40, 0.5%; ATP, 5 mM; MgCl2, 5 mM) or Triton lysis buffer (Tris-HCl [pH 7.4], 50 mM; NaCl, 250 mM; Triton, 1%; ATP, 5 mM; MgCl2, 5 mM), both supplemented with protease and phosphatase inhibitors (leupeptin, 10 μg/ml; aprotinin, 10 μg/ml; phenylmethylsulfonyl fluoride [PMSF], 1 mM; Na3VO4, 0.1 mM; NaF, 50 mM). After clearing was performed by centrifugation at 14,000 rpm for 15 min, extracts were assayed for protein concentration by the Bradford assay (6); 500-μg aliquots were then precleared with either a rabbit preimmune serum or the total mouse IgG fraction (Sigma, St. Louis, Mo.) and protein A-agarose for 2 h at 4°C. After centrifugation at 12,000 rpm, the supernatants were incubated with protein A-agarose or protein G-agarose beads (Pierce) and with the appropriate antibodies for at least 2 to 4 h at 4°C. The immunoprecipitates were washed four times with ice-cold lysis buffer and then resuspended in 2× Laemmli buffer (1× Laemmli buffer is Tris-HCl [pH 6.8], 62.5 mM; SDS, 2%; glycerol, 10%; β-mercaptoethanol, 5%), heat denatured, and run on SDS-polyacrylamide gels.
For Western blotting, whole-cell lysates were prepared by adding warm 2× Laemmli buffer directly to the cell culture plate. The lysates were treated by 10 s of sonication, centrifuged at 14,000 rpm for 15 min, and then separated by SDS-polyacrylamide gel electrophoresis (PAGE).
The proteins were transferred from the gel to a nitrocellulose membrane (Schleicher & Schuell) by semidry electric transfer, and the membrane was blocked in NET buffer (NaCl, 150 mM; Tris-HCl [pH 7.5], 50 mM; EDTA, 5 mM; Triton X-100, 0.05%) containing 0.2% gelatin for 1 h, followed by 1 h of incubation with 4% skim milk powder. Primary and horseradish peroxidase-conjugated secondary antibodies (Cappel) were incubated for 1 h each. Filters were then processed for enhanced chemiluminescence detection (Super Signal; Pierce), according to the manufacturer’s instructions. Records were maintained on Kodak X-OmatS films.
The expression of specific proteins was analyzed by using the following antibodies: clone G3-245 (Pierce), a monoclonal antibody (MAb) specific for pRb; clone 18B6-10, clone 72-13G, and clone PC10 (Santa Cruz Biotechnology, Inc.), MAbs specific for cyclin D3, mouse cyclin D1, and PCNA, respectively; clone IF5D (98), a MAb specific for myogenin; clone MF20 (3), a MAb specific for myosin heavy chain; and sc-528, sc-163, sc-260, sc-596, sc-481, sc-092, and sc-182 (Santa Cruz Biotechnology, Inc.), polyclonal rabbit antibodies, specific for p27, cdk2, cdk4, cyclin A, cyclin E, cyclin D1, and cyclin D3, respectively. The polyclonal rabbit antiserum specific for mouse p21 was kindly provided by C. Schneider.
Immune complex kinase assay.
Cells were suspended in lysis buffer containing 50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) (pH 7.5), 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1 mM DTT, 0.1% Tween 20, 10% glycerol, 1 mM PMSF, 10 μg of leupeptin per ml, 5 μg of aprotinin per ml, 10 mM β-glycerophosphate, 1 mM sodium orthovanadate, and 50 mM NaF (all protease inhibitors were obtained from Sigma Chemicals), followed by a 10-s sonication and clearing by centrifugation at 14,000 rpm for 15 min. Supernatants were assayed for protein concentration as described above; protein samples of 4 mg each were then immunoprecipitated for at least 2 to 4 h at 4°C with protein A-agarose beads precoated with saturating amounts of the appropriate antibody (5 μg). Immunoprecipitated proteins on beads were washed three times with 1 ml of lysis buffer and twice with kinase buffer (50 mM HEPES [pH 7.5], 1 mM DTT, 10 mM MgCl2, plus protease inhibitors, as described above). The beads were resuspended in 50 μl of kinase buffer containing 2 μg of glutathione S-transferase (GST)-pRb (769-921) fusion protein (Santa Cruz Biotechnology, Inc.), 2.5 mM EGTA, 10 mM β-glycerophosphate, 1 mM Na3VO4, 20 μM ATP, and 10 μCi of [γ-32P]ATP (6,000 Ci/mmol; NEN Dupont, Boston, Mass.). After incubation for 30 min at 30°C, the samples were boiled in 2× Laemmli buffer and separated by SDS-PAGE. Phosphorylated proteins were visualized by exposure to phosphorstorage screens. The antibodies used were as follows: for cyclin D1, MAb clone DCS11, kindly provided by J. Bartek; for cyclin D3, MAb clone 18B6-10; and for cdk4 and cdk2, polyclonal rabbit antibodies obtained from Santa Cruz Biotechnology.
RESULTS
The expression of p21, Rb, and cyclin D3 is directly induced by MyoD and requires p300.
Previous work has shown that terminal differentiation of muscle cells is accompanied by transcriptional induction of the retinoblastoma gene (Rb) and the accumulation of the hypophosphorylated (active) form of the Rb protein (pRb) (25, 46, 86). Although the cdk4 and cdk2 pRb kinases are constitutively expressed during myoblast differentiation, their regulatory subunits (cyclin D1, cyclin E, and cyclin A) are down-regulated (33, 71, 83, 91). In contrast, cyclin D3, which is known to function as a cdk4-activating subunit, is greatly induced in differentiating muscle cells (36, 71). The p21 cdk inhibitor is markedly induced upon skeletal muscle differentiation (27, 28, 65), contributing to the decrease of cdk activity (92). Because the increased expression of the Rb, p21, and cyclin D3 mRNAs is a typical feature of muscle cell differentiation, we wished to determine whether these three genes are direct targets of MyoD.
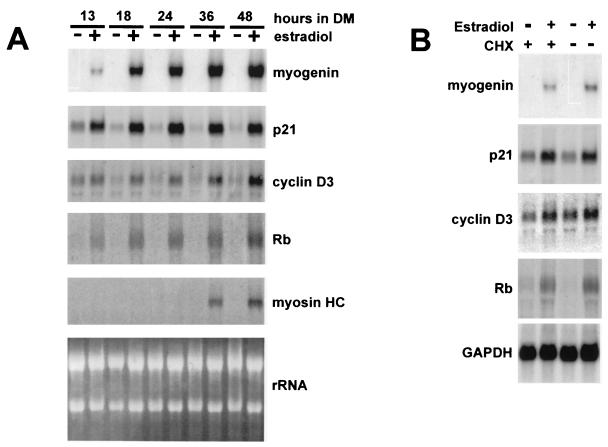
To this end, we established a C3H10T1/2 cell line stably expressing a hormone-inducible MyoD protein, which was created by fusing MyoD to the hormone-binding domain of the estrogen receptor, MyoD-ER (32). C3H10T1/2 fibroblasts were cotransfected with the MyoD-ER construct and pBABEpuro, carrying the puromycin resistance gene; then, puromycin-resistant clones were isolated under hormone-free conditions and selected if they exhibited estrogen-dependent myogenic conversion. One of these clones was used for further studies. Figure 1A shows the Northern blot analysis of total RNA extracted from C3H-MyoD-ER cells placed in differentiation medium, either in the presence or absence of estradiol, for increasing periods of time. The results indicate that p21, Rb, and cyclin D3 were induced upon 13 h of hormone treatment, as early as myogenin, while the induction of myosin heavy chain took place later. The initial induction of p21, Rb, and cyclin D3 required a minimum of 12 h of hormone treatment in differentiation medium; at that time the mRNA levels of p21 were up-regulated approximately threefold, those of cyclin D3 were up-regulated twofold, and those of Rb were up-regulated fourfold relative to untreated cells, as measured by densitometric analysis.
FIG. 1.
Direct activation of p21, Rb, and cyclin D3 by MyoD-ER. (A) Confluent cultures of C3H-ER-MyoD cells were exposed to differentiation medium (DM) for increasing periods of time in the presence or absence of estradiol as indicated. Total RNA was extracted from cells at each stage of differentiation and then subjected to Northern blot analysis. Identical filters were probed for myogenin, p21, cyclin D3, Rb, and myosin heavy chain (myosin HC). Ethidium bromide staining of rRNA, on one of the filters, was photographed with UV light. (B) C3H-ER-MyoD cells were induced to differentiate with estradiol (10−7 M) in the presence or absence of cycloheximide (CHX; 50 μg/ml) for 13 h. The effects of cycloheximide were determined also in C3H-ER-MyoD cells maintained in differentiation medium for 13 h in the absence of estradiol. Total RNA was extracted and then analyzed by Northern blotting, with probes for myogenin, p21, cyclin D3, and Rb. One of the filters was reprobed with a GAPDH probe to normalize the amounts of loaded RNA.
MyoD-ER cell lines have been previously used to determine which myogenic genes were directly activated by MyoD, by inducing the MyoD chimera in the presence of an inhibitor of protein synthesis (cycloheximide) and then monitoring the levels of the mRNA of a number of myogenic markers. These analyses allowed researchers to establish that the endogenous MyoD gene and myogenin are directly induced by MyoD, while the induction of late muscle genes, such as MCK, requires new protein synthesis (32). In order to determine whether MyoD can directly activate p21, Rb, and cyclin D3, total RNA was extracted from C3H-MyoD-ER cells induced to differentiate for 13 h in the presence of estradiol and cycloheximide; the RNA was then subjected to Northern blot analysis. The results shown in Fig. 1B indicate that, as in the case of myogenin, the estrogen-dependent induction of p21, Rb, and cyclin D3 expression was unaffected by cycloheximide, suggesting that the initial induction of these genes directly depends on MyoD activity and does not require the synthesis of new factors. Thus, p21, Rb, and cyclin D3 can be categorized as early differentiation markers. Recently, by expressing MyoD-ER in a glioblastoma cell line, Otten et al. (61) also showed that the p21 gene is a direct target of MyoD.
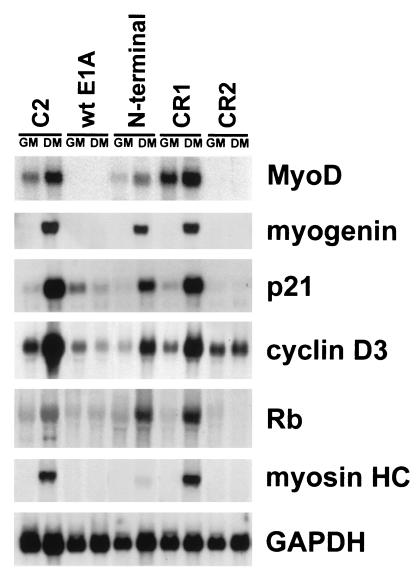
It has been demonstrated that both the transcriptional coactivator p300 and pRb act as cofactors of the MyoD transcriptional activity (see the introduction). To ascertain whether their function is required by MyoD for the induction of Rb, p21, and cyclin D3, we exploited C2 cell clones that stably express either wt E1A or mutant E1A derivatives lacking the conserved domain sequences through which E1A interacts with (and hence inactivates the function of) p300 or pRb family proteins (10). The cellular proteins’ binding properties of such E1A mutants are displayed in Table 1. In previous work, the analysis of these clones allowed us to determine that E1A inhibits muscle differentiation by two different mechanisms as follows: (i) the inhibition of MyoD transcription, which correlates with the ability of E1A to bind p300, and (ii) the inhibition of the MyoD-mediated induction of muscle genes, which correlates with the ability of E1A to bind pRb (10). Figure 2 shows the Northern blot analysis of RNA prepared from C2 parental cells and C2-derived clones constitutively expressing the different E1A constructs, cultured under growth or differentiation conditions for 48 h. The results indicate that terminal differentiation of C2 myoblasts was associated with a strong increase in p21, cyclin D3, and Rb mRNA levels (increases of about 70-, 25-, and 30-fold relative to growing myoblasts). wt E1A and the CR2 E1A mutant (which binds p300 but not pRb) inhibited MyoD transcription and, consequently, the induction of all the differentiation markers. C2 cells expressing the E1A N-terminal mutant (unable to bind p300) allowed MyoD mRNA expression (though to levels approximately fourfold lower than those of parental C2 cells) as well as induction of myogenin, Rb, p21, and cyclin D3 in differentiation medium; by contrast, these cells inhibited the expression of genes activated later during muscle differentiation, such as myosin heavy chain. Finally, the CR1 E1A mutant, which lacks the ability to bind both p300 and pRb, allowed the expression of both early and late differentiation markers.
TABLE 1.
Ability of E1A mutants to coimmunoprecipitate with E1A-associated cellular proteins
| Mutant | Protein
|
|
|---|---|---|
| p300 | pRb | |
| wt E1A | + | + |
| N-terminal (pm563 or RG2) | − | + |
| CR1 (dl646N) | − | − |
| CR2 (dl922-947) | + | − |
FIG. 2.
Levels of p21, cyclin D3, and Rb mRNAs in C2 cells expressing wt E1A and E1A mutant derivatives. Northern blotting was used to analyze the RNA isolated from C2 parental cells and C2-derived stable lines expressing either wt E1A or the E1A mutants described in Table 1. Identical filters were probed for myogenin, MyoD, p21, cyclin D3, Rb, and myosin heavy chain. Shown are results for growing cells (GM) and cells kept for 48 h in differentiation medium (DM). One of the filters was reprobed with a GAPDH probe to normalize the amounts of loaded RNA.
These data indicate that the function of the p300 transcriptional coactivator is required to assist the MyoD-mediated induction of early differentiation markers, while the function of pRb appears to be essential for the induction of late differentiation markers.
Cyclin D3 mediates the interaction of cdk4, p21, and PCNA with pRb in differentiated C2 cells.
The increased levels of pRb, p21, and cyclin D3 during differentiation argued for a role of these proteins in terminally differentiated myotubes. It has been well established that pRb is essential for both cellular growth arrest and myogenic bHLH activity in skeletal muscle cells and that p21 contributes to the mechanism by which differentiating myocytes irreversibly exit the cell cycle (27, 28, 58, 78). By contrast, the significance of cyclin D3 induction still awaits elucidation.
Because D-type cyclins have been shown to bind pRb and PCNA, besides their catalytic partners (15, 20, 34, 47, 99), we sought to determine the differentiation-associated changes in composition and activity of the cdk4 complexes and whether the cyclin D3 subunit might mediate an interaction of these complexes with pRb in differentiated C2 cells.
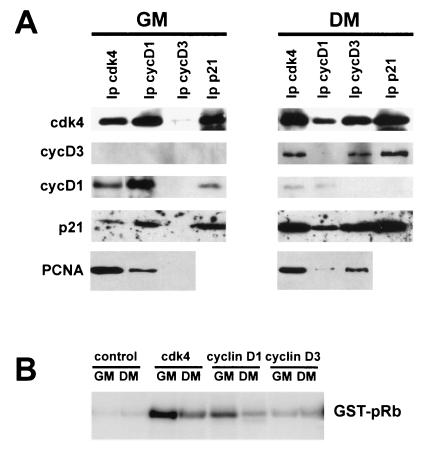
To address this issue, we performed a series of immunoprecipitation-coupled immunoblotting experiments. The analysis of cdk4, cyclin D1, cyclin D3, and p21 immunoprecipitates (Fig. 3A) showed that cdk4 was associated with cyclin D1 and p21 in growing C2 myoblasts, whereas following differentiation, it formed complexes with cyclin D3 and increased amounts of p21. This result was consistent with up-regulation of p21 and cyclin D3 and down-regulation of cyclin D1 in differentiating cells. PCNA was found associated with cdk4 both in growing and in differentiated C2 myoblasts, while the PCNA-associated D-type cyclin switched from cyclin D1 to cyclin D3 as the cells differentiated.
FIG. 3.
Composition and activity of cdk4-cyclin D-p21-PCNA complexes in undifferentiated versus differentiated C2 cells. (A) Cell lysates from C2 myoblasts, either growing (GM) or exposed for 48 h to differentiation medium (DM), were immunoprecipitated by using antibodies against the proteins indicated at the top. The immunoprecipitated proteins were resolved by SDS-PAGE and then analyzed by immunoblotting with antibodies specific for cdk4, cyclin D3, cyclin D1, p21, and PCNA, as indicated on the left. (B) cdk4, cyclin D1, and cyclin D3 were immunoprecipitated from C2 cells either growing (GM) or exposed for 48 h to DM. The protein complexes, collected on protein A-Sepharose beads, were then incubated in the presence of [γ-32P]ATP with a bacterially produced GST-Rb fusion protein as a substrate (as detailed in Materials and Methods). Phosphorylated proteins were resolved on SDS-PAGE gels, and radioactivity was detected with a PhosphorImager (Molecular Dynamics). As a negative control, myoblast and myotube extracts were immunoprecipitated with total mouse IgGs; the Rb-kinase activity of these immunoprecipitates was measured as described above.
Since pRb is found in the hypophosphorylated state in differentiated cells, it could be predicted that in C2 myotubes no kinase activity would be associated with cdk4-cyclin D3, whereas in C2 myoblasts cdk4-cyclin D1 complexes would be active in pRb phosphorylation. To assay for changes in cdk4 kinase activity during myogenic differentiation, extracts prepared from growing and differentiated C2 cells were immunoprecipitated with antibodies specific to cyclin D1, cyclin D3, or cdk4. The kinase activity associated with these immunoprecipitates was assessed by using a bacterially expressed GST-pRb as a substrate. Results shown in Fig. 3B indicate that GST-pRb was phosphorylated by cyclin D1 or cdk4 immunoprecipitated from undifferentiated cells; on the contrary, significantly lower levels of pRb kinase activity were found associated with cyclin D1 or cdk4 immunoprecipitated from differentiated cells. Remarkably, with regard to cyclin D3, almost no associated pRb kinase activity was detected in either condition.
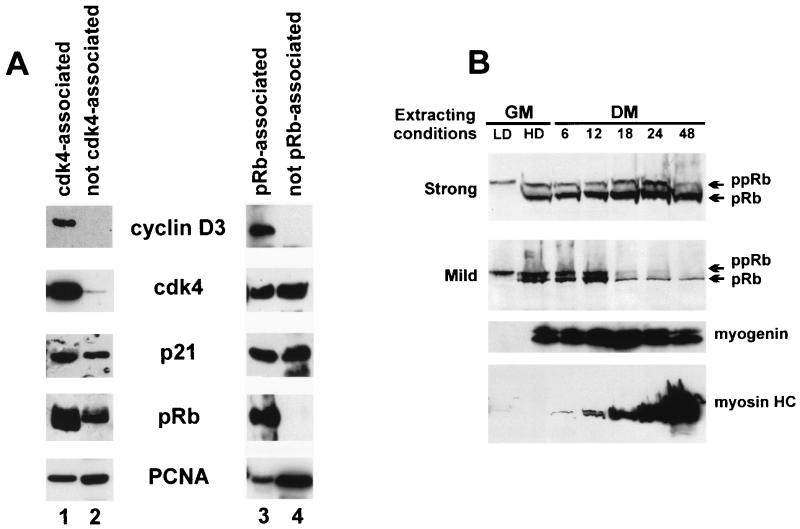
To address the issue of whether cyclin D3 can mediate the interaction of cdk4 complexes with pRb in C2 myotubes, anti-cdk4 and anti-pRb immunoprecipitates were analyzed by Western blotting and compared to those obtained from the supernatants immunodepleted of cdk4 and pRb. Figure 4A shows that cyclin D3, p21, PCNA, and pRb were found in the anti-cdk4 immunoprecipitate, whereas in the cdk4-immunodepleted supernatant p21, PCNA and pRb, but not cyclin D3, were detected. In agreement with this, the anti-pRb immunoprecipitate and the pRb-immunodepleted extract revealed that the bulk of cyclin D3 was associated with pRb, whereas only a fraction of cdk4, PCNA, and p21 were complexed with pRb.
FIG. 4.
(A) Analysis of protein complexes immunoprecipitated with anti-cdk4 or anti-pRb from C2 myotubes. Immunoprecipitation-coupled immunoblot analyses were performed by using extracts prepared from C2 cells harvested after 48 h in differentiation medium. Protein complexes, immunoprecipitated by using anti-cdk4 or anti-pRb antibodies, were resolved by SDS-PAGE and then subjected to immunoblot analyses with anti-cdk4, cyclin D3, p21, pRb, or PCNA antibodies (lanes 1 and 3). Following the first immunoprecipitation, cyclin D3, cdk4, p21, pRb, or PCNA were immunoprecipitated from the cdk4- and the pRb-depleted supernatants and detected by Western blotting by using the specific antibodies (lanes 2 and 4). (B) Time course of pRb protein induction in C2 differentiating cells. C2 myoblasts, cultured at low density (LD; approximately 40% confluent), were allowed to reach confluence (HD; high density) in growth medium (GM) and then transferred to differentiation medium (DM) for the times indicated. The cells, at each stage of differentiation, were extracted either in 0.2% SDS, 2% NP-40, 50 mM NaCl (mild extracting condition), or directly in SDS sample buffer (strong extracting condition). Proteins were separated on SDS-PAGE gels and subjected to immunoblot analysis with the MAb G 245 anti-pRb antibody, as detailed in Materials and Methods. Arrows indicate positions of the underphosphorylated (pRb) and hyperphosphorylated (ppRb) forms of the pRb protein. The staged cell extracts prepared in strong extracting conditions were also monitored for myogenin and myosin heavy chain (myosin HC) expression by using antibodies specific for these proteins.
It has been previously reported that pRb becomes resistant to extraction from nuclei of differentiating muscle cells (86); we examined the time course of pRb accumulation, dephosphorylation, and extractability during the differentiation of C2 myoblasts. Cell lysates were prepared at each stage of differentiation by either extracting directly in SDS sample buffer or in low-salt, low-detergent buffer; immunoblotting analysis was then employed to detect pRb levels and phosphorylation state. The results illustrated in Fig. 4B show that high levels of progressively hypophosphorylated pRb were retrieved when the cells were lysed in SDS sample buffer, whereas in lysates prepared with low-salt, low-detergent buffer the amount of extracted pRb was notably lower, particularly after more than 18 h in differentiation medium. These results indicated that at this stage of differentiation (concomitantly with the appearance of myosin heavy chain) pRb became tightly associated with insoluble components of the nucleus and was thus more resistant to extraction. For the experiments shown in Fig. 4A, C2 cells were lysed in a buffer containing 1% Triton X-100 and 250 mM NaCl (see Materials and Methods), which could recover at least one-third of total pRb; yet, it still conserved the pRb association with the expected proteins. Under these conditions, cdk4, cyclin D3, p21, and PCNA were completely extractable (data not shown).
Taken together, the results of Fig. 3 and 4 indicate that (i) functionally inactive cdk4-cyclinD3-p21-PCNA complexes exist in differentiated C2 cells, (ii) cyclin D3 represents the limiting component of this interaction, (iii) cyclin D3, by its ability to bind pRb, can mediate the interaction of a fraction of cdk4, PCNA, and p21 with pRb, and (iv) hypophosphorylated pRb is able to keep cyclin D3, cdk4, p21, and PCNA complexed into insoluble nuclear structures.
It is worthwhile to mention that differentiated C2 cultures are a mixed population of multinucleated myotubes and quiescent, unfused cells; by means of immunostaining it was verified that cyclin D3 and pRb expression was restricted to the nuclei of multinucleated myotubes, while cdk4 and PCNA were also present in the nuclei of unfused cells (data not shown). Thus, pRb and cdk4 interaction with cyclin D3 detected by the immunoprecipitation-coupled immunoblots is likely to take place in the nuclei of terminally differentiated myotubes.
pRb phosphorylation in E1A-expressing C2 cells.
The sequence motif through which D-type cyclins interact with pRb is similar to the pRb-binding motif of some viral oncoproteins, and cyclin D-pRb complexes are disrupted by E1A and derived peptides (15, 20). Thus, we exploited E1A-expressing C2 cells in order to investigate the effect of the inhibition of the cyclin D3-pRb interaction on pRb phosphorylation.
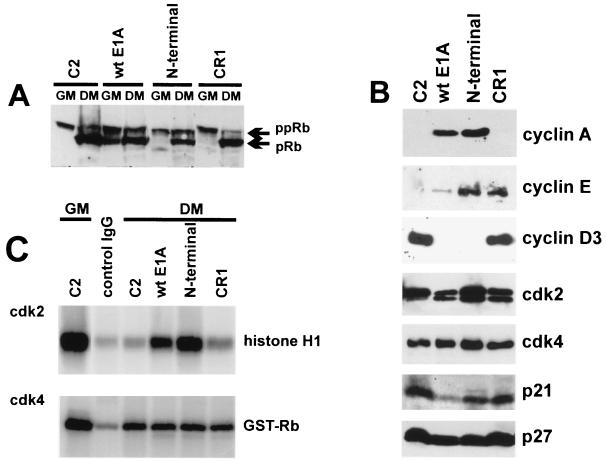
The C2-derived cell lines that were analyzed stably express either wt E1A, the E1A N-terminal mutant (both able to bind pRb), or the E1A CR1 mutant (unable to bind pRb; Table 1). Total cell extracts prepared from each cell clone under growth and differentiation conditions were examined for pRb expression and phosphorylation state by Western blotting. As shown in Fig. 5A, pRb was observed mostly in its slow-migrating, hyperphosphorylated form in all growing cell types (in wt E1A cells a significant fraction of pRb was hypophosphorylated, presumably because these cells must be cultured at a somewhat higher density, to counteract E1A-induced apoptosis). Upon shifting to differentiating conditions, the rapidly-migrating hypophosphorylated form of pRb predominated in the parental C2 and in CR1 cells, whereas in cells expressing wt E1A and the E1A N-terminal mutant, pRb was about equally divided between the active (hypophosphorylated) and inactive (hyperphosphorylated) forms. Thus, the ability of E1A to promote pRb phosphorylation appeared to correlate with the ability of E1A to bind pRb and, hence, with its ability to displace cyclin D3 from pRb.
FIG. 5.
pRb phosphorylation in E1A-expressing C2 cells. (A) Immunoblot analysis of pRb in C2 parental and C2-derived, E1A-expressing cell lines cultured either in growth (GM) or in differentiation (DM) medium for 48 h. For a description of the E1A mutants used to generate the E1A-expressing stable cell lines, see Table 1. Extracts were prepared by lysing cells in SDS sample buffer, proteins were then separated on SDS-PAGE gels and subjected to immunoblot analysis with the MAb G 245 anti-pRb antibody. The hypophosphorylated (pRb) and the hyperphosphorylated (ppRb) forms of the pRb proteins are indicated. (B) Levels of cyclins, cdk’s, and cdk’s in C2-derived stable lines expressing wt or mutant E1A. Cell extracts were prepared from C2 myotubes and from E1A-expressing C2 cells cultured in differentiation medium for 48 h. Levels of cyclin A, cyclin E, cyclin D3, cdk2, cdk4, p21, and p27 were monitored by immunoblot analysis by using antibodies specific for these proteins. (C) Determination of the kinase activity associated with cdk2 and cdk4 in E1A-expressing cells. cdk2 and cdk4 were immunoprecipitated from lysates of C2 myoblasts (growth medium; GM) or myotubes (differentiation medium; DM) or from C2-derived E1A-expressing cell lines cultured for 48 h in DM. Immunocomplexes, coupled to protein A-Sepharose, were incubated in the presence of [γ-32P]ATP with a bacterially expressed GST-Rb fusion protein or with commercial histone H1 as the substrate (see Materials and Methods). The 32P-labelled proteins were then separated on SDS-PAGE gels, and radioactivity was detected with a PhosphorImager (Molecular Dynamics). As a negative control, the C2 myotube extract was immunoprecipitated with total mouse IgGs; the pRb- and histone H1-kinase activities of this immunoprecipitate were measured as described above.
To elucidate the mechanism(s) by which E1A induced pRb phosphorylation, we examined the protein levels of the cdk4 and cdk2 pRb kinases and those of their regulatory subunits in the different E1A-expressing C2 cells, cultured in differentiation medium, and compared them to those of parental C2 cells.
The results shown in Fig. 5B indicate that while cdk4 was expressed to very similar levels in all cell types, cyclin D3 was repressed by wt E1A and the E1A N-terminal mutant but not by the CR1 mutant. The cyclin D3 protein levels in wt E1A and in CR1 cells paralleled the levels of cyclin D3 mRNA in these cells (Fig. 2); in contrast, C2 cells expressing the E1A N-terminal mutant, which did not inhibit the MyoD-mediated induction of cyclin D3 mRNA (Fig. 2), failed to accumulate cyclin D3 as a protein. The absence of cyclin D3 in cells expressing the E1A N-terminal mutant (able to bind pRb) but not in those expressing the CR1 mutant (unable to bind pRb) suggested that E1A, by its ability to interact with pRb, interfered with a mechanism that normally stabilizes cyclin D3 in differentiating C2 cells (see also the results below).
The p21 kinase inhibitor was down-regulated by wt E1A but accumulated nearly to parental levels in cells expressing the E1A N-terminal and CR1 mutants; this was accompanied by augmented mRNA expression (Fig. 2). Cyclin D1 was down-regulated in all cell types (data not shown).
Given the lack of D-type cyclins in cells expressing either wt E1A or the E1A N-terminal mutant and the high levels of p21 in CR1 expressing- and parental C2 cells, the cdk4 kinase activity was expected to be inhibited in all these cells under differentiation conditions. Indeed, as shown in Fig. 5C, very low levels of cdk4-associated pRb kinase activity, comparable to those observed in parental C2 myotubes, were detected in all E1A-expressing cells under differentiation conditions. Thus, cdk4 did not appear to contribute to pRb phosphorylation in these cells.
By contrast, the in vitro kinase activities of cdk2 immunoprecipitates correlated with the changes in pRb phosphorylation observed among the different E1A-expressing cells. As shown in Fig. 5C, the cdk2-associated kinase activity, which was markedly inhibited upon the shifting of C2 cells from growth to differentiation medium, remained inhibited in immunoprecipitates from cells expressing the E1A CR1 mutant, while it was high in cells expressing wt E1A and the E1A N-terminal mutant.
We then determined the levels of cdk2 and its regulatory subunits, cyclins A and E, under differentiation conditions (Fig. 5B). It was found that cdk2 remained at levels comparable to those of parental C2 cells in wt E1A and CR1 cells, while its level was notably increased in N-terminal cells. Cyclins E and A, which completely disappeared in C2 myotubes, were both induced in wt E1A and N-terminal cells, while cyclin E, but not cyclin A, was induced in CR1 cells. The levels of the cdk2 inhibitor p27, which have been shown to be raised upon differentiation of C2 cells (28), were not appreciably modified by E1A expression.
The pattern of expression of the regulatory cyclins A and E and that of the cdk inhibitors p21 and p27 could explain the cdk2 activity observed in the various E1A-expressing cells. Although in CR1 cells cyclin E was induced, the cdk2 activity was antagonized by high levels of p21 and p27, whereas in cells expressing wt E1A and the E1A N-terminal mutant, the induction of cyclin E along with cyclin A reduced the effective inhibitory threshold of p27 and p21. More-direct effects by E1A on p27 and p21 complexes are not excluded, however, as it has been reported, for example, that E1A can bind and inactivate p27 (44) and can interfere with the interaction of p21 with cyclin-cdk complexes (100).
Finally, the persistence of cdk2 activity under differentiating conditions observed in cells expressing wt E1A and the E1A N-terminal mutant correlated with the absence of cyclin D3.
Cyclin D3 is involved in the permanent withdrawal of C2 myotubes from the cell cycle.
It has been previously demonstrated that the expression of E1A in terminally differentiated myotubes, by either adenovirus infection or microinjection, triggers the reactivation of DNA synthesis and that this activity of E1A correlates with its ability to bind the pRb family members (67, 87). These observations and the results shown above indicate that the binding to pRb is required by E1A both to induce DNA synthesis and to suppress cyclin D3, suggesting that these two phenomena might be part of the same mechanism.
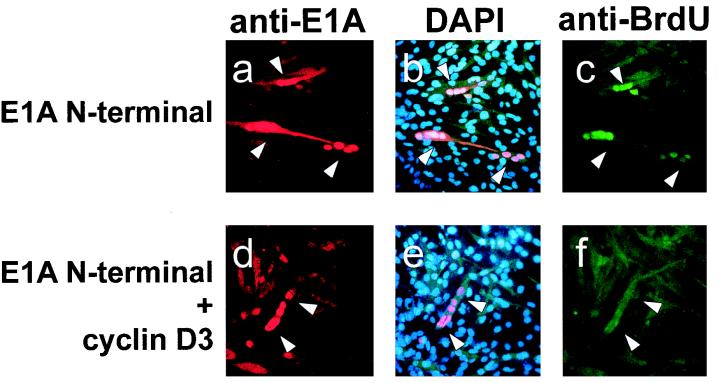
We tried then to assess whether the overexpression of cyclin D3 could counteract the induction of DNA synthesis promoted by E1A microinjection into C2 myotubes. For this experiment we used the E1A N-terminal mutant (capable of binding pRb but not p300) because, unlike wt E1A, it inhibits cyclin D3 expression only at a posttranscriptional level (Fig. 2 and 5B). C2 myoblasts were allowed to differentiate for 3 days, and then cell nuclei were microinjected with the E1A N-terminal encoding plasmid either alone (Fig. 6a, b, and c), or in combination with a cyclin D3 expression construct (Fig. 6d, e, and f). Twenty-four hours postinjection, E1A expression in nuclei was visualized by means of an anti-E1A antibody, and DNA synthesis was assessed by BrdU incorporation. The results show that the reinduction of DNA synthesis triggered by the E1A N-terminal mutant was inhibited by the overexpression of cyclin D3; this suggested that in C2 myotubes cyclin D3 may exert an inhibitory function that must be antagonized by E1A to reactive DNA synthesis.
FIG. 6.
The overexpression of cyclin D3 can counteract the E1A-mediated reactivation of DNA synthesis in terminally differentiated myotubes. C2 myoblasts were allowed to differentiate for three days and then coinjected into the nuclei with 0.2 μg of the E1A N-terminal encoding plasmid in combination with 0.6 μg of either the Rc/CMV-cyclin D3 expression construct (d, e, and f), or 0.6 μg of the Rc/CMV empty vector (a, b, and c). Twelve hours before the injection, myotubes were shifted to 0.1% FCS, and 4 h after the injection BrdU was added for an additional 18 h. Cells were then fixed and stained for nuclear expression of E1A and BrdU incorporation. In panels a and d, E1A expressing nuclei were visualized by using an anti-E1A antibody (M73), followed by incubation with rhodamine-conjugated second-step antimouse antibody. In panels b and e, the nuclei were visualized by DAPI counterstaining. In panels c and f, the nuclei incorporating BrdU were visualized by staining with an anti-BrdU antibody directly conjugated with fluorescein. The injection of a single myotube nucleus resulted in the expression of the injected plasmids in all of the nuclei belonging to the same myotube; one representative field is shown. The results were reproduced in two independent experiments. In each of these experiments at least 50 nuclei positive for the expression of E1A were counted and scored for BrdU staining. When the E1A N-terminal expression construct was injected alone, almost 100% of the E1A-expressing nuclei were also positive for BrdU staining; in contrast, upon coinjection of the cyclin D3 expression vector, only 15% of the E1A-positive nuclei showed significant incorporation of BrdU.
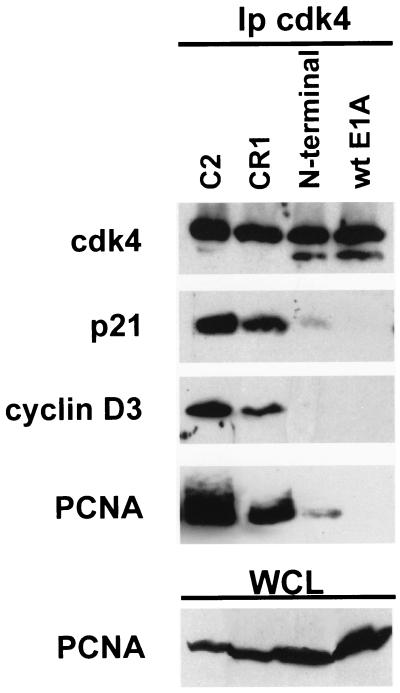
A possible cyclin D3-mediated inhibition might rely upon the ability of D-type cyclins to bind PCNA, the auxiliary factor of DNA polymerases δ and ɛ, required for DNA replication and repair (64). Because PCNA is expressed in differentiated C2 cells and participates in cdk4-cyclin D3-pRb complexes (Fig. 3A and 4A), we asked whether the E1A-mediated inactivation of cyclin D3 might result in PCNA dissociation from these complexes. We thus determined the expression levels and interaction of PCNA with cdk4 in E1A-expressing C2 cells under differentiation conditions (Fig. 7). It was found that in parental C2 myotubes the cdk4 complexes contained p21, cyclin D3, and PCNA, whereas in C2 cells expressing wt E1A or the E1A N-terminal mutant, neither of which accumulated cyclin D3, the interaction of cdk4 with PCNA and p21 was inhibited. Since the PCNA levels were not altered by E1A, these associations appeared to depend on the presence of cyclin D3. Our data indicate that the availability of unbound PCNA increases in wt E1A and N-terminal cells compared to parental C2 cells. We surmise that, in terminally differentiated myotubes, cyclin D3 might function by trapping PCNA into pRb-bound, inactive cdk4 complexes, thus preventing the interaction of PCNA with the DNA synthesis apparatus.
FIG. 7.
Disruption of PCNA-cdk4 complexes in E1A-expressing C2 cells. Cell lysates prepared from C2 or from E1A-expressing C2 cells cultured in differentiation medium for 48 h were immunoprecipitated with the anti-cdk4 antibody. Immunoprecipitated proteins were then resolved by SDS-PAGE and analyzed by immunoblotting by using antibodies specific for p21, cyclin D3, or PCNA. The levels of PCNA in each cell line were detected by immunoblot analysis of whole-cell lysates (WCL).
The function of pRb is required for cyclin D3 stabilization in differentiating muscle cells.
Two findings of the present study let us hypothesize that a pRb-dependent mechanism stabilizes cyclin D3 in differentiating C2 cells. The first finding was that nearly all the cyclin D3 of myotubes was complexed with pRb (Fig. 4A), and the second finding was that the inhibition of cyclin D3 accumulation in C2 cells expressing E1A mutants correlated with the ability of E1A to bind pRb (Fig. 5B). The simplest interpretation of these findings is that cyclin D3 was stabilized by its binding to pRb and that E1A disrupted such interaction and destabilized cyclin D3. If this was correct, the cyclin D3 protein would not be able to accumulate during the differentiation of muscle cells lacking pRb.
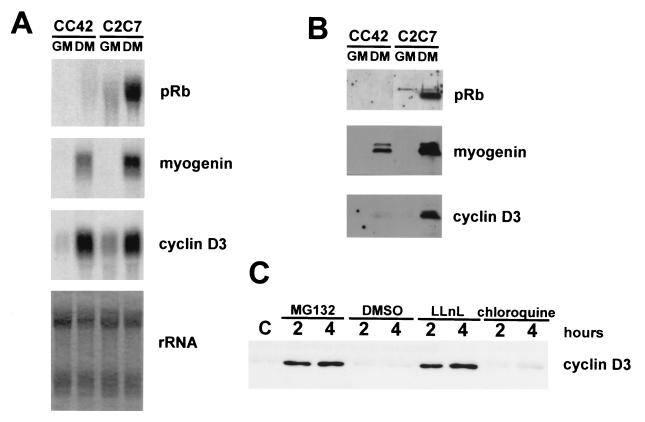
To test this prediction, we determined the expression levels of cyclin D3 in the CC42 Rb−/− myogenic cell line (78). Total RNA and whole-cell protein extracts were prepared from CC42 cells, either proliferating or kept in differentiation medium for 48 h, and analyzed by Northern and Western blotting, respectively (Fig. 8A and B). The results show that cyclin D3 mRNA was induced in differentiating CC42 (Rb−/−) to the same extent as in C2C7 (Rb+/+) cells; in contrast, cyclin D3 protein accumulated only in C2C7 myotubes. The levels of myogenin mRNA were paralleled by the accumulation of similar levels of the myogenin protein in both cell lines. These observations agree with the suggestion that, in the absence of functional pRb, the inherently unstable cyclin D3 is rapidly destroyed.
FIG. 8.
Cyclin D3 is a short-lived protein in Rb−/− myocytes, and its degradation is mediated by the ubiquitin proteasome pathway. (A) Northern blot analysis of total RNA isolated from CC42 and C2 myogenic cells, either proliferating (GM) or maintained in differentiation medium for 48 h (DM). Identical filters were probed for Rb, myogenin, or cyclin D3. Ethidium bromide staining of rRNA on one of the filters was photographed with UV light. (B) Whole-cell extracts were prepared from C2 and CC42 cells, either proliferating (GM) or differentiating (48 h in differentiation medium [DM]). Equal amounts of proteins were separated on SDS-PAGE gels and subjected to immunoblot analysis with antibodies specific for pRb, myogenin, and cyclin D3. (C) Levels of cyclin D3 in differentiating CC42 cells treated with proteasome inhibitors. CC42 cells were exposed to differentiation medium for 24 h and then treated for 2 or 4 h with DMSO solvent alone or with MG132 (10 μM), LLnL (50 μM), or chloroquine (100 μM), as indicated. Whole-cell lysates, normalized for protein concentration, were immunoprecipitated with a cyclin D3 polyclonal antibody (sc182; Santa Cruz Biotechnology). Immunoprecipitated proteins were then resolved by SDS-PAGE and analyzed by immunoblotting by using an anti-cyclin D3 MAb (sc453; Santa Cruz Biotechnology).
Many short-lived regulatory proteins are degraded by a large protease complex, known as the 26S proteasome (23, 31). To determine whether the absence of pRb left cyclin D3 exposed to the action of the proteasome, we explored the effect of specific proteasome inhibitors on the stability of cyclin D3 in differentiating CC42 cells. Confluent cultures were transferred to differentiation medium for 24 h (to induce cyclin D3 transcription) and then treated with either N-acetyl-leucinyl-norleucinal (LLnL) or MG132, both potent inhibitors of the 26S proteasome. The cells treated for 2 or 4 h with 10 μM MG132 or 50 μM LLnL were lysed and immunoprecipitated with an anti-cyclin D3 polyclonal antibody; the immunoprecipitates were then analyzed by Western blotting with a MAb to mouse cyclin D3 (Fig. 8C). Compared with cells treated with the dimethylsulfoxide (DMSO) solvent alone, or with chloroquine, an inhibitor of lysosomal proteolysis, the LLnL or MG132 treatment resulted in a strong increase of the cyclin D3 levels. This result clearly indicates that in the absence of pRb, cyclin D3 is unstable and that its degradation is dependent on the function of the 26S proteasome.
Effect of ectopic expression of cyclin D3 in growing and differentiating myoblasts.
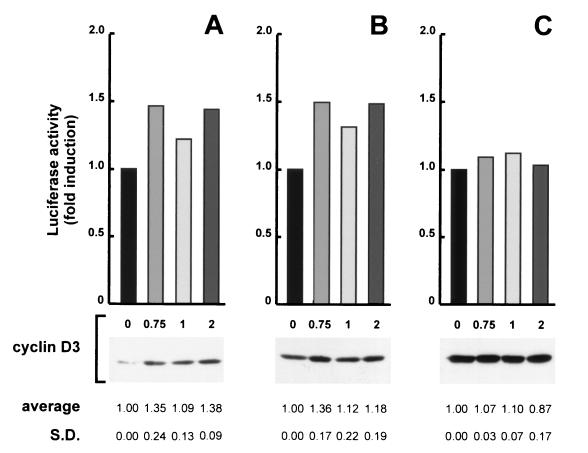
The results reported thus far indicate that muscle differentiation accumulates high levels of cyclin D3 through both transcriptional and posttranscriptional mechanisms and that cyclin D3 contributes a function antagonistic to growth in terminally differentiated myotubes; this must be overcome by E1A to reactivate DNA synthesis in these postmitotic cells. To examine the role of cyclin D3 in differentiation with another, more direct approach, we determined whether the ectopic expression of cyclin D3 could activate the expression of muscle-specific genes in growing C2 myoblasts. We transfected growing C2 myoblasts with a luciferase reporter plasmid containing the MCK promoter-enhancer in the presence or absence of a cyclin D3-expression construct; a CMV-β-galactosidase expression construct was cotransfected to monitor transfection efficiencies. MCK transcriptional activity was then assessed in nonconfluent or mitogen-stimulated confluent cultures as well as in cultures under normal differentiating conditions. As shown in Fig. 9, the MCK promoter activity was slightly enhanced by ectopic cyclin D3 in nonconfluent and confluent cultures in growth medium.
FIG. 9.
Effect of ectopic expression of cyclin D3 on MCK expression in growing and differentiating C2 myoblasts. Proliferating C2 myoblasts were transfected with 0.5 μg of the MCK luc reporter plasmid in the presence of the indicated amount of the cyclin D3 expression construct (Rc/CMV-cycD3); the Rc/CMV expression vehicle without an insert was included to normalize DNA in all transfections. Eighteen hours after transfection, the cells were trypsinized; one half of the cells were plated onto 90-mm-diameter dishes, the other half were plated onto 60-mm-diameter dishes, and refed with growth medium. Parallel transfections were directly transferred to differentiation medium. After 72 h the cells in 90-mm-diameter dishes were subconfluent (A), those in 60-mm-diameter dishes were confluent (B), and those in differentiation medium were fully differentiated (C); at that time cells were collected and assayed for luciferase and β-galactosidase activities. Equal amounts of proteins from each cell lysate were also assayed for cyclin D3 expression by Western blotting. The results shown are from one representative experiment. MCK-luc activity is expressed relative to the levels detected in the absence of cyclin D3. The experiments were done in duplicate and repeated three times. The mean values (expressed as fold induction relative to the baseline values) and the standard deviations (S.D.) of these experiments are shown at the bottom.
We observed, however, that cotransfection with cyclin D3 reproducibly downregulated the expression of CMV-β-galactosidase in all the conditions tested. When corrected for the CMV-β-galactosidase inhibition, the values of induction of MCK promoter activity by ectopic cyclin D3 in mitogen-stimulated myoblasts increased. Ectopic cyclin D3 had no specific effect in the cultures placed in differentiation medium, which exhibited the usual strong induction of endogenous cyclin D3 (Fig. 9). These results indicate that a moderate myogenesis-promoting effect by ectopic cyclin D3 can be appreciated only in C2 myoblasts or, after confluence, before the overt induction of endogenous cyclin D3 takes place. This effect does not appear to be dose dependent, suggesting that the exogenous cyclin D3 cannot accumulate above a given threshold in transfected myoblasts (Fig. 9).
In contrast, previous studies have reported that the overexpression of cyclin D3 in differentiated myocytes either did not specifically inhibit (70, 82) or partly inhibited MCK expression (26). Our results agree with those of Rao et al. and Skapek et al. (70, 82), who found that overexpression of cyclin D3 caused similar reductions of muscle-specific and non-muscle-specific promoter activities. One interpretation of these observations is that the occupation by ectopic cyclin D3 of all the available pRb pockets might preclude the regulatory interaction of pRb with other proteins controlling the transcription of a number of genes.
DISCUSSION
The differentiation of skeletal myoblasts requires that these cells exit the cell cycle. Besides initiating muscle-specific gene expression, the myogenic bHLH factor MyoD has been implicated in promoting the cell cycle arrest, by inducing cell growth repressors, such as pRb and the kinase inhibitor p21. In this study, we show that cyclin D3, whose expression is strongly upregulated in differentiating muscle cells, also plays an important role in the irreversible cell cycle arrest of differentiated myocytes. The analysis of the mechanism of pRb, p21, and cyclin D3 induction during differentiation revealed that these genes are regulated similarly by MyoD.
The expression of p21, Rb, and cyclin D3 is directly regulated by MyoD and requires the function of p300.
By using MyoD-ER cell lines, it has been previously shown that hormone-activated MyoD, in the absence of new protein synthesis, directly induces the expression of the endogenous MyoD gene and that of myogenin but does not induce several downstream muscle genes (32). This finding indicates that the initial activation of MyoD leads to the induction of early differentiation genes, whose function, in turn, is required to activate late muscle genes. We show here that p21, Rb, and cyclin D3 are induced by MyoD (like myogenin) without the requirement of newly synthesized factors, allowing these genes to be categorized as early differentiation markers. The MyoD-mediated induction of these genes is also dependent on p300 function.
Both p300 and pRb have been demonstrated to interact with MyoD and to be required for the completion of the MyoD-mediated transactivation of muscle genes (10, 17, 25, 58, 67, 69, 77). These observations are now refined, as we found that p300 is needed for the MyoD-mediated induction of the early differentiation markers, while pRb is essential for the expression of the late differentiation genes. This conclusion was reached by using C2 cells that stably express either E1A or its mutants that have lost the ability to bind and inactivate p300 and/or the pRb family of cellular proteins.
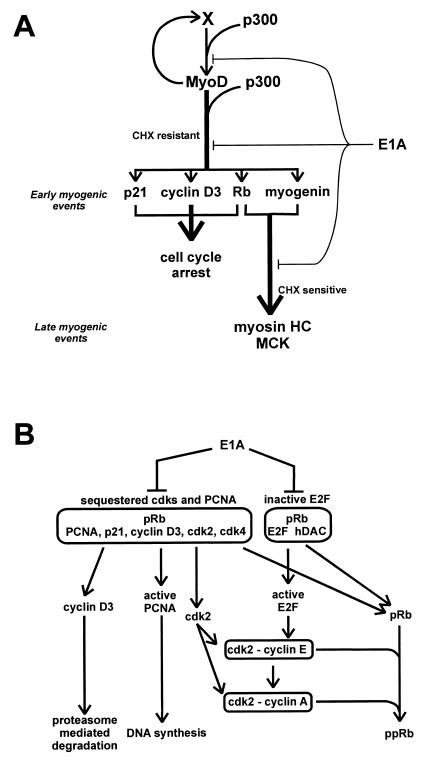
Taken together, the present results allow us propose a model in which MyoD activates its target genes by two sequentially acting molecular mechanisms, the first requiring p300 and the second requiring pRb (Fig. 10A). p300 is essential for the direct induction by MyoD of pRb, p21, cyclin D3, and myogenin. pRb is required for the myogenin-mediated induction of late differentiation genes as well as for the irreversible cell cycle arrest that accompanies terminal differentiation.
FIG. 10.
(A) Schematic model of the multistep process of terminal differentiation. MyoD directly activates its own transcription and that of early differentiation genes through a mechanism that is cycloheximide (CHX) resistant and requires p300. Induction of late differentiation genes is CHX sensitive and requires pRb and myogenin. pRb, p21, and cyclin D3 contribute to cell cycle arrest of differentiating myoblasts. E1A, by targeting both p300 and pRb, interferes with the early and the late steps of muscle differentiation. (B) Schematic model of the E1A-mediated reinduction of DNA synthesis in terminally differentiated myotubes. pRb sequesters cyclin D3 along with inactive cdk’s and PCNA; pRb binds E2F and recruits histone deacetylase (hDAC) to E2F. E1A, by binding the pRb pocket, inhibits the binding of E2F and cyclin D3 to pRb. Consequently, active E2F induces S-phase genes, and cyclin D3 is degraded; active cdk’s and PCNA contribute to S-phase entry.
Regulation of cyclin D3 expression during muscle differentiation.
It has been well established that D-type cyclins, whose expression is induced by serum growth factors (50), promote cellular proliferation by activating cdk4 and cdk6, both of which act as pRb kinases (48, 49, 51). Thus, as myoblasts become committed to terminal differentiation by serum growth factor withdrawal, the expression and/or activity of D-type cyclins would be expected to be down-regulated. Indeed, the activation of the terminal differentiation program in C2 myoblasts leads to the rapid disappearance of cyclin D1 mRNA and protein (33, 71, 91).
With regard to cyclin D3, however, the present results show not only that MyoD induces cyclin D3 mRNA but also that a mechanism stabilizes the cyclin D3 protein in differentiating C2 cells. Such mechanism requires the function of pRb, since Rb−/− muscle cells induced to differentiate fail to accumulate the cyclin D3 protein, despite a normal induction of cyclin D3 mRNA. The extremely low levels of cyclin D3 protein in differentiating Rb−/− myoblasts are due to the intrinsic instability of cyclin D3 when Rb protection is missing; we found that the steady-state level of cyclin D3 increased in the presence of the LLnL and MG132 proteasome inhibitors, which also indicates that the cyclin D3 degradation occurring in the absence of functional pRb is mediated by the ubiquitin proteasome pathway.
The pRb dependence of cyclin D3 stability might either simply require the ability of unphosphorylated pRb to interact physically with cyclin D3 or be an indirect consequence of pRb-mediated repression of unknown genes whose activity leads to cyclin D3 degradation. There is not yet enough information to exclude the latter possibility; the former, however, is supported by two findings. The first finding is that nearly all the cyclin D3 of myotubes is found complexed with pRb. The second finding is that the cyclin D3 protein level in C2 cells expressing E1A mutants correlates with the inability of E1A to sequester pRb. It has been previously reported that E1A, E7, and D-type cyclins, which all possess the LXCXE pRb-binding motif, interact with the pRb pocket region in a competitive manner (15, 20). Taken together, these observations are consistent with the idea that in differentiating muscle cells cyclin D3 gets stabilized by its binding to unphosphorylated pRb, and that E1A promotes cyclin D3 destruction when displacing cyclin D3 from the pRb complexes.
Cyclin D3 complexes in differentiated C2 cells.
The results discussed above indicate that the induction of cyclin D3 mRNA and the stabilization of the cyclin D3 protein are physiological features of muscle differentiation. Interestingly, it has been recently reported that cyclin D3 also accumulates to high levels in differentiating skeletal muscle in vivo during the late stages of mouse fetal development and the first weeks of postnatal life but not in fully differentiated adult skeletal muscle tissues, suggesting a role for cyclin D3 in the induction and/or establishment, rather than in the maintenance, of mammalian skeletal muscle differentiation (4).
We found that the overexpression of ectopic cyclin D3 can augment the transcription from the MCK promoter in growing myoblasts. Such an increase, though relatively modest, is similar in magnitude to that elicited by p21 when overexpressed in mitogen-stimulated myoblasts (reference 82 and data not shown). Moreover, the myogenesis-promoting effect of cyclin D3 was found be dose independent, probably due to the impossibility of ectopic cyclin D3 accumulating to high levels in proliferating myoblasts, in which pRb is hyperphosphorylated and thus unable to protect cyclin D3 from degradation.
Especially intriguing is the observation that cyclin D1 is down-regulated in differentiating muscle cells and that its ectopic expression (unlike that of cyclin D3) prevents muscle gene activation (26, 70, 82). This clearly indicates that in spite of many similarities, cyclin D1 and D3 may have profoundly different roles in muscle cells. The three D-type cyclins exhibit a similar capability of activating cdk4 toward pRb in in vitro kinase assays and in a baculovirus-insect cell overexpression system (34, 49). On the other hand, it should be noted that they do not behave identically with respect to their interaction with pRb, as cyclins D2 and D3 bind to unphosphorylated pRb much more efficiently than cyclin D1 (20, 34). In addition, while all D-type cyclins can form complexes with other catalytic partners besides cdk4/6, such as cdk2, and cdk5 (48, 99), only cyclin D2 and D3 yield an active pRb kinase when allowed to interact with cdk2 in insect cells, whereas cyclin D1 does not (20).
By means of antibody depletion experiments, we have been able to demonstrate that in myotubes cyclin D3 is nearly totally complexed with (inactive) cdk4 as well as with a fraction of unphosphorylated pRb. Others have shown that cyclin D3 also forms complexes with inactive cdk2 in myotubes (83). Considering that the bulk of cyclin D3 is pRb associated and that pRb can form high-order structures by association between the N and C termini (73), it is conceivable that in differentiated C2 cells, cdk4, cdk2, cyclin D3, and pRb participate in multiprotein complexes in which cyclin D3 is the limiting component mediating the interaction of cdk4 and cdk2 with pRb. Neither cdk2, cdk4, nor cyclin D3 immunoprecipitates from differentiated C2 cells had associated pRb kinase activity, presumably due to the presence of high amounts of p21 and p27 in these complexes (present study and references 83 and 92). This failure of the cdk4 and cdk2 subunits to catalyze pRb phosphorylation suggests a mechanism that is able to sequester these kinases in inactive complexes with unphosphorylated pRb in which cyclin D3 may act as a bridge.
The present work also shows that in differentiated C2 cells cyclin D3 forms complexes with PCNA, thus mediating the interaction of a fraction of PCNA with pRb. By analogy with the effect previously reported for the binding of PCNA to cyclin D1 (64), the binding of cyclin D3 is also likely to have an inhibitory effect on PCNA function. During the G1 phase of the cell cycle, the association of cyclin D1 with PCNA negatively regulates PCNA function, whereas in S-phase cells (or following DNA damage) the cyclin D1 down-regulation leads to PCNA release and DNA synthesis (or repair) (64).
Our time course analyses have shown that pRb, progressively dephosphorylated during differentiation, also becomes extraction resistant. pRb is known to be easily extractable from the nuclei of cells in S phase but not in G0 or G1 phase (53), due to a cell cycle-dependent interaction with the nuclear matrix (45). The binding of cyclin D3 to unphosphorylated pRb might provide a docking site allowing the formation of inactive cdk4 and cdk2 complexes, also containing PCNA. Such complexes might keep these kinases, and PCNA, sequestered into structures architecturally organized on the nuclear matrix or at specific nuclear subcompartments.
The function of cyclin D3 is required for the irreversible cell cycle arrest of differentiated C2 cells.
The cyclin D3-mediated associations discussed above delineate a self-sustaining mechanism able to ensure prompt pRb dephosphorylation and an irreversible cell cycle arrest during myogenic differentiation. In this mechanism, the critical role played by cyclin D3 is supported by the effects of E1A interference with the normal cell cycle regulation as follows: (i) the ability of E1A, when stably expressed in C2 cells, to induce pRb phosphorylation correlates with its ability to inhibit cyclin D3 expression, and (ii) E1A targets cyclin D3 to reactivate DNA synthesis in terminally differentiated myotubes.
Concerning the first point, we have shown that E1A with intact ability to bind pRb leads to repression of cyclin D3 and, concomitantly, to the induction of cyclins E and A. Consequently, due to the activation of cdk2, a large pRb fraction is found in its hyperphosphorylated, inactive form when E1A-expressing C2 cells are shifted to differentiation medium. It has been established that the binding of E1A to the pocket region of the pRb family proteins releases the E2F transcription factors, required to induce cyclin E and cyclin A (5, 21, 59, 79, 93), and that the activity of cyclin E/cdk2 is required for the induction of the cyclin A gene (76, 103, 104). The observed correlation between the inhibition of cyclin D3 and the induction of cyclin A upon E1A expression is interesting, as it suggests that cyclin D3 might contribute to maintain cyclin A repressed in differentiated C2 cells. Again, such a mechanism might rely upon the ability of cyclin D3 to keep cdk2 sequestered in inactive complexes with unphosphorylated pRb. This hypothetical mechanism would agree with the observations made by others (35, 83) that cyclin D3 forms complexes with inactive cdk2 in myotubes and with our finding that in these cells nearly all cyclin D3 is pRb associated.
With respect to the second point, we have shown that the ability of microinjected E1A to reactivate DNA synthesis in terminally differentiated myotubes is counteracted by cyclin D3 coinjection, indicating that in these cells cyclin D3 exerts an inhibitory function that must be overcome by E1A to induce DNA synthesis. This function is likely due to the ability of cyclin D3 to trap PCNA into inactive complexes with cdk4 and pRb, as suggested by the finding that by inactivating cyclin D3, E1A causes the dissociation of PCNA from these complexes, thus increasing the level of free, active PCNA.
The above observations together with the recent finding that the overexpression of E2F alone is insufficient to restart DNA synthesis in terminally differentiated myotubes (68), suggest that the ability of E1A to reinduce DNA synthesis in myotubes relies on its ability to release both cyclin D3 and E2F from their interaction with the pocket region of pRb (Fig. 10B). By displacing cyclin D3, E1A also dissociates cdk4, cdk2, and PCNA from pRb, thus releasing these proteins from an inhibitory interaction. Concomitantly, the liberation of E2F from pRb causes activation of S-phase genes, such as cyclin E and cyclin A and DNA polymerase α (5, 13, 21, 76, 93, 104). Cdk2-cyclin E-PCNA and cdk2-cyclin A-PCNA complexes would then assemble, phosphorylate pRb, and become available to the DNA replication apparatus. Several studies have demonstrated a requirement of cyclin A and cdk2 for cell entry into S phase (22, 62, 63, 88, 105) and the colocalization of cyclin A and cdk2 with PCNA at the sites of DNA replication (9).
Although the three-dimensional structure of the pRb pocket region revealed that the binding site for E2F and that for LXCXE proteins are distinct from each other and that E2F and LXCXE peptides can bind concurrently (38), the model in Fig. 10B suggests that cyclin D3 and E2F interact with different pools of pRb molecules. This is based on our finding that in differentiated C2 myotubes the concentration of pRb vastly exceeds that of cyclin D3 and is consistent with the recent discovery that pRb can interact simultaneously with E2F and with a histone deacetylase through the LXCXE-binding site, thus leading to transcription repression at promoters containing E2F binding sites (7, 40, 41).
ACKNOWLEDGMENTS
We are indebted to J. Bartek, M. Eilers, M. Ewen, A. Giordano, E. Moran, L. Kedes, J. Pines, V. Sartorelli, C. Schneider, C. Sherr, and H. Weintraub for providing plasmids and reagents. We thank L. Baron and G. Santarelli for their excellent technical assistance. We acknowledge the help of A. Graessmann, in whose laboratory P.L.P. performed the microinjection experiments. We are grateful to C. Vesco for truly helpful discussions and valuable suggestions during the preparation of the manuscript. We thank F. Tirone for critical reading of the manuscript.
This work was supported by the Associazione Italiana Ricerca sul Cancro (AIRC), Milan, Italy. C.C. received a an AIRC postdoctoral fellowship. L.R. and F.B. were supported by CNR fellowships.
REFERENCES
- 1.Andres V, Walsh K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J Cell Biol. 1996;132:657–666. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 3.Bader D, Masaki T, Fischman D A. Immunochemical analysis of myosin heavy chain during avian myogenesis in vivo and in vitro. J Cell Biol. 1982;95:763–770. doi: 10.1083/jcb.95.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartkova J, Lukas J, Strauss M, Bartek J. Cyclin D3: requirement for G1/S transition and high abundance in quiescent tissues suggest a dual role in proliferation and differentiation. Oncogene. 1998;17:1027–1037. doi: 10.1038/sj.onc.1202016. [DOI] [PubMed] [Google Scholar]
- 5.Botz J, Zerfass-Thome K, Spitkovsky D, Delius H, Vogt B, Eilers M, Hatzigeorgiou A, Jansen-Dürr P. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol Cell Biol. 1996;16:3401–3409. doi: 10.1128/mcb.16.7.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 8.Buckingham M. Making muscle in mammals. Trends Genet. 1992;8:144–148. doi: 10.1016/0168-9525(92)90373-C. [DOI] [PubMed] [Google Scholar]
- 9.Cardoso M C, Leonhardt H, Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- 10.Caruso M, Martelli F, Giordano A, Felsani A. Regulation of MyoD gene transcription and protein function by the transforming domains of the adenovirus E1A oncoprotein. Oncogene. 1993;8:267–278. [PubMed] [Google Scholar]
- 11.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 12.Davis R L, Weintraub H. Acquisition of myogenic specificity by replacement of three amino acid residues from MyoD into E12. Science. 1992;256:1027–1030. doi: 10.1126/science.1317057. [DOI] [PubMed] [Google Scholar]
- 13.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Wet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dowdy S F, Hinds P W, Louie K, Reed S I, Arnold A, Weinberg R A. Physical interaction of the retinoblastoma protein with human D cyclins. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- 16.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A- associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 17.Eckner R, Yao T P, Oldread E, Livingston D M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 18.Emerson C P. Skeletal myogenesis: genetics and embryology to the fore. Curr Opin Genet Dev. 1993;3:265–274. doi: 10.1016/0959-437x(93)90033-l. [DOI] [PubMed] [Google Scholar]
- 19.Endo T, Goto S. Retinoblastoma gene product Rb accumulates during myogenic differentiation and is deinduced by the expression of SV40 large T antigen. J Biochem. 1992;112:427–430. doi: 10.1093/oxfordjournals.jbchem.a123916. [DOI] [PubMed] [Google Scholar]
- 20.Ewen M E, Sluss H K, Sherr C J, Matsushime H, Kato J, Livingston D M. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 21.Geng Y, Eaton E N, Picon M, Roberts J M, Lundberg A S, Gifford A, Sardet C, Weinberg R A. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene. 1996;12:1173–1180. [PubMed] [Google Scholar]
- 22.Girard F, Strausfeld U, Fernandez A, Lamb N J. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell. 1991;67:1169–1179. doi: 10.1016/0092-8674(91)90293-8. [DOI] [PubMed] [Google Scholar]
- 23.Goldberg A L. Functions of the proteasome: the lysis at the end of the tunnel. Science. 1995;268:522–523. doi: 10.1126/science.7725095. [DOI] [PubMed] [Google Scholar]
- 24.Graessmann M, Graessmann A. Microinjection of tissue culture cells. Methods Enzymol. 1983;101:482–492. doi: 10.1016/0076-6879(83)01033-2. [DOI] [PubMed] [Google Scholar]
- 25.Gu W, Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 26.Guo K, Walsh K. Inhibition of myogenesis by multiple cyclin-Cdk complexes. Coordinate regulation of myogenesis and cell cycle activity at the level of E2F. J Biol Chem. 1997;272:791–797. doi: 10.1074/jbc.272.2.791. [DOI] [PubMed] [Google Scholar]
- 27.Guo K, Wang J, Andres V, Smith R C, Walsh K. MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol Cell Biol. 1995;15:3823–3829. doi: 10.1128/mcb.15.7.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halevy O, Novitch B G, Spicer D B, Skapek S X, Rhee J, Hannon G J, Beach D, Lassar A B. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 29.Harlow E, Franza B R, Jr, Schley C. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J Virol. 1985;55:533–546. doi: 10.1128/jvi.55.3.533-546.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harper J W, Elledge S J. Cdk inhibitors in development and cancer. Curr Opin Genet Dev. 1996;6:56–64. doi: 10.1016/s0959-437x(96)90011-8. [DOI] [PubMed] [Google Scholar]
- 31.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 32.Hollenberg S M, Cheng P F, Weintraub H. Use of a conditional MyoD transcription factor in studies of MyoD trans-activation and muscle determination. Proc Natl Acad Sci USA. 1993;90:8028–8032. doi: 10.1073/pnas.90.17.8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jahn L, Sadoshima J, Izumo S. Cyclins and cyclin-dependent kinases are differentially regulated during terminal differentiation of C2C12 muscle cells. Exp Cell Res. 1994;212:297–307. doi: 10.1006/excr.1994.1147. [DOI] [PubMed] [Google Scholar]
- 34.Kato J, Matsushime H, Hiebert S W, Ewen M E, Sherr C J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 35.Kiess M, Gill R M, Hamel P A. Expression and activity of the retinoblastoma protein (pRB)-family proteins, p107 and p130, during L6 myoblast differentiation. Cell Growth Differ. 1995;6:1287–1298. [PubMed] [Google Scholar]
- 36.Kiess M, Montgomery Gill R, Hamel P A. Expression of the positive regulator of cell cycle progression, cyclin D3, is induced during differentiation of myoblasts into quiescent miotubes. Oncogene. 1995;10:159–166. [PubMed] [Google Scholar]
- 37.Lassar A B, Skapek S X, Novitch B. Regulatory mechanisms that coordinate skeletal muscle differentiation and cell cycle withdrawal. Curr Opin Cell Biol. 1994;6:788–794. doi: 10.1016/0955-0674(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 38.Lee J O, Russo A A, Pavletich N P. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 39.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 40.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 41.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 42.Maione R, Amati P. Interdependence between muscle differentiation and cell-cycle control. Biochim Biophys Acta. 1997;1332:M19–M30. doi: 10.1016/s0304-419x(96)00036-4. [DOI] [PubMed] [Google Scholar]
- 43.Maione R, Fimia G M, Holman P, Schaffhausen B, Amati P. Retinoblastoma anti-oncogene is involved in the inhibition of myogenesis by polyomavirus large T antigen. Cell Growth Differ. 1994;5:231–237. [PubMed] [Google Scholar]
- 44.Mal A, Poon R Y, Howe P H, Toyoshima H, Hunter T, Harter M L. Inactivation of p27Kip1 by the viral E1A oncoprotein in TGFβ-treated cells. Nature. 1996;380:262–265. doi: 10.1038/380262a0. [DOI] [PubMed] [Google Scholar]
- 45.Mancini M A, Shan B, Nickerson J A, Penman S, Lee W H. The retinoblastoma gene product is a cell cycle-dependent, nuclear matrix-associated protein. Proc Natl Acad Sci USA. 1994;91:418–422. doi: 10.1073/pnas.91.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martelli F, Cenciarelli C, Santarelli G, Polikar B, Felsani A, Caruso M. MyoD induces retinoblastoma gene expression during myogenic differentiation. Oncogene. 1994;9:3579–3590. [PubMed] [Google Scholar]
- 47.Matsuoka S, Yamaguchi M, Matsukage A. D-type cyclin-binding regions of proliferating cell nuclear antigen. J Biol Chem. 1994;269:11030–11036. [PubMed] [Google Scholar]
- 48.Matsushime H, Ewen M E, Strom D K, Kato J Y, Hanks S K, Roussel M F, Sherr C J. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell. 1992;71:323–334. doi: 10.1016/0092-8674(92)90360-o. [DOI] [PubMed] [Google Scholar]
- 49.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J Y. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsushime H, Roussel M F, Ashmun R A, Sherr C J. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- 51.Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol. 1994;14:2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Missero C, Calautti E, Eckner R, Chin J, Tsai L H, Livingston D M, Dotto G P. Involvement of the cell-cycle inhibitor Cip1/WAF1 and the E1A-associated p300 protein in terminal differentiation. Proc Natl Acad Sci USA. 1995;92:5451–5455. doi: 10.1073/pnas.92.12.5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mittnacht S, Weinberg R A. G1/S phosphorylation of the retinoblastoma protein is associated with an altered affinity for the nuclear compartment. Cell. 1991;65:381–393. doi: 10.1016/0092-8674(91)90456-9. [DOI] [PubMed] [Google Scholar]
- 54.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 55.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mymryk J S, Lee R W, Bayley S T. Ability of adenovirus 5 E1A proteins to suppress differentiation of BC3H1 myoblasts correlates with their binding to a 300 kDa cellular protein. Mol Biol Cell. 1992;3:1107–1115. doi: 10.1091/mbc.3.10.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Norbury C, Nurse P. Animal cell cycles and their control. Annu Rev Biochem. 1992;61:441–470. doi: 10.1146/annurev.bi.61.070192.002301. [DOI] [PubMed] [Google Scholar]
- 58.Novitch B G, Mulligan G J, Jacks T, Lassar A B. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohtani K, DeGregori J, Nevins J R. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olson E N, Klein W H. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- 61.Otten A D, Firpo E J, Gerber A N, Brody L L, Roberts J M, Tapscott S J. Inactivation of MyoD-mediated expression of p21 in tumor cell lines. Cell Growth Differ. 1997;8:1151–1160. [PubMed] [Google Scholar]
- 62.Pagano M, Pepperkok R, Lukas J, Baldin V, Ansorge W, Bartek J, Draetta G. Regulation of the cell cycle by the cdk2 protein kinase in cultured human fibroblasts. J Cell Biol. 1993;121:101–111. doi: 10.1083/jcb.121.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pagano M, Theodoras A M, Tam S W, Draetta G F. Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev. 1994;8:1627–1639. doi: 10.1101/gad.8.14.1627. [DOI] [PubMed] [Google Scholar]
- 65.Parker S B, Eichele G, Zhang P, Rawls A, Sands A T, Bradley A, Olson E N, Harper J W, Elledge S J. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science. 1995;267:1024–1027. doi: 10.1126/science.7863329. [DOI] [PubMed] [Google Scholar]
- 66.Phelps D E, Hsiao K M, Li Y, Hu N P, Franklin D S, Westphal E, Lee E Y, Xiong Y. Coupled transcriptional and translational control of cyclin-dependent kinase inhibitor p18INK4c expression during myogenesis. Mol Cell Biol. 1998;18:2334–2343. doi: 10.1128/mcb.18.4.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Puri P L, Avantaggiati M L, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puri P L, Cimino L, Fulco M, Zimmerman C, La Thangue N B, Giordano A, Graessmann A, Levrero M. Regulation of E2F4 mitogenic activity during terminal differentiation by its heterodimerization partners for nuclear translocation. Cancer Res. 1998;58:1325–1331. [PubMed] [Google Scholar]
- 69.Puri P L, Sartorelli V, Yang X J, Hamamori Y, Ogryzko V V, Howard B H, Kedes L, Wang J Y, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 70.Rao S S, Chu C, Kohtz D S. Ectopic expression of cyclin D1 prevents activation of gene transcription by myogenic basic helix-loop-helix regulators. Mol Cell Biol. 1994;14:5259–5267. doi: 10.1128/mcb.14.8.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rao S S, Kohtz D S. Positive and negative regulation of D-type cyclin expression in skeletal myoblasts by basic fibroblast growth factor and transforming growth factor beta. A role for cyclin D1 in control of myoblast differentiation. J Biol Chem. 1995;270:4093–4100. doi: 10.1074/jbc.270.8.4093. [DOI] [PubMed] [Google Scholar]
- 72.Reed S I. The role of p34 kinases in the G1 to S-phase transition. Annu Rev Cell Biol. 1992;8:529–561. doi: 10.1146/annurev.cb.08.110192.002525. [DOI] [PubMed] [Google Scholar]
- 73.Riley D J, Lee E Y, Lee W H. The retinoblastoma protein: more than a tumor suppressor. Annu Rev Cell Biol. 1994;10:1–29. doi: 10.1146/annurev.cb.10.110194.000245. [DOI] [PubMed] [Google Scholar]
- 74.Rosenthal N. Identification of regulatory elements of cloned genes with functional assays. Methods Enzymol. 1987;152:704–720. doi: 10.1016/0076-6879(87)52075-4. [DOI] [PubMed] [Google Scholar]
- 75.Samuels H H, Stanley F, Casanova J. Depletion of L-3,5,3′-triiodothyronine and l-thyroxine in euthyroid calf serum for use in cell culture studies of the action of thyroid hormone. Endocrinology. 1979;105:80–85. doi: 10.1210/endo-105-1-80. [DOI] [PubMed] [Google Scholar]
- 76.Sanchez I, Dynlacht B D. Transcriptional control of the cell cycle. Curr Opin Cell Biol. 1996;8:318–324. doi: 10.1016/s0955-0674(96)80004-4. [DOI] [PubMed] [Google Scholar]
- 77.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schneider J W, Gu W, Zhu L, Mahdavi V, Nadal-Ginard B. Reversal of terminal differentiation mediated by p107 in Rb−/− muscle cells. Science. 1994;264:1467–1471. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- 79.Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Durr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc Natl Acad Sci USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sherr C J. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 81.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 82.Skapek S X, Rhee J, Spicer D B, Lassar A B. Inhibition of myogenic differentiation in proliferating myoblasts by cyclin D1-dependent kinase. Science. 1995;267:1022–1024. doi: 10.1126/science.7863328. [DOI] [PubMed] [Google Scholar]
- 83.Tedesco D, Baron L, Fischer-Fantuzzi L, Vesco C. Induction of cyclins E and A in response to mitogen removal: a basic alteration associated with the arrest of differentiation of C2 myoblasts transformed by simian virus 40 large T antigen. J Virol. 1997;71:2217–2224. doi: 10.1128/jvi.71.3.2217-2224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tedesco D, Caruso M, Fischer-Fantuzzi L, Vesco C. The inhibition of cultured myoblast differentiation by the simian virus 40 large T antigen occurs after myogenin expression and Rb up-regulation and is not exerted by transformation-competent cytoplasmic mutants. J Virol. 1995;69:6947–6957. doi: 10.1128/jvi.69.11.6947-6957.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thomas P S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci USA. 1980;77:5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thorburn A M, Walton P A, Feramisco J R. MyoD induced cell cycle arrest is associated with increased nuclear affinity of the Rb protein. Mol Biol Cell. 1993;4:705–713. doi: 10.1091/mbc.4.7.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tiainen M, Spitkovsky D, Jansen-Dürr P, Sacchi A, Crescenzi M. Expression of E1A in terminally differentiated muscle cells reactivates the cell cycle and suppresses tissue-specific genes by separable mechanisms. Mol Cell Biol. 1996;16:5302–5312. doi: 10.1128/mcb.16.10.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsai L H, Lees E, Faha B, Harlow E, Riabowol K. The cdk2 kinase is required for the G1-to-S transition in mammalian cells. Oncogene. 1993;8:1593–1602. [PubMed] [Google Scholar]
- 89.Waga S, Hannon G J, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 90.Wang H G, Rikitake Y, Carter M C, Yaciuk P, Abraham S E, Zerler B, Moran E. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J Virol. 1993;67:476–488. doi: 10.1128/jvi.67.1.476-488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang J, Nadal-Ginard B. Regulation of cyclins and p34CDC2 expression during terminal differentiation of C2C12 myocytes. Biochem Biophys Res Commun. 1995;206:82–88. doi: 10.1006/bbrc.1995.1012. [DOI] [PubMed] [Google Scholar]
- 92.Wang J, Walsh K. Inhibition of retinoblastoma protein phosphorylation by myogenesis-induced changes in the subunit composition of the cyclin-dependent kinase 4 complex. Cell Growth Differ. 1996;7:1471–1478. [PubMed] [Google Scholar]
- 93.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 94.Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 95.Weintraub H, Genetta T, Kadesch T. Tissue-specific gene activation by MyoD: determination of specificity by cis-acting repression elements. Genes Dev. 1994;8:2203–2211. doi: 10.1101/gad.8.18.2203. [DOI] [PubMed] [Google Scholar]
- 96.Whyte P, Williamson N M, Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 97.Wigler M, Silverstein S, Lee L S, Pellicer A, Cheg Y C, Axel R. Transfer of purified Herpes virus thymidine kinase gene to cultured mouse cells. Cell. 1977;11:223–232. doi: 10.1016/0092-8674(77)90333-6. [DOI] [PubMed] [Google Scholar]
- 98.Wright W E, Binder M, Funk W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol Cell Biol. 1991;11:4104–4110. doi: 10.1128/mcb.11.8.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xiong Y, Zhang H, Beach D. D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell. 1992;71:505–514. doi: 10.1016/0092-8674(92)90518-h. [DOI] [PubMed] [Google Scholar]
- 100.Xiong Y, Zhang H, Beach D. Subunit rearrangement of the cyclin-dependent kinases is associated with cellular transformation. Genes Dev. 1993;7:1572–1583. doi: 10.1101/gad.7.8.1572. [DOI] [PubMed] [Google Scholar]
- 101.Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 102.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- 103.Zerfass-Thome K, Schulze A, Zwerschke W, Vogt B, Helin K, Bartek J, Henglein B, Jansen-Durr P. p27KIP1 blocks cyclin E-dependent transactivation of cyclin A gene expression. Mol Cell Biol. 1997;17:407–415. doi: 10.1128/mcb.17.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zerfass K, Spitkovsky D, Schulze A, Joswig S, Henglein B, Jansen-Durr P. Adenovirus E1A activates cyclin A gene transcription in the absence of growth factors through interaction with p107. J Virol. 1996;70:2637–2642. doi: 10.1128/jvi.70.4.2637-2642.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zindy F, Lamas E, Chenivesse X, Sobczak J, Wang J, Fesquet D, Henglein B, Brechot C. Cyclin A is required in S phase in normal epithelial cells. Biochem Biophys Res Commun. 1992;182:1144–1154. doi: 10.1016/0006-291x(92)91851-g. [DOI] [PubMed] [Google Scholar]