Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common type of B-cell non-Hodgkin lymphoma in adults and the pathogenesis of DLBCL is multifactorial and complex. Understanding the molecular mechanisms involved in DLBCL is important to identify new therapeutic targets. The present study aimed to screen and identify differentially expressed microRNAs (miRNAs/miRs) between diffuse large B-cell lymphoma (DLBCL) and control [lymph node reactive hyperplasia (LRH)] groups, and to investigate whether miRNAs associated with DLBCL could serve as potential therapeutic targets. In total, 5 DLBCL experimental samples and 5 control samples were obtained from fresh patient tissues. Firstly, the fresh samples were analyzed using miRNA microarray to identify differentially expressed miRNAs. Next, three databases (TargetScan, microRNA.org and PITA) were used to predict by intersection the potential target genes of the 204 differential miRNAs identified, and a Venn diagram of the results was performed. Subsequently, the target genes of differential miRNAs were analyzed by Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis. Finally, to validate the miRNA microarray data, reverse transcription-quantitative PCR (RT-qPCR) was performed for 8 differentially expressed miRNAs (miR-193a-3p, miR-19a-3p, miR-19b-3p, miR-370-3p, miR-1275, miR-490-5p, miR-630 and miR-665) using DLBCL and LRH fresh samples. In total, 204 miRNAs exhibited differential expression, including 105 downregulated and 54 upregulated miRNAs. The cut-off criteria were set as P≤0.05 and fold-change ≥2. A total of 7,522 potential target genes for the 204 miRNAs were predicted. Potential target genes were enriched in the following pathways: ‘Cancer’, ‘MAPK signaling pathway’, ‘regulation of actin cytoskeleton’, ‘focal adhesion’, ‘endocytosis’, ‘Wnt signaling pathway’, ‘axon guidance’, ‘calcium signaling pathway’ and ‘PI3K/AKT signaling pathway’. A total of 8 miRNAs were validated by RT-qPCR, and 4 miRNAs (miR-19b-3p, miR-193a-3p, miR-370-3p and miR-490-5p) exhibited low expression levels in DLBCL (P<0.05), while miR-630 was highly expressed in DLBCL (P<0.05). Overall, the present study screened 204 differentially expressed miRNAs and analyzed the expression levels of 8 differentially expressed miRNAs in DLBCL. These differentially expressed miRNAs may serve as therapeutic targets for improvement of therapeutic efficacy in DLBCL in the future.
Keywords: DLBCL, miRNAs, signaling pathway, therapeutic target
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common type of B-cell non-Hodgkin lymphoma in adults, and is known to be heterogeneous in clinical manifestations, tissue morphology, immune typing and prognosis (1,2). The sub-classification of DLBCL, which is based on cell-of-origin by using the Hans algorithm (3), includes germinal center B-cell like (GCB) and non-GCB DLBCL subtypes. The 5-year overall survival rate for patients with DLBCL is 60–70% with standard chemotherapy of rituximab plus cyclophosphamide, doxorubicin, vincristine and prednisone (4). However, DLBCL has a recurrence rate of 30–50% shortly after treatment, and commonly progresses to the advanced stage (4). Previous studies (5,6) have reported that multiple targets and abnormal signaling pathways lead to DLBCL development, recurrence and drug resistance; thus, potential therapeutic targets associated with signaling pathways are key to improve outcomes for patients. The pathogenesis of DLBCL and effective therapeutic drugs for the disease are currently being investigated.
It is well known that abnormally expressed proteins lead to diseases or tumors. Therefore, targeting abnormal proteins has become a research hotspot. However, without changing the regulation of gene expression, abnormal proteins will still be produced. Since DNA is usually not easy to change, RNA has become the target of regulation. There are numerous non-coding RNAs (ncRNAs), which serve numerous important roles (7).
MicroRNAs (miRNAs/miRs) form a large family of ncRNAs with a length of 19–22 nucleotides, and dysregulation of miRNAs is involved in cancer development and progression (8). Since the first miRNA (lin-4) was identified by Lee et al (9) in 1993, the role of miRNAs in tumors has been widely studied, and it has been reported that miRNAs can regulate ~1/3 of all mammalian expressing genes at the post-transcriptional level, as well as inhibiting translation and/or degrading their targeted mRNAs (8).
The pathogenesis of DLBCL is multifactorial and complex; understanding the molecular mechanisms involved in DLBCL is important to identify new therapeutic targets. The present study aimed to explore the possible pathogenic factors responsible for the occurrence and development of DLBCL at the miRNA, mRNA and protein levels, in order to expand the understanding of the development of DLBCL and provide therapeutic targets for this disease. Our previous study (5) used proteomics methods isobaric tags for relative and absolute quantification (iTRAQ) to explore differentially expressed proteins in DLBCL. A total of 335 differentially expressed proteins were identified. Through Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, it was found that pathways in cancer, PI3K/AKT signaling and alcoholism were significantly changed. The PI3K/AKT signaling pathway was associated with a large number of differentially expressed proteins, suggesting that DLBCL may be associated with PI3K/AKT signaling, which may help to develop a comprehensive treatment of DLBCL and provides new insights into the pathogenesis of the disease (5). Furthermore, formalin-fixed paraffin-embedded DLBCL tissue samples were subjected to immunohistochemistry to verify the expression levels of the differentially expressed proteins identified by iTRAQ and associated with the PI3K/AKT signaling pathway (6). The results revealed that this signaling pathway serves an essential regulatory role in DLBCL (6).
In the current study, microarray methods involving Agilent Human miRNA Array were employed to detect differentially expressed miRNAs between DLBCL and lymph node reactive hyperplasia (LRH) control groups. A total of 10 fresh tissue samples (5 DLBCL and 5 LRH) were analyzed. Databases were used to predict the potential target genes of the differentially expressed miRNAs. The potential target genes were then analyzed by Gene Ontology (GO) and KEGG pathway enrichment analyses. The results of proteomics and miRNA array were comprehensively analyzed, and the expression levels of 8 differentially expressed miRNAs, whose potential target genes may be regulated by the PI3K/AKT signaling pathway, were verified by reverse transcription-quantitative PCR (RT-qPCR). The present study screened and identified differentially expressed miRNAs, and predicted the potential target genes of the differentially expressed miRNAs and their key signaling pathways. The present results provide a basis for further studies on the causes, underlying molecular mechanisms and molecular biomarkers for the diagnosis, prevention and effective treatment of DLBCL.
Materials and methods
Patient samples
A total of 30 fresh tissues were collected from untreated patients who did not receive radiation or chemotherapy prior to surgery at The First Affiliated Hospital of Xinjiang Medical University (Ürümqi, China) between January 2012 and December 2019, including 15 cases of DLBCL (5 GCB and 10 non-GCB) and 15 cases of LRH. There were 21 males and 11 females patients with an age range of 3 to 83 years and a median age of 41.5. The exclusion criteria comprised patients with other lymphoid diseases and patients undergoing chemotherapy. The inclusion criteria were fresh lymph node tissue from patients with DLBCL before any treatment. All fresh lymph node tissues were collected according to standard operating procedures during surgery, and the samples were washed with isotonic saline. The samples were slowly frozen in liquid nitrogen within 8 min, and then stored in a refrigerator at −80°C. The complete clinical and pathological data, hematoxylin and eosin sections and paraffin-embedded tissue samples of all patients were obtained, and the histological diagnosis of the tissues was confirmed and classified by two senior hematologists. The pathological classification was based on the 2016-revised 4th edition of the World Health Organization classification (10), and was further subtyped using the Hans algorithm (3) by senior hematopathologists. In total, 10 fresh frozen tissues from the aforementioned 30 fresh frozen samples were randomly selected and divided into two groups: 5 DLBCL (2 GCB and 3 non-GCB) tissues as the experimental group and 5 LRH tissues as the control group. The samples were subjected to miRNA microarray analysis to screen differential miRNAs. All the aforementioned 30 fresh frozen tissues were divided into two groups: 15 DLBCL (5 GCB and 10 non-GCB) tissues as the experimental group and 15 LRH tissues as the control group. The samples were subjected to RT-qPCR for detecting the expression levels of the differentially expressed miRNAs. Written informed consent was obtained from all patients. All procedures involving human participants in the present study comply with the ethical standards of institutions and/or national research councils, as well as with the Declaration of Helsinki and its subsequent amendments or similar ethical standards. The present study was approved by the Ethics Committee of the Department of Medicine of The First Affiliated Hospital of Xinjiang Medical University (approval no. 20160218-13). The series record GSE173080 provides access to all of the current data (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173080).
Screening and identification of differentially expressed miRNAs RNA extraction
Total RNA was extracted from fresh frozen tissues using the mirVana™ RNA Isolation kit (cat. no. AM1561; Thermo Fisher Scientific Inc.) according to the manufacturer's instructions. Total RNA was quantified by NanoDrop ND-2000 (Thermo Fisher Scientific, Inc.) and the RNA integrity was assessed using Agilent Bioanalyzer 2100 (Agilent Technologies, Inc.).
miRNA microarray
The Agilent Human miRNA Array (cat. no. 8×60 K; Design ID: 070156; Agilent Technologies, Inc.) was used for analysis of the fresh frozen samples. Sample labeling, microarray hybridization and washing were performed according to the manufacturer's standard protocols. Briefly, total RNA was dephosphorylated, denatured and labeled with cyanine-3-CTP(Cy3). After purification, the labeled RNAs were hybridized onto the microarray. After washing, the arrays were scanned with the Agilent Scanner G2505C (Agilent Technologies, Inc.).
miRNA microarray data analysis
Feature Extraction software (version 10.7.1.1; Agilent Technologies, Inc.) was used to analyze array images to obtain the raw data. Next, GeneSpring software (version 12.5; Agilent Technologies, Inc.) was employed for basic analysis of the raw data. First, the raw data were normalized with the quantile algorithm. Probes that had ≥75% of samples in any one condition out of two conditions with flags in ‘Detected’ were selected for further data analysis. Differentially expressed miRNAs were identified by their fold-change and P-value calculated using the unpaired Student's t-test. The thresholds set for upregulated and downregulated genes were fold-change ≥2.0 and P≤0.05 (11). Target genes of differentially expressed miRNAs were the intersection predicted with three databases [TargetScan, www.targetscan.org; microRNA.org, www.microrna.org; and PITA, http://genie.weizmann.ac.il/pubs/mir07/mir07_dyn_data.html). GO and KEGG analyses were applied to determine the roles of these target genes (12). Hierarchical clustering was performed to show the distinguishable miRNA expression pattern among samples.
RT-qPCR
Nine differentially expressed proteins which associated with the PI3K/AKT signaling pathway were screened by the proteomics method in DLBCL in our previous study (5). A total of 204 differentially expressed miRNAs were screened by miRNA microarray in the present study, and miRNA-mRNA prediction was conducted between 204 differentially expressed miRNAs and the aforementioned 9 differentially expressed genes, and it was found that the 8 differential miRNAs had targeting relationships with the 9 differentially expressed genes. Based on the array and miRNA-mRNA prediction results, 8 differentially expressed miRNAs were selected, including miR-193a-3p, miR-19a-3p, miR-19b-3p, miR-370-3p, miR-1275, miR-490-5p, miR-630 and miR-665, whose potential target genes may regulate the PI3K/AKT signaling pathway, were verified by RT-qPCR in DLBCL and LRH tissues. Total RNA was isolated from tissues using TRIzol® reagent (cat. no. 15596026; Thermo Fisher Scientific, Inc.) and reversely transcribed into cDNA using the miRNA First Strand cDNA kit (cat. no. B532451; Sangon Biotech Co., Ltd.) according to the manufacturer's protocol. The sequences of the forward primers for miRNAs are shown in Table I, and the reverse primer was provided in the miRNA Fluorescence Quantitative PCR kit (SYBR Green dye method) (cat. no. B532461; Sangon Biotech Co., Ltd.). qPCR was performed with the Applied Biosystems 7500 Fast PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the miRNA Fluorescence Quantitative PCR kit. U6 was used as an internal control gene (13). The miRNA expression levels were determined by RT-qPCR and calculated using the 2−ΔΔCq method (14,15). The thermocycling conditions for the PCR were 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec.
Table I.
Sequences of the miRNA primers used for reverse transcription-quantitative PCR.
| miRNA | Sequence (5′-3′) |
|---|---|
| hsa-miR-193a-3p-F | GCAGAACTGGCCTACAAAG |
| hsa-miR-19a-3p-F | GCAGTGTGCAAATCTATGCAA |
| hsa-miR-19b-3p-F | AGTGTGCAAATCCATGCAA |
| hsa-miR-370-3p-F | CCTGCTGGGGTGGAA |
| hsa-miR-1275-F | GGTGGGGGAGAGGCT |
| hsa-miR-490-5p-F | GCAGCCATGGATCTCC |
| hsa-miR-630-F | CAGAGTATTCTGTACCAGGGAA |
| hsa-miR-665-F | ACCAGGAGGCTGAGG |
| U6-F | GCTTCGGCAGCACATATACTAAAAT |
F, forward; miR/miRNA, microRNA.
Statistical analysis
All quantitative data are expressed as the mean ± SD of 3 experiments and were analyzed with SPSS 18.0 software (SPSS, Inc.). Unpaired Student's t-test and one-way ANOVA followed by the least significance difference post-hoc test were used for statistical analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Differential miRNAs screening
A total of 2,550 miRNAs were detected by Agilent Human miRNA Array. For the comparison between the experimental group and the control group, the probes marked with 75% samples ‘Detected’ in at least one group were selected for the second step of differential screenings. For the group with biological repeats, fold-change ≥2 and P≤0.05 were used as the screening criteria. In total, 204 differential miRNAs were screened (Table SI), among which, 54 were upregulated and 150 were downregulated in DLBCL compared with in LRH tissues.
Target gene prediction
Target genes of differentially expressed miRNAs were the intersection predicted with three databases (TargetScan, microRNA.org and PITA). A total of 121,174 target genes were predicted only in TargetScan, 13,401 target genes were predicted only in PITA and 117,485 target genes were predicted only in microRNA.org. Additionally, 67,875 overlapping genes (between microRNA.org and TargetScan), 3,523 overlapping genes (between microRNA.org and PITA) and 2,353 overlapping genes (between TargetScan and PITA) were identified. The intersection of the target genes predicted by all three databases consisted of 7,522 target genes (Fig. 1).
Figure 1.
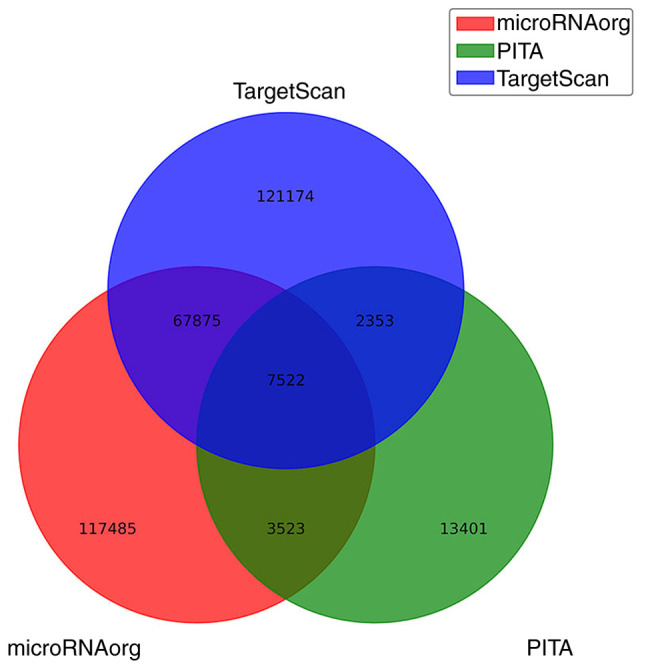
TargetScan, microRNA.org and PITA databases were used to create a Venn diagram, and the intersection predicted 7,522 overlapping target genes of the 204 differential microRNAs.
GO functional enrichment analysis
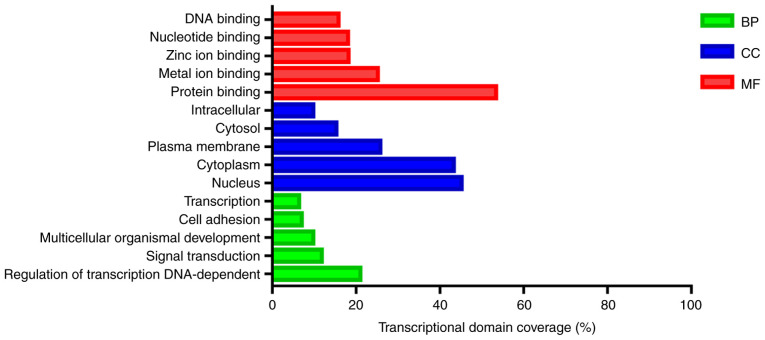
The target genes of differentially expressed miRNAs were particularly enriched in molecular functions, especially in ‘protein binding’, ‘metal ion binding’ and ‘zinc ion binding’ (Table II and Fig. 2). For biological processes, the target genes of differentially expressed miRNAs were enriched in ‘regulation of transcription, DNA-dependent’, ‘signal transduction’ and ‘multicellular organismal development’ (Table II and Fig. 2). In addition, GO cell component analysis indicated that the target genes were enriched in ‘nucleus’, ‘cytoplasm’ and ‘plasma membrane’ (Table II and Fig. 2).
Table II.
GO functional enrichment analysis of target genes.
| A, Biological process | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Rank | ID | Term | List hits | List total | Population hits | Population total | P-value |
| 1 | GO:0006355 | Regulation of transcription, DNA-dependent | 607 | 3,540 | 1,900 | 14,200 | 6.83×10−14 |
| 2 | GO:0007165 | Signal transduction | 341 | 3,540 | 1,146 | 14,200 | 6.18×10−5 |
| 3 | GO:0007275 | Multicellular organismal development | 282 | 3,540 | 913 | 14,200 | 1.49×10−5 |
| 4 | GO:0007155 | Cell adhesion | 204 | 3,540 | 542 | 14,200 | 1.62×10−11 |
| 5 | GO:0006351 | Transcription, DNA-dependent | 203 | 3,540 | 538 | 14,200 | 1.40×10−11 |
| 6 | GO:0006468 | Protein phosphorylation | 186 | 3,540 | 487 | 14,200 | 3.13×10−11 |
| 7 | GO:0045944 | Positive regulation of transcription from RNA polymerase II promoter | 183 | 3,540 | 470 | 14,200 | 6.66×10−12 |
| 8 | GO:0007399 | Nervous system development | 169 | 3,540 | 387 | 14,200 | 2.83×10−16 |
| 9 | GO:0006915 | Apoptosis | 168 | 3,540 | 582 | 14,200 | 1.51×10−2 |
| 10 | GO:0030154 | Cell differentiation | 158 | 3,540 | 496 | 14,200 | 2.40×10−4 |
|
| |||||||
| B, Cellular component | |||||||
|
| |||||||
| Rank | ID | Term | List hits | List total | Population hits | Population total | P-value |
|
| |||||||
| 1 | GO:0005634 | Nucleus | 1,537 | 3,899 | 5,241 | 16,399 | 7.40×10−30 |
| 2 | GO:0005737 | Cytoplasm | 1,471 | 3,899 | 4,954 | 16,399 | 4.45×10−31 |
| 3 | GO:0005886 | Plasma membrane | 876 | 3,899 | 3,398 | 16,399 | 1.18×10−3 |
| 4 | GO:0005829 | Cytosol | 562 | 3,899 | 1,980 | 16,399 | 2.54×10−7 |
| 5 | GO:0005622 | Intracellular | 521 | 3,899 | 1,939 | 16,399 | 4.15×10−4 |
| 6 | GO:0005730 | Nucleolus | 334 | 3,899 | 1,097 | 16,399 | 9.81×10−8 |
| 7 | GO:0005794 | Golgi apparatus | 294 | 3,899 | 885 | 16,399 | 3.72×10−11 |
| 8 | GO:0005783 | Endoplasmic reticulum | 248 | 3,899 | 952 | 16,399 | 4.95×10−2 |
| 9 | GO:0005654 | Nucleoplasm | 240 | 3,899 | 813 | 16,399 | 6.67×10−5 |
| 10 | GO:0005856 | Cytoskeleton | 239 | 3,899 | 773 | 16,399 | 2.03×10−6 |
|
| |||||||
| C, Molecular function | |||||||
|
| |||||||
| Rank | ID | Term | List hits | List total | Population hits | Population total | P-value |
|
| |||||||
| 1 | GO:0005515 | Protein binding | 1,675 | 3,673 | 5,297 | 14,962 | 3.30×10−49 |
| 2 | GO:0046872 | Metal ion binding | 790 | 3,673 | 2,879 | 14,962 | 3.88×10−5 |
| 3 | GO:0008270 | Zinc ion binding | 571 | 3,673 | 1,988 | 14,962 | 2.68×10−6 |
| 4 | GO:0000166 | Nucleotide binding | 569 | 3,673 | 1,985 | 14,962 | 3.65×10−6 |
| 5 | GO:0003677 | DNA binding | 497 | 3,673 | 1,775 | 14,962 | 2.09×10−4 |
| 6 | GO:0005524 | ATP binding | 402 | 3,673 | 1,494 | 14,962 | 1.44×10−2 |
| 7 | GO:0003700 | Sequence-specific DNA binding transcription factor activity | 337 | 3,673 | 936 | 14,962 | 4.15×10−16 |
| 8 | GO:0005509 | Calcium ion binding | 206 | 3,673 | 649 | 14,962 | 1.36×10−5 |
| 9 | GO:0043565 | Sequence-specific DNA binding | 205 | 3,673 | 548 | 14,962 | 6.39×10−12 |
| 10 | GO:0003723 | RNA binding | 170 | 3,673 | 608 | 14,962 | 2.69×10−2 |
‘List hits’ indicates the number of genes annotated by the considered GO category or annotation cluster within the analyzed list of target genes. ‘List total’ indicates the number of genes within the analyzed list of target genes having at least one GO annotation. ‘Population hits’ indicates the number of genes, available on the entire microarray, annotated by the considered GO category or annotation cluster. ‘Population total’ indicates the number of genes available on the entire microarray and having at least one GO annotation. P-value indicates the significance P-value of the gene enrichment of the considered GO category or annotation cluster, calculated with a unilateral Fisher's exact test. GO, Gene Ontology.
Figure 2.
Results of Gene Ontology functional enrichment analysis of the 7,522 target genes. BP, biological process; CC, cellular component; MF, molecular function.
KEGG pathway functional enrichment analysis
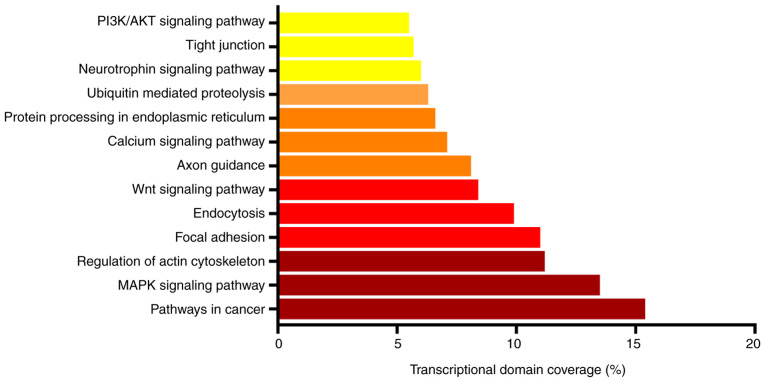
According to the results of KEGG enrichment analysis, the target genes of differentially expressed miRNAs were enriched in ‘pathways in cancer’, ‘MAPK signaling pathway’, ‘regulation of actin cytoskeleton’, ‘focal adhesion’, ‘endocytosis’, ‘Wnt signaling pathway’, ‘axon guidance’, ‘calcium signaling pathway’ and ‘PI3K/AKT signaling pathway’ (Table III and Fig. 3).
Table III.
Kyoto Encyclopedia of Genes and Genomes pathway functional enrichment analysis.
| Rank | ID | Term | List hits | List total | Population hits | Population total | P-value |
|---|---|---|---|---|---|---|---|
| 1 | 5200 | Pathways in cancer | 131 | 1,399 | 327 | 5,981 | 3.51×10−12 |
| 2 | 4010 | MAPK signaling pathway | 115 | 1,399 | 268 | 5,981 | 3.68×10−13 |
| 3 | 4810 | Regulation of actin cytoskeleton | 95 | 1,399 | 214 | 5,981 | 4.37×10−12 |
| 4 | 4510 | Focal adhesion | 94 | 1,399 | 200 | 5,981 | 8.62×10−14 |
| 5 | 4144 | Endocytosis | 84 | 1,399 | 203 | 5,981 | 5.40×10−9 |
| 6 | 4310 | Wnt signaling pathway | 71 | 1,399 | 151 | 5,981 | 1.02×10−10 |
| 7 | 4360 | Axon guidance | 69 | 1,399 | 130 | 5,981 | 1.27×10−13 |
| 8 | 4020 | Calcium signaling pathway | 60 | 1,399 | 177 | 5,981 | 8.32×10−4 |
| 9 | 4141 | Protein processing in endoplasmic reticulum | 56 | 1,399 | 167 | 5,981 | 1.63×10−3 |
| 10 | 4120 | Ubiquitin mediated proteolysis | 53 | 1,399 | 139 | 5,981 | 5.85×10−5 |
| 11 | 4722 | Neurotrophin signaling pathway | 51 | 1,399 | 127 | 5,981 | 1.56×10−5 |
| 12 | 4530 | Tight junction | 48 | 1,399 | 133 | 5,981 | 5.75×10−4 |
| 13 | 4151 | PI3K/AKT signaling pathway | 46 | 1,399 | 138 | 5,981 | 4.63×10−3 |
| 14 | 4514 | Cell adhesion molecules (CAMs) | 42 | 1,399 | 136 | 5,981 | 2.61×10−2 |
| 15 | 4660 | T cell receptor signaling pathway | 42 | 1,399 | 108 | 5,981 | 2.03×10−4 |
‘List hits’ indicates the number of genes annotated by the considered KEGGpathwaycategory or annotation cluster within the analyzed list of target genes. ‘List total’ indicates the number of genes within the analyzed list of target genes having at least one KEGG pathwayannotation. ‘Population hits’ indicates the number of genes, available on the entire microarray, annotated by the considered KEGG pathway category or annotation cluster. ‘Population total’ indicates the number of genes available on the entire microarray and having at least one KEGG pathway annotation. P-value indicates the significance P-value of the gene enrichment of the considered KEGG pathwaycategory or annotation cluster, calculated with a unilateral Fisher's exact test. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 3.
Results of Kyoto Encyclopedia of Genes and Genomes pathway functional enrichment analysis of the 7,522 target genes.
Hierarchical clustering
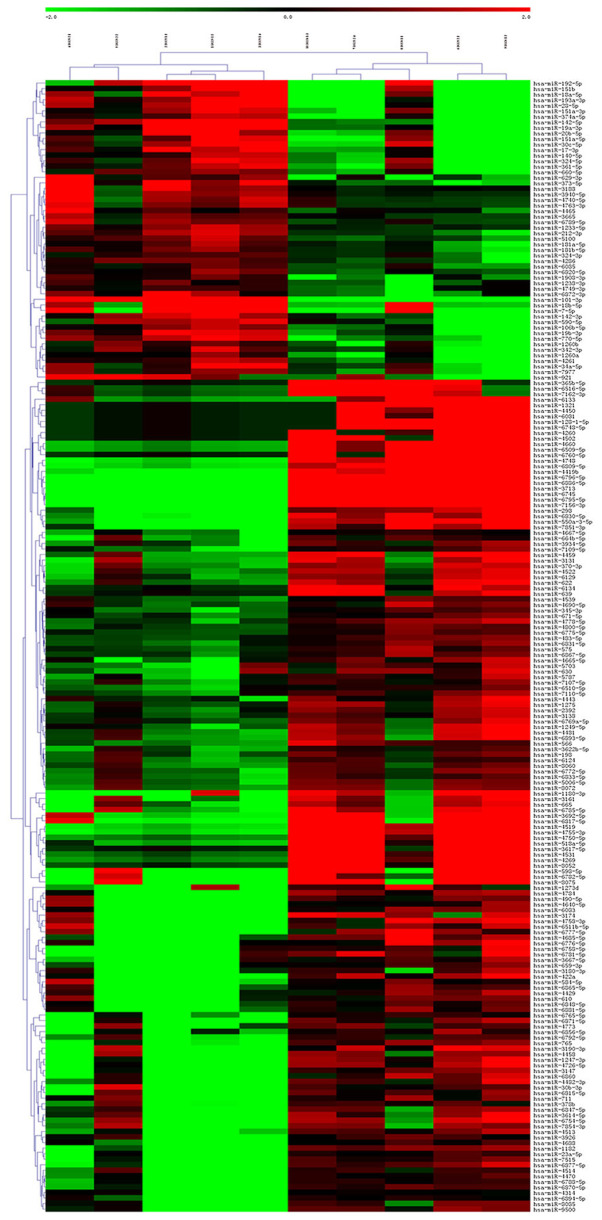
Through hierarchical clustering analysis, it was found that the same samples can appear in the same cluster, and miRNAs clustered in the same cluster may have similar biological functions (Fig. 4).
Figure 4.
Results of unsupervised hierarchical cluster analysis of the 204 differentially expressed microRNAs as shown by a heat map. The color scale indicates the change in gene expression from relatively low (green) to relatively high (red) levels. The row name represents the sample name, while the column name represents the probe name.
Validation of the differentially expressed miRNAs
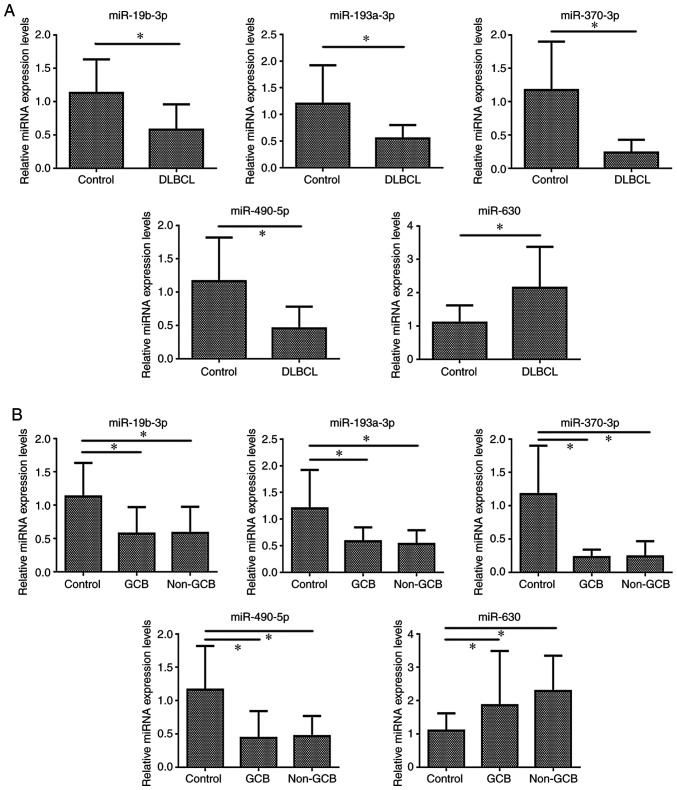
Only statistically significant different miRNAs RT-qPCR results were shown. Compared with those in the control group, the expression levels of miR-19b-3p, miR-193a-3p, miR-370-3p and miR-490-5p were significantly lower in the DLBCL group (P<0.05), while miR-630 expression was significantly higher in DLBCL (P<0.05) (Table IV and Fig. 5A).
Table IV.
Analysis of miRNA expression levels in DLBCL vs. control groups (mean±SD).
| Groups | miR-19a-3p | miR-19b-3p | miR-193a-3p | miR-370-3p | miR-1275 | miR-490-5p | miR-630 | miR-665 |
|---|---|---|---|---|---|---|---|---|
| Control (LRH) (n=15) | 1.134±0.576 | 1.132±0.503 | 1.200±0.722 | 1.172±0.728 | 1.186±0.788 | 1.164±0.655 | 1.095±0.524 | 1.085±0.430 |
| DLBCL (n=15) | 0.882±0.648 | 0.578±0.382 | 0.548±0.252 | 0.236±0.192 | 1.299±0.753 | 0.456±0.326 | 2.145±1.235 | 1.440±0.948 |
| T value | 1.126 | 3.397 | 3.299 | 4.817 | −0.401 | 3.742 | −3.030 | −1.319 |
| P-value | 0.270 | 0.002 | 0.004 | 0.000 | 0.692 | 0.001 | 0.007 | 0.198 |
DLBCL, diffuse large B-cell lymphoma; LRH, lymph node reactive hyperplasia; miR/miRNA, microRNA.
Figure 5.
Reverse transcription-quantitative PCR results of the expression levels of different miRNAs. (A) Expression levels of different miRNAs in DLBCL compared with those in LRH (control) (*P<0.05). (B) Different miRNA expression levels in the GCB DLBCL, non-GCB DLBC and LRH groups (*P<0.05). miRNA, microRNA; DLBCL, diffuse large B-cell lymphoma; LRH, lymph node reactive hyperplasia; GCB, germinal center B-cell like; miR/miRNA, microRNA.
As shown in Table V, the GCB and non-GCB groups exhibited significantly lower expression levels of miR-19b-3p, miR-193a-3p, miR-370-3p and miR-490-5p (P<0.05), and significantly higher expression levels of miR-630 compared with the LRH group. Additionally, there was no significant differences in miRNA expression between the GCB and non-GCB groups (Fig. 5B).
Table V.
Analysis of miRNA expression levels in GCB, non-GCB DLBCL subtypes and control groups (mean±SD).
| Groups | miR-19a-3p | miR-19b-3p | miR-193a-3p | miR-370-3p | miR-1275 | miR-490-5p | miR-630 | miR-665 |
|---|---|---|---|---|---|---|---|---|
| Control (LRH) (n=15) | 1.134±0.576 | 1.132±0.503 | 1.269±0.904 | 1.172±0.728 | 1.186±0.788 | 1.164±0.655 | 1.095±0.524 | 1.085±0.430 |
| GCB (n=5) | 0.682±0.308 | 0.573±0.397a | 0.582±0.263a | 0.230±0.111a | 0.975±0.568 | 0.437±0.406a | 1.860±1.628a | 0.949±0.372 |
| Non-GCB (n=10) | 0.982±0.760 | 0.580±0.396a | 0.531±0.259a | 0.239±0.228a | 1.461±0.807 | 0.466±0.303a | 2.287±1.061a | 1.685±1.067 |
P<0.05 vs. control. DLBCL, diffuse large B-cell lymphoma; LRH, lymph node reactive hyperplasia; GCB, germinal center B-cell like; miR/miRNA, microRNA.
Discussion
DLBCL is a common lymphoid malignancy in adults. Despite the improvements in therapeutic options and survival of patients, treatment resistance is a major clinical challenge in DLBCL, and ~40% of patients have refractory disease or relapse (16). Therefore, it is urgent to find novel treatment targets and effective therapeutic drugs to improve the survival of patients with DLBCL.
Microarray analysis has become a widely used tool for generating gene expression data on a genomic scale and has emerged as a promising and efficient tool for screening significant genetic or epigenetic alterations in carcinogenesis (17). miRNAs are known as one of the main regulators of gene expression in important biological and physiological processes, such as cell proliferation, apoptosis, proliferation, differentiation, cell motility and angiogenesis, as well as in disease initiation and progression, being particularly important in cancer cell invasion, migration and metastasis (18). Additionally, miRNAs are involved in B-cell development, B-cell receptor, B-cell migration/adhesion and production of follicles, plasma cells and memory B cells (19). Therefore, miRNAs are expected to receive increased attention as molecular targets for the diagnosis, prediction of prognosis and treatment of DLBCL. Beheshti et al (20) identified a circulating miRNA signature in a Smurf2-deficient mouse model that spontaneously develops DLBCL. By using cut-points from recursive partitioning analysis, they derived a 5-miRNA signature (let-7b, let-7c, miR-18a, miR-24, and miR-15a) with a classification rate of 91% for serum from patients with DLBCL vs. normal controls. These circulating miRNAs were able to distinguish between DLBCL subtypes and disease characteristics for clinicopathological diagnosis. Ting et al (21) found that miR-155, miR-17/92, miR-21, miR-224 and mir-146b-5p have value in predicting treatment response to chemotherapy in DLBCL. It has been suggested that miRNAs can be employed as an indicator to predict relapse or refractoriness after treatment of DLBCL (21). In the present study, 204 differential miRNAs were screened in DLBCL, which may provide a basis for researchers to identify the pathogenesis of DLBCL, and may serve in the future as reliable biomarkers for precise diagnosis and as therapeutic targets for improvement of treatment efficacy in DLBCL.
miRNAs are important regulators of gene expression, since they eventually lead to a decrease in the observed mRNA expression levels of target genes (22). Therefore, the present study predicted the target genes of the 204 differentially expressed miRNAs with three databases (TargetScan, microRNA.org and PITA). In total, 7,522 overlapping target genes of the 204 differential miRNAs were identified. The current results may provide a theoretical basis for future studies on the occurrence and development of DLBCL.
miRNAs regulate target genes or themselves to activate or inhibit signaling pathways, and have become a research hotspot for tumor development and therapeutic targets in DLBCL (23). In the present study, KEGG analysis was implemented to determine the roles of these target genes. It was found that the target genes were enriched in ‘pathways in cancer’, ‘MAPK signaling pathway’, ‘regulation of actin cytoskeleton’, ‘focal adhesion’, ‘endocytosis’, ‘Wnt signaling pathway’, ‘axon guidance’, ‘calcium signaling pathway’ and ‘PI3K/AKT signaling pathway’. Shim et al (24) has reported that miR-124 acts as a tumor suppressor by targeting NF-κB p65 in B-cell lymphoma. Zhao et al (25) has indicated that SMAD5 antisense RNA 1 inhibits DLBCL cell proliferation by sponging miR-135b-5p to upregulate adenomatous polyposis coli expression and inactivate the classic Wnt/β-catenin signaling pathway. Yoon et al (23) found that the PI3K/AKT signaling pathway is strongly enriched with targets of miR-29 in DLBCL. In the present study, certain signaling pathways, such as pathways in cancer, and the MAPK, Wnt and PI3K/AKT signaling pathways, were identified, which is consistent with the results of previous studies on DLBCL (5,23). In addition, a number of novel signaling pathways were identified in the present study, such as regulation of actin cytoskeleton, focal adhesion, endocytosis, axon guidance and the calcium signaling pathway, which may therefore be associated with the occurrence and development of DLBCL. The role of these novel signaling pathways in DLBCL needs to be further studied.
According to our previous studies (5,6) the PI3K/AKT signaling pathway serves an important regulatory role in the occurrence and progression of DLBCL. A previous proteomic study demonstrated that there were 9 differential proteins in DLBCL that regulated the PI3K/AKT signaling pathway (5), hence miRNA-mRNA prediction was conducted with these 9 miRNAs in the present study. Thus, eight differentially expressed miRNAs in DLBCL and LRH, whose potential target genes may regulate the PI3K/AKT signaling pathway, were verified by RT-qPCR. It was revealed that miR-19b-3p, miR-193a-3p, miR-370-3p and miR-490-5p exhibited low expression in DLBCL (P<0.05), while miR-630 expression in DLBCL was high (P<0.05). These differentially expressed miRNAs serve an important regulatory role in a variety of tumors; however, there are limited studies on DLBCL.
miR-19b-3p has been reported to be associated with favorable or unfavorable events in several types of cancer. Its role is controversial depending on the tumor, and it may be a good non-invasive biomarker for cancer detection. Tang et al (26) has reported that miR-19b-3p promotes intrahepatic cholangiocarcinoma (ICC) cell proliferation and epithelial-mesenchymal transition (EMT), and inhibits apoptosis, while knockdown of coiled-coil domain containing 6 (CCDC6) reverses these effects. These results suggest that serum miR-19b-3p expression may be an important biomarker for ICC diagnosis, and targeting the miR-19b-3p-CCDC6 axis may be a promising strategy for ICC treatment (26). Park et al (27) has reported that miR-19b-3p can inhibit the migration of breast cancer cells through exosome-mediated delivery by targeting aquaporin-5. Marcuello et al (28) has demonstrated that a plasma 6-miRNA signature (miR-15b-5p, miR-18a-5p, miR-29a-3p, miR-335-5p, miR-19a-3p and miR-19b-3p) can be used to distinguish between colorectal cancer (CRC) or advanced adenoma and healthy individuals. The aforementioned serum 6-miRNA signature may be a useful strategy to improve the diagnostic performance of current CRC screening programs. Although miR-19b-3p has been implicated in certain types of cancer, its role in cancer remains controversial, and there are no relevant studies on its role in DLBCL.
Numerous studies (29,30) have revealed the crucial role of the miR-193 family, which comprises miR-193a-3p, miR-193a-5p, miR-193b-3p and miR-193b-5p, in health and disease-associated biological processes by interacting with specific target genes and signaling pathways, mainly acting as a tumor suppressor. Wang et al (29) has demonstrated that miR-193a-3p is a tumor suppressor gene that serves an important role in the biology of health and disease by interacting with specific targets and signals, and it also inhibits the proliferation and facilitates the apoptosis of hepatocellular carcinoma (HCC) cells. It has been suggested that miR-193a-3p may be used as a promising biomarker for the diagnosis of HCC and as a therapeutic target for HCC in the future (29). Lin et al (30) has revealed that the expression levels of miR-193a-3p in human CRC cell lines are significantly decreased compared with those in a normal colonic epithelium cell line. Overexpression of miR-193a-3p inhibits the proliferation, migration and angiogenesis of CRC cells, while forced expression of plasminogen activator urokinase rescues these inhibitory effects (30). Chen et al (31) has demonstrated that miR-193a-3p expression in pancreatic ductal adenocarcinoma (PDAC) tissues is significantly lower than that in non-cancerous tissues. When overexpressing miR-193a-3p in PDAC cells, their proliferative ability was significantly inhibited, the apoptosis rate was accelerated, and the cell cycle was blocked in the G1 and G2/M phases (31). In the present study, it was found that miR-193a-3p exhibited low expression in DLBCL (P<0.05). It was speculated that the functional role of this miRNA in DLBCL may be considered as a tumor suppressor. However, to the best of our knowledge, there are no relevant studies on the role of miR-193a-3p in DLBCL.
miR-370-3p serves an important regulatory role in a variety of tumors, and increasing evidence has suggested that it is downregulated and acts as a tumor suppressor in numerous types of cancer. Several studies (32–34) have indicated that it serves a regulatory role in tumors through the regulation of target genes, it can increase the sensitivity to chemotherapy drugs and it can be used as a biomarker or a therapeutic target in tumors. Li et al (32) has found that miR-370-3p inhibits chronic myeloid leukemia cell proliferation and induces apoptosis by suppressing PDZ and LIM domain protein 1/Wnt/β-catenin signaling. Nadaradjane et al (33) has indicated that miR-370-3p/temozolomide (TMZ) treatment is 2-fold more efficient than TMZ treatment alone in decreasing the volume of glioblastoma multiforme in mice. It has been suggested that miR-370-3p may be used as a therapeutic tool for anti-glioblastoma multiforme therapy (33). Leivonen et al (34) has profiled the miRNAs of matched primary and relapsed DLBCL by next-generation sequencing and has revealed that miR-370-3p is markedly downregulated in the majority of relapsed DLBCL samples, while overexpression of miR-370-3p regulates the target genes MAP3K8, PIK3R1, PIK3CG, PI3KCD and SYK, resulting in downregulated mRNA expression levels. It has been demonstrated that miR-370-3p downregulates genes involved in the PI, MAPK and BCR signaling pathways, and enhances the chemosensitivity of DLBCL cells in vitro (34). The present study indicated that miR-370-3p expression was downregulated in DLBCL, suggesting that it may inhibit the occurrence and development of DLBCL. Research on miR-370-3p in patients with DLBCL is rare; thus, further studies are required.
Recent studies have found that miR-490-5p is associated with the occurrence and development of tumors, and serves an important role in a variety of tumors. Wang et al (35) has revealed that miR-490-5p expression is markedly downregulated in neuroblastoma (NB) tissues and cell lines. Overexpression of miR-490-5p suppresses cell proliferation, migration and invasion, and induces cell cycle G0/G1 arrest and apoptosis in NB cell lines (35). The aforementioned results have demonstrated for the first time that miR-490-5p may function as a tumor suppressor in NB by targeting a myeloma overexpressed gene (35). Xiang et al (36) has demonstrated that miR-490-5p promotes the proliferation of bladder cancer cells and inhibits their apoptosis. In the present study, low miR-490-5p expression was observed in DLBCL. It was speculated that miR-490-5p may be involved in the occurrence and development of DLBCL, but no relevant studies on its regulatory mechanism in DLBCL have been conducted thus far.
Previous evidence has demonstrated that miR-630 is involved in multiple processes during cancer development and progression. Valera et al (37) has performed miRNA profiling in young patients with prostate cancer (PCa) and compared the results with those of PCa in older men. Their results have revealed that, compared with its expression in PCa and its normal counterpart, miR-630 expression is significantly upregulated in PCa (37). Differentially expressed miRNAs provide insights into the molecular mechanisms involved in this PCa subtype. Pan et al (38) has provided the first evidence that miR-630 inhibits papillary thyroid carcinoma (PTC) cell growth, metastasis and EMT by suppressing the JAK2/STAT3 signaling pathway, and indicated that a potential therapeutic strategy through enhancing miR-630 expression may benefit patients with PTC. The present study indicated that miR-630 was a differentially expressed miRNA with high expression in DLBCL. It was hypothesized that miR-630 may be involved in the occurrence or development of DLBCL, but no relevant studies have been conducted on DLBCL to date. However, since it serves an important role in other tumors, it is worth further studying the role of miR-630 in DLBCL.
In summary, the present study screened 204 miRNAs that exhibited differential expression between DLBCL and control groups via miRNA microarray. Altered expression levels of five miRNAs, including miR-19b-3p, miR-193a-3p, miR-370-3p, miR-490-5p and miR-630, may contribute to the development of DLBCL. The current findings provide valuable information to understand the pathogenesis of DLBCL, and may lead to the development of therapeutic strategies involving the use of miRNAs for the treatment of patients with DLBCL. Further exploration of the biological role of these differentially expressed miRNAs in DLBCL will strengthen the conclusions of the present study. For example, future studies should be performed to analyze these differentially expressed miRNAs in DLBCL cell lines.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- DLBCL
diffuse large B-cell lymphoma
- LRH
lymph node reactive hyperplasia
- miRNAs/miRs
microRNAs
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- RT-qPCR
reverse transcription-quantitative PCR
- GCB
germinal center B-cell like
- ncRNAs
non-coding RNAs
- iTRAQ
isobaric tags for relative and absolute quantification
- ICC
intrahepatic cholangiocarcinoma
- CCDC6
coiled-coil domain containing 6
- EMT
epithelial-mesenchymal transition
- HCC
hepatocellular carcinoma
- CRC
colorectal cancer
- PDAC
pancreatic ductal adenocarcinoma
- TMZ
temozolomide
- NB
neuroblastoma
- PCa
prostate cancer
- PTC
papillary thyroid carcinoma
Funding Statement
The present study was supported by the Chinese Society of Clinical Oncology (CSCO)-Roche Solid Tumor Research Fund (grant no. Y-Roche2019/2-0061) and the Non-profit Central Research Institute Fund of the Chinese Academy of Medical Sciences (grant no. 2020-PT330-003).
Funding
The present study was supported by the Chinese Society of Clinical Oncology (CSCO)-Roche Solid Tumor Research Fund (grant no. Y-Roche2019/2-0061) and the Non-profit Central Research Institute Fund of the Chinese Academy of Medical Sciences (grant no. 2020-PT330-003).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus repository (GSE173080) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173080).
Authors' contributions
All authors participated in the preparation of the manuscript. HXG wrote the first draft of the manuscript and edited the original figures and tables. XXL conceived and designed the study. HXG, SJL and MBW conducted the experiments and contributed to data analysis. XXL and HXG confirmed the authenticity of all the raw data. XXL, WZ and WLC conceived and designed the study and revised and reviewed this article for important intellectual content. SFY, ZPM, JX and WS collected the paraffin-embedded tissue samples, fresh tissue samples and clinicopathological information. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all patients. All procedures involving human participants in the present study comply with the ethical standards of institutions and/or national research councils, as well as with the Declaration of Helsinki and its subsequent amendments or similar ethical standards. The present study was approved by the Ethics Committee of the Department of Medicine of The First Affiliated Hospital of Xinjiang Medical University (Ürümqi, China) (approval no. 20160218-13).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Li S, Young KH, Medeiros LJ. Diffuse large B-cell lymphoma. Pathology. 2018;50:74–87. doi: 10.1016/j.pathol.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Roschewski M, Staudt LM, Wilson WH. Diffuse large B-cell lymphoma-treatment approaches in the molecular era. Nat Rev Clin Oncol. 2014;11:12–23. doi: 10.1038/nrclinonc.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Müller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 4.Coiffier B, Sarkozy C. Diffuse large B-cell lymphoma: R-CHOP failure-what to do? Hematology Am Soc Hematol Educ Program. 2016;2016:366–378. doi: 10.1182/asheducation-2016.1.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao HX, Nuerlan A, Abulajiang G, Cui WL, Xue J, Sang W, Li SJ, Niu J, Ma ZP, Zhang W, Li XX. Quantitative proteomics analysis of differentially expressed proteins in activated B-cell-like diffuse large B-cell lymphoma using quantitative proteomics. Pathol Res Pract. 2019;215:152528. doi: 10.1016/j.prp.2019.152528. [DOI] [PubMed] [Google Scholar]
- 6.Gao HX, Li SJ, Niu J, Ma ZP, Nuerlan A, Xue J, Wang MB, Cui WL, Abulajiang G, Sang W, et al. TCL1 as a hub protein associated with the PI3K/AKT signaling pathway in diffuse large B-cell lymphoma based on proteomics methods. Pathol Res Pract. 2020;216:152799. doi: 10.1016/j.prp.2019.152799. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Wang Y, Shu S, Cai J, Tang C, Dong Z. Non-coding RNAs in kidney injury and repair. Am J Physiol Cell Physiol. 2019;317:C177–C188. doi: 10.1152/ajpcell.00048.2019. [DOI] [PubMed] [Google Scholar]
- 8.Baronti L, Guzzetti I, Ebrahimi P, Friebe Sandoz S, Steiner E, Schlagnitweit J, Fromm B, Silva L, Fontana C, Chen AA, Petzold K. Base-pair conformational switch modulates miR-34a targeting of Sirt1 mRNA. Nature. 2020;583:139–144. doi: 10.1038/s41586-020-2336-3. [DOI] [PubMed] [Google Scholar]
- 9.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 10.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, et al., editors. 4th edition. IARC Press; Lyon: 2017. WHO classification of tumours of haematopoetic and lymphoid tissues. [Google Scholar]
- 11.Felix TF, Lopez Lapa RM, de Carvalho M, Bertoni N, Tokar T, Oliveira RA, M Rodrigues MA, Hasimoto CN, Oliveira WK, Pelafsky L, et al. MicroRNA modulated networks of adaptive and innate immune response in pancreatic ductal adenocarcinoma. PLoS One. 2019;14:e0217421. doi: 10.1371/journal.pone.0217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue J, Gao HX, Sang W, Cui WL, Liu M, Zhao Y, Wang MB, Wang Q, Zhang W. Identification of core differentially methylated genes in glioma. Oncol Lett. 2019;18:6033–6045. doi: 10.3892/ol.2019.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu S, Zheng Q, Wu H, Wang C, Liu T, Zhou W. miR-532 promoted gastric cancer migration and invasion by targeting NKD1. Life Sci. 2017;177:15–19. doi: 10.1016/j.lfs.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 14.Kakurina GV, Kolegova ES, Shashova EE, Cheremisina OV, Choynzonov EL, Kondakova IV. Relationship between the mRNA expression levels of calpains 1/2 and proteins involved in cytoskeleton remodeling. Acta Naturae. 2020;12:110–113. doi: 10.32607/actanaturae.10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Due H, Brøndum RF, Young KH, Bøgsted M, Dybkær K. MicroRNAs associated to single drug components of R-CHOP identifies diffuse large B-cell lymphoma patients with poor outcome and adds prognostic value to the international prognostic index. BMC Cancer. 2020;20:237. doi: 10.1186/s12885-020-6643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Lei Q, Zhang S, Kong L, Qin B. Screening and identification of key biomarkers in hepatocellular carcinoma: Evidence from bioinformatic analysis. Oncol Rep. 2017;38:2607–2618. doi: 10.3892/or.2017.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khordadmehr M, Shahbazi R, Sadreddini S, Baradaran B. miR-193: A new weapon against cancer. J Cell Physiol. 2019;234:16861–16872. doi: 10.1002/jcp.28368. [DOI] [PubMed] [Google Scholar]
- 19.Musilova K, Mraz M. MicroRNAs in B-cell lymphomas: How a complex biology gets more complex. Leukemia. 2015;29:1004–1017. doi: 10.1038/leu.2014.351. [DOI] [PubMed] [Google Scholar]
- 20.Beheshti A, Stevenson K, Vanderburg C, Ravi D, McDonald JT, Christie AL, Shigemori K, Jester H, Weinstock DM, Evens AM. Identification of circulating serum multi-MicroRNA signatures in human DLBCL models. Sci Rep. 2019;9:17161. doi: 10.1038/s41598-019-52985-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ting CY, Liew SM, Price A, Gan GG, Bee-Lan Ong D, Tan SY, Bee PC. Clinical significance of aberrant microRNAs expression in predicting disease relapse/refractoriness to treatment in diffuse large B-cell lymphoma: A meta-analysis. Crit Rev Oncol Hematol. 2019;144:102818. doi: 10.1016/j.critrevonc.2019.102818. [DOI] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 23.Yoon S, Nguyen HCT, Jo W, Kim J, Chi SM, Park J, Kim SY, Nam D. Biclustering analysis of transcriptome big data identifies condition-specific microRNA targets. Nucleic Acids Res. 2019;47:e53. doi: 10.1093/nar/gkz139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shim H, Nam J, Kim SW. NF-κB p65 represses microRNA-124 transcription in diffuse large B-cell lymphoma. Genes Genomics. 2020;42:543–551. doi: 10.1007/s13258-020-00922-y. [DOI] [PubMed] [Google Scholar]
- 25.Zhao CC, Jiao Y, Zhang YY, Ning J, Zhang YR, Xu J, Wei W, Kang-Sheng G. Lnc SMAD5-AS1 as ceRNA inhibit proliferation of diffuse large B cell lymphoma via Wnt/β-catenin pathway by sponging miR-135b-5p to elevate expression of APC. Cell Death Dis. 2019;10:252. doi: 10.1038/s41419-019-1479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Y, Yang J, Wang Y, Tang Z, Liu S, Tang Y. MiR-19b-3p facilitates the proliferation and epithelial-mesenchymal transition, and inhibits the apoptosis of intrahepatic cholangiocarcinoma by suppressing coiled-coil domain containing 6. Arch Biochem Biophys. 2020;686:108367. doi: 10.1016/j.abb.2020.108367. [DOI] [PubMed] [Google Scholar]
- 27.Park EJ, Jung HJ, Choi HJ, Jang HJ, Park HJ, Nejsum LN, Kwon TH. Exosomes co-expressing AQP5-targeting miRNAs and IL-4 receptor-binding peptide inhibit the migration of human breast cancer cells. FASEB J. 2020;34:3379–3398. doi: 10.1096/fj.201902434R. [DOI] [PubMed] [Google Scholar]
- 28.Marcuello M, Duran-Sanchon S, Moreno L, Lozano JJ, Bujanda L, Castells A, Gironella M. Analysis of A 6-mirna signature in serum from colorectal cancer screening participants as non-invasive biomarkers for advanced adenoma and colorectal cancer detection. Cancers (Basel) 2019;11:1542. doi: 10.3390/cancers11101542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang SS, Huang ZG, Wu HY, He RQ, Yang LH, Feng ZB, Dang YW, Lu HP, Fang YY, Chen G. Downregulation of miR-193a-3p is involved in the pathogenesis of hepatocellular carcinoma by targeting CCND1. PeerJ. 2020;8:e8409. doi: 10.7717/peerj.8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin M, Zhang Z, Gao M, Yu H, Sheng H, Huang J. MicroRNA-193a-3p suppresses the colorectal cancer cell proliferation and progression through downregulating the PLAU expression. Cancer Manag Res. 2019;11:5353–5363. doi: 10.2147/CMAR.S208233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen ZM, Yu Q, Chen G, Tang RX, Luo DZ, Dang YW, Wei DM. MiR-193a-3p inhibits pancreatic ductal adenocarcinoma cell proliferation by targeting CCND1. Cancer Manag Res. 2019;11:4825–4837. doi: 10.2147/CMAR.S199257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li LM, Luo FJ, Song X. MicroRNA-370-3p inhibits cell proliferation and induces chronic myelogenous leukaemia cell apoptosis by suppressing PDLIM1/Wnt/β-catenin signaling. Neoplasma. 2020;67:509–518. doi: 10.4149/neo_2020_190612N506. [DOI] [PubMed] [Google Scholar]
- 33.Nadaradjane A, Briand J, Bougras-Cartron G, Disdero V, Vallette FM, Frenel JS, Cartron PF. miR-370-3p is a therapeutic tool in anti-glioblastoma therapy but is not an intratumoral or cell-free circulating biomarker. Mol Ther Nucleic Acids. 2018;13:642–650. doi: 10.1016/j.omtn.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leivonen SK, Icay K, Jäntti K, Siren I, Liu C, Alkodsi A, Cervera A, Ludvigsen M, Hamilton-Dutoit SJ, d'Amore F, et al. MicroRNAs regulate key cell survival pathways and mediate chemosensitivity during progression of diffuse large B-cell lymphoma. Blood Cancer J. 2017;7:654. doi: 10.1038/s41408-017-0033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Zhang X, Yao H, Le Y, Zhou W, Li J, Lu L, Chen M, Li X. MiR-490-5p functions as tumor suppressor in childhood neuroblastoma by targeting MYEOV. Hum Cell. 2020;33:261–271. doi: 10.1007/s13577-019-00302-z. [DOI] [PubMed] [Google Scholar]
- 36.Xiang M, Yuan W, Zhang W, Huang J. Expression of miR-490-5p, miR-148a-3p and miR-608 in bladder cancer and their effects on the biological characteristics of bladder cancer cells. Oncol Lett. 2019;17:4437–4442. doi: 10.3892/ol.2019.10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valera VA, Parra-Medina R, Walter BA, Pinto P, Merino MJ. microRNA expression profiling in young prostate cancer patients. J Cancer. 2020;11:4106–4114. doi: 10.7150/jca.37842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan XM, He XY, Yang YL, Jia WJ, Yang ZQ, Yan D, Ma JX. MiR-630 inhibits papillary thyroid carcinoma cell growth, metastasis, and epithelial-mesenchymal transition by suppressing JAK2/STAT3 signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:2453–2460. doi: 10.26355/eurrev_201903_17392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus repository (GSE173080) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173080).