Abstract
Background
High-sensitivity severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigen assays are desirable to mitigate false negative results. Limited data are available to quantify and track SARS-CoV-2 antigen burden in respiratory samples from different populations.
Methods
We developed the Microbubbling SARS-CoV-2 Antigen Assay (MSAA) with smartphone readout, with a limit of detection of 0.5 pg/mL (10.6 fmol/L) nucleocapsid antigen or 4000 copies/mL inactivated SARS-CoV-2 virus in nasopharyngeal (NP) swabs. We developed a computer vision and machine learning–based automatic microbubble image classifier to accurately identify positives and negatives and quantified and tracked antigen dynamics in intensive care unit coronavirus disease 2019 (COVID-19) inpatients and immunocompromised COVID-19 patients.
Results
Compared to qualitative reverse transcription−polymerase chain reaction methods, the MSAA demonstrated a positive percentage agreement of 97% (95% CI 92%–99%) and a negative percentage agreement of 97% (95% CI 94%–100%) in a clinical validation study with 372 residual clinical NP swabs. In immunocompetent individuals, the antigen positivity rate in swabs decreased as days-after-symptom-onset increased, despite persistent nucleic acid positivity. Antigen was detected for longer and variable periods of time in immunocompromised patients with hematologic malignancies. Total microbubble volume, a quantitative marker of antigen burden, correlated inversely with cycle threshold values and days-after-symptom-onset. Viral sequence variations were detected in patients with long duration of high antigen burden.
Conclusions
The MSAA enables sensitive and specific detection of acute infections and quantification and tracking of antigen burden and may serve as a screening method in longitudinal studies to identify patients who are likely experiencing active rounds of ongoing replication and warrant close viral sequence monitoring.
Keywords: SARS-CoV-2, viral antigen, longitudinal NP swab samples, microbubbling assay
Introduction
A substantial challenge in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has been the development of sensitive, specific, and easily accessible diagnostics for the early identification of infections and for monitoring infection progress to guide appropriate isolation and infection control procedures. The current diagnostic gold standard is qualitative real-time reverse transcription−polymerase chain reaction (rRT-PCR), with various methods demonstrating limit of detection (LOD) from 102 to 105 copies/mL according to manufacturer package inserts and reference panels (1–3). While many nucleic acid-based methods are sensitive enough to detect acute infections, persistent nucleic acid positivity after symptom resolution and disease recovery makes it challenging to determine the right level of infection control measures during patient care, especially for immunocompromised patients with long periods of nucleic acid positivity and diverse presentations (4, 5). Positive SARS-CoV-2 nucleic acid results do not always predict infectivity. Studies have suggested that infectious viruses are absent from most specimens taken 8 days after symptom onset, despite measurable viral RNA loads (6–9). In a golden hamster SARS-CoV-2 model, although viral RNA was present in nasal washes up to 14 days postinoculation, the detection of infectious viruses and the communicable period were much shorter (10). Antigen results were shown to correlate better with viral culture results than nucleic acid determinations (11), suggesting positive antigen results may provide good risk prediction of transmissibility. However, the challenge of most rapid antigen tests is their low sensitivity and low positive percentage agreement in high viral load cases as compared to rRT-PCR [e.g., (12–14)]. Antigen tests with improved sensitivity are desirable to minimize false negatives and correctly identify truly infectious cases (13).
In a few recent studies (15–18), the single molecule array (SIMOA) technology was used to develop an ultrasensitive antigen test to track changes of antigen and antibodies to SARS-CoV-2 in plasma, capillary blood and saliva, with an LOD of 0.2 pg/mL nucleocapsid (N) antigen. Besides the 2 cases in which saliva N protein levels were tracked using the SIMOA assay for about 12 days (18), we are not aware of large-scale studies tracking antigen dynamics in respiratory samples using a highly sensitive antigen test.
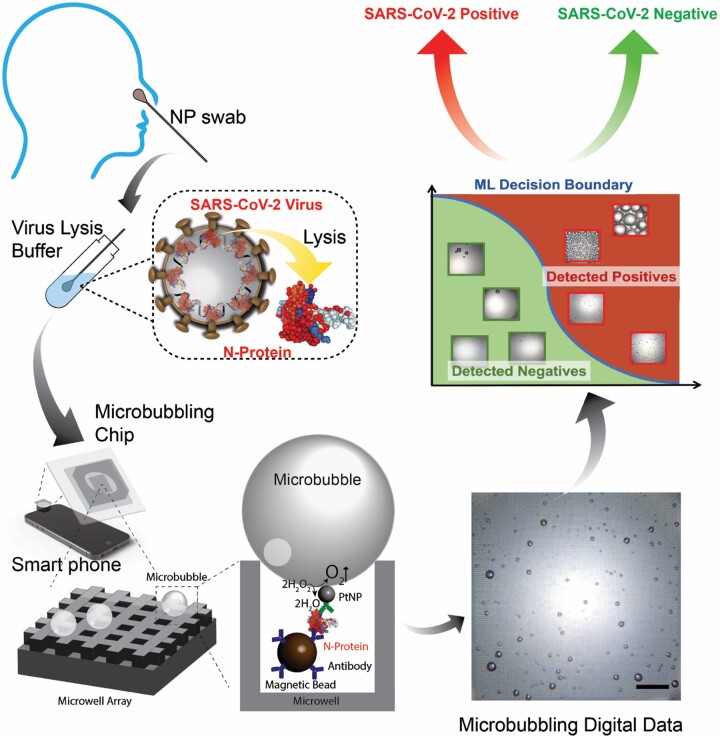
In this study, we developed and validated a highly sensitive and specific SARS-CoV-2 antigen assay using the microbubbling digital assay with smartphone readout (19) for acute SARS-CoV-2 infection detection. This system incorporates computer vision image recognition and machine learning (ML) classification to enable antigen burden quantification and tracking. In the microbubbling digital assay (19), individual sandwich complexes formed between magnetic bead/N antigen/platinum nanoparticle (PtNP) are distributed in microwells in a microchip. Bright field images of oxygen microbubbles generated through PtNP catalysis of H2O2 decomposition are captured using a smartphone camera enabling facile and accurate signal readout (19). Building on the features of the microbubbling digital assay including directly visible signals, femtomolar analytical sensitivity, digital quantitation capability and compatibility with smartphone and artificial intelligence, we developed the Microbubbling SARS-CoV-2 Antigen Assay (MSAA) for the detection of SARS-CoV-2 N antigen. The performance of this assay was assessed in clinical swab samples. Using this assay, we quantified and tracked antigen dynamics in an intensive care unit coronavirus disease 2019 (COVID-19) inpatient population and an immunocompromised COVID-19 patient population.
Methods
Detailed methods are in the online Data Supplement.
Clinical Swab Samples
Residual clinical swab samples were used in the study with the approval of the Institutional Review Board at the University of Pennsylvania. Refer to the online Data Supplement for details.
Microbubbling SARS-CoV-2 Antigen Assay
Clinical nasopharyngeal (NP) swab eluant samples (200 µL) were mixed with 2 µL of 10% Tween 20 and 2 µL of protease inhibitor cocktail for viral lysis and incubated at room temperature for 30 min. The lysed sample was then processed using the centrifugal filter (Pierce™ protein concentrators PES, 100K MWCO) by centrifuging at 12 000 g for 10 min collecting the filtrate for the following assays. The protocol for the microbubbling digital assay was followed as previously published (19), with the exception that the incubation time lengths were 30 min each, which were optimized to reduce assay time without sacrificing performance. Details of the assay steps are described in the online Data Supplement.
Imaging and Automatic Microbubble Detection and Counting
Microbubbles on the microbubbling microchips were imaged using an iPhone 11 or an iPad with the uHandy mobile phone microscope (9×, 5 mm focusing length; Aidmics Biotechnology Co.). A computer vision algorithm was designed to process these images to detect microbubbles, through 2 parallel approaches. The first approach employs Canny edge detection to find microbubble contours (20). The second approach applies the Hough Circle transform to improve detection of larger bubbles (21). To avoid double counting bubbles detected through both approaches, those bubbles detected by the second approach that had large overlap with the bubbles detected by the first approach were removed.
A ML-based classifier was developed to classify images. In each image, we counted the number of detected bubbles whose radius was above and below a set threshold (about 50 microns, corresponding to 8 pixels in the images). These big bubble and small bubble counts formed the 2-dimensional feature representation of each image, and we trained a linear separator using the linear support vector ML algorithm. Link to the codes for the computer vision and ML pipeline is available in the online Data Supplement.
Results
Microbubbling SARS-CoV-2 Antigen Assay Performance
The design of MSAA is shown in Fig. 1. A monoclonal capture antibody is conjugated to the surface of magnetic microbeads, and a polyclonal detection antibody is biotinylated to bind to avidin coated PtNPs. When N protein is present, immunosandwich complexes form between the magnetic microbeads and PtNPs. After washing, the immunocomplexes are pulled down into a microwell array in the microbubbling chip via an external magnetic field, where microbubbles are generated through degradation of H2O2 catalyzed by PtNPs. The microbubble images, captured using a smartphone camera and a mobile microscope, are analyzed by computer vision and ML algorithms, which generate quantitative outputs (bubble size, number, total bubble volume) and classify the images as either SARS-CoV-2 positive or negative.
Fig. 1.
Schematic of the Microbubbling SARS-CoV-2 Antigen Assay. NP swab eluant is first treated with lysis buffer to release N proteins from SARS-CoV-2 viruses. N protein is then detected by the smartphone-based microbubbling digital assay. The microbubble images are quantified and classified as positive or negative by computer vision and ML algorithms. Scale bar, 500 μm.
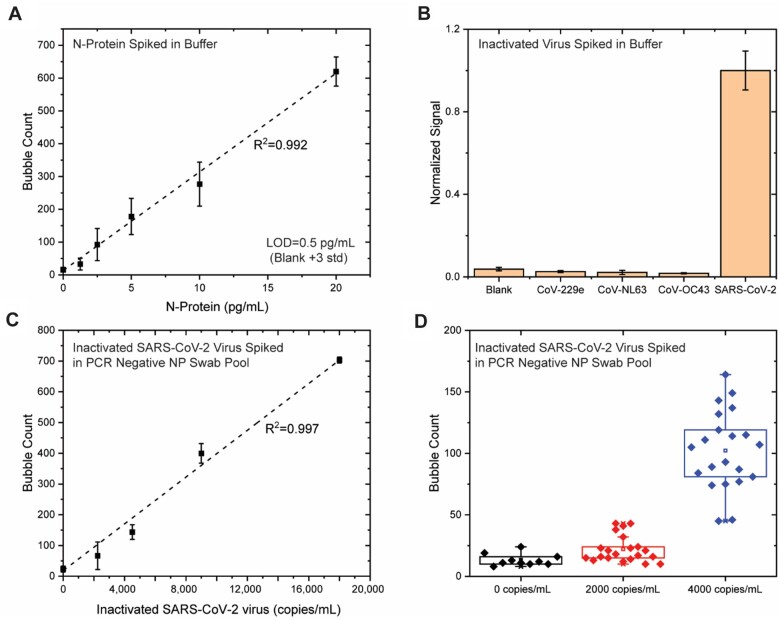
To identify the best antibody pair to use in MSAA, we screened different antibody pairs that bind the SARS-CoV-2 N protein with various capture/detection combinations (online Supplemental Fig. 1). The pair (N1 as capture antibody, Np as detection antibody) that generated the highest analytical sensitivity was chosen (Fig. 1). To assess the intrinsic analytical sensitivity of the assay, different amounts of recombinant N protein were spiked into PBS buffer and tested by the MSAA. As shown in Fig. 2, A, the number of microbubbles increased linearly with the concentration of N protein between 0 and 20 pg/mL, with the LOD for recombinant N protein at 0.5 pg/mL (10.6 fmol/L, blank + 3 SD, n = 10).
Fig. 2.
Performance of the Microbubbling SARS-CoV-2 Antigen Assay. (A) Dose–response curve of recombinant N protein in buffer. Mean ± 3 SD; n = 10 at 0, n = 3 at other points. (B) Specificity test. Mean ± 3 SD; n = 3. (C) Dose–response curve of inactivated SARS-CoV-2 viruses in rRT-PCR–negative NP swab pool. Mean ± 3 SD; n = 10 at 0, n = 3 at other points. (D) LOD following the FDA antigen template guideline.
To assess the analytical specificity of the MSAA toward different strains of coronaviruses, we challenged the assay with 3 human coronaviruses: CoV-229e, CoV-NL63, and CoV-OC43. Human coronaviruses OC43, 229E, and NL63 cause common cold and bronchiolitis and are the pathogens that are likely to be present in respiratory samples and possibly cross-react in the MSAA based on sequence homology. As shown in Fig. 2, B, no significant difference in signal was observed between the blank and the 3 coronaviruses (1 × 105 pfu/mL), while an over 10 times signal increase was observed for SARS-CoV-2 (1 × 105 copies/mL or 1.4 × 104 pfu/mL).
We then evaluated the analytical performance of the assay in swab samples. Mucosal swab samples contain catalase and peroxidase, which may interfere with the MSAA by degrading the signaling reagent, H2O2. To assess this possibility, recombinant N protein was spiked into a negative pool of NP swab samples (confirmed by rRT-PCR to be negative for SARS-CoV-2) and tested in the MSAA with and without PtNPs as catalyzing reagent. As shown in online Supplemental Fig. 2, bubbles were only observed when PtNPs were present, indicating catalase and peroxidase from the mucosal matrix had been eliminated during washing steps, and did not interfere in the MSAA.
We subsequently challenged the assay with 10 rRT-PCR-negative NP swab samples. As shown in online Supplemental Fig. 3, A, the background signals of the 10 samples varied. To investigate if the background signal was specific to the MSAA, a representative sample (sample 3, with a medium background signal) was tested in an ELISA using 96-well plate bottom to immobilize the N1 capture antibody and luciferase as the reporter enzyme labeled onto the Np detection antibody. As shown in Supplemental Fig. 3, B, a medium background signal was also observed for sample 3, but not for buffer control, in the ELISA assay. This indicates that the background was assay format-agnostic and intrinsic to the swab matrix. Mucins have been reported as the major cause of nonspecific bindings and background signals in many immunoassays using mucosal samples (21), due to their ability to bind to a variety of solid surfaces (22, 23, 24). We postulated that mucins were the source of the background signal in our assay. To eliminate the background signal, we took advantage of the high molecular weight of mucins and used a filter with molecular weight cutoff between mucins (200 kDa∼200 MDa) and N protein (47.08 kDa) (Pierce™ protein concentrator PES, 100K MWCO) to remove mucins and retain filtrate for testing. As shown in Supplemental Fig. 3, C, background signals in the rRT-PCR–negative NP swab pool were removed by filtering the sample through the previously described concentrator, while specific signals from SARS-CoV-2 were retained.
We compared the analytical sensitivity of the assay for inactivated SARS-CoV-2 spiked in buffer (without filtration) vs in negative NP swab pool (with filtration). As shown in online Supplemental Fig. 4, the analytical sensitivity in buffer (without filtration) was about 10 times higher than that in negative NP swab pool.
To determine the analytical sensitivity of the MSAA for viruses in NP swabs, different amounts of inactivated SARS-CoV-2 viruses were spiked into an rRT-PCR-negative NP swab pool, lysed, filtered, and tested using the MSAA. As shown in Fig. 2, C, the number of microbubbles increased linearly with the concentrations of inactivated SARS-CoV-2 viruses between 0 and 18 000 copies/mL [virus genome RNA concentration, determined by RT-quantitative PCR (RT-qPCR)] using a primer set specific to nsp14, with detailed methods in the online Data Supplemental). To determine the LOD following the Food and Drug Administration (FDA) antigen assay template (25), we tested 10 rRT-PCR-negative NP swab pools, 21 swab pools spiked with inactivated SARS-CoV-2 viruses at 2000 copies/mL, and 21 pools at 4000 copies/mL. As shown in Fig. 2, D, at a concentration of 2000 copies/mL, the signals of 17 out of the 21 samples were above the mean of blank signal; at a concentration of 4000 copies/mL, the signals of all 21 samples were above the blank mean + 3 SD. Therefore, 4000 copies/mL was determined as the LOD of the MSAA for SARS-CoV-2 in swabs. This LOD translates to 400 virus copies/reaction (100 μL sample volume) in the assay.
We tested deidentified residual clinical NP swab samples (at least 50 samples in each category) using the MSAA and compared the results to clinical testing results using FDA emergency use authorization (EUA)-approved rRT-PCR methods (Table 1; list of rRT-PCR assays can be found in the online Data Supplement). Characteristics of the clinical samples, including presence of symptoms, days after symptom onset, and clinical locations were also listed in Table 1. Compared to rRT-PCR, the positive percentage agreement (PPA) was 97% (95% CI 92%–99%) in symptomatic individuals within 7 days of symptom onset and positive nucleic acid results (n = 128), and the negative percentage agreement (NPA) was 97% (95% CI 94%–100%) in symptomatic and asymptomatic individuals with negative nucleic acid results (n = 73). The percentage of antigen-positive samples decreased in individuals at 7 to 12 days and >12 days after symptom onset or initial COVID-19 diagnosis, despite positive rRT-PCR results from the same samples.
Table 1.
Clinical performance of the Microbubbling SARS-CoV-2 antigen assay.
| rRT-PCR results | Presence of symptoms and days after symptom onset or initial diagnosis | N | Clinical location | Microbubbling SARS-CoV-2 antigen results |
PPA and NPA (95% CI) | |
|---|---|---|---|---|---|---|
| Positive | Negative | |||||
| Positive | Symptom onset <7 days | 128 | Emergency department, obstetrics department, preoperative evaluation, ICUa | 124 | 4 | PPA = 97% (92% to 99%) |
| Symptom onset 7–12 days | 51 | 27 | 24 | PPA = 53% (38% to 67%) | ||
| Symptom onset or initial diagnosis >12 days | 58 | 15 | 43 | PPA = 26% (15% to 39%) | ||
| Asymptomatic | 62 | 28 | 34 | PPA = 45% (32% to 58%) | ||
| Negative | Symptomatic and asymptomatic | 73 | 2 | 71 | NPA = 97% (94% to 100%) | |
Intensive care unit.
In the asymptomatic but rRT-PCR-positive group (n = 62), the PPA was 45% (95% CI 32%–58%). In this asymptomatic group, the MSAA was able to detect a presymptomatic case, in which both antigen and nucleic acid results were positive in the swab sample collected 1 day before the patient developed cough and other respiratory symptoms.
Computer Vision and ML Algorithm for MSAA Readout
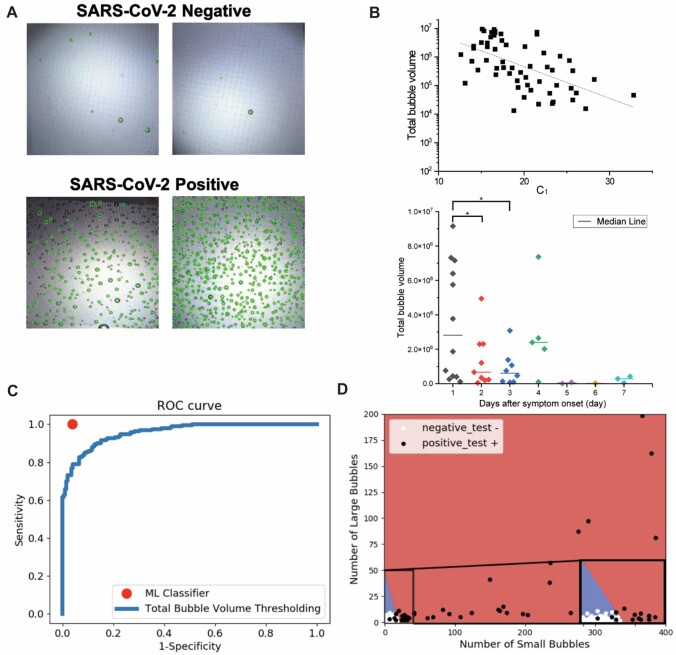
A computer vision algorithm was developed to detect microbubbles of varying sizes in each image. Figure 3, A, shows microbubble detection with green circles overlaid on the original images. Having automatically detected microbubble locations and sizes, we calculate the volumes of all bubbles to estimate the total bubble volume as a quantitative readout. As shown in Fig. 3, B, log-transformed total bubble volume correlated inversely with cycle threshold (Ct) values, and total bubble volume decreased with days-after-symptom-onset. This indicates that total bubble volume is a potential quantitative marker for antigen burden.
Fig. 3.
(A) Examples from clinical NP swab samples. (B) Upper: log-transformed total bubble volume correlated inversely with Ct by Pearson linear correlation. Lower: total bubble volume decreased with days-after-symptom-onset. (C) ROC curve comparing classification performance of 2 approaches. (D) Linear decision boundaries from the ML algorithm that learns to accurately classify images as negatives or positives in the validation data set.
We then compared 2 approaches (ML algorithm vs total bubble volume thresholding) for automatic classification of microbubble images as positives or negatives. We first randomly sampled 168 images (from 168 unique samples) to use as the data set to train a linear separator using the linear support vector ML algorithm and then evaluated the performance on the remaining 168 images (from another set of 168 samples). Next, we varied the threshold on the estimated total bubble volume to generate a total bubble volume-based classifier and evaluate its performance on the same 168 images. Figure 3, C, shows the performance for both classifiers. The ML classifier performs with much higher sensitivity (100%) and specificity (95%) than any point on the bubble volume thresholding ROC curve. The decision boundaries identified by the ML classifier are shown in Fig. 3, D. These results were very stable across 3 random 50–50 splits of the full data set (336 images) into training and testing data. Overall, this demonstrates that fully automated image analysis and ML are able to produce antigen burden quantitation and accurate classification from the MSAA.
Antigen Dynamics Tracking
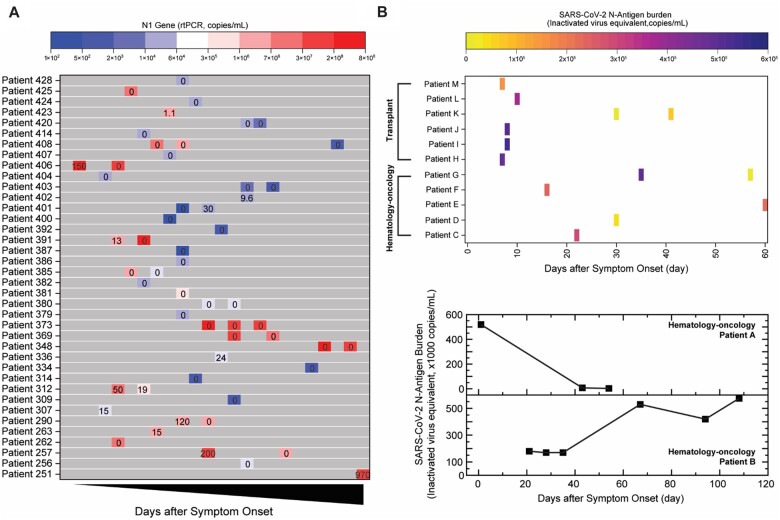
In the clinical validation group (Table 1), antigen and nucleic acid results showed limited correlation in individuals after the acute infection phase (>7 days). To further investigate this phenomenon longitudinally, we quantified antigen burden using the MSAA in serial NP/oropharyngeal swabs collected from 38 intensive care unit patients hospitalized due to COVID-19 (Fig. 4, A). To compare nucleic acid with antigen dynamics, both N1 gene copy number quantified using RT-qPCR and antigen burden were plotted in Fig. 4, A. Consistent with what was observed in Table 1, nucleic acid and antigen results did not always correlate with each other in this patient cohort. Many serial samples did not have detectable N antigens despite abundant copy numbers of the N1 gene (≫4000 copies/mL). Antigen burden in several patients (e.g., #406, #391, #257) decreased considerably despite N1 gene copies either increasing or remaining stable and above LOD of the MSAA. This suggests the RT-qPCR may be detecting N1 gene fragments that were not producing intact N protein with the epitope for antibody recognition. On the other hand, for patient #401, N1 gene copy number increase correlated with the increase in antigen burden, possibly indicating active virus replication during this period.
Fig. 4.
Tracking antigen burden using Microbubbling SARS-CoV-2 Antigen Assay in (A) inpatients in intensive care unit and (B) immunocompromised patients (rRT-PCR persistently positive) with either hematological malignancies or transplants. Numbers in boxes in (A) indicate N antigen burden quantitated by MSAA (inactivated virus equivalent, ×1000 copies/mL). Heatmap colors in (A) indicate N1 gene copy number. Heatmap colors in (B) indicate N antigen burden quantitated by MSAA.
Based on recent observations in immunocompromised patients, immunodeficiency plays a critical role in prolonged viral shedding, replication, and possibly mutation (5, 26–30). With persistently positive RNA results, the challenge is to determine at which point these patients are no longer contagious (5). In our validation group (Table 1), immunocompromised individuals also remained antigen positive for longer periods of time. We tracked antigen burden at various days-after-symptom-onset in a group of 13 immunocompromised patients with either hematological malignancies or transplants (Fig. 4, B, upper panel). Nucleic acid was detected in all samples for these patients, while antigen burden varied over time. In Fig. 4, B (lower panel), antigen burden dynamics are shown for 2 patients with hematological malignancies, demonstrating profiles distinct from each other. The furthest timepoint with antigen result in this group was from a patient (patient B in Fig. 4, B, lower panel) who had active multiple myeloma, a high antigen burden at 108 days and had received multiple rounds of convalescent plasma and remdesivir. In this patient, we detected multiple sequence variations at 3 time points (online Supplemental Fig. 5 and Table 3), with the highest number of variants in the spike protein sequence. Such variations were not detected in patient A (Fig. 4, B, lower panel) or patients with short duration of antigen detected. This suggests that prolonged high antigen burden may be a surrogate for high levels of viral replication and mutation. Thus, the MSAA could serve as a screen to identify patients who warrant close viral sequence monitoring.
Discussion
We have demonstrated that the MSAA can detect the N antigen with an analytical sensitivity of 0.5 pg/mL (10.6 fmol/L) and high specificity. Our observed LOD is 100 times better than the stated LOD of commercially available antigen assays and comparable to the SIMOA N antigen assay (18) (Supplemental Table 4). The LOD in swabs (4000 copies/mL) is comparable to many FDA EUA-approved rRT-PCR assays (102–105 copies/mL), better than current EUA-approved lateral flow antigen tests and the majority of emerging SARS-CoV-2 antigen assays (Supplemental Table 4). Clinical validation comparing to EUA-approved rRT-PCR methods showed excellent PPA and NPA in both symptomatic and asymptomatic individuals for acute infection detection (<7 days). Residual NP swabs in 3 mL saline tubes were used for convenience during our clinical validation. We expect higher PPA using swabs directly reconstituted using a lower volume of extraction reagents, which can be tested in future studies. These findings suggest that the MSAA may be a valuable tool in detecting acute SARS-CoV-2 infections. The gradual decrease in PPA in samples after 7 days of symptom onset is consistent with seroconversion (31) and clearance of antigen from the body. It has been suggested that infectivity of individuals with high Ct values and symptom onset to testing >8 days is low (8). Our data in Table 1 and Fig. 4, A, are consistent with this hypothesis. The fact that the Microbubbling Antigen Assay was able to detect a presymptomatic infection case indicates that the assay has the required sensitivity to detect presymptomatic antigen levels. The low PPA in the asymptomatic group is likely due to the heterogeneous nature of this group, in which individuals may present for testing at various stages of their infections (SARS-CoV-2 testing was conducted as part of presurgery evaluation, prenatal care, or emergency department visits with nonrespiratory complaints), sometimes late enough that antigens have been cleared by immune system while viral genomic fragments remain.
We have also demonstrated that total bubble volume, a quantitative output from the MSAA, correlated inversely with Ct values and days-after-symptom-onset, and may serve as a potential marker for antigen burden. This allows us to probe and monitor the level and dynamics of virus antigens at different stages of infection and to explore the correlation between antigen burden and disease severity/prognosis in future studies, which may be valuable in advancing understanding of the SARS-CoV-2 virus. With the exception of some reports focusing on antigens in blood (16, 18, 32) and some antigen data in saliva (18), the burden and dynamics of SARS-CoV-2 antigens in respiratory samples have not been thoroughly characterized. Our data demonstrated that in immunocompetent individuals, antigen is often cleared much faster than nucleic acid in respiratory samples. On the other hand, we also showed that immunocompromised individuals remained antigen positive for much longer periods of time and cleared antigen at various timepoints. In general, antigen dynamics did not correlate well with that of nucleic acids outside of the acute infection window. Notably, we detected many variants in longitudinal samples from a patient with prolonged high antigen burden. This suggests that patients who have long duration of high antigen burden in the MSAA are likely experiencing active rounds of ongoing replication and are at higher risk for viral mutation. Therefore, the MSAA could serve as a screen in longitudinal studies to identify patients who warrant close viral sequence monitoring.
We expect that the MSAA will be able to mitigate concerns for false negatives using antigen tests. However, we did not compare the MSAA with current lateral flow antigen tests in a side-by-side manner. Another limitation of the study is that the data of days-since-symptom-onset came from limited documentation in the medical records based on patient self-reporting. These data may not always be accurate or complete. Ct values were not available from some rRT-PCR methods used in this study and are in general not well-standardized across platforms (33). This may lead to heterogeneity in the nucleic acid data presented. Finally, due to the poor culturability of most clinical specimens, we did not use viral culture to assess the infectivity of antigen positive samples.
In future studies, the MSAA can be automated and multiplexed with other infectious disease antigens and applied to other sample types. With the convenient computer vision and ML-based algorithms on smartphones for automated microbubble detection, this assay also has the potential to be used at the point-of-care for frequent and repeated testing. Finally, in an integrated system, the microbubble results could also be conveniently uploaded into a database that integrates other clinical parameters and molecular/serology results, etc. to provide a comprehensive picture of the disease in the population, enabling disease tracking and future predictive algorithm development.
In summary, we have demonstrated that the MSAA can be applied to the detection of SARS-CoV-2 antigen with high sensitivity and specificity. The MSAA demonstrated high PPA and NPA with rRT-PCR methods in symptomatic and asymptomatic individuals early in the infection course. We have developed computer vision and ML algorithms to quantify and classify the image output and also shown that the assay can be a valuable tool in providing insights into antigen dynamics in various patient populations.
Supplemental Materials
Supplemental material is available at Clinical Chemistry online.
Supplementary Material
Nonstandard Abbreviations:
- NP
nasopharyngeal
- LOD
limit of detection
- N antigen
nucleocapsid antigen
- RT-qPCR
reverse transcription−quantitative polymerase chain reaction
- SIMOA
single molecule array
- ML
machine learning
- PtNP
platinum nanoparticle
- COVID-19
coronavirus disease 2019
- PSA
prostate specific antigen
- βhCG
beta human chorionic gonadotropin
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- CV
coefficient of variation
- rRT-PCR
real-time reverse transcription−polymerase chain reaction
- PPA
positive percent agreement
- NPA
negative percent agreement
- MSAA
Microbubbling SARS-CoV-2 Antigen Assay
Author Contributions
All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
Authors’ Disclosures or Potential Conflicts of Interest
Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership
P. Wang, Clinical Chemistry, AACC, Instanosis Inc.
Consultant or Advisory Role
D. Vogl, Karyopharm Therapeutics, GSK, Oncopeptides, Takeda, Amgen, Celgene Corporation, Active Biotech; A. Garfall, Tmunity, Amgen, Surface Oncology; P. Wang, Truvian Sciences.
Stock Ownership
None declared.
Honoraria
A. Garfall, Janssen, GlaxoSmithKline; P. Wang, AACC.
Research Funding
H. Chen, Z. Li, and P. Wang have received support from National Institute of Health grants R01DA035868, R01EB029363 and National Science Foundation grant 1928334. S.R. Weiss has received support from National Institute of Health grant R01AI40442 and Penn Center for Research on Coronaviruses and Other Emerging Pathogens. We thank the RADx-Tech Program, Penn Center for Precision Medicine, Penn Health-Tech and Penn Center for Innovation & Precision Dentistry for providing funding for this project. This work was carried out in part at the Singh Center for Nanotechnology, part of the National Nanotechnology Coordinated Infrastructure Program, which is supported by the National Science Foundation grant NNCI-2025608. D. Jayaraman, Amazon Research, General Electric, NEC Laboratories America; D. Vogl, Takeda, Active Biotech; A. Garfall, Janssen, Novartis, Tmunity, and CRISPR Therapeutics to institution; R.G. Collman, OraSure, Inc; J.S. Lee, Bill & Melinda Gates Foundation INV-018479, 5R01AI140539-02; S. Cherry, NIH 3R01AI150246-01S1, 5R01AI140539, Burroughs Wellcome Fund, Bill & Melinda Gates Foundation INV-018479, Mercatus Fast Grant.
Expert Testimony
None declared.
Patents
A. Garfall, 15/757,123, 16/768,260, 16/746,459; P. Wang, PCT, US and EU patents pending; Z. Li, U.S. provisional application no 621730,719.
Role of Sponsor
The funding organizations played a direct role in the design of study, review and interpretation of data, and preparation of manuscript. The funding organizations played no role in the choice of enrolled patients or final approval of manuscript.
References
- 1. Wang X, Yao H, Xu X, Zhang P, Zhang M, Shao J, et al. Limits of detection of 6 approved RT-PCR kits for the novel SARS-coronavirus-2 (SARS-CoV-2). Clin Chem 2020;66:977–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shi J, Han D, Zhang R, Li J, Zhang R. Molecular and serological assays for SARS-CoV-2: Insights from genome and clinical characteristics. Clin Chem 2020;66:1030–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. SARS-CoV-2 reference panel comparative data. https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-reference-panel-comparative-data (Accessed June 2021).
- 4. Henderson DK, Weber DJ, Babcock H, Hayden MK, Malani A, Wright SB, et al. ; SHEA Board of Trustees. The perplexing problem of persistently PCR-positive personnel. Infect Control Hosp Epidemiol 2021;42:203–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abbasi J. Researchers tie severe immunosuppression to chronic COVID-19 and virus variants. JAMA 2021;325:2033–5. [DOI] [PubMed] [Google Scholar]
- 6. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465–9. [DOI] [PubMed] [Google Scholar]
- 7. La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis 2020;39:1059–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis 2020;71:2663–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gniazdowski V Morris CP, Wohl S, Mehoke T, Ramakrishnan S, Thielen P, et al. Repeated Coronavirus Disease 2019 Molecular Testing: Correlation of Severe Acute Respiratory Syndrome Coronavirus 2 Culture With Molecular Assays and Cycle Thresholds. Clin Infect Dis 2021;73(4):e860–9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sia SF, Yan L-M, Chin AWH, Fung K, Choy K-T, Wong AYL, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020;583:834–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pekosz A, Parvu V, Li M, Andrews JC, Manabe YC, Kodsi S, et al. Antigen-based testing but not real-time polymerase chain reaction correlates with severe acute respiratory syndrome coronavirus 2 viral culture. [Epub ahead of print] Clin Infect Dis January 20, 2021. as doi:10.1093/cid/ciaa1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mak GC, Cheng PK, Lau SS, Wong KK, Lau CS, Lam ET, et al. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol 2020;129:104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lanser L, Bellmann-Weiler R, Ottl KW, Huber L, Griesmacher A, Theurl I, Weiss G. Evaluating the clinical utility and sensitivity of SARS-CoV-2 antigen testing in relation to RT-PCR CT values. Infection 2021;49:555–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coronavirus (COVID-19) update: FDA authorizes first antigen test to help in the rapid detection of the virus that causes covid-19 in patients. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-antigen-test-help-rapid-detection-virus-causes (Accessed June 2021)
- 15. Norman M, Gilboa T, Ogata AF, Maley AM, Cohen L, Busch EL, et al. Ultrasensitive high-resolution profiling of early seroconversion in patients with COVID-19. Nat Biomed Eng 2020;4:1180–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ogata AF, Maley AM, Wu C, Gilboa T, Norman M, Lazarovits R, et al. Ultra-sensitive serial profiling of SARS-CoV-2 antigens and antibodies in plasma to understand disease progression in covid-19 patients with severe disease. Clin Chem 2020;66:1562–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shan D, Johnson JM, Fernandes SC, Mendes M, Suib H, Holdridge M, et al. SARS-coronavirus-2 nucleocapsid protein measured in blood using a SIMOA ultra-sensitive immunoassay differentiates covid-19 infection with high clinical sensitivity. medRxiv 2020. 10.1101/2020.08.14.20175356. [DOI]
- 18. Shan D, Johnson JM, Fernandes SC, Suib H, Hwang S, Wuelfing D, et al. N-protein presents early in blood, dried blood and saliva during asymptomatic and symptomatic SARS-CoV-2 infection. Nat Commun 2021;12:1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen H, Li Z, Zhang L, Sawaya P, Shi J, Wang P. Quantitation of femtomolar-level protein biomarkers using a simple microbubbling digital assay and bright-field smartphone imaging. Angew Chem Int Ed Engl 2019;58:13922–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Canny J. A computational approach to edge detection. IEEE Trans Pattern Anal Mach Intell 1986;8:679–98. [PubMed] [Google Scholar]
- 21. Duda RO, Hart PE. Use of the Hough transformation to detect lines and curves in pictures. Commun ACM 1972;15:11–5. [Google Scholar]
- 22. Helton KL, Nelson KE, Fu E, Yager P. Conditioning saliva for use in a microfluidic biosensor. Lab Chip 2008;8:1847–51. [DOI] [PubMed] [Google Scholar]
- 23. McColl J, Yakubov GE, Ramsden JJ. Complex desorption of mucin from silica. Langmuir 2007;23:7096–100. [DOI] [PubMed] [Google Scholar]
- 24. Song D, Cahn D, Duncan GA. Mucin biopolymers and their barrier function at airway surfaces. Langmuir 2020;36:12773–83. [DOI] [PubMed] [Google Scholar]
- 25.Coronavirus disease 2019 (COVID-19) emergency use authorizations for medical devices/in vitro diagnostics EUAS. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas (Accessed June 2021).
- 26. Baang JH, Smith C, Mirabelli C, Valesano AL, Manthei DM, Bachman MA, et al. Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis 2021;223:23–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tarhini H, Recoing A, Bridier-Nahmias A, Rahi M, Lambert C, Martres P, et al. Long-term severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectiousness among three immunocompromised patients: from prolonged viral shedding to SARS-CoV-2 superinfection. J Infect Dis 2021;223:1522–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aydillo T, Gonzalez-Reiche AS, Aslam S, van de Guchte A, Khan Z, Obla A, et al. Shedding of viable SARS-CoV-2 after immunosuppressive therapy for cancer. N Engl J Med 2020;383:2586–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kemp SA, Collier DA, Datir RP, Ferreira I, Gayed S, Jahun A, et al. ; COVID-19 Genomics UK (COG-UK) Consortium. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021;592:277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abdul-Jawad S, Baù L, Alaguthurai T, Barrio DMDI, Laing AG, Hayday TS, et al. Acute immune signatures and their legacies in severe acute respiratory syndrome coronavirus-2 infected cancer patients. Cancer Cell 2021;39:257–75.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang AT, Garcia-Carreras B, Hitchings MDT, Yang B, Katzelnick LC, Rattigan SM, et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun 2020;11:4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hingrat QL, Visseaux B, Laouenan C, Tubiana S, Bouadma L, Yazdanpanah Y, et al. Detection of SARS-CoV-2 n-antigen in blood during acute covid-19 provides a sensitive new marker and new testing alternatives. Clin Microbiol Infect 2020;27:e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rhoads D, Peaper DR, She RC, Nolte FS, Wojewoda CM, Anderson NW, Pritt BS. College of American Pathologists (CAP) microbiology committee perspective: caution must be used in interpreting the cycle threshold (CT) value. Clin Infect Dis 2021;72:e685–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.