Abstract
Background
We studied risk factors, antibodies, and symptoms of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in a diverse, ambulatory population.
Methods
A prospective cohort (n = 831) previously undiagnosed with SARS-CoV-2 infection underwent serial testing (SARS-CoV-2 polymerase chain reaction, immunoglobulin G [IgG]) for 6 months.
Results
Ninety-three participants (11.2%) tested SARS-CoV-2-positive: 14 (15.1%) asymptomatic, 24 (25.8%) severely symptomatic. Healthcare workers (n = 548) were more likely to become infected (14.2% vs 5.3%; adjusted odds ratio, 2.1; 95% confidence interval, 1.4–3.3) and severely symptomatic (29.5% vs 6.7%). IgG antibodies were detected after 79% of asymptomatic infections, 89% with mild-moderate symptoms, and 96% with severe symptoms. IgG trajectories after asymptomatic infections (slow increases) differed from symptomatic infections (early peaks within 2 months). Most participants (92%) had persistent IgG responses (median 171 days). In multivariable models, IgG titers were positively associated with symptom severity, certain comorbidities, and hospital work. Dyspnea and neurologic changes (including altered smell/taste) lasted ≥ 120 days in ≥ 10% of affected participants. Prolonged symptoms (frequently more severe) corresponded to higher antibody levels.
Conclusions
In a prospective, ethnically diverse cohort, symptom severity correlated with the magnitude and trajectory of IgG production. Symptoms frequently persisted for many months after infection.
Clinical Trials Registration. NCT04336215.
Keywords: COVID-19, humoral immunity, longitudinal data analysis, postacute sequelae of COVID-19, prospective cohort, risk factors, SARS-CoV-2 infection, symptoms
In a diverse, ambulatory cohort (548 healthcare workers; 283 nonhealthcare workers), 11.2% tested positive for SARS-CoV-2 over 6-month follow-up. COVID-19 symptom severity correlated with magnitude and trajectory of IgG production. Symptoms lasting ≥ 30 days afflicted one-third of infected participants.
As the coronavirus disease 2019 (COVID-19) pandemic continues to surge, as of early May 2021, the United States has recorded the most cases (>32 million) and deaths (>580 000) of any country [1]. Approximately one-third of infections are estimated to be asymptomatic [2–4] and are considered important drivers of viral transmission [5]. Nonetheless, asymptomatic infections may be accompanied by subclinical abnormalities in laboratory tests and lung imaging [6]. Important questions remain about long-term clinical and immunologic consequences of asymptomatic infections.
Most persons infected by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) develop antibodies against the virus [7]. However, immune responses vary considerably, with a minority of infected people not producing detectable antibodies [8]. The magnitude of humoral immune responses may be proportional to illness severity [9, 10]. The duration and trajectory of humoral immunity also remain unclear; some studies report substantial declines in antibody responses within a few months [11, 12] while others report persistent responses over many months [8, 13, 14]. One challenge in interpreting these studies is differences in study populations: most studies have focused on hospitalized, convalescent, and referred patients previously diagnosed with SARS-CoV-2 infection, raising questions about selection bias and generalizability. Few prospective studies have systematically evaluated long-term antibody trends and associated factors among diverse, previously undiagnosed populations of individuals across a spectrum of illness severity, including asymptomatic infections [15].
We characterized the incidence of and risk factors for SARS-CoV-2 infection in a prospective cohort of ambulatory, previously undiagnosed healthcare workers (HCWs) and non-HCWs recruited early in the US pandemic and followed over 6 months. The study was conducted in New Jersey (NJ), an ethnically diverse state hit particularly hard by the spring 2020 COVID-19 surge [1, 16]. We further examined dynamics and correlates of anti-SARS-CoV-2 antibodies and persistence of symptoms up to 6 months postinfection.
METHODS
Study Design and Population
As described [17], the Rutgers Corona Cohort is a prospective, university-based observational cohort of HCWs and non-HCW comparators recruited and consented 24 March to 7 April 2020, across 2 campuses (Newark and New Brunswick/Piscataway). Eligibility criteria included: (1) age ≥ 20 years; (2) not pregnant or breastfeeding; (3) no recent (prior 30 days) urgent care or emergency department visits, hospitalizations, operations, or changes in prescribed medicines; (4) no previously diagnosed SARS-CoV-2 infection/COVID-19; and (5) no fever at the baseline visit. Eligibility for HCWs required: (1) ≥ 20 hours of weekly hospital work; (2) roles with regular patient exposure (eg, physicians, nurses, technicians, respiratory therapists); and (3) regular direct patient contact (≥3 patients/shift). Eligibility for a comparator group of non-HCWs required: (1) work as faculty, staff, or students at Rutgers for ≥ 20 hours weekly; and (2) no patient contact. Hospital-based employees without direct patient care responsibilities were not eligible for enrollment. All study activities were approved by the Rutgers Institutional Review Board (Pro2020000679) and all participants provided electronic informed consent prior to engaging in study activities.
Study Activities and Data
Study visits took place at baseline, 2, 4, 8, 16, and 26 weeks. At each visit, study staff in personal protective equipment (PPE) measured body temperature and collected oropharyngeal swabs immersed in phosphate-buffered saline [18] and blood using serum separator tubes. For the first 2 study months, participants recorded an early evening body temperature using a study-issued oral thermometer. Participants completed questionnaires at baseline, weekly for 2 months, then every other week. Questionnaire items included demographics, comorbidities, lifestyle, occupation, COVID-19 exposures and diagnoses, recent symptoms, and, for HCWs, unit locations, patient contacts, and PPE use. Participant zip code was used to classify residence in areas with high COVID-19 rates, defined as > 2% of residents with confirmed infections as of 20 August 2020.
Additional information about SARS-CoV-2–positive participants was obtained through follow-up surveys, telephone calls, and medical chart review; symptoms were assessed using all available data sources. Among symptomatic participants, overall symptom severity was assessed using the question: “Please consider any past or present COVID-19 symptoms when answering the following question: Overall, when these symptoms were at their worst, how bad or bothersome were they?” Responses options were: mild, moderate, severe, and very severe. Based on the distribution of responses, we categorized symptom severity as asymptomatic, mild-moderate, or severe. All study data were managed using REDCap electronic data capture tools hosted at Rutgers Robert Wood Johnson Medical School [19].
SARS-CoV-2 Assays
SARS-CoV-2 assays were conducted at all study visits under Food and Drug Administration-approved Emergency Use Authorization number 200090 at Infinity Biologix (Piscataway, NJ), as described [17, 18]. In brief, total RNA was extracted from oropharyngeal swabs using nucleic acid-binding paramagnetic beads (Chemagic Viral DNA/RNA 300 Kit H96). Reverse transcriptase-polymerase chain reaction (RT-PCR) was performed in triplicate for 3 SARS-CoV-2 genomic regions: nucleocapsid (N), spike protein (S), and ORF1ab. Positive and negative assay controls were used.
SARS-CoV-2 Antibody Testing
We used an in-house developed enzyme-linked immunosorbent assay (ELISA) platform for antibody binding to 2 portions of SARS-CoV-2 spike protein (S1 subunit receptor-binding domain [RBD], full-length S2 subunit) [20]. Detection of antigen-bound antibodies used combined alkaline phosphatase-conjugated anti-human immunoglobulin A (IgA), IgM, and IgG secondary antibodies, or anti-human IgG antibody alone, at 1:2000 dilution (Supplementary Methods and Supplementary Table 1). For participants with optical density 450 (OD405) ≥1 positive IgM/G/A (total) antibody test and/or positive PCR, anti-RBD IgG titers were determined. Seropositivity was defined as total antibody levels OD405 ≥0.7 across ≥ 2 time points or ≥ 1.0 once; or IgG titers ≥ 1:80 across ≥ 2 time points or ≥ 1:320 once. We chose RBD as our solid-phase antigen in our assay because it is a preferred target of neutralizing antibodies [21].
Routine Chemistries and Blood Counts
Comprehensive metabolic panels were analyzed on baseline plasma samples, and cell counts were analyzed on whole-blood samples from all visits, using standard clinical assays (Beckman Coulter).
Statistical Analyses
Study outcomes were defined by SARS-CoV-2 positivity with PCR and/or antibody testing (IgM/G/A or IgG only). Comparisons of characteristics between SARS-CoV-2–infected and –uninfected participants and between HCWs and non-HCWs used χ 2 or Fisher exact testing (categorical data) and t tests or Wilcoxon rank sum testing (continuous data), as appropriate. Trends were evaluated across levels of symptom severity using Cochran-Armitage tests (categorical data) and Jonckheere-Terpstra tests (continuous data).
To identify explanatory baseline and early exposure characteristics associated with likelihood of SARS-CoV-2 infection, multivariable logistic regression models were fitted with elastic net penalty for regularization, permitting selection from many variables (Table 1) while avoiding overfitting due to penalties on regression coefficients. Separate models were applied to all participants and to HCWs, the latter including HCW-specific variables (eg, role, PPE use) (Table 1). Models accounted for time-varying exposures (eg, sick contacts, patient care metrics) over the first study month, during the first surge’s peak [22]; data after SARS-CoV-2 diagnosis were excluded to limit bias from factors resulting from infection.
Table 1.
Characteristics of Rutgers Corona Cohort Study Participants Stratified by SARS-CoV-2 Test Resultsa
| Characteristics | All (n = 831) | Ever Positive (n = 93, 11.2%) | Never Positive (n = 738, 88.8%) | P Valueb |
|---|---|---|---|---|
| Female | 533 (64.1) | 66 (71.0) | 467 (63.3) | .15 |
| Age, y | .52 | |||
| 20–39 | 430 (51.7) | 50 (53.8) | 380 (51.5) | |
| 40–59 | 315 (37.9) | 31 (33.3) | 284 (38.5) | |
| ≥60 | 86 (10.3) | 12 (12.9) | 74 (10.0) | |
| Race | .03 | |||
| White | 483 (58.6) | 53 (58.9) | 430 (58.6) | |
| Asian | 171 (20.8) | 10 (11.1) | 161 (21.9) | |
| Black | 90 (10.9) | 15 (16.7) | 75 (10.2) | |
| Other | 80 (9.71) | 12 (13.3) | 68 (9.26) | |
| Hispanic/Latino ethnicity | 101 (12.2) | 19 (20.4) | 82 (11.1) | .01 |
| Residence in high-risk zip codec | 101 (12.6) | 6 (6.59) | 95 (13.3) | .18 |
| Child < 18 y in the home | 326 (39.2) | 36 (38.7) | 290 (39.3) | .91 |
| Smoking | .21 | |||
| Current | 37 (4.47) | 4 (4.30) | 33 (4.49) | |
| Former | 230 (27.8) | 33 (35.5) | 197 (26.8) | |
| Chronic illness | 376 (45.2) | 51 (54.8) | 325 (44.0) | .05 |
| Obesity | 188 (22.8) | 31 (33.3) | 157 (21.5) | .01 |
| Diabetes mellitus | 48 (5.84) | 2 (2.15) | 46 (6.31) | .11 |
| Hypertension | 125 (15.2) | 20 (21.7) | 105 (14.3) | .06 |
| Cardio/cerebrovascular disease | 20 (2.41) | 3 (3.23) | 17 (2.30) | .58 |
| Chronic respiratory disorderd | 113 (13.6) | 10 (10.8) | 103 (14.0) | .40 |
| Autoimmune disease or immunosuppressant use | 40 (4.81) | 5 (5.38) | 35 (4.74) | .79 |
| HCW | 548 (65.9) | 78 (83.9) | 470 (63.7) | <.001 |
| Attending physician | 113 (13.6) | 8 (8.60) | 105 (14.2) | <.001 |
| Resident or fellow physician | 98 (11.8) | 8 (8.60) | 90 (12.2) | |
| Nurse | 225 (27.1) | 45 (48.4) | 180 (24.4) | |
| Other role | 112 (13.5) | 17 (18.3) | 95 (12.9) | |
| Work location | <.001 | |||
| Newark | 342 (41.2) | 54 (58.1) | 288 (39.0) | |
| New Brunswick/Piscataway | 489 (58.8) | 39 (41.9) | 450 (61.0) | |
| Exposure over the first monthe | ||||
| Worked on site, ever | 752 (90.8) | 86 (95.6) | 666 (90.2) | .10 |
| Stayed home as much as possible when not working | 502 (60.5) | 63 (68.5) | 439 (59.5) | .10 |
| Avoided others as much as possible when not at work | 514 (61.9) | 67 (72.0) | 447 (60.6) | .03 |
| Mask use outside the homef | .51 | |||
| None | 510 (61.4) | 52 (55.9) | 458 (62.1) | |
| Sometimes | 225 (27.1) | 29 (31.2) | 196 (26.6) | |
| Always | 95 (11.4) | 12 (12.9) | 83 (11.3) | |
| Unprotected COVID-19 exposure at home | 112 (14.4) | 16 (42.1) | 96 (13.0) | <.001 |
| Unprotected COVID-19 exposure at work | 543 (67.0) | 66 (90.4) | 477 (64.6) | <.001 |
| Unprotected COVID-19 exposure outside home/work | 103 (13.3) | 12 (34.3) | 91 (12.3) | <.001 |
| Unprotected COVID-19 exposure at home or outside home/work | 188 (24.1) | 25 (61.0) | 163 (22.1) | <.001 |
| Average level of patient contact | <.001 | |||
| Non-HCW | 283 (34.2) | 15 (16.3) | 268 (36.4) | |
| Patient contact below median | 268 (32.4) | 60 (65.2) | 208 (28.3) | |
| Patient contact at or above median | 277 (33.5) | 17 (18.5) | 260 (35.3) | |
| HCWs only | ||||
| Worked in emergency department | 311 (38.9) | 43 (69.4) | 268 (36.3) | <.001 |
| Worked on medical floor | 246 (31.5) | 22 (50.0) | 224 (30.4) | .01 |
| Worked in operating room | 147 (18.8) | 24 (52.2) | 123 (16.7) | <.001 |
| Worked in intensive care unit | 262 (33.7) | 19 (47.5) | 243 (32.9) | .06 |
| Worked in designated COVID-19 unit | 242 (31.2) | 17 (44.7) | 225 (30.5) | .06 |
| Average % patients for whom used PPE per shift | .86 | |||
| <25% | 42 (7.79) | 6 (7.89) | 36 (7.78) | |
| 25%–49% | 48 (8.91) | 8 (10.5) | 40 (8.64) | |
| ≥50% | 449 (83.3) | 62 (81.6) | 387 (83.6) | |
| Average % time in PPE using N95 mask per shift | <.001 | |||
| <25% | 75 (14.0) | 22 (28.9) | 53 (11.5) | |
| 25%–49% | 51 (9.51) | 8 (10.5) | 43 (9.35) | |
| ≥50% | 410 (76.5) | 46 (60.5) | 364 (79.1) | |
| Average number of patients with COVID-19 per shift | .12 | |||
| 0 | 41 (8.17) | 10 (13.3) | 31 (7.26) | |
| 1–4 | 170 (33.9) | 20 (26.7) | 150 (35.1) | |
| ≥5 | 291 (58.0) | 45 (60.0) | 246 (57.6) |
Data are No. (%).
Abbreviations: COVID-19, coronavirus disease 2019; HCW, healthcare worker; PPE, personal protective equipment; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
aPCR- or antibody-positive for SARS-CoV-2 were classified as positive.
b P values were computed using χ 2 or Fisher exact testing, as appropriate.
cHigh-risk zip code defined as having confirmed SARS-CoV-2 infections in > 2% of residents as of 20 August 2020.
dAsthma, chronic obstructive pulmonary disease, or other chronic lung disease.
eExcluding any values after diagnosis of SARS-CoV-2 infection.
fLowest reported value in the first month of participation.
Antibody curves were estimated for (1) different levels of symptom severity and (2) different durations of symptoms, with spline for time and random effects set to 0. To identify factors associated with IgG titers at each visit, we fitted generalized additive mixed effects models. We included all SARS-CoV-2–positive participants (PCR positive or antibody positive) except individuals newly positive by PCR at the final (26-week) visit (n = 2), who had not yet mounted an antibody response. Time of positivity was anchored by the date of the first positive test (PCR or antibody), defined as time 0. Given the nonlinear changes in antibody levels expected over time, models included a spline function for time. A random intercept accounted for within-subject correlation over time. Model variables included symptom severity (none, mild-moderate, severe), preselected baseline chemistries (glomerular filtration rate, alanine aminotransferase, albumin), time-updating cell counts (lymphocytes, neutrophils, platelets, hemoglobin), and variables listed in Table 1. Missing data were imputed using multiple imputation with chained equations based on 50 imputed data sets [23]. Presence and persistence of symptoms over time was graphed using Kaplan-Meier plots and summarized by the median, 75th and 90th percentiles. Analyses were performed using SAS 9.4, R 4.0.3, and Stata 16.1.
RESULTS
We enrolled 831 participants (548 HCWs, 283 non-HCWs; Supplementary Figure 1); 722 (86.9%) completed a 26-week visit and 758 (91.2%) completed at least 5 of 6 study visits. Overall, 71% of participants completed at least 12 of 16 follow-up questionnaires. Two-thirds of participants were female, and half were < 40 years old (Table 1). The cohort was racially and ethnically diverse (58.6% white, 20.8% Asian, 10.9% black, 9.7% other race, and 12.2% Hispanic/Latino). Nearly half (45.3%) of participants had at least 1 comorbidity, most commonly obesity (22.8%). Within 1 month after enrollment, 23.8% reported exposure to someone outside of home/work suspected or confirmed to have COVID-19. Most HCWs (91.8%) reported close contact with ≥ 1 patient with suspected or confirmed COVID-19 within 1 month of enrollment. Compared to non-HCWs, HCWs were younger, more racially diverse, more likely Newark-based, and more likely to report unprotected COVID-19 exposures at and outside work before diagnosis (Supplementary Table 2). Compared to eligible persons who did not enroll, enrolled participants were more likely to be of Hispanic/Latino ethnicity, have certain comorbidities (eg, respiratory, autoimmune), be a HCW, and be recruited at the Newark campus, and less likely to be female and a HCW caring for patients with COVID-19 (Supplementary Table 3).
Ultimately, 93 participants (11.2%) tested positive for virus and/or antibodies, 86 (92.5%) within the first 2 months of the study, echoing trends more broadly in NJ and participating hospitals (Supplementary Figure 2). Five participants tested positive at the final visit (3 by PCR, 2 by antibody), during the second COVID-19 surge in NJ in late 2020. These included 1 late asymptomatic PCR-positive infection following early asymptomatic infection in April, with sustained low-titer IgG and 5 negative PCRs until late September.
Among infected participants, 62 (66.7%) tested positive by both PCR and antibodies, 12 (12.9%) tested positive by PCR only, and 19 (20.4%) tested positive by antibody only. Of infected participants, 24 (25.8%) reported severe symptoms (including the 5 hospitalized participants, none in intensive care units [ICUs]), 55 (59.1%) reported mild to moderate symptoms, and 14 (15.1%) reported no symptoms. Only 13 (14.0%) infected participants received pharmacologic treatment. Despite being less likely to live in high-risk zip codes (10.3% vs 17.1%, P = .02), HCWs were significantly more likely to test positive for virus or antibodies (14.2% vs 5.3%, P < .001). Among infected individuals, HCWs were more likely to have severe symptoms (29.5% vs 6.7%, P = .04) and require hospitalization (6.4% vs 0%, P = .31). Self-reported severity correlated with symptom burden, settings of care, treatments received, and laboratory values (Supplementary Table 4 and Supplementary Figure 3). More-symptomatic participants had lower hemoglobin and absolute lymphocyte and neutrophil counts during follow-up, especially while infected (Supplementary Figure 4).
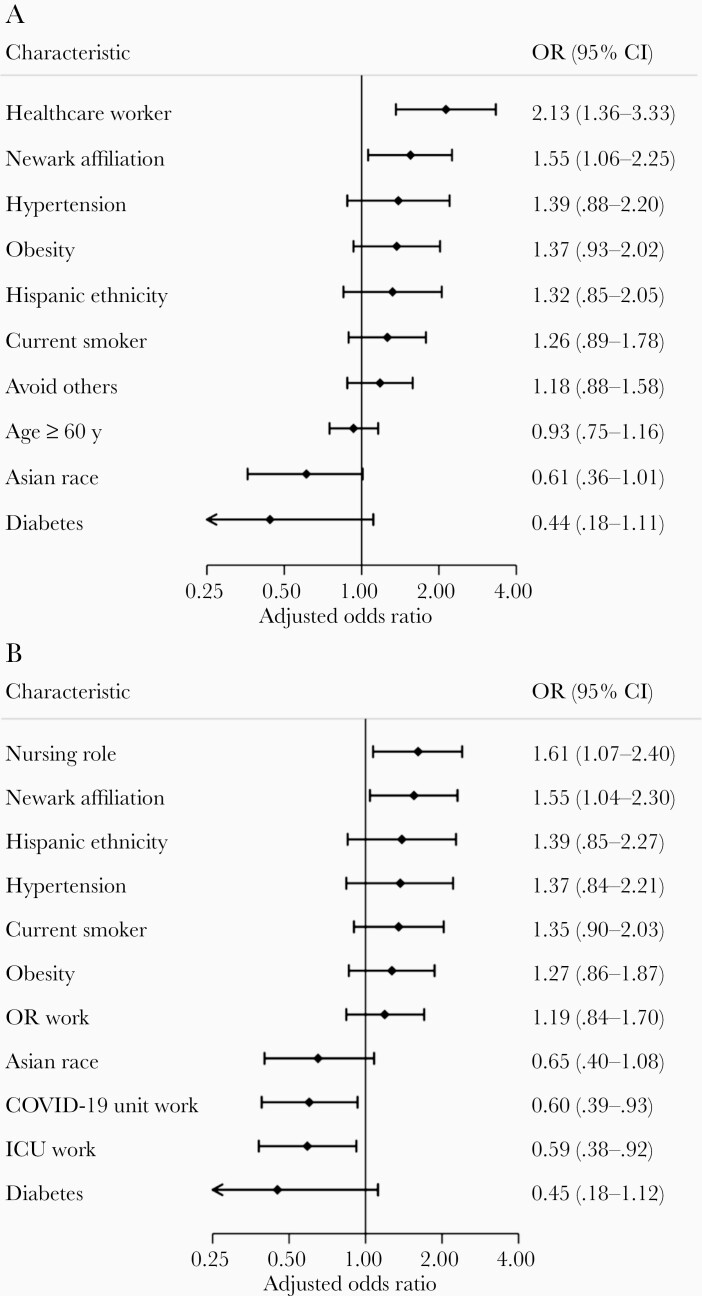
Among all participants, the factors most strongly associated with infection were HCW status (adjusted odds ratio [aOR], 2.13; 95% confidence interval [CI], 1.36–3.33) and Newark affiliation (aOR, 1.55; 95% CI, 1.06–2.25) (Figure 1A). Among HCWs, nursing role and Newark affiliation were most positively associated with infection, whereas work in ICUs or COVID-19 units was negatively associated with infection (Figure 1B). More-extensive N95 use was reported among SARS-CoV-2–negative participants (Table 1).
Figure 1.
Factors associated with SARS-CoV-2 infection. Forest plots show factors associated with infection in (A) Rutgers Corona Cohort participants (n = 831) and (B) the subset of healthcare workers (n = 548) as measured by positive SARS-CoV-2 PCR or antibody testing. Results reflect aORs from multivariable logistic regression models fitted with elastic net penalty for regularization and variable selection from among variables listed in Table 1. Reference groups included: age < 40 years (versus ≥ 60 years), white race (versus Asian race), and attending physician (vs nursing). Abbreviations: aOR, adjusted odds ratio; CI, confidence interval; ICU, intensive care unit; OR, operating room; PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
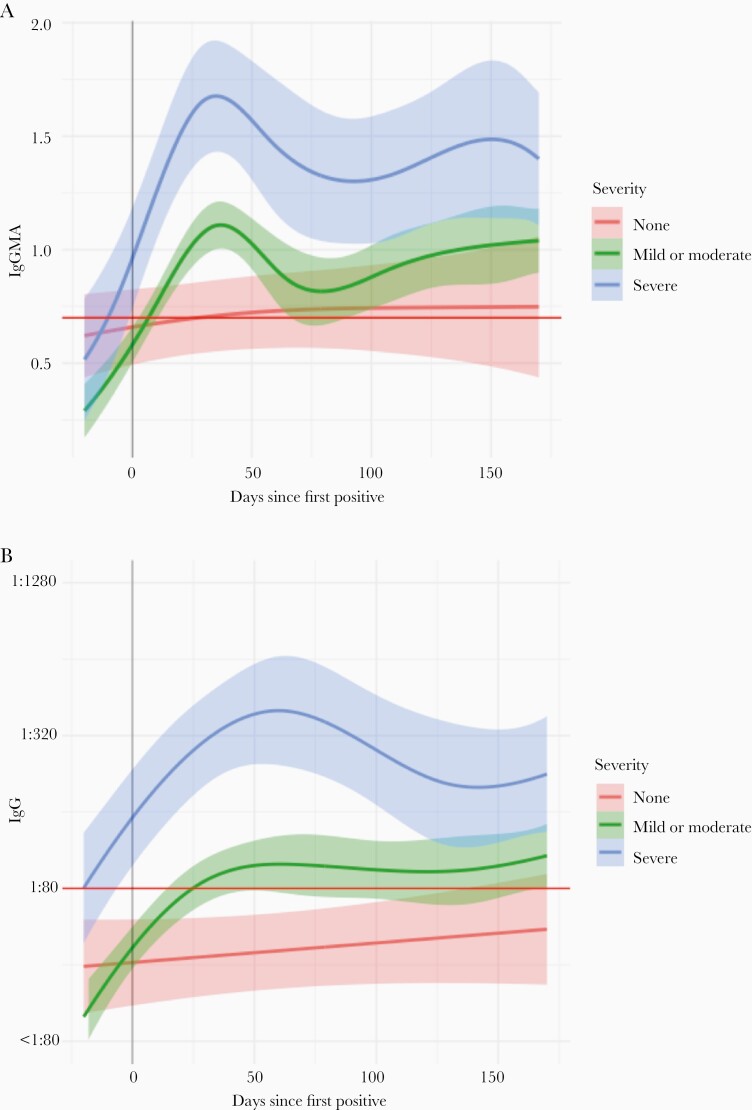
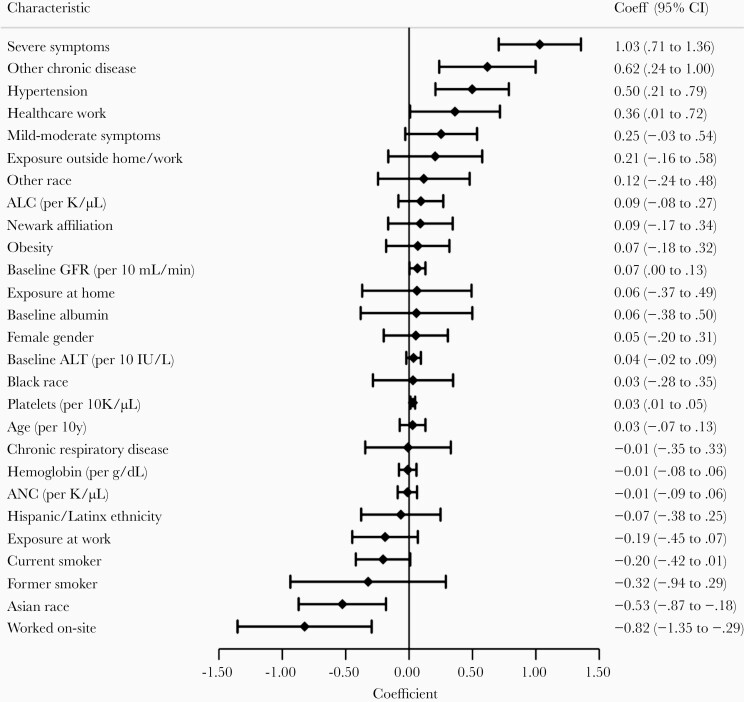
Median follow-up after diagnosis was 171 days (interquartile range, 158–180). Not including 2 participants newly PCR-positive at week 26, overall seropositivity was lower among asymptomatic participants (IgG 79%) compared to mildly-moderately symptomatic (IgG 89%) and severely symptomatic participants (IgG 96%) (Supplementary Table 4). At the final visit, IgG antibody prevalences among previously infected participants were: asymptomatic, IgG 69%; mildly-moderately symptomatic, IgG 83%; severely symptomatic, IgG 91%. Among antibody-positive participants (by total Ig or IgG) infected in the first wave with available samples at month 6, detectable antibodies persisted in most (67/73, 92%) participants, irrespective of symptom severity. Severe symptomatic illness was most strongly associated with higher IgG titers over time (coefficient 1.03; 95% CI, .71–1.36; Figure 2, Figure 3, Supplementary Table 4, and Supplementary Figure 5). Other factors associated with higher IgG titers included several comorbidities and HCW status; factors associated with lower IgG titers included Asian race, smoking, and working on site within 1 month after enrollment (Figure 3).
Figure 2.
Average antibody levels over time among SARS-CoV-2–infected Rutgers Corona Cohort participants, stratified by symptom severity. Plots show estimated average levels of (A) total antibody and (B) IgG over time with 95% confidence intervals based on symptom severity. Curves and 95% confidence bands were estimated for different levels of symptom severity by fitting a model with a spline function for time and a random intercept to account for repeated measures. Abbreviations: Ig, immunoglobulin; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Figure 3.
Factors associated with IgG titer among SARS-CoV-2–infected Rutgers Cohort participants (n = 81). Estimates reflect coefficients for factors in association with log-transformed IgG titer over time from a generalized additive mixed model, fitted with a spline for time. Factors reflect baseline values except disease severity (global assessment), cell counts (updated over time), and selected variables reflecting exposure in the first month of follow-up but excluding any values after SARS-CoV-2–positive testing (unprotected exposures to infected persons, worked on site). See “Methods” for details. Reference groups not shown are no symptoms, white race, and never smoker. Other chronic disease includes diabetes mellitus, cardio/cerebrovascular disease, cancer, chronic kidney disease, autoimmune disease, or immunosuppressant use. Chronic respiratory disease includes asthma, chronic obstructive pulmonary disease, or other chronic lung disease. Abbreviations: ALC, absolute lymphocyte count; ALT, alanine transaminase; ANC, absolute neutrophil count; CI, confidence interval; GFR, glomerular filtration rate; IgG, immunoglobulin G; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
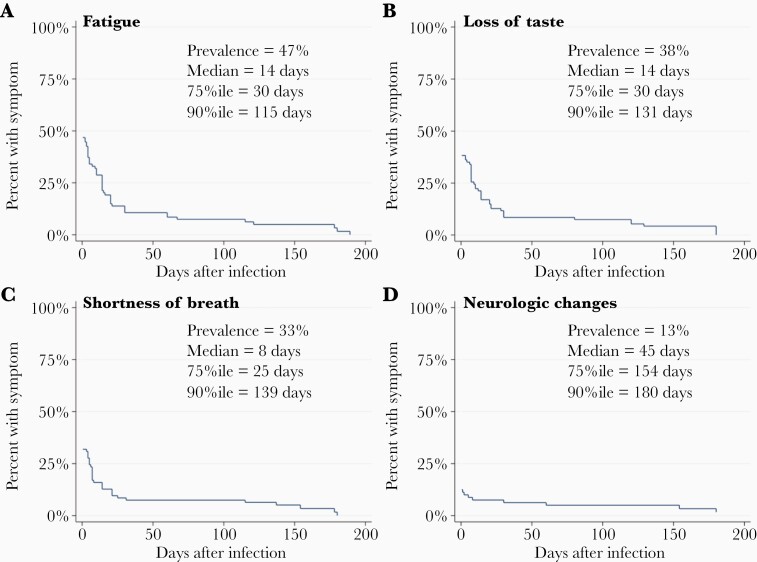
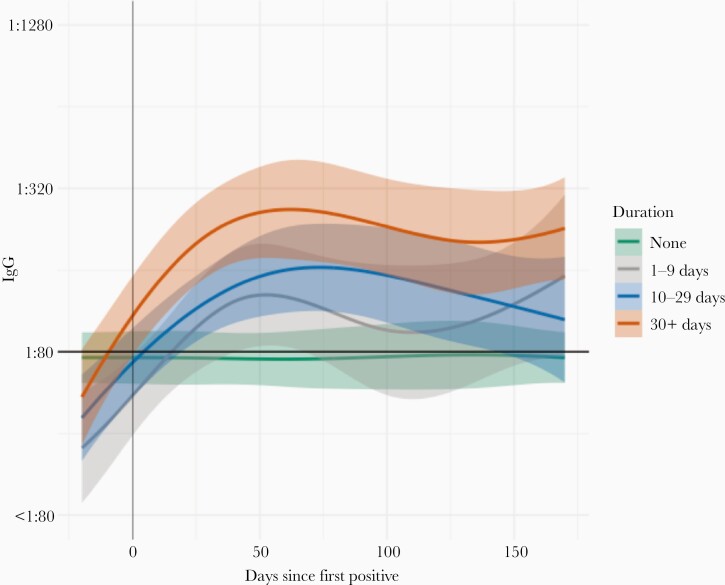
Among symptomatic infected participants, the median duration of most symptoms was ≤ 2 weeks except for neurologic changes besides altered smell and taste (eg, brain fog, memory problems, visual disturbances; median 45 days), which were least prevalent among 15 symptoms measured (changes reported in 12% of participants) (Figure 4 and Supplementary Figure 6). Nonetheless, multiple symptoms were reported in ≥ 25% of affected individuals for ≥ 30 days, and ≥ 10% of affected individuals reported having ≥ 120 days of shortness of breath, chest congestion, loss of small and/or taste, and other neurologic changes (Figure 4 and Supplementary Figure 6). About one-third (33/93, 35%) reported symptoms lasting 30 days or longer. Not surprisingly, symptom duration was correlated with symptom severity (r = 0.26, P = .03) and antibody titer (P = .03; Figure 5).
Figure 4.
Duration of selected symptoms in infected RCC participants. Kaplan-Meier plots show prevalence and time course (in days) of 4 selected symptoms among infected RCC participants from infection to resolution: (A) fatigue; (B) loss of taste; (C) shortness of breath; and (D) neurologic changes besides altered taste or smell, eg, altered cognition or visual changes. Symptoms are shown in decreasing order of overall prevalence (A–D). Median, 75th, and 90th percentiles are indicated for each symptom among those who reported the symptom. Abbreviations: %ile, percentile; RCC, Rutgers Corona Cohort.
Figure 5.
Average antibody levels over time among SARS-CoV-2–infected Rutgers Corona Cohort participants, stratified by symptom duration. Plots show estimated average levels of IgG over time with 95% confidence intervals based on symptom duration. Curves and 95% confidence bands were estimated for different durations of symptoms by fitting a model with a spline function for time and a random intercept to account for repeated measures. Abbreviations: IgG, immunoglobulin G; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
DISCUSSION
This study represents a 6-month prospective cohort study of risk factors, humoral responses, and symptoms in ambulatory, previously undiagnosed, at-risk individuals, recruited from a diverse professional community affected early in the US pandemic. Over 1 in 10 participants were SARS-CoV-2 infected, and most were followed for 5–6 months with excellent cohort retention. HCWs were more likely to become infected and have more severe illness. Among hospital workers, nurses were at greater risk for infection, whereas ICU and COVID-19 unit workers were at lower risk. Symptom severity and duration were associated with magnitude and trajectory of antibody responses. In contrast, most demographic characteristics, comorbidities (except hypertension), and laboratory criteria were not associated with antibody responses. Persons with asymptomatic infections had few changes in cell counts, lower seroconversion rates, and lower antibody levels. Multiple symptoms lasted 1 month or longer in at least 25% of participants; neurologic changes besides altered smell or taste were less frequent (approximately 1/8) but generally long lasting in those reporting them.
We and others have reported increased risks of SARS-CoV-2 among HCWs [17, 22, 24]. In our cohort, professional role (eg, nursing) was associated with greater risk. We also observed differences in illness severity between HCWs and others: HCWs were more likely to have severe symptoms and more robust antibody responses, consistent with other research suggesting higher risk of hospitalization among infected HCWs, particularly those with patient-facing roles, including nurses [25, 26]. In contrast to some [26] but consistent with other [27] studies, we observed lower infection rates among ICU workers and even those on COVID-19 units, which may have related to more rigorous N95-mask usage. Unlike in other studies [28], HCWs in our cohort were less likely to live in areas with higher rates of local transmission, and the increased rates observed were not well explained by outside exposures. The excess rates of infection in Newark may have resulted from later implementation of universal masking in that hospital versus in New Brunswick [29].
Our longitudinal cohort study contributes new insights into several aspects of the humoral response to SARS-CoV-2 infection. One is the correlation between rates of seropositivity and presence or severity of symptoms. Some studies have reported high rates of seropositivity after even asymptomatic or mild infections [6, 13], while others have found a correlation between illness severity and seropositivity [9, 30]. Such distinctions may relate to differences in recruitment, because studies that recruited previously diagnosed or self-referred infected volunteers are subject to selection bias by excluding those with asymptomatic or milder, undiagnosed infection. This was not an issue with our prospective cohort study design, which minimized the influence of selection bias and enhanced the internal validity of our findings. Second, we observed a strong positive correlation between antibody levels and symptom severity, which might be explained by stronger B-cell activation in the context of excessive inflammation typically associated with severe COVID-19 [31]. Because our cohort comprised an ambulatory population with mostly mild infections not requiring hospitalization, our work expands on previous observations of positive correlations between strength of antibody responses and COVID-19 severity, which were obtained in studies including only hospitalized patients or clinically diagnosed convalescent cases [32–34]. Moreover, while our study does not address cellular immunity, it allows for indirect inferences about immune status, because no or weak anti-SARS-CoV-2 antibody responses are accompanied by low frequencies of antigen-specific T cells [35]. Third, prior reports of anti-SARS-CoV-2 antibody trajectories have been conflicting, with some studies reporting antibody declines and loss of detectable antibodies, particularly following asymptomatic infections [6, 11, 12], and others showing antibody responses persisting for several months [8, 13, 14, 36]. In our study, most participants had sustained IgG up to 6 months after infection, irrespective of symptom severity. We also found severity-related differences in antibody trajectories, with slow, steady increases following asymptomatic infections compared to sharper rises and declines after symptomatic infections. These findings echo previous findings of sustained, nondeclining antibody responses in those with milder infections [36].
We cannot extrapolate our results to SARS-CoV-2 variants of concern (VOCs) because our cohort was recruited and followed in 2020 during earlier phases of the pandemic, prior to the emergence of several VOCs, including the highly transmissible Delta variant [37, 38]. Furthermore, given the relative reduction of protective immune responses against the Delta variant among previously infected persons [39, 40], our data do not support changes in current recommendations for vaccinating those with history of prior infection.
Our data on the duration of symptoms complement other reports of prolonged abnormalities in studies of previously infected subjects [41–44]. Notably, we studied an ambulatory population with generally mild illnesses, most not requiring hospitalization, over a 6-month timeframe that bracketed 2 infection surges in NJ. Of infected participants, about one-third reported symptoms lasting at least 30 days, and greater than 10% had persistent symptoms lasting for months, including fatigue, altered smell and taste, and shortness of breath. Notably, in contrast to other research [45], we found that participants with longer duration of symptoms also had higher levels of antibodies over time, perhaps reflecting the correlation between symptom duration and symptom severity, although further study is warranted on the immune profiles among those with prolonged symptoms. Nonetheless, because mild illnesses are much more common than illnesses that required hospitalization, the frequency of prolonged symptoms in our cohort raises a cautionary note about post–SARS-CoV-2 sequelae.
Our study had multiple strengths. The prospective inclusion of generally healthy, ethnically diverse, previously undiagnosed participants followed longitudinally from the early phases of the US pandemic, with exceptional retention, captured a range of clinical responses, including asymptomatic infections. Most participants received no medical intervention for their illness (86%), allowing us to characterize the natural history of disease and biomarker trajectories in a predominantly untreated cohort. Compared to results from hospitalized or convalescent cohorts, our findings may be more generalizable to the broader population of people with mild or asymptomatic infections [3, 4], often undiagnosed and contributing to viral transmission [5]. Following participants at 6 time points over 6 months, with ≥ 5 months of follow-up data for most SARS-CoV-2–infected participants, enabled study of antibody responses and symptoms longitudinally in relation to disease severity and other factors. The high levels of subject participation and retention increased confidence in the validity of our findings.
This study also had limitations. The rapid enrollment of a highly motivated convenience cohort may have preferentially enriched the study population with persons who perceived themselves at higher risk for infection, such as people with underlying respiratory diseases (although not itself a risk factor in our analysis). Enrolled participants and eligible participants who did not enroll also differed in certain respects related to the populations recruited at each campus location (eg, more HCWs and Hispanic/Latino participants in Newark). Nonetheless, the enrollment of persons not previously diagnosed with infection remains a strength of this study, and we do not believe that the enrollment procedures substantially affected the internal validity of our findings. Certain analyses were limited in statistical power due to smaller sample sizes of infected individuals. Less-frequent sampling during periods of lower transmission may have missed some asymptomatic infections without seroconversion. Disease severity was based on self-report, but this classification correlated well with symptom burden, levels of care and treatments received, and antibody responses. Sources of infectious exposure among participants could not be known with certainty; some infected participants who reported infected household members before their diagnosis may still have been the source of infections for other household members. Finally, given the differences in infection risk and severity between HCWs and non-HCWs, findings from our study population may not fully generalize to all populations.
In summary, in our prospective, ethnically diverse cohort of ambulatory, previously undiagnosed participants recruited early in the COVID-19 pandemic in the United States, levels and trajectories of antibody responses correlated with disease severity more so than any other factor. Asymptomatic infections led to lower seroconversion rates and antibody levels. One-third of infected participants had symptoms lasting 1 month or longer. Fatigue, respiratory, and neurologic symptoms lasted for months in at least 10% of affected individuals. Participants with prolonged symptoms, who were generally more severely symptomatic, also tended to have higher antibody levels over time. Going forward, this cohort of uninfected and infected, seropositive and seronegative, participants will allow further investigation of postacute sequelae of SARS-CoV-2 infection, risks factors for reinfection, and relationships between infection and vaccine responses.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank the Rutgers Corona Cohort (RCC) participants serving on the frontlines of the pandemic in New Jersey. We acknowledge the work of the entire RCC study team, all based at Rutgers University, for the following contributions: conducting study visits, Kathleen Black, PhD, MPH, Deborah McCloskey, RN, BSN, Taylor Black, MPH, Adriana De Resende, Alicia Legard, Christie Lyn Costanza, MPH, Christina Dalliani, Eric Asencio, Jared Khan, MPH, Valorie Cadorett, Ariana Alcaide, MPH, Kathleen Bott, RN, Randall Teeter, Juan Diego Ramirez, Adriana Hemans, Eliana Obando-Jaramillo, Yanille Taveras, MS, Marisol Rivera, Lisa Cerracchio, RNC, Halina Malveaux, RN, Jessica Kirby-Smith, Fei Chen, RN, Christina Varghese, William Russell, RPFT, Marie Macor, RN, Anthony Helena, Karen Dragert, RN, Aura Velasco, LPN, Susette Coyle, RN, MSN, Ami Patel, and Elizabeth George, RPH, BCOP; providing supplies, Joseph Barone, PharmD, and Daniel Notterman, MD; facilitating implementation of the study at the participating clinical sites, Brian Buckley, PhD, Debra Chew, MD, MPH, Mark Einstein, MD, MS, Shereef Elnahal, MD, Cecile Feldman, DMD, MBA, John Gantner, MBA, CPA, Brian Strom, MD, MPH, Stanley Z. Trooskin, MD, and Helmut Zarbl, DCS, PhD; assistance with data collection, Nicole Brennan, BS and Angel Chen, MPA; assistance with NJ state data, Panos Georgopoulos, PhD, Zhongyuan Mi, MPH, and Xiang Ren, MS; administrative assistance, Helaine Novek, Judith Argon, MS, Matthew Leonardelli; assistance with literature review, Michael Hoven; and assistance with ELISA work Parth Patel, Mennat Elsayed, and Valentina Guerrini. We also thank Charles Hevi and Dana Garbolino, Infinity Biologix for support with laboratory testing. In-kind logistics support was provided by Marken/UPS and dfYoung.
Disclaimer . The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial support. This work was supported by the National Institutes of Health (grant numbers 3U01AI122285-05S1, U01HL133817, UL1TR003017, 3UL1TR003017-02S1, 3UL1TR003017-02S2, UH3AI122309, P30ES005022, R01HL149450, R01HL149450-02S1, K23AR070286, and R61HD105619).
Potential conflicts of interest. D. B. H. reports grant support from Danisco USA Inc. S. S. reports advisory board membership, research grant support, and speaker fees from Gilead Sciences. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. New York Times. Coronavirus map: tracking the global outbreak. https://www.nytimes.com/interactive/2020/world/coronavirus-maps.html. Accessed 18 December 2020.
- 2. Oran DP, Topol EJ. The proportion of SARS-CoV-2 infections that are asymptomatic: a systematic review. Ann Intern Med 2021; 174:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yanes-Lane M, Winters N, Fregonese F, et al. Proportion of asymptomatic infection among COVID-19 positive persons and their transmission potential: a systematic review and meta-analysis. PLoS One 2020; 15:e0241536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buitrago-Garcia D, Egli-Gany D, Counotte MJ, et al. Occurrence and transmission potential of asymptomatic and presymptomatic SARS-CoV-2 infections: a living systematic review and meta-analysis. PLoS Med 2020; 17:e1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 2020; 368:489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Long QX, Tang XJ, Shi QL, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 2020; 26:1200–4. [DOI] [PubMed] [Google Scholar]
- 7. Wajnberg A, Mansour M, Leven E, et al. Humoral response and PCR positivity in patients with COVID-19 in the New York City region, USA: an observational study. Lancet Microbe 2020; 1:e283–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gudbjartsson DF, Norddahl GL, Melsted P, et al. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med 2020; 383:1724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hansen CB, Jarlhelt I, Pérez-Alós L, et al. SARS-CoV-2 antibody responses are correlated to disease severity in COVID-19 convalescent individuals. J Immunol 2021; 206:109–17. [DOI] [PubMed] [Google Scholar]
- 10. Crawford KHD, Dingens AS, Eguia R, et al. Dynamics of neutralizing antibody titers in the months after severe acute respiratory syndrome coronavirus 2 infection. J Infect Dis 2021; 223:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patel MM, Thornburg NJ, Stubblefield WB, et al. Change in antibodies to SARS-CoV-2 over 60 days among health care personnel in Nashville, Tennessee. JAMA 2020; 324:1781–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ibarrondo FJ, Fulcher JA, Goodman-Meza D, et al. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild Covid-19. N Engl J Med 2020; 383:1085–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wajnberg A, Amanat F, Firpo A, et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 2020; 370:1227–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Noh JY, Kwak JE, Yang JS, et al. Longitudinal assessment of anti-SARS-CoV-2 immune responses for six months based on the clinical severity of COVID-19. J Infect Dis 2021; 224:754–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trieu MC, Bansal A, Madsen A, et al. ; Bergen COVID-19 Research Group. SARS-CoV-2-specific neutralizing antibody responses in Norwegian health care workers after the first wave of COVID-19 pandemic: a prospective cohort study. J Infect Dis 2021; 223:589–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. New Jersey Department of Health. New Jersey COVID-19 dashboard. https://www.nj.gov/health/cd/topics/covid2019_dashboard.shtml. Accessed 29 December 2020.
- 17. Barrett ES, Horton DB, Roy J, et al. Prevalence of SARS-CoV-2 infection in previously undiagnosed health care workers in New Jersey, at the onset of the U.S. COVID-19 pandemic. BMC Infect Dis 2020; 20:853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Radbel J, Jagpal S, Roy J, et al. Detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is comparable in clinical samples preserved in saline or viral transport medium. J Mol Diagn 2020; 22:871–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Datta P, Ukey R, Bruiners N, et al. Highly versatile antibody binding assay for the detection of SARS-CoV-2 infection. medRxiv, doi: 10.1101/2021.07.09.21260266, 14. July 2021, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Premkumar L, Segovia-Chumbez B, Jadi R, et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol 2020; 5:eabc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barrett ES, Horton DB, Roy J, et al. Risk factors for SARS-CoV-2 infection in hospital workers: results from a screening study in New Jersey, US in Spring 2020. Open Forum Infect Dis 2020; 7:ofaa534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. White IR, Royston P, Wood AM. Multiple imputation using chained equations: issues and guidance for practice. Stat Med 2011; 30:377–99. [DOI] [PubMed] [Google Scholar]
- 24. Chou R, Dana T, Buckley DI, Selph S, Fu R, Totten AM. Epidemiology of and risk factors for coronavirus infection in health care workers: a living rapid review. Ann Intern Med 2020; 173:120–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kambhampati AK, O’Halloran AC, Whitaker M, et al. COVID-19–associated hospitalizations among health care personnel—COVID-NET, 13 states, March 1–May 31, 2020. Morb Mort Wkly Rep 2020; 69:1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shah ASV, Wood R, Gribben C, et al. Risk of hospital admission with coronavirus disease 2019 in healthcare workers and their households: nationwide linkage cohort study. BMJ 2020; 371:m3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grant JJ, Wilmore SMS, McCann NS, et al. Seroprevalence of SARS-CoV-2 antibodies in healthcare workers at a London NHS Trust. Infect Control Hosp Epidemiol 2021; 42:212–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jacob JT, Baker JM, Fridkin SK, et al. Risk factors associated with SARS-CoV-2 seropositivity among US health care personnel. JAMA Netw Open 2021; 4:e211283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang X, Ferro EG, Zhou G, Hashimoto D, Bhatt DL. Association between universal masking in a health care system and SARS-CoV-2 positivity among health care workers. JAMA 2020; 324:703–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rijkers G, Murk JL, Wintermans B, et al. Differences in antibody kinetics and functionality between severe and mild severe acute respiratory syndrome coronavirus 2 infections. J Infect Dis 2020; 222:1265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol 2020; 20:355–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robbiani DF, Gaebler C, Muecksch F, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020; 584:437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020; 26:845–8. [DOI] [PubMed] [Google Scholar]
- 34. Lynch KL, Whitman JD, Lacanienta NP, et al. Magnitude and kinetics of anti-severe acute respiratory syndrome coronavirus 2 antibody responses and their relationship to disease severity. Clin Infect Dis 2021; 72:301–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 2020; 183:158–68.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Y, Zuiani A, Fischinger S, et al. Quick COVID-19 healers sustain anti-SARS-CoV-2 antibody production. Cell 2020; 183:1496–507.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alizon S, Haim-Boukobza S, Foulongne V, et al. Rapid spread of the SARS-CoV-2 Delta variant in some French regions, June 2021. Euro Surveill 2021; 26:2100573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Centers for Disease Control and Prevention. COVID data tracker, variant proportions. https://covid.cdc.gov/covid-data-tracker/#variant-proportions. Accessed 30 July 2021.
- 39. Liu C, Ginn HM, Dejnirattisai W, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell 2021; 184:4220–36.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021; 596:276–80. [DOI] [PubMed] [Google Scholar]
- 41. Nehme M, Braillard O, Alcoba G, et al. ; COVICARE TEAM. COVID-19 symptoms: longitudinal evolution and persistence in outpatient settings. Ann Intern Med 2021; 174:723–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Carfì A, Bernabei R, Landi F; Gemelli Against COVID-19 Post-Acute Care Study Group. Persistent symptoms in patients after acute COVID-19. JAMA 2020; 324: 603–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Willi S, Lüthold R, Hunt A, et al. COVID-19 sequelae in adults aged less than 50 years: a systematic review. Travel Med Infect Dis 2021; 40:101995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nalbandian A, Sehgal K, Gupta A, et al. Post-acute COVID-19 syndrome. Nat Med 2021; 27:601–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gerhards C, Thiaucourt M, Kittel M, et al. Longitudinal assessment of anti-SARS-CoV-2 antibody dynamics and clinical features following convalescence from a COVID-19 infection. Int J Infect Dis 2021; 107:221–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.