Abstract
Background
Households are hot spots for severe acute respiratory syndrome coronavirus 2 transmission.
Methods
This prospective study enrolled 100 coronavirus disease 2019 (COVID-19) cases and 208 of their household members in North Carolina though October 2020, including 44% who identified as Hispanic or non-White. Households were enrolled a median of 6 days from symptom onset in the index case. Incident secondary cases within the household were detected using quantitative polymerase chain reaction of weekly nasal swabs (days 7, 14, 21) or by seroconversion at day 28.
Results
Excluding 73 household contacts who were PCR-positive at baseline, the secondary attack rate (SAR) among household contacts was 32% (33 of 103; 95% confidence interval [CI], 22%–44%). The majority of cases occurred by day 7, with later cases confirmed as household-acquired by viral sequencing. Infected persons in the same household had similar nasopharyngeal viral loads (intraclass correlation coefficient = 0.45; 95% CI, .23–.62). Households with secondary transmission had index cases with a median viral load that was 1.4 log10 higher than those without transmission (P = .03), as well as higher living density (more than 3 persons occupying fewer than 6 rooms; odds ratio, 3.3; 95% CI, 1.02–10.9). Minority households were more likely to experience high living density and had a higher risk of incident infection than did White households (SAR, 51% vs 19%; P = .01).
Conclusions
Household crowding in the context of high-inoculum infections may amplify the spread of COVID-19, potentially contributing to disproportionate impact on communities of color.
Keywords: SARS-CoV-2, household transmission, viral load, secondary attack rate, living density
Households are hot spots for severe acute respiratory syndrome coronavirus 2 transmission. In the United States, household crowding in the context of high-inoculum infections may amplify the spread of COVID-19, potentially contributing to disproportionate impact on communities of color.
Households are hot spots for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission [1]. Person-to-person transmission is difficult to control in shared living spaces. For those who isolate at home with young children in small living spaces, following guidelines to physically distance and use separate sleeping, eating, and lavatory facilities is difficult [2]. Because the period of peak infectiousness starts prior to the onset of symptoms, spread can occur before these measures are taken [3–5].
Meta-analyses of the secondary household attack rate (SAR) in the first 6 months of the pandemic ranged from 15% to 20% [6, 7], but these analyses incorporated both retrospective and prospective analyses [6, 7]. Prospective testing of household contacts (HCs) regardless of symptom status is required to capture all secondary cases. Previously identified risk factors associated with increased transmission include the presence of symptoms [6–8] and high viral load in the index case [9]. Among HCs, spouses and those aged >18 years (ie, adults compared with children) are more likely to acquire infection [6, 7, 10]. Measuring secondary household attack rates in vulnerable communities and identifying risk factors for transmission in these communities are critical.
The University of North Carolina (UNC) COVID-19 Household Transmission Study (CO-HOST) is the largest, single-site, observational, household cohort in the United States to date and the most ethnically and racially diverse. Weekly sampling for quantitative viral loads combined with SARS-CoV-2 antibody testing at baseline and at 1 month provided an extended period to evaluate transmission. Our objective in this study was to measure the incident SAR in a setting where infected individuals were asked to quarantine at home and given standard guidance. Household and individual demographics as well as daily symptoms and weekly viral loads were collected to identify risk factors and timing of household transmission.
METHODS
Study Design and Enrollment
In the CO-HOST study, we evaluated SARS-CoV-2 acquisition among persons undergoing quarantine in their home after a care-seeking household member (the index case) tested positive for SARS-CoV-2. Recruitment occurred between April 2020 and October 2020, prior to the introduction of SARS-CoV-2 vaccines in the United States. Inclusion criteria for the index cases were age >18 years with a positive qualitative nasopharyngeal (NP) swab for SARS-CoV-2 RNA using polymerase chain reaction (PCR) performed at the UNC hospital clinical laboratory, willingness to self-isolate at home for a 14-day period, living with at least 1 HC who was also willing to participate, and living within driving distance (<1 hour) from the study site. Inclusion criteria for HCs of index patients included age >1 year and currently living in the same home as the index case without plans to live elsewhere during the 28-day study. All participants (or their parents/guardians) gave written, informed consent. Minors aged >7 years provided assent.
As shown in Supplementary Figure 1, all households were visited on day 1 by the study team. NP and nasal midturbinate (NMT) swabs were collected for SARS-CoV-2 PCR testing, and blood samples were collected for serology. All study participants received instruction on self-collection of NMT swabs and completed baseline questionnaires that included basic demographic and household information, abbreviated medical history, symptoms, recent travel history, and exposure to confirmed cases of coronavirus disease 2019 (COVID-19).
All participants received a daily symptom questionnaire via email until no symptoms were reported for 2 consecutive days. HCs who remained asymptomatic received the questionnaire for 21 days to monitor for new symptoms. On days 7, 14, and 21, a study staff member conducted home visits to collect self-collected NMT swabs. At the final study visit (day 28), participants were asked about COVID-19–related care-seeking and underwent serologic testing.
Laboratory Analyses
Details for all laboratory methods are found in the Supplementary Materials. Quantitative PCR testing for SARS-CoV-2 was performed using a Centers for Disease Control and Prevention reverse-transcription quantitative-PCR protocol authorized by the US Food and Drug Administration for emergency use, as previously described [11]. Serology was performed using enzyme-linked immunoassay (ELISA) that detects antibodies to the receptor binding domain of the spike protein with high sensitivity and specificity [12]. When ELISA results were not available (ie, in children who did not undergo venipuncture), results from a BioMedomics COVID-19 immunoglobulin (Ig) M/IgG Rapid Test [13–15] were used.
To determine the prevalence of the 614G variant, which predominated in North Carolina at the time of the study, in our study samples, we developed a real-time PCR assay to target a 107 bp region encompassing the D614G mutation in the S1 segment of the SARS-CoV-2 spike protein.
To determine whether secondary cases were acquired outside the household rather than due to household transmission, we performed high-density amplicon sequencing on NP/NMT swab samples from households with late secondary cases to assign SARS-CoV-2 clades and determine the genetic relatedness of viral isolates within and between households.
Statistical Analyses
We summarized baseline demographic characteristics and underlying conditions of index cases and HCs, as well as their household demographics. We determined if baseline NP SARS-CoV-2 viral loads were correlated within households by the intraclass correlation coefficient (ICC), which compares within vs between household variation.
We evaluated the secondary household attack rate among household members of persons who tested positive for SARS-CoV-2. For each household, if multiple participants were positive at enrollment, the index case was defined as the person with the earliest onset of infection based on reported onset of symptoms and known date(s) of PCR test positivity. If this was ambiguous, baseline antibody positivity was used as evidence of less recent infection. We calculated the SAR as the risk of incident infection among HCs, defined as the proportion of contacts who were PCR-negative at baseline but then developed SARS-CoV-2 infection during the 28-day study follow-up, confirmed by either PCR or antibody seroconversion. Those with evidence of prior infection (antibody-positive and PCR-negative) or HCs who reported the same COVID-19 exposure outside the household as the index case were excluded from the analysis. To avoid misclassification of asymptomatic infection, HCs who tested PCR-negative by weekly nasal swabs (days 7, 14, and 21) but without day 28 antibody results were also excluded. A 95% confidence interval (CI) for the SAR was calculated using a robust variance estimation for the intercept term in a logistic regression model to account for outcome dependence within a household.
Potential risk factors for secondary transmission within the household, including characteristics of index cases, households, and HCs, were examined. These risk factors and details of symptom severity evaluation are described in the Supplementary Materials. Statistical significance was tested using either the Fisher exact test or χ2 test for categorical variables and the Mann-Whitney test for continuous variables. To determine if the association between index race-ethnicity and SAR was related to housing density or viral load (which were correlated with race-ethnicity and/or transmission in unadjusted analyses), we calculated the odds ratio of the race-ethnicity–specific SAR, with the 95% CI estimated using the same robust variance estimation described above, then added the other risk factors as covariates in the logistic regression model to calculate adjusted odds ratios.
Analyses were performed using R 4.0.2 (R Core Team, Vienna, Austria). All hypothesis tests were 2-sided at a significance level of 0.05 with no adjustments for multiple comparisons.
The University of North Carolina Institutional Review Board approved the study.
RESULTS
Study Enrollment
Between April 2020 and October 2020, the UNC CO-HOST study enrolled 102 households across the central Piedmont Region of North Carolina (Supplementary Figure 2). After excluding participants who did not complete follow-up, had evidence of prior infection, or those who were possibly infected at the same time as the index case based on a common exposure, 91 households were included in the baseline analysis (Figure 1). Households were enrolled a median of 6 days (interquartile range [IQR], 4–7) after symptom onset of the designated index case, which was reassigned in 11 households. Baseline characteristics of the 91 index cases and 176 HCs are listed in Table 1. The median index case age was 37 years (IQR, 23–49), while the age of HCs ranged from 2 to 77 years with 34% aged <18 years. Overall, 44% of participants identified as other than White, non-Hispanic race-ethnicity, and 33% of adults were obese (body mass index >30 kg/m2; Table 1, other comorbidities in Supplementary Table 1).
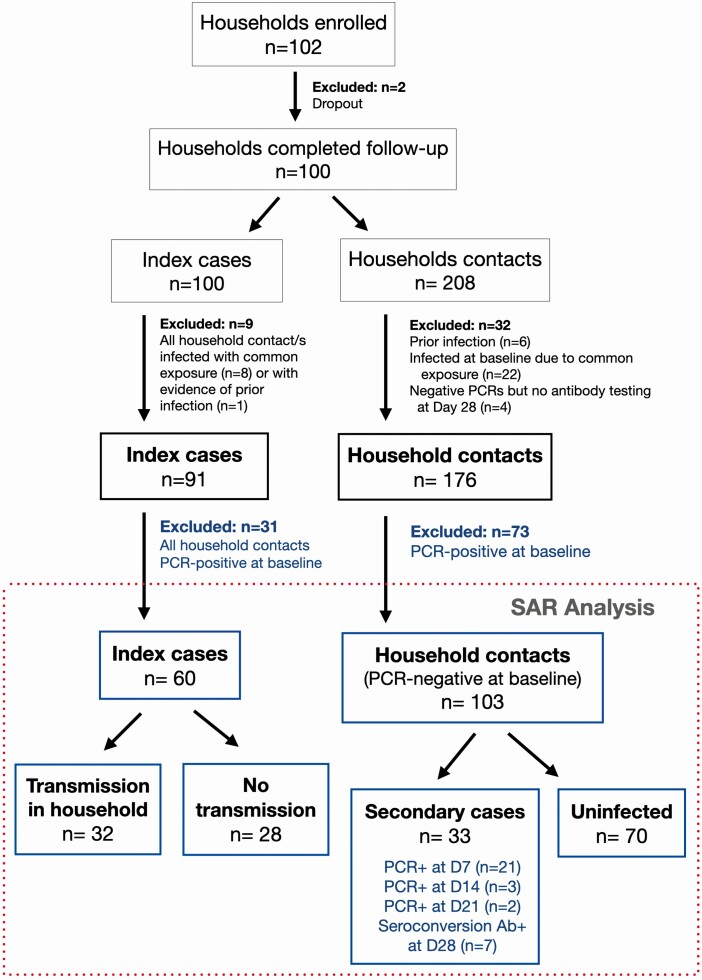
Figure 1.
COVID-19 Household Transmission Study enrollment and SAR. Among 100 households that completed the 28-day follow-up, household contacts were excluded if they had evidence of prior infection (negative PCR and positive antibody test at enrollment), were possibly infected at the same time as the index case based on a common exposure event, or negative PCR testing could not be confirmed with a negative antibody test at day 28. Of the remaining 176 household contacts of 91 index cases, 41% (73) were already PCR-positive at baseline and thus excluded from the primary SAR analysis. During study follow-up, 33 incident severe acute respiratory syndrome coronavirus 2 cases were identified, yielding a SAR of 32% (33 of 103). Among the 33 secondary cases, 22 were identified by both PCR and seroconversion from day 1 to day 28, 4 were identified by PCR only, and 7 were identified based on seroconversion. Abbreviations: Ab+, antibody positive; D, day; PCR, polymerase chain reaction; SAR, secondary household attack rate.
Table 1.
Demographics of Study Participants
| Characteristic | Index Cases, n (%) | Household Contacts, n (%) | ||
|---|---|---|---|---|
| 91 | 176 | |||
| Male | 43 | (47) | 87 | (49) |
| Female | 48 | (53) | 89 | (51) |
| Race/Ethnicity | ||||
| White, non-Hispanic | 52 | (57) | 96 | (55) |
| Non-White | 39 | (43) | 77 | (44) |
| Black or African American | 10 | (11) | 17 | (9.7) |
| Hispanic/Latinx | 26 | (29) | 58 | (33) |
| Other, non-Hispanic | 3 | (3.3) | 2 | (1.1) |
| Unknown | 0 | (0.0) | 3 | (1.7) |
| Spanish speaking | ||||
| Yes | 13 | (14) | 28 | (16) |
| No | 78 | (86) | 148 | (84) |
| Age, years | ||||
| 0–12 | 2 | (2.2) | 38 | (22) |
| 13–17 | 6 | (6.6) | 22 | (13) |
| 18–24 | 20 | (22) | 23 | (13) |
| 25–49 | 41 | (45) | 56 | (32) |
| 50–64 | 18 | (20) | 27 | (15) |
| >65 | 4 | (4.4) | 10 | (5.7) |
| Education (excluding those aged <18 years) | ||||
| Total responses for adults aged >18 years | 80 | 113 | ||
| High school or lower | 36 | (45) | 54 | (48) |
| College degree | 23 | (29) | 34 | (30) |
| Graduate degree | 21 | (26) | 25 | (22) |
| Occupation (excluding those aged <18 years) | ||||
| Total responses for adults aged >18 years | 83 | 116 | ||
| Education | 3 | (3.6) | 6 | (5.2) |
| Healthcare worker | 11 | (13) | 9 | (7.8) |
| Retail/Hospitality/Other frontline worker | 19 | (23) | 22 | (19) |
| Student | 7 | (8.4) | 12 | (10) |
| White collar worker | 23 | (28) | 34 | (29) |
| Other (trade and arts) | 6 | (7.2) | 6 | (5.2) |
| Not working outside the home | 14 | (17) | 27 | (23) |
| Comorbidities (excluding those aged <18 years) | ||||
| Diabetes | 4 | (4.8) | 9 | (7.8) |
| High blood pressure | 12 | (15) | 24 | (21) |
| Body mass index, kg/m2 | ||||
| >30 | 28 | (34) | 38 | (33) |
| 25–29.9 | 24 | (29) | 31 | (27) |
| >30 and 1 or more comorbidity | ||||
| Adults aged >18 years (n = 83 index, 116 HC) | 16 | (19) | 22 | (19) |
| Adults aged >50 years (n = 22 index, 37 HC) | 7 | (32) | 12 | (32) |
Abbreviation: HC, household contact.
Household Characteristics
The median size of households was 4 persons (Supplementary Table 2). However, in 38% of households, at least 1 household member chose not to participate. Households with a non-White index case were more likely to live in a home less than 2000 square feet in size (71% vs 43%, P = .01). This led to a higher “living density” for non-White households: 43% had more than 3 household members living in a home with fewer than 6 rooms compared with 8% of White households (P < .001).
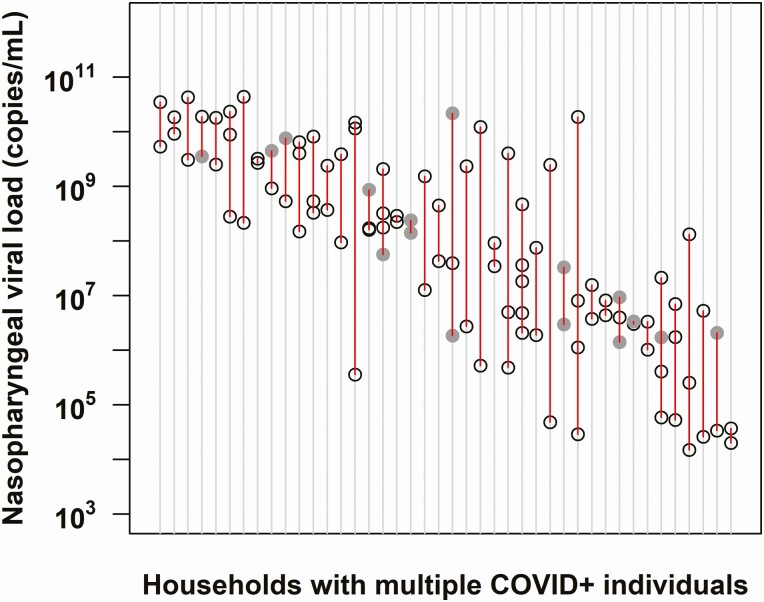
SARS-CoV-2 viral burden was correlated within households (Figure 2). When comparing the baseline NP viral load within vs between households, viral burden showed significant clustering within households (ICC = 0.45; 95%, CI 0.23–0.62; P < .001). Differences in viral load are not attributable to D614G mutation in the viral spike protein that has been associated with increased viral load and infectivity [16], as 90 of 92 (98%) of genotyped SARS-CoV-2 isolates contained the 614G mutant, while only 2 of 92 were wild type at this locus.
Figure 2.
Severe acute respiratory syndrome coronavirus 2 viral burden is correlated within families. The viral load obtained at enrollment from nasopharyngeal (NP) swabs in households with multiple COVID-19–positive household members is shown (n = 42 households). Each vertical row in red depicts an individual household, with circles delineating the log viral load of each member within the household. Gray-shaded circles represent values derived from a nasal midturbinate swab if NP sampling was not performed. This was based on a linear regression equation generated from more than 100 study participants with positive viral load from both NP and nasal midturbinate swabs [11]. Households are depicted across the x-axis in order of decreasing viral load. Data drawn from 148 participants. The intraclass correlation coefficient = 0.45; 95% confidence interval, .23–.62; P value < .001. Abbreviation: COVID-19, coronavirus disease 2019.
Secondary Attack Rate Among Household Contacts
The incident SAR among HCs was 32% (33 of 103; 95% CI, 22%–44%). Among 91 households, 73 of 176 (41%) HCs tested PCR-positive at baseline and were excluded from the primary SAR analysis (Figure 1, Supplementary Table 3 for demographics of baseline infected cases). Among the remaining 103 HCs of 51 index cases, 33 incident SARS-CoV-2 infections were observed during the 28-day study follow-up (SAR = 32%; 33 of 103). Of these 33 secondary cases, 22 were identified by both PCR and seroconversion from day 1 to day 28, 4 were identified by PCR only, and 7 were identified by seroconversion alone. The majority of secondary cases in the household experienced symptoms (27 of 33), while 18% (n = 6) remained asymptomatic.
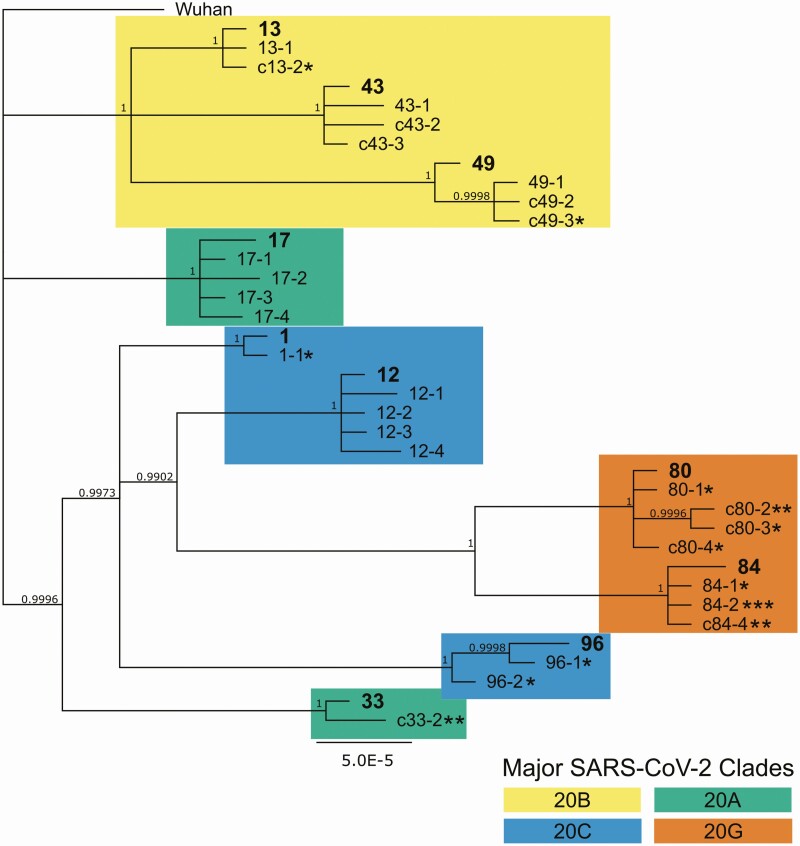
Secondary household transmission occurred early, within the first week following enrollment for the majority of cases (n = 21 of 26 for those identified by PCR; Figure 1). Five cases were detected by PCR after the first week of enrollment, at day 14 or day 21. Four of these late secondary cases occurred in households of 5 or more (including 2 from the same household), which suggests the possibility of sequential transmission within the household. High-density amplicon sequencing of viral isolates from these late secondary cases and others in their household confirmed that 4 of 5 were indeed due to household transmission (1 isolate failed sequencing) and not community-acquired (Figure 3).
Figure 3.
Bayesian phylogeny showing high relatedness within household infections, indicating household transmission. High-density amplicon sequencing was performed on all available viral isolates from 10 households with secondary infections to assess relatedness between infections. Whole-genome sequences were assembled according to the Wuhan reference genome, assigned to major clades, and then used for Bayesian phylogeny reconstruction. Index cases within each numbered household are in bold. Household contacts are numbered sequentially starting with the index case number, eg, X-1, X-2. Minors are indicated with the letter “c” prior to the case number. Each asterisk indicates 1 study week preceding a positive quantitative polymerase chain reaction (PCR) test, ie, * indicating a D7 positive test, ** indicating a D14 positive test, and *** indicating a D21 positive test. Household contacts without asterisks were PCR-positive at baseline. Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Risk Factors for Household Transmission
At the household level, 44% of households (40 of 91) had at least 1 infected household member at enrollment in addition to the index case, rising to 69% (63 of 91) of households 1 month later. Sixty households contained susceptible HCs at enrollment and were thus included in the risk factor analysis.
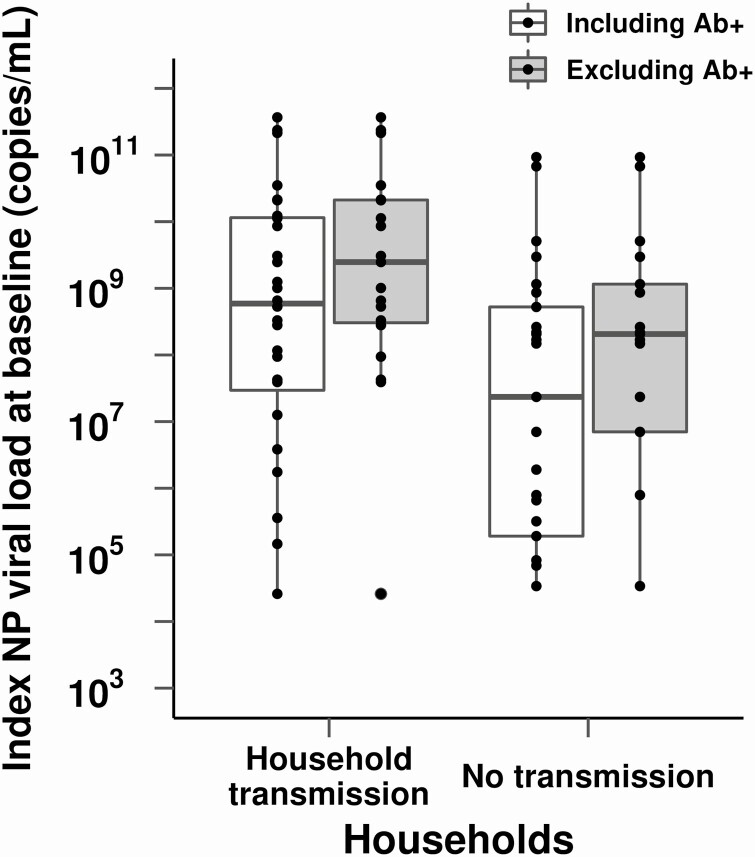
Secondary transmission in the household was associated with a higher NP viral load in index cases at enrollment. The median NP viral load among index cases was 1.4 log10 higher in households with secondary cases detected during the study vs those with no incident cases in the household (P = .03; Table 2). This difference persisted when the analysis was restricted to index cases who were still antibody-negative and thus more recently infected [16, 18] (Figure 4). Symptom severity was not associated with household transmission, though secondary transmission did occur in households of the 4 index cases who were hospitalized (Table 2).
Table 2.
Potential Risk Factors for Severe Acute Respiratory Syndrome Coronavirus 2 Transmission From Index Cases
| Index Cases | All Indexes, n (%) | Household Transmission, n (%) | No Transmission, n (%) | P Value |
|---|---|---|---|---|
| 60 (100) | 32 (53) | 28 (47) | — | |
| Age, years | ||||
| <18 | 7 (12) | 4 (13) | 3 (11) | NS |
| 18–50 | 45 (75) | 25 (78) | 20 (71) | |
| >50 | 8 (13) | 3 (9) | 5 (18) | |
| Sex | ||||
| Female | 33 (55) | 15 (47) | 18 (64) | NS |
| Male | 27 (45) | 17 (53) | 10 (36) | |
| Mask wearing at home prior to enrollment (missing n = 3) | 10 (18) | 4 (13) | 6 (22) | NS |
| Race-Ethnicity | ||||
| White, non-Hispanic | 34 (57) | 13 (41) | 21 (75) | .01a |
| Black or African American | 7 (12) | 5 (16) | 2 (7) | |
| Other, non-Hispanic | 4 (7) | 4 (12) | 0 (0) | |
| Hispanic/Latinx | 15 (25) | 10 (31) | 5 (18) | |
| Symptom severity | ||||
| Mild | 12 (21) | 4 (14) | 8 (29) | .07 |
| Moderate/Severe | 41 (72) | 21 (72) | 20 (71) | |
| Hospitalized | 4 (7) | 4 (14) | 0 (0) | |
| Duration of symptoms at enrollment, median (IQR), days | 5 (4–7) | 5 (4–7) | 6 (4–7) | NS |
| Nasopharyngeal viral load (log10 copies/mL) at enrollment (missing n = 6), median (IQR) | 8.3 (5.9–9.5) | 8.8 (7.3–10.1) | 7.4 (5.3–8.7) | .03 |
| Comorbidities for adults aged >18 years (missing n = 1 for diabetes, n = 3 for obesity) | ||||
| Diabetes | 0 (0) | 0 (0) | 0 (0) | NS |
| Obesity, body mass index >30 kg/m2 | 21 (42) | 13 (50) | 8 (33) | NS |
| Education for adults aged >18 years (missing n = 3) | ||||
| High school or less | 21 (42) | 13 (52) | 8 (32) | NS |
| College degree | 17 (34) | 9 (36) | 8 (32) | |
| Graduate degree | 12 (24) | 3 (12) | 9 (36) |
P values only reported if ≤ .10, otherwise noted as NS.
Abbreviations: IQR, interquartile range; NS, not significant.
aCompares White, non-Hispanic vs all other categories.
Figure 4.
Association of index NP viral load and transmission in the household. Households with secondary cases were more likely to have index cases with high NP viral load compared with households without secondary transmission (median NP viral load log 8.8 vs 7.4 copies/mL, respectively; P = .03). Index cases who were not yet antibody-positive at enrollment, as a marker of more recent infection, are depicted to the right in gray. Abbreviations: Ab+, antibody positive; NP, nasopharyngeal.
Households with non-White index cases were more likely to experience incident transmission in the household (Table 2), despite there being no difference in index case viral loads by race-ethnicity (median NP viral load for White vs non-White: 8.3 vs 8.3 log10 copies/mL). This corresponds to a SAR of 51% (95% CI, 33%–69%) in households with a non-White or Hispanic index case compared with 19% (95% CI, 10%–35%) in White, non-Hispanic households (P = .008). Higher living density, defined as more than 3 household members living in a home with fewer than 6 rooms, was associated with a greater odds of transmission (OR, 3.3; 95% CI, 1.02–10.9; P = .047; Table 3); a greater proportion of non-White/Hispanic households met this definition of high living density (42%, 11 of 26) compared with White, non-Hispanic households (12%, 4 of 34; P = .01). However, after adjusting for viral load and living density, Hispanic/non-White race-ethnicity remained associated with secondary household transmission (Table 4).
Table 3.
Potential Household-Level Risk Factors for Severe Acute Respiratory Syndrome Coronavirus 2 Transmission
| Households | All Households, n (%) | Infected, n (%) | Uninfected, n (%) | P Value |
|---|---|---|---|---|
| 60 (100) | 32 (53) | 28 (47) | - | |
| Household size, mean | 3.8 | 4.2 | 3.4 | .03 |
| Living space (missing n = 3), sq ft | ||||
| <2000 | 29 (51) | 18 (60) | 11 (41) | NS |
| >2000 | 28 (49) | 12 (40) | 16 (59) | |
| Number of roomsa | ||||
| 2 or fewer | 5 (8) | 2 (6) | 3 (10) | .06 |
| 3–5 | 27 (45) | 19 (59) | 8 (29) | |
| 6 or more | 28 (47) | 11 (34) | 17 (61) | |
| More than 3 people and fewer than 6 rooms | 15 (25) | 13 (41) | 2 (7) | .003 |
| Home ownership (missing n = 1) | ||||
| Renting apartment | 4 (7) | 2 (7) | 2 (7) | NS |
| Renting home | 17 (29) | 11 (36) | 6 (21) | |
| Own home | 38 (64) | 18 (58) | 20 (71) |
P values only reported if ≤ .10, otherwise noted as NS.
Abbreviation: NS, not significant.
aNumber of rooms includes bedrooms, kitchen, and common rooms but not bathrooms or garage.
Table 4.
Risk Factors for Severe Acute Respiratory Syndrome Coronavirus 2 Transmission in the Household
| Index or Household Risk Factor | Susceptible Household Contacts | Incident Secondary Cases | Secondary Household Attack Rate (95% CI) | Odds Ratio (95% CI) | Adjusted Odds Ratio (95% CI) |
|---|---|---|---|---|---|
| Non-White | 41 | 21 | 51% (33–69) | 4.4 (1.5–13.0) | 4.8 (1.5–15.4) |
| White, non-Hispanic | 62 | 12 | 19% (10–35) | - | |
| Higher index nasopharyngeal viral loada | - | - | - | 3.5 (1.5–8.1) | 3.6 (1.5–8.5) |
| High living densityb | 29 | 15 | 52% (27–75) | 3.3 (1.02–10.9) | 1.4 (.4–4.6) |
| Not high living density | 74 | 18 | 24% (15–37) | - |
Adjusted odds ratio adjusts for viral load as a continuous variable and race-ethnicity and living density as dichotomous variables.
Abbreviation: CI, confidence interval.
aOdds ratio for 3 log10 increase in the index viral load (VL). For example, the odds of transmission in a household where the index VL is 1 × 109 copies/mL is 3.5 times greater than in a household where the index VL is 1 × 106 copies/mL.
bDefined as more than 3 people occupying fewer than 6 rooms.
Among susceptible HCs, those who shared a bathroom with the index case were at higher risk of acquiring infection (Supplementary Table 4). Obesity and being female were also associated with a higher risk of incident infection, though these associations were not statistically significant. Though a slightly greater percentage of participants in households without secondary transmission reported wearing a mask at home in the week prior to enrollment (22% vs 13% for index cases and 30% vs 20% in HCs), these differences were not statistically significant.
Discussion
Household transmission is one of the main drivers of the SARS-CoV-2 pandemic. By incorporating timely recruitment of index cases, prospective sampling to 21 days regardless of symptom status, and confirmatory viral sequencing in a subset of households, we show that household transmission occurs in a substantial proportion of COVID-19–positive households, with racial-ethnic disparities in secondary attack rates and higher risk of infection in more crowded households. Our data also suggest that those infected with a high viral load are not only more likely to transmit virus to other household members but they may seed other high viral load infections, putting the entire household at higher risk for more severe illness.
The incident SAR in this study was 32%, rising to 51% in minority households. While a meta-analysis of household transmission studies conducted in the first 6 months of the pandemic (prior to circulation of new variants) found a much lower overall household SAR of 17% (95% CI, 14%–19%), it noted significant heterogeneity between studies (ranging from 4% to 45%) and combined both retrospective studies based on contact tracing data and prospective analyses [6]. As expected, prospective studies with increased frequency of testing regardless of symptom status generally show higher infection rates [7]. In the United States, a retrospective study in New York that included household testing offered regardless of symptom status reported a SAR of 38% [17], while 2 prospective studies following households in Utah and Wisconsin (58 households, SAR 29%) [18] and Tennessee and Wisconsin (101 households, SAR 53%) [10] also reported higher SARs. Altogether, these studies document high secondary household attack rates within US households.
To our knowledge, this is the first study to show increased transmission in non-White US households. Though they experience similar case fatality rates, African American/Black and Hispanic populations in the United States experience disproportionately higher rates of SARS-CoV-2 infection [19–21]. These racial disparities are likely due to differences in healthcare access and exposure risk that are driven by systemic societal inequities rather than individual biological or behavioral characteristics [22–25]. Our limited findings are consistent with this explanation. While the sample size precluded full investigation of drivers of increased transmission in minority households, we found that high living density/household crowding was more common in non-White households, while viral load and reported masking in the home did not differ by race-ethnicity.
We also found that SARS-CoV-2 viral burden was correlated within households. Increased viral load increases infectivity in vivo [26]. A study of 282 clusters in Spain showed an increased risk of transmission with shorter time to onset of symptoms among contacts as viral load of the index cases increased [9]. Since greater viral burden (high viral load or lower cycle threshold values by PCR) is associated with disease severity [27–30], our findings imply that when a person is hospitalized, others in the same household may be at a higher risk for a similar outcome than would be predicted based on their individual risk factors alone. An inoculum effect may underlie this finding [31] and also explain why secondary cases in households appear to be overdispersed, with either most or all members infected, or none at all [6, 32, 33].
This study has several limitations. First, although we enrolled most households within 24 to 48 hours of a positive SARS-CoV-2 test result, delays in testing meant that it was common for others in the household to be PCR-positive at enrollment. While 33 HCs met our end point as incident SARS-CoV-2 cases, 73 HCs were infected at study baseline and hence could not be categorized definitively as due to household transmission and were excluded from the analysis. Thus, we likely underestimated the true SAR, and the resultant small sample size was not sufficient to investigate all drivers of household transmission. We were similarly limited in our ability to do adjusted analyses beyond a simplistic exploration of whether living density might account for the observed racial disparity in SAR. In the households with multiple infected household members at baseline, we cannot be certain that the designated index case was the source of infection for all infected household members. This may have affected our evaluation of index case risk factors associated with transmission.
Additionally, we were unable to adequately assess the effects of age, mask-wearing, and the presence of symptoms on transmission. We recruited adult index cases, and in 38% of households, at least 1 household member (most often young children) declined to participate. While mask use was queried, mask use prior to any COVID-19 diagnosis in the household was not specifically elicited. All index cases except 1 were tested because they were symptomatic.
In conclusion, SARS-CoV-2 transmits early and often among household members. While masking, physical distancing, and quarantining the whole household may reduce or prevent transmission beyond the household, these strategies are less effective within the household, especially in the setting of high viral load infections and crowded living spaces. Frequent point-of-care testing and post-exposure prophylaxis in those at risk for severe illness [34] and ultimately widespread and equitable distribution of vaccines [35] are needed to lessen the impact of COVID-19 within households and vulnerable communities.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the wonderful COVID-19 Household Transmission Study participants, Moby and the HIV Clinical Trials Unit, and the University of North Carolina (UNC) Respiratory Diagnostic Center team. Thanks to Michelle Berrey, JoAnn Kuruc, and Dania Munson for help with protocol writing and submission; Oksana Kharabora, Maureen Furlong, Amy James Loftis, Tia Belvin, and Dana Swilley for help with study preparation and implementation; and Joe Eron, Billy Fischer, and Ada Adimora for their input and support.
Financial support. Research was supported by funds and charitable contributions from the UNC Department of Medicine (emergency funds to principal investigator [PI] J. T. L., UNC School of Medicine), UNC COVID-19 Response Fund/Health Foundation (via UNC Health Foundation to PI J. T. L., UNC School of Medicine), a Gillings Innovations Laboratory Award funded by the 2007 Gillings Gift to UNC–Chapel Hill’s Gillings School of Global Public Health (to co-PIs K. A. P. and J. T. L.), and the National Center for Advancing Translational Sciences, National Institutes of Health, through grant award UL1TR002489 (NC TrACs Pilot Funding Award to PI J. T. L.). L. P. reports grants as the co-investigator for NCI-U54 CA260543 and NC Collaboratory Fund. Trainees were supported by the National Institute of Allergy and Infectious Diseases (NIAID; T32A1007151, K. T.) and the Infectious Diseases Society of America (T. R.). Rapid antibody tests were provided by BioMedomics Inc, Morrisville, NC.
Potential conflicts of interest. K. R. M. received grant support from Ridgeback Biotherapeutics LP (2020–2021) and has human immunodeficiency virus collaborations unrelated to this study with Gilead Sciences (ongoing). L. P. reports grants or contracts as co-investigator for NIAID (U01AI151788). All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
REFERENCES
- 1. Report of the WHO-China Joint Commission on Coronavirus Disease 2019 (COVID-19). Available at: https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf. Accessed 16 February 2021.
- 2. Centers for Disease Control and Prevention. Public health guidance for community-related exposure. 2020. Available at: https://www.cdc.gov/coronavirus/2019-ncov/php/public-health-recommendations.html. Accessed 16 February 2021.
- 3. He X, Lau EHY, Wu P, et al. . Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 2020; 26:672–5. [DOI] [PubMed] [Google Scholar]
- 4. Tindale LC, Stockdale JE, Coombe M, et al. . Evidence for transmission of COVID-19 prior to symptom onset. Elife 2020; 9. doi: 10.7554/eLife.57149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benefield AE, Skrip LA, Clement A, Althouse RA, Chang S, Althouse BM. SARS-CoV-2 viral load peaks prior to symptom onset: a systematic review and individual-pooled analysis of coronavirus viral load from 66 studies. bioRxiv [Preprint]. 2020. Available from: http://medrxiv.org/lookup/doi/10.1101/2020.09.28.20202028.
- 6. Madewell ZJ, Yang Y, Longini IM Jr, Halloran ME, Dean NE. Household transmission of SARS-CoV-2: a systematic review and meta-analysis. JAMA Netw Open 2020; 3:e2031756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fung HF, Martinez L, Alarid-Escudero F, et al. . The household secondary attack rate of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a rapid review. Clin Infect Dis 2020. Available at: https://academic.oup.com/cid/advance-article-abstract/doi/10.1093/cid/ciaa1558/5921151. Accessed 21 February 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buitrago-Garcia D, Egli-Gany D, Counotte MJ, et al. . Occurrence and transmission potential of asymptomatic and presymptomatic SARS-CoV-2 infections: a living systematic review and meta-analysis. PLoS Med 2020; 17:e1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marks M, Millat-Martinez P, Ouchi D, et al. . Transmission of COVID-19 in 282 clusters in Catalonia, Spain: a cohort study. Lancet Infect Dis 2021; doi: 10.1016/S1473-3099(20)30985-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grijalva CG, Rolfes MA, Zhu Y, et al. . Transmission of SARS-COV-2 infections in households—Tennessee and Wisconsin, April-September 2020. MMWR Morb Mortal Wkly Rep 2020; 69:1631–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Muller MS, Chhetri SB, Basham C, et al. . Practical strategies for SARS-CoV-2 RT-PCR testing in resource-constrained settings. medRxiv [Preprint]. 2021. doi: 10.1101/2021.02.18.21251999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Premkumar L, Segovia-Chumbez B, Jadi R, et al. . The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol 2020; 5. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Z, Yi Y, Luo X, et al. . Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol 2020; 92:1518–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. COVID-19 IgM/IgG rapid test—BioMedomics Inc. Available at: https://www.biomedomics.com/products/infectious-disease/covid-19-rt/. Accessed 17 February 2021.
- 15. Naranbhai V, Chang CC, Beltran WFG, et al. . High seroprevalence of anti-SARS-CoV-2 antibodies in Chelsea, Massachusetts. J Infect Dis 2020; 222:1955–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Korber B, Fischer WM, Gnanakaran S, et al. . Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020; 182:812–27.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rosenberg ES, Dufort EM, Blog DS, et al. . COVID-19 testing, epidemic features, hospital outcomes, and household prevalence, New York State—March 2020. Clin Infect Dis 2020; 71:1953”9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lewis NM, Chu VT, Ye D, et al. . Household transmission of SARS-CoV-2 in the United States. Clin Infect Dis 2021; 73:e1805”13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mackey K, Ayers CK, Kondo KK, et al. . Racial and ethnic disparities in COVID-19-related infections, hospitalizations, and deaths: a systematic review. Ann Intern Med 2020; doi: 10.7326/M20-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lopez CA, Cunningham CH, Pugh S, et al. . Disparities in SARS-CoV-2 seroprevalence among individuals presenting for care in central North Carolina over a six-month period. medRxiv [Preprint]. 2021. doi: 10.1101/2021.03.25.21254320. [Google Scholar]
- 21. Brandt K, Goel V, Keeler C, et al. . SARS-CoV-2 testing in North Carolina: racial, ethnic, and geographic disparities. Health Place 2021; 69:102576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karmakar M, Lantz PM, Tipirneni R. Association of social and demographic factors with COVID-19 incidence and death rates in the US. JAMA Netw Open 2021; 4:e2036462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poteat T, Millett GA, Nelson LE, Beyrer C. Understanding COVID-19 risks and vulnerabilities among black communities in America: the lethal force of syndemics. Ann Epidemiol 2020; 47:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Holmes L, Enwere M, Williams J, et al. . Black–White risk differentials in COVID-19 (SARS-COV2) transmission, mortality and case fatality in the United States: translational epidemiologic perspective and challenges. Int J Environ Res Public Health 2020; 17:4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rogers TN, Rogers CR, VanSant-Webb E, Gu LY, Yan B, Qeadan F. Racial disparities in COVID-19 mortality among essential workers in the United States. World Medi Health Policy 2020; 12:311–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hou YJ, Chiba S, Halfmann P, et al. . SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 2020; 370:1464–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maltezou HC, Raftopoulos V, Vorou R, et al. . Association between upper respiratory tract viral load, comorbidities, disease severity and outcome of patients with SARS-CoV-2 infection. J Infect Dis 2021; doi: 10.1093/infdis/jiaa804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Magleby R, Westblade LF, Trzebucki A, et al. . Impact of SARS-CoV-2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clin Infect Dis 2021; 73:e4197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Y, Yan LM, Wan L, et al. . Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis 2020; 20:656–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fajnzylber J, Regan J, Coxen K, et al. ; Massachusetts Consortium for Pathogen Readiness. . SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun 2020; 11:5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Silvia Munoz-Price L, Rivera F, Ledeboer N. Air contamination of households versus hospital inpatient rooms occupied by SARS-CoV-2 positive patients. Infect Control Hosp Epidemiol 2021; 1–5. doi: 10.1017/ice.2021.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ladhani SN, Andrews N, Aiano F, et al. . Secondary attack rate and family clustering of SARS-CoV-2 infection in children of healthcare workers with confirmed COVID-19. Clin Infect Dis 2021; 73:e260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paul LA, Daneman N, Brown KA, et al. . Characteristics associated with household transmission of SARS-CoV-2 in Ontario, Canada: a cohort study. Clin Infect Dis 2021; 73:1840–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hurt AC, Wheatley AK. Neutralizing antibody therapeutics for COVID-19. Viruses 2021; 13. doi: 10.3390/v13040628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harris RJ, Hall JA, Zaidi A, Andrews NJ, Dunbar JK, Dabrera G. Effect of vaccination on household transmission of SARS-CoV-2 in England. N Engl J Med 2021; doi: 10.1056/NEJMc2107717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.