Abstract
Background
The detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in patient samples is of critical importance in the management of patients and monitoring transmission in the population. However, data on the analytical performance characteristics for detection of SARS-CoV-2 in clinical specimens between individual targets within the same platform, and among different analytical platforms, are limited.
Methods
Here we evaluated the performance of 6 different sample-to-answer SARS-CoV-2 detection methods—Roche cobas 6800, Cepheid GeneXpert, Diasorin Simplexa, Luminex Aries emergency use authorization (EUA), Luminex Aries research use only (RUO), and bioMérieux BioFire—in clinical specimens with a range of viral loads.
Results
The positive percentage agreement between the Roche cobas 6800 and GeneXpert was 100%, Diasorin 95%, Aries EUA 74%, Aries RUO 83%, and BioFire 97%. Notably, in samples with cycle threshold (Ct) values below 30 for the E gene on the Roche cobas 6800 platform, we found 100% positive agreement among all platforms. Given these results, we examined the distribution of over 10 000 Ct values of all positive specimens from individuals at our institution on the Roche cobas platform. Nearly 60% of specimens from asymptomatic individuals had a PCR Ct value >30 as measured using the cobas 6800 assay E gene.
Conclusions
Our results demonstrate performance characteristics between different platforms by Ct value and provide data regarding the distribution of viral RNA present in positive specimens.
Keywords: COVID -19, SARS-CoV-2, PCR, diagnostics
Introduction
Impact Statement
We determined the analytic performance of 6 different sample-to-answer severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection platforms. Our results show that below cycle threshold (Ct) values of 30, all platforms demonstrate 100% positive agreement, while this is varied at Ct values above 30. For clinical context, we analyzed the distribution of Ct values in over 10 000 consecutive clinical specimens obtained from both symptomatic and asymptomatic patients since the initial phase of the pandemic. We found that samples obtained from 15% of symptomatic patients demonstrated Ct values greater than 30, while in asymptomatic patients, this approached 60%. This data are important in the interpretation of SARS-CoV-2 results across molecular detection platforms in asymptomatic individuals.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiologic agent of the global coronavirus disease 2019 (COVID-19) pandemic, belongs to the beta-coronavirus family, which includes SARS-CoV, Middle East respiratory syndrome coronavirus, and the endemic OC43 and HKU1 strains (1). Although infection with SARS-CoV-2 can lead to severe respiratory symptoms, pneumonia, and death, a significant proportion of individuals have mild disease or remain asymptomatic (2). While presymptomatic and symptomatic individuals are known to harbor infectious virus, transmission by asymptomatic individuals is also thought to occur (3). Thus, the testing of asymptomatic individuals has resulted in substantial burden to the clinical laboratory due to demand, fueled by a variety of factors including pretravel testing, preprocedure testing, and workplace testing. Moreover, turnaround time is of critical importance as asymptomatic individuals who test positive for SARS-CoV-2 can engage in additional precautions. To ensure timely results in the setting of unpredictable supply chain disruptions or equipment downtime, most clinical laboratories require redundant methods for this analyte to support the clinical demand. This raises an important challenge, as it remains unclear how results generated across different platforms compare when tested with the same specimen; this may have consequences for result interpretation and patient management.
Reverse-transcriptase PCR of patient specimens obtained from the nasopharynx, oropharynx, or lower respiratory tract is utilized for SARS-CoV-2 detection. Many assays with Food and Drug Administration emergency use authorization (EUA) status have been designed to detect at least 2 targets (e.g., E gene, N gene, orf1a/b) within the SARS-CoV-2 genome. While these assays are designed to be qualitative, they produce a cycle threshold (Ct) value, which can be used as a proxy to estimate viral load and is inversely related to the number of viral copies present in the samples. Thus, the Ct value could demonstrate clinical utility (e.g., by predicting disease severity and/or transmission risk) (4, 5). However, as each gene and platform produces nonstandardized Ct values, this specific information is typically absent from clinical reports. Several studies have examined the concordance between different instruments using control nucleic acid and pooled positive specimens; however, how performance correlates with viral load is unclear (6–11). Understanding the relationship between reported Ct value and clinical outcomes and/or the ability to transmit virus may have an impact in the management of this pandemic. In addition, the distribution of Ct values in positive patients will help inform the use of rapid antigen detection assays, which could relieve stress on the clinical laboratory.
Here, we sought to determine the cross-platform concordance of SARS-CoV-2 detection across 6 different commercial platforms with clinical specimens. Given that the viral load in positive specimens is highly variable, we analyzed the agreement between instruments based on Ct value in clinical specimens. To give context to the performance of each platform in positive patients, we analyzed the Ct values of over 10 000 specimens on the Roche cobas 6800. Finally, we compared the reported Ct values in the same specimens across 6 different platforms to determine cross-platform correlations.
Methods
Specimens Evaluated for Cross-Platform Comparison
To perform a cross-platform comparison, we utilized deidentified nasopharyngeal (NP) and oropharyngeal specimens submitted to Barnes-Jewish Hospital Laboratory for SARS-CoV-2 detection from March 2020 to May 2020. Specimens were collected using flocked nylon swabs and eluted into either universal transport media, viral transport medium, or Eswab liquid Amies medium. The 39 positive and 40 negative specimens were selected randomly from a subset of all samples processed at Barnes-Jewish Hospital Laboratory that had adequate residual volume for additional analysis. These specimens were initially identified as positive or negative using the Quidel Lyra SARS-CoV-2 assay with the Qiagen Rotor-Gene Q PCR instrument. Residual specimen was diluted 1:10 in the original specimen matrix (i.e., universal transport media, viral transport medium, or Amies) and frozen in aliquots at −80 °C until testing.
Distribution of Ct Values on All Specimens Analyzed on Roche Cobas 6800
All positive NP specimens obtained at Barnes-Jewish Hospital from May 1, 2020, through December 31, 2020 were included in this analysis. Absence or presence of symptoms (defined as 1 or more of the following: new or worsening fever, chills, cough, dyspnea, supplemental oxygen requirement, myalgia, sore throat, headache, anosmia, or pulmonary infiltrate on imaging) was recorded by the ordering clinician at the time of testing for SARS-CoV-2. These data were retrieved from the Epic electronic medical record after approval from the Washington University in St. Louis Medical Center Human Research Protection Office (IRB #202005002). Orf1a/b gene and E gene Ct values were retrieved directly from the cobas 6800 instrument. All data visualizations were performed with GraphPad Prism 9.
Instrumentation and Analysis
Each commercial sample-to answer assay was performed according to manufacturer’s instructions, except for the Aries research use only (RUO), which are detailed in the following discussion. In brief, the Roche cobas 6800 SARS-CoV-2 test (https://www.fda.gov/media/136049/download) is a high-throughput dual target assay for orf1a/b (Target 1) and E gene (Target 2); 0.6 mL of specimen is required. The Cepheid Xpert Xpress SARS-CoV-2 test (https://www.fda.gov/media/136314/download) is a rapid dual target assay for N2 gene (Target 1) and E gene (Target 2); 0.3 mL of specimen is required. The Diasorin Simplexa COVID-19 Direct (https://www.fda.gov/media/136286/download) is a platform that uses a 8-well Direct Amplification Disc. Fifty microliters of sample is loaded onto the disc, and 2 different targets are assayed S gene, and orf1a/b. The Aries EUA SARS-CoV-2 Assay (https://www.fda.gov/media/136693/download) detects orf1a/b and N gene targets; 0.2 mL of sample is used. The BioFire COVID-19 test is a qualitative test on the FilmArray 2.0 or Torch systems (12). The assay detects the orf1a/b and orf8 genes in 3 simultaneous PCR reactions, and 0.3 mL of sample is analyzed. Ct values are not generated for the BioFire assay.
The Aries RUO assay was performed using Assay 2 oligonucleotides targeting SARS-CoV-2 orf1a/b (primers 5′-CCCTGTGGGTTTTACACTTAA-3′ and 5′-ACGATTGTGCATCAGCTGA-3′, probe 56-FAM/CCGTCTGCG/ZEN/GTATGTGGAAAGGTTATGG/3IABkFQ), N (primers 5′-GGGGAACTTCTCCTGCTAGAAT-3′ and 5′-CAGACATTTTGCTCTCAAGCTG-3′, probe/5TexRD-XN/ TTGCTGCTGCTTGACAGATT/3IAbRQSp) and RNase P (Primers 5′-AGATTTGGACCTGCGAGCG-3′ and 5′-GAGCGGCTGTCTCCACAAGT-3′ and probe/5TYE665/TTCTGACCTGAAGGCTCTGCGCG/3IAbRQSp). Reverse-transcriptase PCR was performed with the following cycling protocol: 1 cycle for 7 min at 50 °C, 1 cycle for 2 min a t 95 °C, and 45 cycles of 95 °C for 15 sec and 60 °C for 30 sec (https://www.luminexcorp.com/eu/targeted-molecular-testing-solutions/); 0.2 mL of sample is used.
The reference method for our analysis was the Quidel Lyra assay (https://www.fda.gov/media/136820/download). In brief, the Lyra assay targets pp1ab of SARS-CoV-2 from nucleic acid samples prepared using either the bioMérieux NucliSENS® easyMAG® system or EMAG® system, which use 0.18 mL of sample as input.
Statistical Analysis
All statistical analyses and data visualizations were performed with GraphPad Prism 9. The Wilson/Brown method was performed to obtain the 95% confidence interval (CI) for percent agreement.
Results
Cross-Platform Performance in Clinical Specimens
To directly compare the performance of different SARS-CoV-2 detection assays, we tested 39 positive and 40 negative clinical specimens obtained following routine clinical testing in March and April 2020. These samples were all processed using the Lyra Quidel platform, one of the first assays used for SARS-CoV-2 detection at our institution and thus served as a reference comparator for our study. These samples were diluted 1:10, aliquoted and frozen at −80 °C prior to cross-platform analysis. The diluted samples were then analyzed on the Roche cobas 6800, Cepheid GeneXpert, Diasorin Simplexa, Aries EUA, Aries RUO, and BioFire assays. Due to limited residual sample, only a subset of the samples were processed on the Aries EUA, Aries RUO, and BioFire platforms (n = 72, 44, and 44, respectively).
The positive percentage agreement (PPA) compared to Quidel Lyra assay was as follows: cobas 6800 100% (91%–100% CI), GeneXpert 100% (91%–100% CI), Diasorin 95% (83%–99% CI), Aries EUA 74% (59%–85% CI), Aries RUO 83% (66%–93% CI), and BioFire 97% (83%–100% CI) (Table 1). The negative percentage agreement was 100% for all platforms, except the Diasorin (98%).
Table 1.
Positive percentage agreement, negative percentage agreement, and overall percentage agreement between SARS-CoV-2 detection platforms.
| Xpert | Agreement (%) | 95% CI (%) | |
|---|---|---|---|
| PPA | 39/39 | 100 | 91–100 |
| NPA | 40/40 | 100 | 91–100 |
| OPA | 79/79 | 100 | 95–100 |
| Roche | |||
| PPA | 39/39 | 100 | 91–100 |
| NPA | 40/40 | 100 | 91–100 |
| OPA | 79/79 | 100 | 95–100 |
| Diasorin | |||
| PPA | 37/39 | 95 | 83–99 |
| NPA | 39/40 | 98 | 87–100 |
| OPA | 76/79 | 96 | 89–99 |
| Aries EUA | |||
| PPA | 29/39 | 74 | 59–85 |
| NPA | 33/33 | 100 | 90–100 |
| OPA | 62/72 | 86 | 76–92 |
| Aries RUO | |||
| PPA | 25/30 | 83 | 66–93 |
| NPA | 14/14 | 100 | 78–100 |
| OPA | 39/44 | 89 | 76–95 |
| BioFire | |||
| PPA | 29/30 | 97 | 83–100 |
| NPA | 14/14 | 100 | 78–100 |
| OPA | 43/44 | 98 | 88–100 |
Ct values obtained from Roche cobas E gene result. Roche, Roche cobas SARS-CoV-2 assay; Xpert, Cepheid GeneXpert Xpress SARS-CoV-2 assay; Diasorin, Diasorin Simplexa COVID-19 assay; BioFire, BioFire COVID-19 test; Aries RUO, Luminex Aries non-EUA COVID-19 assay; Aries EUA, Luminex Aries EUA COVID-19 assay; NPA, negative percentage agreement; OPA, overall percentage agreement.
Given the importance of viral load in sensitivity for detection of viral detection, we analyzed the PPA in samples with higher viral load (Ct <30 cycles by Roche E gene) and lower viral load (Ct >30 cycles by Roche E gene). Importantly, in samples with higher viral load, we found 100% PPA on all platforms tested (Table 2). Consistent with decreased sensitivity in lower viral load specimens, the PPA between different platforms in samples with Ct > 30 was 100% (84%–100% CI) for GeneXpert, 90% (70%–98% CI) for Diasorin, 50% (19%–51% CI) for Aries EUA, 69% (44%–86% CI) for Aries RUO, and 94% (72%–100% CI) for the BioFire. Thus, the differences in clinical performance between the platforms is observed when the specimens have lower viral concentrations.
Table 2.
PPA between platforms by Ct value.
| <30 Ct agreement |
>30 Ct agreement |
|||
|---|---|---|---|---|
| Ratio, n/n | % (95% CI) | Ratio, n/n | % (95% CI) | |
| Xpert | 19/19 | 100 (83–100) | 20/20 | 100 (91–100) |
| Roche | 19/19 | 100 (83–100) | 20/20 | 100 (91–100) |
| Diasorin | 19/19 | 100 (83–100) | 18/20 | 90 (70–98) |
| Aries EUA | 19/19 | 100 (83–100) | 10/20 | 50 (30–70) |
| Aries RUO | 14/14 | 100 (78–100) | 11/16 | 69 (48–86) |
| BioFire | 14/14 | 100 (78–100) | 15/16 | 94 (72–100) |
Ct values obtained from Roche cobas E gene result. Roche, Roche cobas SARS-CoV-2 assay; Xpert, Cepheid GeneXpert Xpress SARS-CoV-2 assay; Diasorin, Diasorin Simplexa COVID-19 assay; BioFire, BioFire COVID-19 test; Aries RUO, Luminex Aries non-EUA COVID-19 assay; Aries EUA, Luminex Aries EUA COVID-19 assay.
Distribution of Ct Values in Clinical Specimens
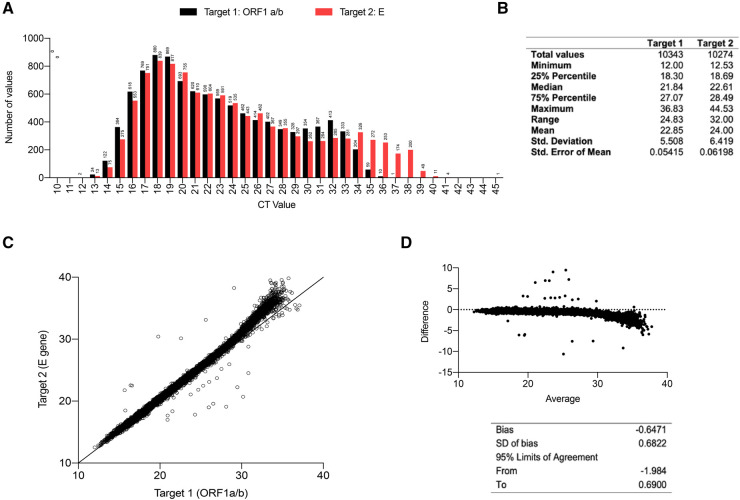
As our results indicated that low viral load specimens could result in false negatives, we analyzed the distribution in Ct values present in positive specimens from our institution on our primary instrument, the Roche cobas 6800. Our institution is a tertiary care academic medical center with samples obtained from both asymptomatic and symptomatic patients. We analyzed all 10 453 positive samples (including presumptive positives) processed on the Roche cobas 6800 platform between May 1, 2020 and December 31, 2020. Not all specimens were positive for both targets. The Ct values ranged from 12.00 to 36.83 for Target 1 (orf1a/b) and from 12.53 and 44.53 for Target 2 (E gene) with a mean of 22.84 and 24.00, respectively (Fig. 1, A and B). A total of 20.7% of positive samples showed Ct values above 30 for Target 2 (E gene) (Figs. 1, A, and 3). When we plotted the Ct value of Target 1 and Target 2 against each other, we found a very strong correlation below Ct value of 30, which deviated toward higher Ct values of Target 2 above this value (Fig. 1, C). The deviation above a Ct of 30 was also evident by Bland-Altman analysis (Fig. 1, D).
Fig. 1.
Analysis of SARS-CoV-2 positive NP specimens processed on the Roche cobas 6800. (A) Ct values for Target 1 (orf1a/b, black) and Target 2 (E, red) in all positive specimens processed on the Roche cobas 6800. Numbers above bars indicate number of positive specimens. (B) Table showing distribution of Ct values for positive samples processed on Roche cobas 6800. (C) Plot of Target 1 (orf1a/b) and Target 2 (E gene) in all positive samples processed on the Roche Cobas 6800. Each point reflects a Ct values generated for an individual patient. (D) Bland-Altman analysis of paired Target 1 and Target 2 Ct values.
Cross-Platform Comparison of Ct Values in Clinical Specimens
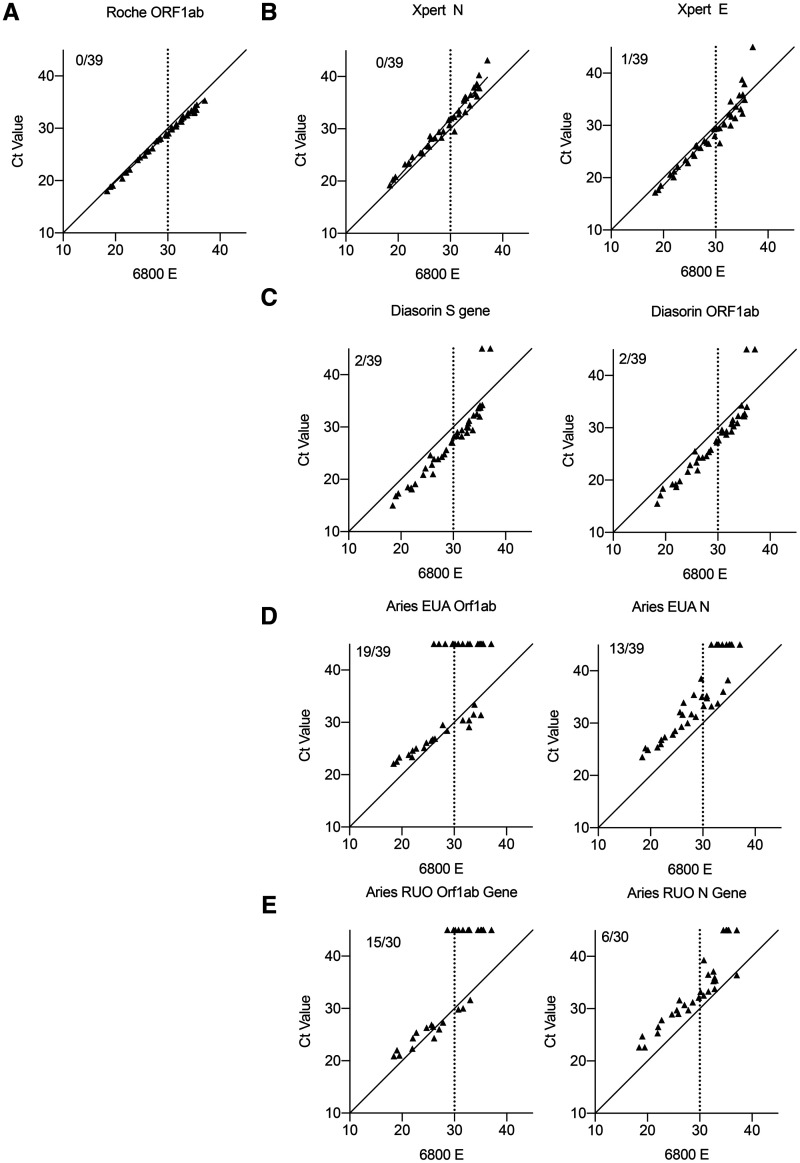
To determine interassay precision and commutability, we extracted the Ct values from the 39 positive samples run on the 6 platforms (online Supplemental Table 1). We found a linear relationship in Ct value between Roche cobas 6800 and Cepheid Xpress systems, which slightly deviated at high Ct value (Fig. 2, A and B). However, the Diasorin reported Ct values were lower across a broad range of viral loads compared to the Roche (Fig. 2, C). The Aries platform (both EUA and RUO) generally reported higher Ct values than the Roche, which accounts for the decreased PPA with Ct > 30 (Fig. 2D and E). These data demonstrate slight biases in reported Ct value for each instrument, which in theory can be standardized based on calculated bias for clinical use.
Fig. 2.
Correlation of Ct values between 5 different platforms on the same clinical specimens by Roche cobas 6800 E gene. (A) Graph showing the correlation of Ct value by Roche cobas 6800 E gene and orf1a/b. Inset reflects number of samples that were not detected by orf1a/b. Graph showing the correlation of Ct value by Roche cobas 6800 E gene and indicated target on Cepheid GeneXpert (B), Diasorin Simplexa (C), Aries EUA (D), and Aries RUO (E). Inset reflects number of samples that were not detected by the given target.
Distribution of Ct Values in Asymptomatic and Symptomatic COVID-19 Patients
The large burden placed on the clinical laboratory due the COVID-19 pandemic is in part due to the testing of both symptomatic and asymptomatic individuals, which is not generally performed for other respiratory viruses. We thus sought to determine the distribution of Ct values among asymptomatic and symptomatic individuals. To answer this question, we categorized all positive patient specimens processed on the Roche cobas 6800 platform by physician-reported symptom status (see Methods). An important caveat of this categorization is that we are unable to discriminate between asymptomatic and presymptomatic individuals as we did not follow symptom evolution. Nevertheless, we found 58.1% of asymptomatic and 15.0% of symptomatic individuals to have Ct values above 30 (Fig. 3). While asymptomatic patients only constituted 7.5% of all positive tests, the vast majority had high Ct values. A limitation of our study design is that we are unable to identify repeat results or distinguish the validity of repeat testing in the appropriate clinical context. However, a limited reanalysis in which 9.2% of the results were excluded due to repeat testing demonstrated no significant changes in the Ct value distribution (data not shown). These results highlight (a) asymptomatic individuals with specimens containing viral loads correlating with Ct values greater than 30 may not be detected using less sensitive methods and (b) the majority of symptomatic individuals will continue to test positive across all platforms assessed here.
Fig. 3.
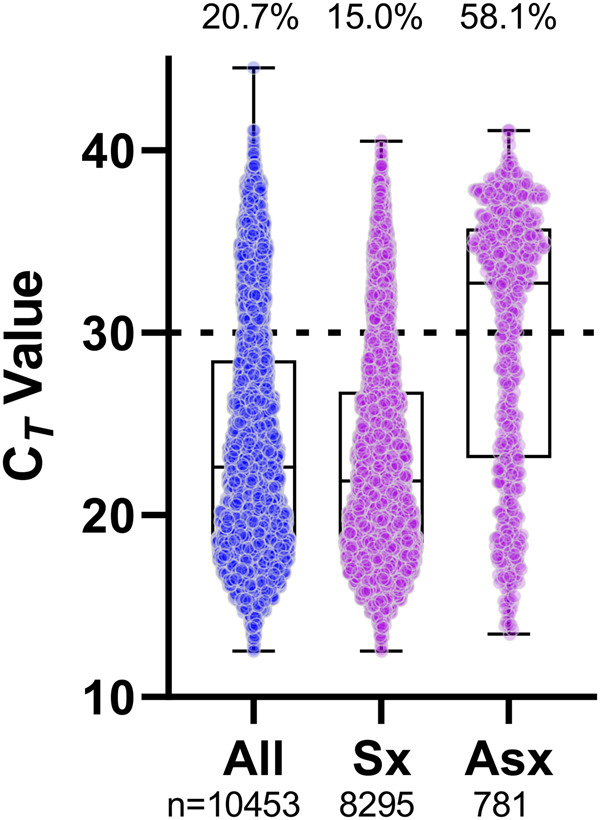
Distribution of SARS-CoV-2 E gene Ct values from positive NP specimens grouped by symptom status and detected on the Roche cobas 6800. All data points are shown in the first column and subsequently grouped by symptom status, if indicated on the order. The number of specimens is shown under the column. The percentage of specimens with Ct values greater than 30 (dashed line) is shown at the top of the column. Standard box and whisker plots show the median and interquartile range (box) and full range (whiskers) of data. Sx, symptomatic; Asx, asymptomatic.
Discussion
The detection SARS-CoV-2 in clinical specimens forms the backbone for the management of the COVID-19 pandemic. In March 2020, the Food and Drug Administration granted EUA for several detection platforms; however, the performance of these platforms in clinical specimens has not been clearly defined. In this work we report the concordance between different platforms with the clinical specimens. Moreover, we have analyzed the distribution of reported Ct values in all positive specimens (>10 000) run on our primary diagnostic platform, which provides clinical context for the performance profiles of the various platforms.
The COVID-19 pandemic has placed substantial strain on the clinical laboratory with regard to testing volume, rapid turnaround time, and accurate performance. While each manufacturer reports a theoretical limit of detection (LoD), the performance in clinical specimens is not yet well-established. Given that clinical laboratories possess multiple platforms for SARS-CoV-2 detection, understanding their limitations is critical for accurate reporting. In this regard, several studies have compared the concordance of results among different platforms using diluted control material, and pooled patient samples. The Cepheid GeneXpert and Roche cobas 6800 systems have been tested by others who report sensitivity and PPA compared to a reference standard to be >98% (7, 9). We confirm and extend these findings as we find excellent overall agreement with the Roche cobas 6800, Cepheid GeneXpert, and Diasorin Simplexa, and Biofire platforms. Importantly, we find that in high viral load, low Ct value samples, all platforms tested including Aries EUA and RUO also demonstrate excellent concordance. Importantly, the Food and Drug Administration has recently released data regarding performance of SARS-CoV-2 detection platforms using a reference panel (https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-reference-panel-comparative-data). The sensitivity of each assay is approximated by the reported LoD based on dilution of a positive specimen with known concentration. The Roche cobas SARS-CoV-2 assay demonstrates a LoD of 1800 RNA nucleic acid amplification testing–detectable units/mL (NDU/mL) while the Cepheid GeneXpert, Biofire Defense, and Diasorin Simplexa show a LoD of approximately 6000 NDU/mL. However, the LoD of the ARIES EUA platform was 180 000 NDU/mL. These data are similar to the PPA we observe in our study. Specifically, we observe decreased PPA with higher Ct, corresponding to lower viral loads. Overall, our study provides information that is critical for rapid maneuvering in the setting of instrument downtime or supply-line shortages for an individual platform.
Given that the differences in platform performance occur in low viral load samples, we analyzed the distribution of all positive samples performed on our primary analytical platform at our institution. The Ct values were distributed homogenously across the range of positive Ct values. Since our patient population was selected from both symptomatic and asymptomatic individuals, this data set likely reflects the true distribution of Ct values in SARS-CoV-2–positive patients. Using the Roche cobas E gene as a reference standard, we found approximately 21% of positive samples to have a Ct value above 30. Our results are similar to that of Buchan et al., who found the Ct values of symptomatic patients to be 3× greater than the LoD (13). Importantly, nearly 60% of specimens from asymptomatic patients had a Ct value above 30. These specimens could be reported as false negatives if run on other less sensitive platforms. These observed differences likely reflect variation in extraction, volume of specimen assayed, thermocycling, and primer-probe efficiency. A key issue regarding SARS-CoV-2 testing is the extent to which these differences matter both clinically and in the management of transmission chains. The main strain on the clinical laboratory is the testing of asymptomatic patients in the community and prior to admission to the hospital. A perfect test in these scenarios would report infectivity rather than presence of SARS-CoV-2 RNA. Prior work has shown cultures with Ct values above 34 to have low likelihood of carrying infectious virus (14). While more data are clearly required to establish conditions in which an individual is infectious, it could be acceptable to utilize less sensitive platforms in asymptomatic individuals to determine if they are infectious. This will be of importance with the approval of SARS-CoV-2 antigen detection platforms (BiNAX NOW and BD Veritor) as these are likely to be less sensitive, but might be better suited for the testing of asymptomatic individuals to assess transmission risk. Thus, SARS-CoV-2 detection platforms should take patient status into consideration both for assay evaluation and implementation.
The role of viral load in patient management is important in the management of several viral infections. While accurate quantitation is difficult to achieve with NP and oropharyngeal specimens due to variability in acquisition of the sample and thus loss of normalization, Ct values still have the potential to inform care. This could be important in individuals with low Ct values as it could reflect poor control of the virus, and thus necessitate more frequent clinical assessments. The standardization of Ct value reporting would be critical in this regard, and our work indicates that this could be feasible as we observed strong linearity in reported Ct values between different platforms, especially between the cobas 6800, GeneXpert, and Simplexa assays.
In summary, our analysis of the performance 6 sample-to-answer platforms using the same clinical specimens as opposed to synthetic genetic material is directly applicable to the accurate interpretation of respiratory samples collected from patients. However, a limitation of our analysis is that samples were diluted and frozen prior to cross-platform analysis. In addition, while we analyzed the distribution of Ct values in over 10 000 patients, there could be bias in our data set as it is mainly derived from patients from a single tertiary academic medical center. Specifically, the data regarding the Ct value distribution in asymptomatic and symptomatic patients might not be generalizable to different patient populations. An additional limitation is that our categorization of patients by symptomology relied upon nonstandardized clinical assessment by the ordering clinician. Nevertheless, our study brings context to Ct values produced by SARS-CoV-2 detection platforms a key step in its use in patient management.
Supplemental Material
Supplemental material is available at The Journal of Applied Laboratory Medicine online.
Supplementary Material
Nonstandard Abbreviations
SARS-CoV-2evere acute respiratory syndrome coronavirus 2COVID-19coronavirus disease 2019EUAemergency use authorizationCtcycle thresholdNPnasopharyngealRUOresearch use onlyPPApositive percentage agreementLoDlimit of detection
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:Employment or Leadership: None declared. Consultant or Advisory Role: C.-A.D. Burnham is a consultant for Cepheid. N.W. Anderson is a member of the Scientific Advisory Board for Diasorin Molecular. Stock Ownership: None declared. Honoraria: C.-A.D. Burnham has received honoraria from BioFire and Roche. N.W. Anderson has received honoraria from Biomerieux. Research Funding: C.-A.D. Burnham, grants to institution from Cepheid, bioMerieux, and Luminex, BioFire. Expert Testimony: None declared. Patents: None declared.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
Acknowledgments: We are grateful to the laboratory staff at Barnes-Jewish Hospital for their ongoing efforts for our patients and in support of the COVID-19 pandemic response.
REFERENCES
- 1.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF.. The proximal origin of SARS-CoV-2. Nat Med 2020;26:450–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long Q-X, Tang X-J, Shi Q-L, Li Q, Deng H-J, Yuan J, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 2020;26:1200–4. [DOI] [PubMed] [Google Scholar]
- 3.Furukawa NW, Brooks JT, Sobel J.. Evidence supporting transmission of severe acute respiratory syndrome coronavirus 2 while presymptomatic or asymptomatic. Emerg Infect Dis 2020;26. Available from: 10.3201/eid2607.201595 (Accessed April 2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westblade LF, Brar G, Pinheiro LC, Paidoussis D, Rajan M, Martin P, et al. SARS-CoV-2 viral load predicts mortality in patients with and without cancer who are hospitalized with COVID-19. Cancer Cell 2020;38:661–671.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magleby R, Westblade LF, Trzebucki A, Simon MS, Rajan M, Park J, et al. Impact of SARS-CoV-2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease. Clin Infect Dis [Epub ahead of print 2020. Jun 30] as 10.1093/cid/ciaa851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith E, Zhen W, Manji R, Schron D, Duong S, Berry GJ.. Analytical and clinical comparison of three nucleic acid amplification tests for SARS-CoV-2 detection. J Clin Microbiol 2020;58:e01134–20. 10.1128/JCM.01134-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou H, Chen J, Wang Y, Lu Y, Zhu Y, Zhang B, et al. Multi-center evaluation of the Cepheid Xpert Xpress SARS-CoV-2 assay for the detection of SARS-CoV-2 in oropharyngeal swab specimens. J Clin Microbiol 2020;58:e00926–20. 10.1128/JCM.01288-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore NM, Li H, Schejbal D, Lindsley J, Hayden MK.. Comparison of two commercial molecular tests and a laboratory-developed modification of the CDC 2019-nCoV RT-PCR assay for the detection of SARS-CoV-2. J Clin Microbiol 2020;58:e00938–20. 10.1128/JCM.00938-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broder K, Babiker A, Myers C, White T, Jones H, Cardella J, et al. Test agreement between Roche Cobas 6800 and Cepheid GeneXpert Xpress SARS-CoV-2 assays at high cycle threshold ranges. J Clin Microbiol 2020;58:e01187–20. 10.1128/JCM.01187-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fung B, Gopez A, Servellita V, Arevalo S, Ho C, Deucher A, et al. Direct comparison of SARS-CoV-2 analytical limits of detection across seven molecular assays. J Clin Microbiol 2020;58:e01535–20. 10.1128/JCM.01535-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhen W, Smith E, Manji R, Schron D, Berry GJ.. Clinical evaluation of three sample-to-answer platforms for the detection of SARS-CoV-2. J Clin Microbiol 2020; 58:e00783–20. 10.1128/JCM.00783-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liotti FM, Menchinelli G, Marchetti S, Morandotti GA, Sanguinetti M, Posteraro B, et al. Evaluating the newly developed BioFire COVID-19 test for SARS-CoV-2 molecular detection. Clin Microbiol Infect 2020;26:1699–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchan BW, Hoff JS, Gmehlin CG, Perez A, Faron ML, Munoz-Price LS, et al. Distribution of SARS-CoV-2 PCR Cycle Threshold values provide practical insight into overall and target-specific sensitivity among symptomatic patients. Am J Clin Pathol 2020;154:479–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tom MR, Mina MJ.. To interpret the SARS-CoV-2 test, consider the cycle threshold value. Clin Infect Dis 2020;71:2252–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.