Abstract
The SARS-CoV-2 variant is rapidly spreading across the world and causes to resurge infections. We previously reported that CT-P59 presented its in vivo potency against Beta variants, despite its reduced activity in cell experiments. Yet, it remains uncertain to exert the antiviral effect of CT-P59 on Gamma, Delta and its associated variants (L452R). To tackle this question, we carried out cell tests and animal studies. CT-P59 showed neutralization against Gamma, Delta, Epsilon, and Kappa variants in cells, with reduced susceptibility. The mouse challenge experiments with Gamma and Delta variants substantiated in vivo potency of CT-P59 showing symptom remission and virus abrogation in the respiratory tract. Collectively, cell and animal studies showed that CT-P59 is effective against Gamma and Delta variants infection, hinting that CT-P59 has therapeutic potential for patients infected with Gamma, Delta and its associated variants.
Keywords: SARS-CoV-2 variant, Therapeutic antibody, CT-P59, Regdanvimab, Gamma, Delta
1. Introduction
On November 2020 in Brazil, SARS-CoV-2 infection rapidly increased again in Manaus, where previously showed high sero-prevalences and the newly emerging virus was designated as P.1 or Gamma [1,2]. Gamma variant possesses 12 mutations in the spike protein of SARS-CoV-2. In particular, three mutations (K417T, E484K, and N501Y) in RBD (Receptor Binding Domain) are common in Alpha (N501Y) and Beta (K417 N, E484K, and N501Y) which are associated with increased transmissibility, immune escape and pathogenicity [3,4].
New lineage B.1.617 is considered to cause a steep increase in infection cases and death in India; Kappa (B.1.617.1) and Delta (B.1.617.2). In particular, highly transmissible Delta rapidly spread and replaced globally dominant Alpha even in most vaccinated countries such as the United Kingdom and the United States [[5], [6], [7], [8]]. The Delta has L452R, T478K and P681R in the spike protein (Table 1 ) which may contribute to increased infectivity, pathogenicity and immune evasion, resulting in re-infection or resistance to vaccine-elicited antibody and therapeutic antibodies [9,10]. One of Delta-associated variants, B.1.427 and B.1.429 also known as Epsilon have L452R mutation, causing reduction in antibody neutralization [11]. In addition, B.1.1.519 has common mutation sites with Delta at T478 and P681 residues, in common with Alpha variant [12]. T478K was a previously predominant variant in Mexico and located at the interface of RBD and ACE2 (Angiotensin-Converting Enzyme 2) interaction, thereby potentially impacting viral infection [13]. Delta and Kappa have a P681R mutation at furin cleavage sites (681PRRAR/S686). Recent studies showed that P681R enhances spike cleavage and fusion for viral entry, suggesting augmented transmissibility and pathogenicity [14,15]. Thus, it is important to assess the impact of mutations within the spike on COVID-19 therapeutics.
Table 1.
Mutations in spike protein of variants.
| SARS-CoV-2 Variant |
Origin | S1 |
S2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N-Terminal Domain | Receptor Binding Domain | – | Furin Cleavage Site | – | |||||||
| Delta (B.1.617.2) | India | T19R | G142D | Δ156-157 | R158G | L452R | T478K | – | D614G | P681R | D950 N |
| Epsilon (B.1.427) | US | – | – | – | – | L452R | – | – | D614G | – | – |
| Epsilon (B.1.429) | US | S13I | G142D | – | W152C | L452R | – | – | D614G | – | – |
| Kappa (B.1.617.1) | India | – | – | E154K | – | L452R | – | E484Q | D614G | P681R | Q1071H |
| B.1.1.519 | Mexico | – | – | – | – | – | T478K | – | D614G | P681H | T732A |
| Gamma (P.1) | Brazil | L18F T20 N P26S |
D138Y | – | R190S | K417T | – | E484K N501Y |
D614G H655Y |
– | T1027I V1176F |
The CT-P59 human monoclonal antibody (Regdanvimab) specifically binds to spike of SARS-CoV-2, blocking interaction with ACE2 for viral entry. We have demonstrated that CT-P59 has neutralization and protection against the original and Beta viruses in in vitro and in vivo studies [16,17]. However, it remains uncertain whether CT-P59 has the therapeutic effect on Gamma, Delta and its associated variants. To this end, we examined the sensitivity of CT-P59 against Gamma, Delta, Epsilon and Kappa variants by cell-based assays, and animal test using Gamma and Delta-infected mice.
2. Materials and methods
2.1. Viruses
Gamma (hCoV-19/Japan/TY7-503/2021) were obtained from BEI Resources. Gamma (hCoV-19/Korea/KDCA95637/2021), Delta (hCoV-19/Korea/KDCA5439/2021), Epsilon (hCoV-19/Korea/KDCA1792/2020, and hCoV-19/Korea/KDCA1793/2020), Kappa (hCoV-19/Korea/KDCA2950/2021) were obtained through National Culture Collection for Pathogens. Pseudoviruses for Gamma, Delta, Epsilon, Kappa, L452R, T478K, and P681H, K417T, E484K, and N501Y were produced.
2.2. Biolayer interferometry (BLI)
Binding affinity of CT-P59 to wild type and mutant RBDs were evaluated by the Octet QKe system (ForteBio). Mutant RBDs (K417T/E484K/N501Y, L452R/T478K, and L452R) were purchased from Sino Biological [16,17].
2.3. Plaque reduction neutralization test (PRNT)
PRNT were carried out as previously described [17]. Briefly, pre-incubated mixture of variant and CT-P59 was inoculated to VeroE6 cells. After incubation, the neutralization was determined as plaque counting.
2.4. Microneutralization (ViroSpot) assay
ViroSpot assay was performed as previously described [16]. Briefly, the mixture of virus and CT-P59 was inoculated to VeroE6. The infected cells were probed by treatment with anti-nucleocapsid antibody and TrueBlue staining. Neutralization were assessed by Immunospot analyzer.
2.5. Pseudovirus assay
Luciferase-based pseudovirus assay was performed using wild-type and mutant spike-expressing lentiviruses. Mutations in spike gene were introduced by gene cloning and its protein expression was confirmed by Western blot. Pseudoviruses were produced by transfection with luciferase reporter plasmid along with Gag-Pol, Rev, and Spike expression plasmids and then the copy number of pseudoviruses was quantitated by qPCR. Pseudoviruses were mixed with diluted antibodies ranging either from 100 to 0.005 ng/mL or 1000 to 0.05 ng/mL. The inocula infected ACE2-expressing HEK293T cells. After 72 h, luciferase activities were measured and IC50 values were calculated with Prism.
2.6. Animal experiments
8-week-old female human ACE2 transgenic mice, tg(K18-ACE2)2Prlmn (The Jackson Laboratory), were housed in a certified A/BSL-3 facility at International Vaccine Institute (IVI) and Korea Disease Control and Prevention Agency (KDCA) for Gamma and Delta variants studies, respectively. All procedures were approved by the Institutional Animal Care and Use Committee at IVI (IACUC Approval No. 2020-021) and KDCA (No. KDCA-IACUC-21-026) for Gamma and Delta studies, respectively.
2.7. Mouse study
hACE2 transgenic (TG) mice were intranasally inoculated with 30 μL of Gamma or Delta variant (1 × 104 PFU) under anesthesia for Gamma (n = 11/group) and Delta (n = 8–9/group) variants studies, respectively. CT-P59 was administered via a single intraperitoneal IP injection at 8 h post-inoculation at 5, 20, 40, and 80 mg/kg. Lung tissues and nasal washes were collected at scheduled necropsy at 3 dpi (days post-infection) and 6 dpi. The nasal washes were collected by flushing with 50 μL of PBS twice through the nasal cavity in the Gamma variant study only. The lungs were aseptically removed and washed with Hank's balanced salt solution containing 1% penicillin/streptomycin (ThermoFisher Scientific), then homogenized and strained through a 70-μm cell strainer (Becton Dickinson). Virus titration was determined by plaque assay in VeroE6 cells. The body weight and mortality were monitored daily until 6 dpi for Delta and until 10 dpi for Gamma variants, respectively. Animals with 25% (Delta) or 30% (Gamma) loss of body weight were euthanized according to each facility's standard and excluded from statistics.
3. Results
3.1. In vitro potency of CT-P59 against Gamma, Delta and its associated variants in cells
To investigate the therapeutic efficacy of CT-P59 against Gamma, Delta and Epsilon variants, we first evaluated the binding affinity of CT-P59 against mutant RBDs (K417T/E484K/N501Y, L452R/T478K and L452R) by using Bio-Layer interferometry. The equilibrium dissociation constant (KD) of CT-P59 against RBD of Gamma, Delta and Epsilon variants was reduced by 9–12-fold compared to that against wild-type (Supplementary Table 1). Next, we carried out two types of in vitro assays with live or pseudotyped viruses to assess the susceptibility of Gamma, Delta and its associated variants to CT-P59. The live virus microneutralization and pseudovirus assays showed approx. 61∼138-fold reduced susceptibility against Gamma variants, compared to that against wild type SARS-CoV-2 (Table 2 and Supplementary Fig. 2). E484K (8.66-fold) and N501Y (5.49-fold) was less than 10-fold susceptible to CT-P59, but CT-P59 showed lower IC50 value (0.7-fold) against K417T. CT-P59 showed reduced susceptibility against Delta, Epsilon and Kappa variants with IC50 of 1,237, 365.6–499.5 and 161.5 ng/mL compared to that against wild type (6.76 ng/mL) in PRNT. Approx. 183-, 54–74-, 24-fold reduced susceptibility against Delta, Epsilon and Kappa were observed compared to wild type SARS-CoV-2, respectively (Table 2 and Supplementary Fig. 1A). In addition, CT-P59 showed approx. 98-, 31-, 50-fold reduced neutralizing activity against Delta, Epsilon and Kappa pseudotyped virus compared to that of wild type, respectively (Table 2 and Supplementary Fig. 1B). CT-P59 was less susceptible to L452R, but retained its own neutralizing effect against T478K, and P681H (Table 2 and Supplementary Fig. 1B). Thus, we found that CT-P59 can neutralize Gamma, Delta and Epsilon variants despite reduced binding affinity and antiviral ability in in vitro experiments.
Table 2.
Neutralization effect of CT-P59 against variant live viruses and pseudoviruses.
| Viruses | IC50 (ng/mL) | Fold change (reduction) | |
|---|---|---|---|
| Live virus | Gamma (P.1) | 275.75 (WT: 2.0) | 137.86 |
| Delta (B.1.617.2) | 1237 (WT: 6.76) | 182.99 | |
| Epsilon (B.1.427) | 499.5 (WT: 6.76) | 73.89 | |
| Epsilon (B.1.429) | 365.6 (WT: 6.76) | 54.08 | |
| Kappa (B.1.617.1) | 161.5 (WT: 6.76) | 23.89 | |
| Pseudovirus | Gamma (P.1) | 13.45 (D614G: 0.219) | 61.42 |
| Delta (L452R/T478K/P681R) | 21.52 (D614G: 0.219) | 98.26 | |
| Epsilon (S13I/W152C/L452R) | 6.819 (D614G: 0.219) | 31.14 | |
| K417T | 0.154 (D614G: 0.219) | 0.70 | |
| E484K | 3.273 (D614: 0.378) | 8.66 | |
| N501Y | 1.202 (D614G: 0.219) | 5.49 | |
| L452R (Epsilon) | 13.22 (D614: 0.378) | 34.97 | |
| T478K | 0.213 (D614G: 0.219) | 0.97 | |
| P681H | 0.378 (D614: 0.378) | 1.00 | |
3.2. In vivo potency of CT-P59 against Gamma and Delta variants in mice
To evaluate whether CT-P59 has an ability to lower viral burden against Gamma and Delta variants in in vivo setting, hACE2 TG mice were treated with clinically relevant dosages in consideration of the 40 mg/kg of clinical dose of CT-P59. The dose of 80 mg/kg is considered as the equivalent exposure in clinical study (data not shown), and lower doses of 5 and 20 mg/kg were selected to investigate the lower exposure in the clinical situation.
Survival rate of the Gamma variant-infected placebo group resulted in 0% at 8 dpi, having two mice euthanized which showed more than 30% body weight loss at 7 dpi and one mouse died at 8 dpi. In contrast, there was no lethality in all CT-P59 treatment groups (Fig. 1 A). CT-P59 treatment delayed body weight loss and showed a statistically significant difference compared to the placebo group from 3 dpi to 6 dpi. Mean body weight of CT-P59 treatment groups began to revitalize from 7 dpi and fully recovered at 10 dpi. In contrast, the placebo group showed an average body weight loss of 27.3% at 6 dpi, and showed 28.4% reduction for one mouse while two euthanized mice excluded from statistical analysis at 7 dpi (Fig. 1 B). Delta variant-infected placebo group showed the survival rate of 75% at 5 dpi and 0% at 6 dpi, respectively. In contrast, there was no lethality in all CT-P59 treatment groups (Fig. 2 A). CT-P59 treatment also prevented the weight loss of the mice infected with Delta variant. 5 and 20 mg/kg of treatment groups showed the delay in weight loss until 6 dpi, and 40 and 80 mg/kg of treatment showed statistically significant difference compared to the placebo group at 4 dpi and 5 dpi, respectively (Fig. 2 B).
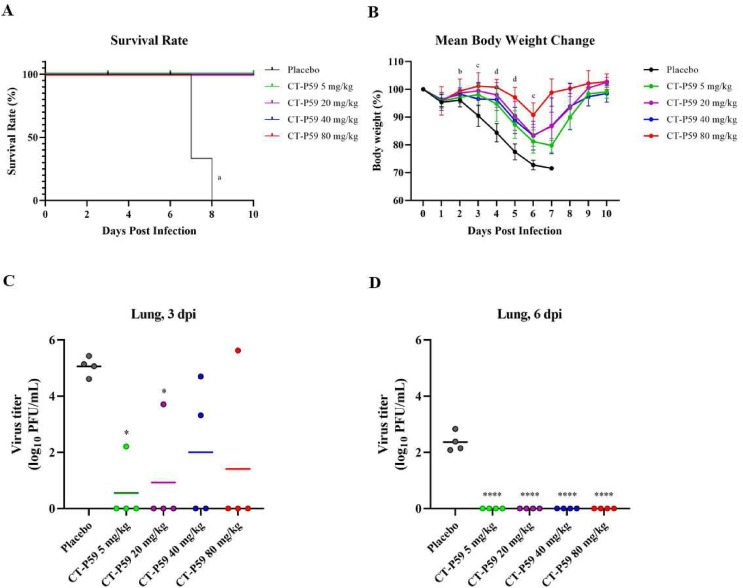
Fig. 1.
In vivo efficacy of CT-P59 against Gamma variant in mice. Vehicle and 5, 20, 40 and 80 mg/kg of CT-P59 were administered intraperitoneally 8 h after virus inoculation. Body weights and survival rates were monitored daily (A, B). The weight per mouse on day 0 was used as a baseline, and weight change was determined relative to baseline. 30% or higher of weight loss was considered to be dead. Four animals each were euthanized for virus titration at 3 dpi and 6 dpi, respectively. The virus titers from lung tissue was measured (C, D) using plaque assay. Alphabets and asterisks indicates statistical significance between the virus only and each treatment group as determined by one-way ANOVA followed by a Dunnett's post-hoc test. a denotes p < 0.0001 between control and CT-P59 treatment groups (pooled). b denotes p < 0.01 to p < 0.05 between control and CT-P59 treatment group at 80 mg/kg c denotes p < 0.0001 to p < 0.01 between control and CT-P59 treatment groups at 5, 20, 40 and 80 mg/kg d denotes p < 0.0001 to p < 0.001 between control and CT-P59 treatment groups at 5, 20, 40 and 80 mg/kg ∗ indicates P < 0.05, and ∗∗∗∗ indicates P < 0.0001.
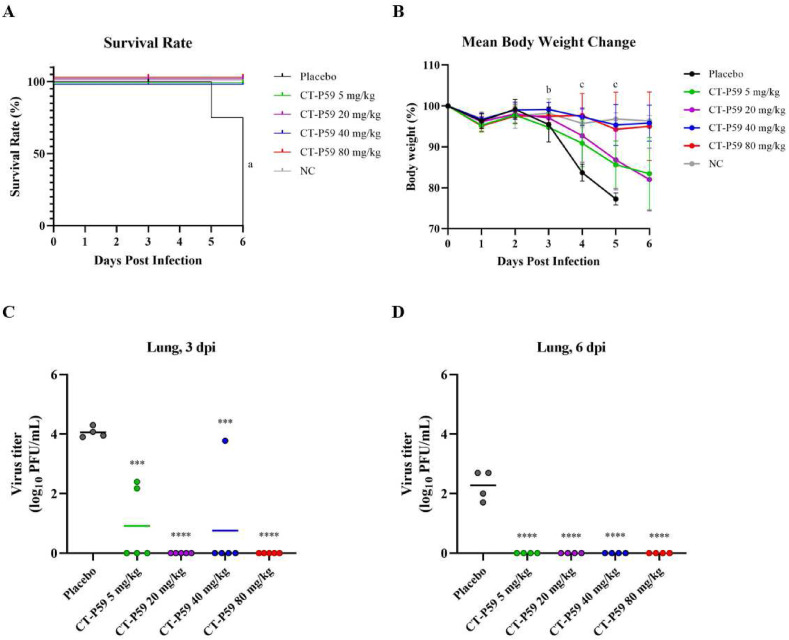
Fig. 2.
In vivo efficacy of CT-P59 against Delta variant in mice. Negative control (NC) indicates naïve mice without virus infection. Vehicle and 5, 20, 40 and 80 mg/kg of CT-P59 were administered intraperitoneally 8 h after virus inoculation. Body weights and survival rates were monitored daily (A, B). The weight per mouse on day 0 was used as a baseline, and weight change was determined relative to baseline. 25% or higher of weight loss was considered to be dead. Five and four animals each were euthanized for virus titration at 3 dpi and 6 dpi, respectively. The virus titers from lung tissue was measured (C, D) using plaque assay. Alphabets and asterisks indicates statistical significance between the virus only and each treatment group as determined by one-way ANOVA followed by a Dunnett's post-hoc test. a denotes statistically significant difference (p < 0.05) between virus only and each treatment group (5, 20, 40 and 80 mg/kg). b denotes statistically significant difference (p < 0.05) between virus only and 40 mg/kg c denotes statistically significant difference (p < 0.05) between virus only and each treatment group (40 and 80 mg/kg). ∗∗∗ indicates P < 0.001, and ∗∗∗∗ indicates P < 0.0001 between virus only and each treatment group.
CT-P59 significantly inhibited the virus replication in the respiratory tract. In the Gamma variant study, the mean virus titer in lung tissues reached 5.1 log10 PFU/mL at 3 dpi and declined to 2.4 log10 PFU/mL at 6 dpi in the placebo group. On the contrary, all CT-P59-treated groups showed considerably reduced viral titers at 3 and 6 dpi. At 6 dpi, complete eradication of virus in all CT P59-treated groups were shown (Fig. 1. C and D). From the nasal washes, there were no statistical differences between placebo and CT-P59 treated groups, where complete eradication of virus was shown from all mice except for a single mice from placebo group at both 3 and 6 dpi (Supplementary Fig. 3. A and B). CT-P59 also significantly abrogated the virus replication from lungs in the study with Delta variant. In the placebo group, the mean virus titer in lung tissues reached 4.1 log10 PFU/mL at 3 dpi and declined to 2.3 log10 PFU/mL at 6 dpi. In contrast, CT-P59 treatment showed substantial reduction in viral titers at 3 dpi and 6 dpi. At 3 dpi, infectious viral titers were 3.1 and 3.3 log reduced in 5 and 40 mg/kg groups, respectively, while complete eradication was observed in 20 and 80 mg/kg groups. At 6 dpi, no viral titers were detected from all CT P59-treated mice (Fig. 2C and D).
4. Discussion
Here, we tested and confirmed in vitro neutralizing ability of CT-P59 against Gamma, Delta, and its associated variants in authentic and pseudo viral cell-based assays which are generally used to determine antiviral effects. However, in vitro cell-based evaluation is limited to determine the in vivo antiviral efficacy of therapeutics since IC50 indicates its antiviral ability and does not reflect the effectiveness of treatment dosage and in vivo environments.
In accordance with the in vivo ferret study against Beta variant [16], CT-P59 reduced viral loads and also ameliorated clinical symptoms in Gamma or Delta variant-infected mice. Recently, Chen et al., showed that a monoclonal antibody with partial loss of in vitro neutralizing activity comparably confers the in vivo protections from variant infections in mice [18]. This notion was supported by our results, suggesting that clinical dosage of CT-P59 could overcome a certain level of reduction in in vitro antiviral activity, as long as it is not a complete loss in neutralizing activity.
In addition, the biodistribution of monoclonal antibody to target sites i.e., respiratory tracts is important to translate the in vitro antiviral activity to the in vivo efficacy in terms of pharmacokinetics (PK) of patients, for COVID-19 [19]. Given that a lung to serum concentration ratio is 15%, predicted lung concentration during the treatment period could provide the rationale for the therapeutic efficacy with administered dosages [[20], [21], [22]]. Moreover, CT-P59 is predicted to achieve sufficient lung epithelial lining fluid concentrations above the IC90 values against Gamma, Delta and its associated variants during the treatment period based on the clinical PK. Hence, the biodistribution of CT-P59 supports that expected sufficient concentration in lung could clinically compensate for the reduced activity of CT-P59 against Gamma, Delta and its associated variants in the patients. In addition, The therapeutic ability of Regdanvimab (40 mg/kg dosage and 6.76 ng/mL IC50) could come to approx. 70-fold over that of Sotrovimab (500 mg dosage and 100 ng/mL IC50) [23]. Importantly, the findings that CT-P59 ameliorated clinical symptoms in Gamma and Delta-infected mice which is reported to represent severe symptoms model [24], supports the potency of CT-P59 in mild to moderate patients by preventing from progressing to severe COVID-19. The human therapeutic effectiveness needs to be further investigated in patients infected with the Gamma and delta variants.
5. Conclusion
CT-P59 retained antiviral activity against Gamma, Delta, Epsilon, and Kappa variants in cells, and showed in vivo substantial protection in Gamma and Delta variant-infected mice, suggesting that the use of CT-P59 could be a curable option to COVID-19 patients infected with these variants and other potential variants having identical mutation sites.
Declaration of competing interest
Authors D.-K Ryu, B. Kang, H. Noh, J.-I Kim, J.-M Seo, C. Kim, M. Kim, K.-S Kwon and S.-Y Lee declare that they were employed by Celltrion when the study was conducted and the manuscript compiled.
Acknowledgements
This study was supported by Korea Disease Control and Prevention Agency for efficacy evaluation (PRNT and animal testing) and Korea National Institute of Health fund (2020-ER5311-00).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2021.09.023.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., Hulswit R.J.G., Franco L.A.M., Ramundo M.S., de Jesus J.G., Andrade P.S., Coletti T.M., Ferreira G.M., Silva C.A.M., Manuli E.R., Pereira R.H.M., Peixoto P.S., Kraemer M.U.G., Gaburo N., Jr., Camilo C.D.C., Hoeltgebaum H., Souza W.M., Rocha E.C., de Souza L.M., de Pinho M.C., Araujo L.J.T., Malta F.S.V., de Lima A.B., Silva J.D.P., Zauli D.A.G., Ferreira A.C.S., Schnekenberg R.P., Laydon D.J., Walker P.G.T., Schluter H.M., Dos Santos A.L.P., Vidal M.S., Del Caro V.S., Filho R.M.F., Dos Santos H.M., Aguiar R.S., Proenca-Modena J.L., Nelson B., Hay J.A., Monod M., Miscouridou X., Coupland H., Sonabend R., Vollmer M., Gandy A., Prete C.A., Jr., Nascimento V.H., Suchard M.A., Bowden T.A., Pond S.L.K., Wu C.H., Ratmann O., Ferguson N.M., Dye C., Loman N.J., Lemey P., Rambaut A., Fraiji N.A., Carvalho M., Pybus O.G., Flaxman S., Bhatt S., Sabino E.C. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buss L.F., Prete C.A., Jr., Abrahim C.M.M., Mendrone A., Jr., Salomon T., de Almeida-Neto C., Franca R.F.O., Belotti M.C., Carvalho M., Costa A.G., Crispim M.A.E., Ferreira S.C., Fraiji N.A., Gurzenda S., Whittaker C., Kamaura L.T., Takecian P.L., da Silva Peixoto P., Oikawa M.K., Nishiya A.S., Rocha V., Salles N.A., de Souza Santos A.A., da Silva M.A., Custer B., Parag K.V., Barral-Netto M., Kraemer M.U.G., Pereira R.H.M., Pybus O.G., Busch M.P., Castro M.C., Dye C., Nascimento V.H., Faria N.R., Sabino E.C. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science. 2021;371:288–292. doi: 10.1126/science.abe9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dejnirattisai W., Zhou D., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., Wang B., Lopez-Camacho C., Slon-Campos J., Walter T.S., Skelly D., Costa Clemens S.A., Naveca F.G., Nascimento V., Nascimento F., Fernandes da Costa C., Resende P.C., Pauvolid-Correa A., Siqueira M.M., Dold C., Levin R., Dong T., Pollard A.J., Knight J.C., Crook D., Lambe T., Clutterbuck E., Bibi S., Flaxman A., Bittaye M., Belij-Rammerstorfer S., Gilbert S.C., Carroll M.W., Klenerman P., Barnes E., Dunachie S.J., Paterson N.G., Williams M.A., Hall D.R., Hulswit R.J.G., Bowden T.A., Fry E.E., Mongkolsapaya J., Ren J., Stuart D.I., Screaton G.R. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184:2939–2954. doi: 10.1016/j.cell.2021.03.055. e2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L., Ho D.D. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29:747–751. doi: 10.1016/j.chom.2021.04.007. e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell F., Archer B., Laurenson-Schafer H., Jinnai Y., Konings F., Batra N., Pavlin B., Vandemaele K., Van Kerkhove M.D., Jombart T., Morgan O., le Polain de Waroux O. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarah Cherian V.P., Jadhav Santosh, Yadav Pragya, Gupta Nivedita, Das Mousmi, Rakshit Partha, Singh Sujeet, Abraham Priya, Panda Samiran, NIC team . The Second Wave of COVID-19 in Maharashtra. BioRxiv; India: 2021. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.England P.H. 2021. Variants: Distribution of Case Data. [Google Scholar]
- 8.Prevention C.f.D.C.a. 2021. Variant Proportions. [Google Scholar]
- 9.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Velay A., Le Guen J., Kassis-Chikhani N., Edriss D., Belec L., Seve A., Courtellemont L., Pere H., Hocqueloux L., Fafi-Kremer S., Prazuck T., Mouquet H., Bruel T., Simon-Loriere E., Rey F.A., Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 10.Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCallum M., Bassi J., De Marco A., Chen A., Walls A.C., Di Iulio J., Tortorici M.A., Navarro M.J., Silacci-Fregni C., Saliba C., Sprouse K.R., Agostini M., Pinto D., Culap K., Bianchi S., Jaconi S., Cameroni E., Bowen J.E., Tilles S.W., Pizzuto M.S., Guastalla S.B., Bona G., Pellanda A.F., Garzoni C., Van Voorhis W.C., Rosen L.E., Snell G., Telenti A., Virgin H.W., Piccoli L., Corti D., Veesler D. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021;373:648–654. doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lubinski B., Tang T., Daniel S., Jaimes J.A., Whittaker G.R. Functional evaluation of proteolytic activation for the SARS-CoV-2 variant B.1.1.7: role of the P681H mutation. bioRxiv. 2021 doi: 10.1016/j.isci.2021.103589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maison D.P., Ching L.L., Shikuma C.M., Nerurkar V.R. Genetic characteristics and phylogeny of 969-bp S gene sequence of SARS-CoV-2 from Hawaii reveals the worldwide emerging P681H mutation. Hawaii J Health Soc Welf. 2021;80:52–61. [PMC free article] [PubMed] [Google Scholar]
- 14.Akatsuki Saito H.N., Uriu Keiya, Kosugi Yusuke, Irie Takashi, Shirakawa Kotaro, Sadamasu Kenji, Kimura Izumi, Ito Jumpei, Wu Jiaqi, Ozono Seiya, Tokunaga Kenzo, Butlertanaka Erika P., Yuri L., Tanaka, Shimizu Ryo, Shimizu Kenta, Fukuhara Takasuke, Kawabata Ryoko, Sakaguchi Takemasa, Yoshida Isao, Asakura Hiroyuki, Nagashima Mami, Yoshimura Kazuhisa, Kazuma Yasuhiro, Nomura Ryosuke, Horisawa Yoshihito, Takaori-Kondo Akifumi. bioRxiv; 2021. The Genotype to Phenotype Japan (G2P-Japan) Consortium, So Nakagawa, Terumasa Ikeda, Kei Sato, SARS-CoV-2 Spike P681R Mutation Enhances and Accelerates Viral Fusion. [Google Scholar]
- 15.Thomas C.M.S., Peacock P., Brown Jonathan C., Goonawardane Niluka, Zhou Jie, Whiteley Max, PHE Virology Consortium, Thushan I., de Silva, Barclay Wendy S. 2021. The SARS-CoV-2 Variants Associated with Infections in India, B.1.617, Show Enhanced Spike Cleavage by Furin, bioRxiv. [Google Scholar]
- 16.Ryu D.K., Song R., Kim M., Kim Y.I., Kim C., Kim J.I., Kwon K.S., Tijsma A.S., Nuijten P.M., van Baalen C.A., Hermanus T., Kgagudi P., Moyo-Gwete T., Moore P.L., Choi Y.K., Lee S.Y. Therapeutic effect of CT-P59 against SARS-CoV-2 South African variant. Biochem. Biophys. Res. Commun. 2021;566:135–140. doi: 10.1016/j.bbrc.2021.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim C., Ryu D.K., Lee J., Kim Y.I., Seo J.M., Kim Y.G., Jeong J.H., Kim M., Kim J.I., Kim P., Bae J.S., Shim E.Y., Lee M.S., Kim M.S., Noh H., Park G.S., Park J.S., Son D., An Y., Lee J.N., Kwon K.S., Lee J.Y., Lee H., Yang J.S., Kim K.C., Kim S.S., Woo H.M., Kim J.W., Park M.S., Yu K.M., Kim S.M., Kim E.H., Park S.J., Jeong S.T., Yu C.H., Song Y., Gu S.H., Oh H., Koo B.S., Hong J.J., Ryu C.M., Park W.B., Oh M.D., Choi Y.K., Lee S.Y. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12:288. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen R.E., Winkler E.S., Case J.B., Aziati I.D., Bricker T.L., Joshi A., Darling T.L., Ying B., Errico J.M., Shrihari S., VanBlargan L.A., Xie X., Gilchuk P., Zost S.J., Droit L., Liu Z., Stumpf S., Wang D., Handley S.A., Stine W.B., Jr., Shi P.Y., Davis-Gardner M.E., Suthar M.S., Knight M.G., Andino R., Chiu C.Y., Ellebedy A.H., Fremont D.H., Whelan S.P.J., Crowe J.E., Jr., Purcell L., Corti D., Boon A.C.M., Diamond M.S. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature. 2021;596:103–108. doi: 10.1038/s41586-021-03720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia R., Wang H. How could in vitro antiviral activity Be applied to optimize the dosing regimens of candidates for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)? Clin. Infect. Dis. 2021;73:352–353. doi: 10.1093/cid/ciaa1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.FDA . 2020. Fact Sheet for Health Care Providers Ememgency Use Authorization (EUA) of Casirivimab and Imdevimab. [Google Scholar]
- 21.FDA . 2021. Fact Sheet for Healthcare Providers Emegency Use Authorization (EUA) of Sotrovimab. [Google Scholar]
- 22.Shah D.K., Betts A.M. Antibody biodistribution coefficients: inferring tissue concentrations of monoclonal antibodies based on the plasma concentrations in several preclinical species and human. mAbs. 2013;5:297–305. doi: 10.4161/mabs.23684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrea C.H.-D., Cathcart L., Lempp Florian A., Ma Daphne, Schmid Michael A., Agostini Maria L., Guarino Barbara, Julia Di iulio, Rosen Laura E., Tucker Heather, Dillen Joshua, Subramanian Sambhavi, Sloan Barbara, Bianchi Siro, Pinto Dora, Saliba Christian, Wojcechowskyj Jason A., Julia Noack, Zhou Jiayi, Kaiser Hannah, Chase Arthur, Montiel-Ruiz Martin, Dellota Exequiel, Jr., Park Arnold, Spreafico Roberto, Anna Sahakyan, Lauron Elvin J., Czudnochowski Nadine, Cameroni Elisabetta, Ledoux Sarah, Adam Werts, Colas Christophe, Soriaga Leah, Telenti Amalio, Purcell Lisa A., Hwang Seungmin, Gyorgy Snell, Virgin Herbert W., Corti Davide, Hebner Christy M. 2021. The Dual Function Monoclonal Antibodies VIR-7831 and VIR-7832 Demonstrate Potent in Vitro and in Vivo Activity against SARS-CoV-2. [Google Scholar]
- 24.Winkler M.S., Skirecki T., Brunkhorst F.M., Cajander S., Cavaillon J.M., Ferrer R., Flohe S.B., Garcia-Salido A., Giamarellos-Bourboulis E.J., Girardis M., Kox M., Lachmann G., Martin-Loeches I., Netea M.G., Spinetti T., Schefold J.C., Torres A., Uhle F., Venet F., Weis S., Scherag A., Rubio I., Osuchowski M.F. Bridging animal and clinical research during SARS-CoV-2 pandemic: a new-old challenge. EBioMedicine. 2021;66:103291. doi: 10.1016/j.ebiom.2021.103291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.